Abstract
The Helicobacter pylori protein CagA may undergo tyrosine phosphorylation following its entry into human gastric epithelial cells with downstream effects on signal transduction. Disruption of the gp130 receptor that modulates the balance of the SHP2/ERK and JAK/STAT pathways enhanced peptic ulceration and gastric cancer in gp130 knock-out mice. In this study, we evaluated the effect of translocated CagA in relation to its tyrosine phosphorylation status on the gp130-mediated signal switch between the SHP2/ERK and JAK/STAT3 pathways. We showed that in the presence of CagA, SHP2 was recruited to gp130. Phosphorylated CagA showed enhanced SHP2 binding activity and ERK1/2 phosphorylation, whereas unphosphorylated CagA showed preferential STAT3 activation. These findings indicate that the phosphorylation status of CagA affects the signal switch between the SHP2/ERK and JAK/STAT3 pathways through gp130, providing a novel mechanism to explain H. pylori signaling.
Keywords: Bacteria, Bacterial Genetics, Cell Metabolism, Epithelial Cell, JAK Kinase, MAP Kinases (MAPKs), Gastric Physiology, STAT, Stomach
Introduction
Helicobacter pylori frequently colonizes the human stomach (1), and hosts infected by cagA-positive strains are at increased risk of gastric cancer and peptic ulceration (2–10). H. pylori injects the CagA protein into host gastric epithelial cells via a type IV secretion system (11–16). The injected CagA is tyrosine-phosphorylated by Src family protein-tyrosine kinases and binds SHP2 (Src homology 2 domain-containing Src homology tyrosine phosphatase) (17–21); the CagA-SHP2 complex has been detected in human gastric mucosa (22, 23).
The IL6/gp130/STAT3 (interleukin-6/glycoprotein 130/signal transducer and activation of transcription 3) pathway has been shown to play a role in the development of gastric cancer (24, 25). IL6 exerts its biological activities through the receptor subunit gp130 (26). At least two functional modules of gp130 have been characterized; one encompasses four membrane-distal Tyr(P) binding sites for the Src homology 2 domain of the latent transcription factors, STAT1 and STAT3. The other comprises the membrane-proximal Tyr(P)757 residue responsible for the engagement of cytoplasmic SHP2 (26, 27).
IL6 induces recruitment and homodimerization of gp130, potentially leading to balanced signaling through both the JAK/STAT and SHP2/Ras/ERK signaling pathways (28, 29). However, disrupting this balance in the gp130 “knock-in” mouse induced premalignant lesions, including atrophy, intestinal metaplasia, dysplasia, and ultimately gastric cancer (24). Similarly, when the IL6 cytokine family signaling pathway is disrupted, increased STAT3 signaling may favor development of gastric adenomas, whereas increased SHP2/ERK2 signaling may lead to mucosal inflammation (25, 30).
That cellular factors that are up-regulated in response to H. pylori could modulate SHP2/ERK or JAK/STAT signaling pathways suggests that H. pylori persistence and its consequent pathologies could be influenced by gp130-mediated signal transduction through these pathways (30). In this study, we examined the role of CagA tyrosine phosphorylation status in the activation of the SHP2/ERK and JAK/STAT pathways downstream of the gp130 receptor. Our studies indicate that the tyrosine phosphorylation status of CagA produced by H. pylori directs the traffic of signal transduction through the two pathways.
EXPERIMENTAL PROCEDURES
Bacteria, Cell Culture, and Co-incubation with H. pylori
A pair of naturally occurring isogenic cagA strains (147C, with a 3′ tyrosine phosphorylation motif, and 147A, without the motif) isolated simultaneously from the same host have been described (31–33). As controls, H. pylori strains 60190 (cagPAI+), 8822 (cagPAI−), and ΔcagA (an isogenic mutant of 60190 lacking cagA (ATCC 49503) were used. H. pylori strains were cultured on agar plates containing 10% horse serum at 37 °C in a microaerobic atmosphere, using the Campy Container System (BBL, Sparks, MD). Human (AGS) gastric epithelial cells were cultured in RPMI 1640 medium (Invitrogen), containing 10% FBS (Invitrogen). The cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. On the day of experimentation, confluent cells were incubated overnight in fresh serum- and antibiotic-free media. AGS cells also were co-incubated with H. pylori up to multiplicities of infection of 100:1 for varying times.
Immunoblotting and Antibodies
Whole cell extracts were prepared with lysis buffer containing 50 mm Tris (pH 7.5), 5 mm EDTA, 100 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors (Roche Applied Science). Lysates were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. Immunodetection was performed using an electrochemiluminescence reagent (Intron, Seoul, Korea), according to the manufacturer's instructions. In experiments using kinase inhibitors, cells were incubated with PP2 (Calbiochem), UO126 (Promega, Madison, WI), or AG490 (Calbiochem) for 1 h before co-incubation with H. pylori. Anti-SHP2, anti-pY99, anti-phospho-STAT3, anti-STAT3, anti-gp130, anti-phospho-JAK2, anti-JAK2, and anti-c-Myc were from Santa Cruz Biotechnology, Inc. (Santa Cruz CA), anti-ERK1/2 and anti-phospho-ERK1/2 were from Cell Signaling (Danvers, MA), and anti-CagA antibody (HPP-5003-9) was from ASTRAL Biologicals (San Ramon, CA). Anti-human IL6R monoclonal neutralizing antibody and gp130 were obtained from R&D Systems (Minneapolis, MN). AGS cells were cultured overnight with monoclonal antibodies to IL6R or gp130 (R&D Systems) in a serum-free medium and co-incubated with H. pylori at 37 °C in 5% CO2. Whole cell extracts were prepared with lysis buffer and analyzed by immunoblotting with antibodies specific for anti-phospho-STAT3 antibody. Equal loading of lanes was controlled by comparison with lanes that had been resolved using anti-STAT3 antibody.
Co-immunoprecipitation
For immunoprecipitation, whole cell extracts prepared with lysis buffer were incubated overnight with appropriate antibodies for 4 h at 4 °C, and immune complexes were trapped on protein G-Sepharose beads (Amersham Biosciences). Beads were washed five times with cold lysis buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and protease inhibitors (Roche Applied Science). The cell lysates and immunoprecipitated materials were subjected to SDS-PAGE, and then proteins were transferred to polyvinylidene difluoride membranes and incubated with antibodies, including anti-SHP2, anti-CagA, or anti-gp130, and then visualized using a chemiluminescence reagent (Intron).
Expression Vectors
SHP2-specific small interfering RNA (siRNA) (kindly provided by Prof. M. Hatakeyama) was used to silence the expression of SHP2 (34). To selectively knock down human SHP2 expression, AGS cells were co-transfected with pSUPER-SHP2 and pBabePuro. After 12 h, cells were incubated in RPMI 1640, 10% fetal bovine serum containing 0.3 μg/ml puromycin (Sigma) to select appropriate transfectants; SHP2 expression of the cloned cells was confirmed by immunoblotting.
Immunofluorescence Microscopy
AGS cells co-incubated with H. pylori were washed twice with cold PBS, fixed in methanol for 10 min, washed with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 4 min. Nonspecific binding was blocked with 3% bovine serum albumin in 0.1% Triton X-100/PBS for 30 min, followed by incubation with anti-phospho-STAT3 mouse monoclonal antibodies in 2% bovine serum albumin, 0.1% Triton X-100/PBS at 4 °C overnight. After washing with PBS, phospho-STAT3 was visualized by treatment with fluorescein isothiocyanate-conjugated goat anti-mouse monoclonal secondary antibodies for 60 min at room temperature. Cells were washed with PBS, mounting medium for fluorescence was added, and slides were sealed with coverslips and examined for immunofluorescence using a confocal laser-scanning microscope (LSM 510; Carl Zeiss, Thornwood, NY).
Nuclear Protein Extraction and Immunoblotting
AGS cells co-cultured with H. pylori were harvested and separated into cytoplasmic and nuclear fractions using CEB buffer (10 mm Tris-Cl, pH 8, 60 mm KCl, 1 mm EDTA, 1 mm dithiothreitol), containing 0.5% Nonidet P-40 and protease inhibitors (Roche Applied Science). The cytoplasmic fraction was clarified by centrifugation at 1,200 × g for 5 min, and the nuclear pellet was washed with CEB buffer containing 0.2% Nonidet P-40 to remove cytoplasmic contamination. The nuclear pellet was lysed with NEB buffer (20 mm Tris-Cl, pH 8, 0.4 m NaCl, 1.5 mm MgCl2, 1.5 mm EDTA, 1 mm dithiothreitol) containing 0.5% Nonidet P-40 and protease inhibitors (Roche Applied Science). Nuclear proteins were analyzed by immunoblotting as described above.
Real Time PCR
Total cellular RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's protocol. First-strand complementary DNA was synthesized from 1 μg of total cellular RNA using an RNA PCR kit (Intron) with random primers. Thereafter, real time RT-PCR analysis also was performed using a PCR mixture containing 1 μmol/liter of each primer and SYBR Green master mix (Applied Biosystems, Foster City, CA) using the ABI PRISM 7000 Quantitative PCR system (Applied Biosystems). Each sample was examined in triplicate, and the amounts of the PCR products produced were normalized with respect to β-actin, as an internal control. The following primer pairs were used: c-myc, 5′-TGC TCC ATG AGG AGA CAC C and 5′-CTC TGA CCT TTT GCC AGG AG; β-actin, 5′-TTG CCG ACA GGA TGC AGA AGA and 5′-AGG TGG ACA GCG AGG CCA GGA T.
Electromobility Shift Assay (EMSA)
AGS cells co-incubated with H. pylori were harvested and separated into cytoplasmic and nuclear fractions using CEB buffer containing 0.5% Nonidet P-40 and protease inhibitor, as described above. The oligonucleotides used as probes for the EMSA were as follows: c-myc E2F, 5′-GACGCTTGGCGGGAAAAAG and 5′-GGCTTTTTCCCGCCAAG (35). Oligonucleotides based on c-myc E2F were end-labeled with [γ-32P]ATP by T4 polynucleotide kinase (Invitrogen). For binding reactions, 1 × 105 cpm of labeled oligonucleotide probes were incubated with 10 μg of nuclear extracts and 1 μg of poly(dI-dC) in binding buffer (4% (v/v) glycerol, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm dithiothreitol, 50 mm NaCl, 10 mm Tris-HCl, pH 7.5) at room temperature for 20 min. Protein-DNA complexes were separated by electrophoresis in a 5% non-denaturing polyacrylamide gel in 0.5× TBE buffer. The gels were dried and exposed to film (Agfa) at −70° using an intensifying screen.
Cell Migration Assay
Cell migration assays were performed using μ-Dish 35-mm culture inserts (Ibidi) according to the manufacturer's protocols. In brief, AGS cells were seeded into each well of culture inserts and incubated at 37 °C in a humidified atmosphere with 5% CO2. On the day of experimentation, confluent cells were refed overnight with RPMI 1640 supplemented with 0.5% FBS. After appropriate cell attachment, the culture inserts were gently removed by using sterile tweezers, and the dish was filled with fresh RPMI 1640 supplemented with 0.5% FBS. AGS cells also were co-incubated for 24 h with H. pylori to a multiplicity of infection of 50.
Statistical Analysis
Significance in differences between experimental conditions was determined by Student's t test; p values of <0.05 were considered significant in all analyses.
RESULTS
Role of CagA Tyrosine Phosphorylation Status and SHP2 Binding
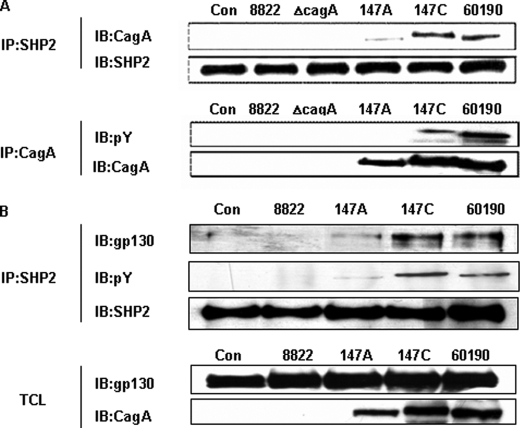
The tyrosine phosphorylation status of CagA has been shown to strongly influence CagA binding affinity to SHP2 (23, 36). We began our experiments by asking whether the H. pylori strains that we were studying would show similar activities. As expected, both of the H. pylori strains (147C and 60190) that express CagA and that have tyrosine phosphorylation sites in their EPIYA domain (C-EPIYA+) showed CagA binding to SHP2, whereas the CagA PAI-negative 8822 and ΔcagA strains showed no detectable SHP2 binding (Fig. 1A). Differences in the mobility of the CagA protein reflect the previously observed microheterogeneity in the cagA open reading frame (31, 32). These results confirm that the CagA molecules we study bind to SHP2, as determined by tyrosine phosphorylation status.
FIGURE 1.
Relationship between the C-EPIYA status of H. pylori CagA and its interaction with SHP2 and gp130. A, lysates of AGS cells co-incubated with H. pylori were immunoprecipitated (IP) with anti-SHP2 or anti-CagA antibody and then immunoblotted (IB) with the converse antibodies. Con, control. B, lysates of AGS cells co-incubated with H. pylori strains for 1 h were immunoprecipitated with anti-SHP2 antibody. The immunoprecipitates and total cell lysates (TCL) were immunoblotted with anti-phosphotyrosine, anti-gp130, anti-CagA, or anti-SHP2 antibody.
Interaction between SHP2 and gp130 in Relation to CagA Status
SHP2 activation is critical for IL6 induction of the mitogen-activated protein kinase (MAPK) pathway (34, 37). IL6 levels in gastric tissue are elevated in H. pylori+ hosts and then fall after H. pylori eradication (38). To examine whether H. pylori affects SHP2 recruitment to gp130, a central constituent of the IL6 receptor complex (37), cellular lysates from AGS cells co-incubated with H. pylori were immunoprecipitated and immunoblotted using antibodies to CagA, gp130, or SHP2. For AGS cells co-incubated with C-EPIYA+ H. pylori strain 147C or 60190, immunoprecipitation showed that SHP2 was associated with both gp130 and with CagA. However, no association with gp130 was detected in the AGS cells that were co-incubated with C-EPIYA− H. pylori strain 147A or cagA− strain 8822 (Fig. 1B). These data indicate that the injected phosphorylated C-EPIYA+ CagA plays a role in the recruitment of SHP2 to gp130.
Influence of CagA Tyrosine Phosphorylation Status on H. pylori-mediated gp130 Receptor Phosphorylation
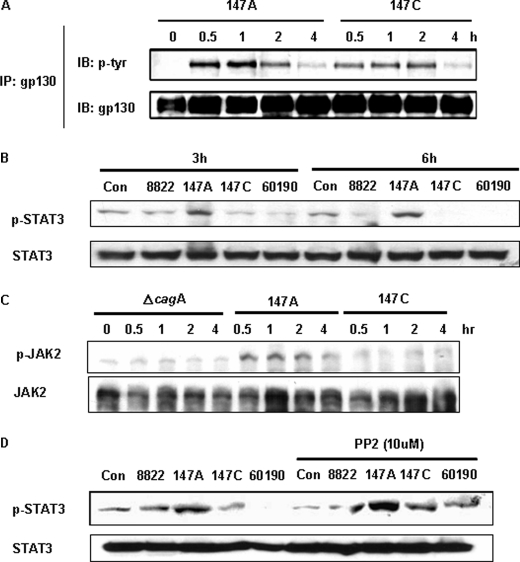
Next we compared the isogenic cagA+ isogenic C-EPIYA− 147A and C-EPIYA+ 147C H. pylori strains by co-incubating them with AGS cells to determine whether the AGS cell gp130 receptors become activated. After co-incubation, the AGS cells were subjected to immunoprecipitation with anti-gp130 antibodies. Both of the H. pylori strains induced activation of the gp130 receptor, indicating its independence from CagA tyrosine phosphorylation status.
Influence of CagA Tyrosine Phosphorylation Status on JAK/STAT3 Activation
Once it became clear that the H. pylori strains we studied activated gp130, confirming the results of Bronte-Tinkew et al. (39), we next examined the downstream JAK/STAT activation. First, to analyze STAT3 activation in relation to the CagA tyrosine phosphorylation status, AGS cells were co-incubated for 3 or 6 h with H. pylori strains of varying cagA genotype and then immunoblotted using anti-phospho-STAT3. STAT3 was phosphorylated in AGS cells that were co-incubated with C-EPIYA− strain 147A (Fig. 2B), but compared with the absence of H. pylori (control), essentially no phosphorylation was observed with two C-EPIYA+ strains (147C and 60190) or with strain 8822. Following cytokine stimulation, STAT3 is activated by the phosphorylation of a single tyrosine residue (Tyr705) by JAK2 and dimerizes via a reciprocal Src homology 2 phosphotyrosine interaction (40). To examine whether the H. pylori STAT3 phosphorylation is mediated by JAK2 activation, AGS cells were co-incubated with H. pylori strains. We found that C-EPIYA− strain 147A induced JAK2 activation, which suggests that the STAT3 activation mediated by non-phosphorylated CagA is dependent on JAK2 activation (Fig. 2C).
FIGURE 2.
Relation of gp130 receptor phosphorylation to H. pylori CagA and STAT3 phosphorylation. A, the gp130 receptor was immunoprecipitated (IP) from the cell lysates with anti-gp130 antibody, and phosphorylated gp130 receptor was detected by immunoblotting (IB) with anti-phosphotyrosine antibody (PY99). The extent of gp130 receptor precipitated then was assessed by reprobing the blot with an anti-gp130 receptor antibody. B, after co-incubation with H. pylori strains for 3 or 6 h, STAT3 status was determined by immunoblotting using antibody recognizing STAT3 phosphorylated at tyrosine 705 (p-STAT3). The same blot was stripped and reprobed with anti-STAT3 antibody to assess for gel loading. C, AGS cells were co-incubated with cells of H. pylori strains for the indicated times, and whole cell lysates were subjected to immunoblot analysis using anti-phospho-JAK2 antibody. The membrane was reprobed with an anti-JAK2 antibody. D, AGS cells were co-incubated with cells of H. pylori strains with or without PP2 (10 μm) pretreatment and were examined for STAT3 phosphorylation by immunoblotting. As a loading control, blots were stripped and reprobed with anti-STAT3 antibody. Con, control.
To address that hypothesis, because CagA is phosphorylated by the Src family of protein-tyrosine kinases (15, 17, 18), we asked whether pretreatment with PP2, a specific Src family kinase inhibitor, would affect the CagA-induced STAT3 activation. AGS gastric epithelial cells were pretreated with PP2 for 1 h prior to co-incubation, and STAT3 activation was examined. In the presence of PP2, STAT activation by C-EPIYA+ strains 147C and 60190 was observed (Fig. 2D). That PP2 inhibition of Src kinase (thus inhibiting CagA phosphorylation) restores STAT3 activation provides further evidence that non-phosphorylated CagA is essential for the observed (39) H. pylori-induced STAT3 activation.
Influence of gp130 and JAK2 Inhibition on CagA-induced STAT3 Phosphorylation
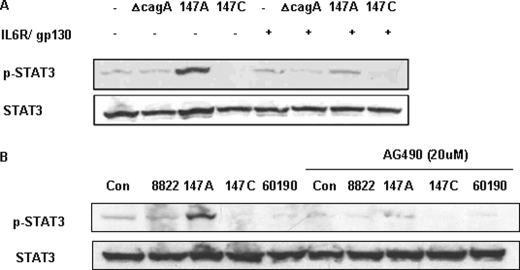
The IL6 receptor complex is composed of a ligand-binding receptor (IL6R) and its signal transducer, gp130 (41). Monoclonal antibodies to IL6R and gp130, that function as specific receptor antagonists, abrogating activation of the IL6/IL6R/gp130 complex (42), were used to examine whether H. pylori-induced STAT3 activation in AGS cells is mediated through the gp130 receptor. AGS cells were preincubated overnight with the neutralizing monoclonal antibodies to IL6R and gp130 in serum-free medium and then co-incubated with H. pylori. We found that the antibody treatment substantially inhibited the STAT3 activation in AGS cells that had been induced by strain 147A, which indicated that the predominant signal by its C-EPIYA− CagA involves the gp130 receptor (Fig. 3A). Next, to determine whether the C-EPIYA− CagA-induced STAT3 phosphorylation is mediated via JAK2, AGS cells were pretreated with AG490, a JAK2-preferential JAK inhibitor, and then co-incubated with H. pylori (Fig. 3B). Importantly, the addition of AG490 had no effect on the viability of the cells (data not shown). AGS cells pretreated with AG490 showed greatly reduced 147A-mediated STAT3 phosphorylation. In total, these results showed that both gp130 and JAK2 activation are involved in the C-EPIYA− CagA-induced STAT3 activation.
FIGURE 3.
Effect of inhibition of gp130 or JAK2 on CagA-induced STAT activation. A, the AGS cells pretreated overnight in serum-free medium with anti-human IL6R and gp130 neutralizing monoclonal antibodies were co-incubated with H. pylori cells. Whole cell lysates were subjected to immunoblot analysis using phospho-STAT3 (p-STAT3) antibody. Immunoblotting of total STAT3 served as the control. B, AGS cells were pretreated (or not) with 20 μm JAK2 inhibitor (AG490) for 1 h and then co-incubated with H. pylori strains for 3 h. STAT3 phosphorylation was analyzed by immunoblotting using anti-p-STAT3 antibody. Con, control.
Influence of C-EPIYA CagA Tyrosine Phosphorylation Status on SHP2/ERK Activation
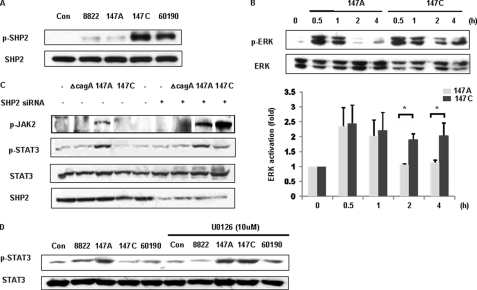
The gp130 receptor is responsible for engagement of cytoplasmic SHP2 and subsequent activation of the Ras-ERK pathway (26, 27). To investigate whether CagA C-EPIYA status affects the SHP2/ERK pathway downstream of the gp130 receptor, AGS cells were co-incubated with the defined H. pylori strains. As expected, SHP2 was phosphorylated in cells that were co-incubated with C-EPIYA+ 147C or 60190 and not with C-EPIYA− 147C or with cagA− 8822 (Fig. 4A). There was early ERK activation by both 147A and 147C, but activation was more sustained for 147C (Fig. 4B). In the absence of H. pylori, AGS control cells showed no detectable levels of phosphorylated ERK1/2. Together with our prior studies (33), these data confirm that C-EPIYA+ CagA facilitates signal transduction through SHP2/ERK to a greater extent than does C-EPIYA− CagA.
FIGURE 4.
SHP2/ERK activation according to H. pylori CagA status. A, lysates from AGS cells co-incubated with H. pylori strains were immunoblotted with anti-p-SHP2, and then the membrane was stripped and analyzed with anti-SHP2 antibody to standardize gel loading. B, AGS cells were co-incubated with cells of H. pylori strain 147A or 147C. The phosphorylated ERK1/2 was assessed by immunoblotting. Blots were reprobed with α-ERK antibody to control for differences in ERK1/2 loading (top). Densitometric ratios between p-ERK and ERK immunoblotting were shown (bottom). Each bar indicates the mean ± S.E. (error bars) of three separate experiments. *, p < 0.05, comparing 147A and 147C. C, AGS cells were transfected with SHP2 siRNA or control empty vector for 12 h and primarily selected for 3 days in RPMI 1640 medium with 10% FBS containing 0.3 μg/ml puromycin. The cells were incubated with control vector alone (−) or SHP siRNA(+) and H. pylori. Total cell lysates were immunoblotted with anti-phospho-JAK2 (p-JAK2), anti-phospho-STAT3 (p-STAT3), or anti-SHP2 antibody. D, AGS cells were pretreated with U0126 (10 μm) for 1 h prior to co-incubation with H. pylori. Phosphorylation of STAT3 was analyzed by immunoblotting. Con, control.
To determine why C-EPIYA+ CagA does not induce STAT3 activation and because SHP2 is known to be an important negative regulator of JAK2/STAT3 signaling in other cells (42), we next investigated the specific involvement of SHP2 in JAK2/STAT3 activation in AGS cells co-incubated with H. pylori. SHP2 activity correlates with its own phosphorylation, which can catalyze tyrosine phosphorylation of JAKs, receptors, or other cellular proteins (41). Because we showed that C-EPIYA+ strain 147C phosphorylates SHP2 in AGS cells (Fig. 4A), but JAK2 is not activated (Fig. 2C), and because the SHP2 recruitment site within gp130 often is involved in negative regulation of IL6-induced STAT3 activation (43, 44), we selectively inhibited SHP2 expression using SHP2 siRNA pretreatment. As expected, treatment with SHP2 siRNA substantially lowered SHP2 levels (Fig. 4C). In AGS cells that were co-incubated with C-EPIYA− 147A, STAT3 activation was not affected by the SHP2 siRNA pretreatment. In contrast, siRNA pretreatment of AGS cells co-incubated with C-EPIYA+ 147C increased JAK2 and STAT3. These studies provide further evidence that phosphorylated SHP2 inhibits JAK2/STAT3 signaling pathways after its interaction with C-EPIYA+ CagA molecules. Thus, phosphorylation-competent (C-EPIYA+) and incompetent (C-EPIYA−) forms of CagA have opposing cellular functions.
ERK kinase activity also can down-regulate STAT3 function via several mechanisms, including inhibition of upstream kinases, such as JAK family members (45), dephosphorylation of STAT3 phosphotyrosine (46), and formation of an ERK-STAT3 complex (47). Because we hypothesized that enhanced ERK activation in cells co-incubated with C-EPIYA+ CagA+ H. pylori cells also might lead to STAT3 inhibition, we next investigated in AGS cells co-incubated with strain 147C whether the ERK pathway downstream from SHP2 was involved in the STAT3 dephosphorylation. Pretreatment of AGS cells with the ERK inhibitor U0126 before H. pylori co-incubation clearly enhanced STAT3 activation by 147C or 60190, indicating that C-EPIYA+ CagA suppressed STAT3 activation via an ERK-dependent mechanism (Fig. 4D).
Relation of C-EPIYA CagA Status to STAT3 Nuclear Translocation
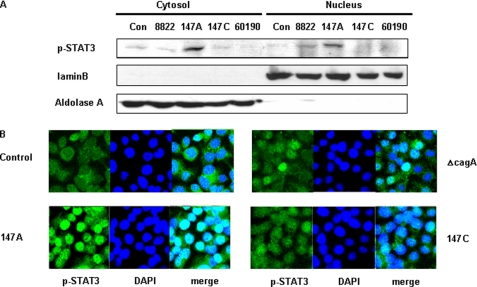
Dimeric STAT3 translocates into the cell nucleus, where it binds to defined elements within promoter regions of target genes, activating their transcription (48–50). Using immunoblotting of cytosolic and nuclear fractions, we investigated effects on STAT3 translocation in AGS cells that were incubated with H. pylori strains of varying CagA status. Co-incubation with C-EPIYA− strain 147A induced greater STAT3 nuclear translocation than the other strains tested (Fig. 5A). Immunofluorescence analysis of AGS cells was used to determine the STAT3 subcellular localization after co-incubation with H. pylori. AGS cells co-incubated with 147A induced greater phospho-STAT3 nuclear translocation than did the cells with either 147C or the CagA-negative strain (Fig. 5B), consistent with the immunoblotting results.
FIGURE 5.
STAT3 phosphorylation and nuclear localization according to the C-EPIYA status of H. pylori CagA. A, AGS cells were co-incubated with H. pylori strains for 3 h, and then their cytoplasmic and nuclear fractions were separated. The nuclear proteins were analyzed by immunoblotting with anti-phospho-STAT3 (p-STAT3). To verify complete separation of the cytosolic and nuclear fractions, cytosolic and nuclear extracts were immunoblotted for aldolase A and lamin B. B, AGS cells were co-incubated with H. pylori cells and examined using confocal microscopy, with anti-phospho-STAT3 (p-STAT3) stained in green (magnification, ×100), and DNA stained with 4′,6-diamidino-2-phenylindole.
Role of STAT3 on the gp130-mediated Activation of c-myc
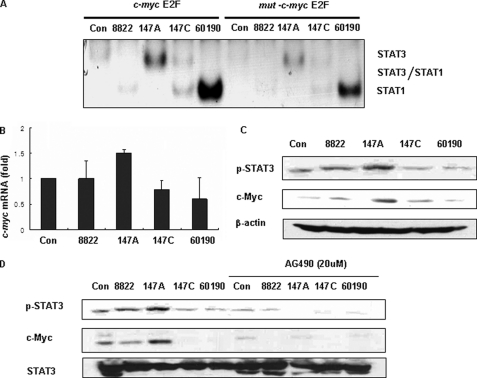
Next, we examined the consequences of STAT3 activation, which is mediated by C-EPIYA− unphosphorylated CagA, by monitoring the protein levels of c-myc, a target gene of STAT3. The c-myc product is an effector molecule responsible for cell proliferation and apoptosis (48, 51–54), and STAT3 is known to be involved in the rapid c-myc activation at least partly by binding to a site (overlapping the c-myc E2F binding site) within the c-myc P2 promoter (55, 56). We first performed EMSAs with oligonucleotide probes containing the STAT3 c-myc E2F binding site (TTGGCGGGAAA) and/or mut-c-myc E2F, which has a mutated c-myc E2F site (TTGGAAGTTAA). We examined nuclear extracts from AGS cells co-incubated with H. pylori to resolve the binding specificity of the STAT proteins for the c-myc E2F site. In the IL6 cytokine family-stimulated (control) cells, nuclear extracts contain STAT3, STAT3/STAT1, and STAT1 (55). However, our studies showed that STAT3 or STAT1 homodimers were strongly detected in AGS cells co-incubated with H. pylori. Nuclear extracts from AGS cells co-incubated with 147A had STAT3 homodimers bound to the c-myc E2F binding site, whereas cells co-incubated with 147C or 60190 had binding of the STAT1 homodimer (Fig. 6A). By Western blotting, phospho-STAT1 was induced preferentially in AGS cells co-incubated with 147C or 60190, consistent with the EMSA results (supplemental Fig. 1).
FIGURE 6.
Relation of STAT3 activation to the induction of c-myc. A, nuclear extracts from AGS cells co-incubated with H. pylori were incubated with 32P-labeled oligonucleotide containing the c-myc promoter E2F site and then subjected to electrophoresis and autoradiography. B, AGS cells were co-incubated with H. pylori for 6 h and expression of c-myc mRNA was analyzed by real-time PCR. C, AGS cells were co-incubated with H. pylori, and cell lysates were analyzed by Western blot using phospho-STAT3 (p-STAT3) or c-Myc antibody. The blot was stripped and reprobed with β-actin antibody to confirm equal loading. D, cell lysates prepared from AGS cells co-incubated with H. pylori in the presence or absence of AG490 were subjected to Western blot using phospho-STAT3 or c-Myc antibody. The blot was stripped and reprobed with STAT3 antibody. Con, control. Error bars, S.E.
Next, we examined c-myc mRNA and c-Myc protein levels by real-time PCR and Western blotting, respectively. As expected, c-myc mRNA and c-Myc protein expression were induced by C-EPIYA− CagA, paralleling the EMSA findings. However, c-myc mRNA and c-Myc protein both were down-regulated in AGS cells co-incubated with C-EPIYA+ strain 147C or 60190 (Fig. 6, B and C). To ask whether c-Myc induction was STAT3-mediated, we used AG490 to specifically inhibit STAT3 activation. Pretreatment of AGS cells with AG490 before co-incubation with H. pylori affected c-Myc induction, indicating that STAT3 activation is involved in the H. pylori-induced c-Myc response (Fig. 6D).
The C-EPIYA− CagA-mediated JAK/STAT Pathway Induces Cell Migration
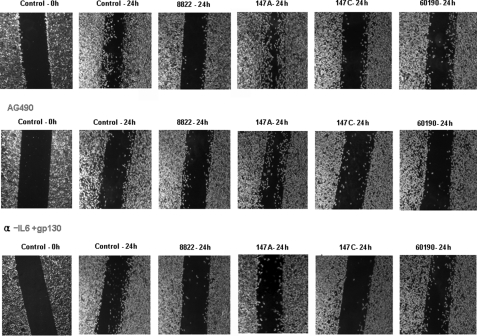
IL6 has an important role in the initial phase of intestinal wound healing, and gp130-mediated STAT1/3 signaling has a protective effect on intestinal epithelium, consistent with STAT3 function in epithelial migration during epidermal wound healing (57). To investigate the involvement of the JAK/STAT pathway in healing of wounded epithelium in vitro in relation to the CagA status of H. pylori, migration assays were performed in the presence of H. pylori. There was significant inhibition of cell migration by H. pylori strains with C-EPIYA+ CagA, whereas cell migration was induced by H. pylori with C-EPIYA− CagA. To determine whether C-EPIYA− CagA-mediated STAT3 activation induces cell migration, we also used the JAK2-specific inhibitor AG490, or IL6/gp130-neutralizing antibody to determine their effects on cell migration in AGS cells co-incubated with H. pylori (Fig. 7). Cell migration induced by H. pylori cells with C-EPIYA− CagA was abolished by inhibition of either JAK or the gp130 receptor. These data indicate that the JAK/STAT pathway that is induced by C-EPIYA− CagA may participate in the regulation of cell migration.
FIGURE 7.
Effect of STAT3 activation on the migration of AGS cells co-incubated with H. pylori. The cell migration assay was performed as described under “Experimental Procedures.” AGS cells were co-incubated with H. pylori in each well of culture inserts in the presence or absence of AG490 or IL6 + gp130 neutralizing antibody and examined using Olympus IX70 microscopy (magnification, ×100).
DISCUSSION
Gastric colonization with cagA-positive H. pylori strains increases the risk for atrophic gastritis, peptic ulcer disease, and gastric cancer (2–6) and may decrease the risk for asthma and other allergic disorders (58). However, the molecular mechanisms involved are incompletely understood. In this study, we used a pair of isogenic strains (147A and 147C) with identical CagA proteins, differing only by the presence of a single C-EPIYA motif (31–33) to elucidate the mechanisms by which tyrosine-phosphorylated CagA affects signaling within gastric epithelial cells. Because the C-EPIYA+ (phosphorylated) CagA (from 147C) has greater binding affinity to SHP2, as shown previously (23, 34, 36) and confirmed in our co-immunoprecipitation experiments, the extent of this interaction could affect downstream signaling pathways. For example, SHP2 positively regulates ERK activity, which is necessary for CagA induction of the hummingbird phenotype (23). Consistent with this pathway, we have shown that phosphorylated C-EPIYA+ 147C CagA induced greater ERK1/2 activation than did unphosphorylated C-EPIYA− 147A CagA, confirming our prior observations (33).
However, the IL6 receptor gp130 has bifunctional domains; thus, its activation can lead to signaling through either the SHP2/ERK or JAK/STAT pathways (26, 27). We asked whether signaling through these competing transduction pathways is affected by CagA in a tyrosine phosphorylation (EPIYA)-dependent manner. We first showed that the gp130 receptor is phosphorylated by CagA, regardless of its C-EPIYA status. Thus, in addition to gp130 induction by IL6 in the human gastric mucosa (38), H. pylori also affects gp130-regulated signal transduction from the interior of epithelial cells. Next, we found that C-EPIYA− CagA induced substantially more STAT3 than did C-EPIYA+ CagA, and we confirmed the inverse relationship for ERK activation.
Because STAT3 is activated via the JAK2 signaling pathway (40), we sought to determine whether C-EPIYA− CagA-induced STAT3 activation is mediated through the gp130 receptor. The use of the JAK2 inhibitor AG490 showed that STAT3 activation by 147A requires JAK2 activation, and the use of PP2, a specific inhibitor of Src family kinases, which blocks CagA phosphorylation, restored induction of STAT3 phosphorylation by 147C or 60190 CagA. Both of these studies indicate that STAT3 activation is dependent on C-EPIYA− CagA. That monoclonal antibodies to IL6R and to gp130 significantly inhibited STAT3 phosphorylation in the presence of unphosphorylated CagA confirmed the central role of gp130 in CagA-induced STAT3 activation. Our findings consistently indicate that the gp130/STAT3 pathway is preferentially activated by C-EPIYA− CagA, whereas as expected (11, 23), C-EPIYA+ CagA preferentially activates the SHP2/ERK pathway. Thus, the relative proportion of the two forms of CagA within an H. pylori population may determine the balance of signaling pathways in the underlying epithelial cells.
Next, we examined why C-EPIYA+ CagA failed to activate the STAT3 pathway. ERK kinase activity can down-regulate STAT3 activation by dephosphorylation of STAT3 phosphotyrosine (46), and SHP2 activity can dephosphorylate gp130 and associated factors, including JAKs and STATs (42). The negative feedback induced by C-EPIYA+ CagA on the JAK-related signal pathway can be explained by down-regulation of the JAK/STAT signal transduction pathway by several families of proteins, including ERK, or by SHP2 suppression of the JAK/STAT pathway. The prolonged STAT3 activity induced by C-EPIYA− CagA also can be explained by prolonged STAT3 phosphorylation through receptor-associated kinases that are not suppressed by SHP2 as well as by reduced dephosphorylation of receptor-recruited JAKs or STATs by SHP2 (59).
Upon stimulation of the IL6 receptor (gp130), STAT3 mediates the rapid activation of c-myc. STAT3 binds to a region overlapping the E2F site in the c-myc promoter, a locus critical for c-myc transcriptional activation by IL6 or gp130 signals (55). To understand the linkage between STAT3 and c-myc induction, we performed EMSA to analyze whether, in the context of H. pylori co-incubation, STAT3 binds to the c-myc promoter and performed real time PCR to evaluate STAT3 c-myc induction. The data show that STAT3 induced in AGS cells by C-EPIYA− CagA binds preferentially to c-myc E2F, whereas c-myc was not induced in AGS cells co-incubated with C-EPIYA+ CagA because STAT1 bound to the c-myc promoter is likely to interact with a co-repressor. A likely candidate is MBP-1, which represses c-myc expression when bound to the E2F site (56). IL6 and HGF activate STAT3 and can promote epithelial cell growth or migration in vivo (60, 61). In keratinocyte-specific STAT3-null mice, slower skin wound healing and migration defects were observed compared with control mice (57). In parallel, we provide evidence that STAT3 activation facilitates gastric epithelial wound healing.
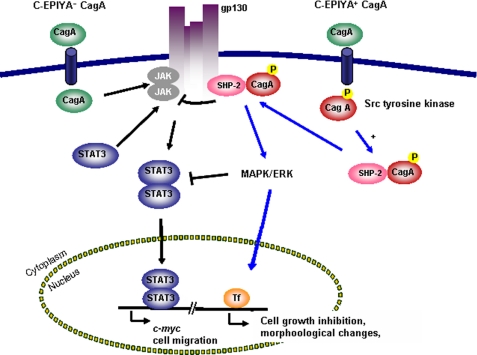
In total, these findings indicate that regulation of human signal transduction pathways induced by translocated CagA vary depending on its tyrosine phosphorylation status, with C-EPIYA− CagA playing a novel role. Variation in CagA C-EPIYA status thus may be responsible for differential gastric epithelial cell phenotypes. Overall, the tyrosine phosphorylation status of CagA in H. pylori in part determines the traffic flows of two opposing pathways, gp130-activated SHP2/ERK and JAK/STAT signaling, in gastric epithelial cells (Fig. 8).
FIGURE 8.
Proposed role of CagA in signal switching between the JAK/STAT3 and SHP2/ERK pathways via the gp130 receptor in AGS cells. AGS cells incubated with CagA+ EPIYA− H. pylori have preferential activation of the JAK/STAT3 pathway, via the IL6 receptor gp130, whereas AGS cells incubated with CagA+ EPIYA+ H. pylori first tyrosine-phosphorylate CagA and then have preferential activation of the SHP2/ERK pathway. (The interaction of phosphorylated CagA with SHP2 results in inhibition of gp130-mediated JAK activation.) Activated JAK recruits STAT3 to the cell membrane gp130 complex, which then signals the nucleus via the STAT pathway. STAT3 translocated to the nucleus induces c-myc expression and facilitates cell migration, where the SHP2/ERK pathways, which are predominantly mediated by phosphorylated CagA, induce cell morphological changes and cell growth inhibition.
H. pylori populations in a host are dynamic (62) and can gain or lose cagA-encoded EPIYA motifs (62) through recombination involving repetitive DNA sequences (31). Such changes, which may be stochastic with selection determined by host and environmental circumstances (63), affect epithelial cell phenotypes that can promote prolonged colonization. The active role of C-EPIYA− CagA in the JAK/STAT pathway induction now explains the utility to H. pylori of being able to modify CagA tyrosine phosphorylation status, with the polar (EPIYA− and EPIYA+) forms inducing separate and inhibitory pathways. In this way, the H. pylori population biology with respect to CagA status, creates a “rheostat,” under selective pressure, by which the divergent forms affect host cell phenotypes and ultimately the downstream risk of disease.
Supplementary Material
Acknowledgment
We thank Prof. M. Hatakeyama for sharing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM63270. This work was also supported by the Korea Research Foundation and Korean government (Ministry of Education and Human Resources Development, Basic Research Promotion Fund) Grant KRF-2004-015-C00444.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ERK
- extracellular signal-regulated kinase
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- FBS
- fetal bovine serum
- siRNA
- small interfering RNA
- PBS
- phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1.Huang J. Q., Sridhar S., Chen Y., Hunt R. H. (1998) Gastroenterology 114, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 2.Suerbaum S., Michetti P. (2002) N. Engl. J. Med. 347, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 3.Tummuru M. K., Cover T. L., Blaser M. J. (1993) Infect. Immun. 61, 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser M. J., Perez-Perez G. I., Kleanthous H., Cover T. L., Peek R. M., Chyou P. H., Stemmermann G. N., Nomura A. (1995) Cancer Res. 55, 2111–2115 [PubMed] [Google Scholar]
- 5.Nomura A. M., Lee J., Stemmermann G. N., Nomura R. Y., Perez-Perez G. I., Blaser M. J. (2002) J. Infect. Dis. 186, 1138–1144 [DOI] [PubMed] [Google Scholar]
- 6.Nomura A. M., Pérez-Pérez G. I., Lee J., Stemmermann G., Blaser M. J. (2002) Am. J. Epidemiol. 155, 1054–1059 [DOI] [PubMed] [Google Scholar]
- 7.Parsonnet J., Friedman G. D., Orentreich N., Vogelman H. (1997) Gut 40, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe T., Tada M., Nagai H., Sasaki S., Nakao M. (1998) Gastroenterology 115, 642–648 [DOI] [PubMed] [Google Scholar]
- 9.Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R. J. (2001) N. Engl. J. Med. 345, 784–789 [DOI] [PubMed] [Google Scholar]
- 10.Covacci A., Censini S., Bugnoli M., Petracca R., Burroni D., Macchia G., Massone A., Papini E., Xiang Z., Figura N. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5791–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., Backert S. (2007) Nature 449, 862–866 [DOI] [PubMed] [Google Scholar]
- 12.Segal E. D., Cha J., Lo J., Falkow S., Tompkins L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14559–14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asahi M., Azuma T., Ito S., Ito Y., Suto H., Nagai Y., Tsubokawa M., Tohyama Y., Maeda S., Omata M., Suzuki T., Sasakawa C. (2000) J. Exp. Med. 191, 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backert S., Ziska E., Brinkmann V., Zimny-Arndt U., Fauconnier A., Jungblut P. R., Naumann M., Meyer T. F. (2000) Cell Microbiol. 2, 155–164 [DOI] [PubMed] [Google Scholar]
- 15.Odenbreit S., Püls J., Sedlmaier B., Gerland E., Fischer W., Haas R. (2000) Science 287, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 16.Stein M., Rappuoli R., Covacci A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbach M., Moese S., Hauck C. R., Meyer T. F., Backert S. (2002) J. Biol. Chem. 277, 6775–6778 [DOI] [PubMed] [Google Scholar]
- 18.Stein M., Bagnoli F., Halenbeck R., Rappuoli R., Fantl W. J., Covacci A. (2002) Mol. Microbiol. 43, 971–980 [DOI] [PubMed] [Google Scholar]
- 19.Feng G. S., Hui C. C., Pawson T. (1993) Science 259, 1607–1611 [DOI] [PubMed] [Google Scholar]
- 20.Freeman R. M., Jr., Plutzky J., Neel B. G. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11239–11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad S., Banville D., Zhao Z., Fischer E. H., Shen S. H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2197–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki S., Yamakawa A., Ito Y., Ohtani M., Higashi H., Hatakeyama M., Azuma T. (2003) J. Infect. Dis. 187, 334–337 [DOI] [PubMed] [Google Scholar]
- 23.Higashi H., Nakaya A., Tsutsumi R., Yokoyama K., Fujii Y., Ishikawa S., Higuchi M., Takahashi A., Kurashima Y., Teishikata Y., Tanaka S., Azuma T., Hatakeyama M. (2004) J. Biol. Chem. 279, 17205–17216 [DOI] [PubMed] [Google Scholar]
- 24.Ohtani T., Ishihara K., Atsumi T., Nishida K., Kaneko Y., Miyata T., Itoh S., Narimatsu M., Maeda H., Fukada T., Itoh M., Okano H., Hibi M., Hirano T. (2000) Immunity 12, 95–105 [DOI] [PubMed] [Google Scholar]
- 25.Tebbutt N. C., Giraud A. S., Inglese M., Jenkins B., Waring P., Clay F. J., Malki S., Alderman B. M., Grail D., Hollande F., Heath J. K., Ernst M. (2002) Nat. Med. 8, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 26.Heinrich P. C., Behrmann I., Müller-Newen G., Schaper F., Graeve L. (1998) Biochem. J. 334, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taga T., Kishimoto T. (1997) Annu. Rev. Immunol. 15, 797–819 [DOI] [PubMed] [Google Scholar]
- 28.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamimura D., Ishihara K., Hirano T. (2003) Rev. Physiol. Biochem. Pharmacol. 149, 1–38 [DOI] [PubMed] [Google Scholar]
- 30.Wang T. C., Goldenring J. R. (2002) Nat. Med. 8, 1080–1082 [DOI] [PubMed] [Google Scholar]
- 31.Aras R. A., Lee Y., Kim S. K., Israel D., Peek R. M., Jr., Blaser M. J. (2003) J. Infect. Dis. 188, 486–496 [DOI] [PubMed] [Google Scholar]
- 32.Kim S. Y., Lee Y. C., Kim H. K., Blaser M. J. (2006) Cell Microbiol. 8, 97–106 [DOI] [PubMed] [Google Scholar]
- 33.Pillinger M. H., Marjanovic N., Kim S. Y., Lee Y. C., Scher J. U., Roper J., Abeles A. M., Izmirly P. I., Axelrod M., Pillinger M. Y., Tolani S., Dinsell V., Abramson S. B., Blaser M. J. (2007) J. Biol. Chem. 282, 18722–18731 [DOI] [PubMed] [Google Scholar]
- 34.Higuchi M., Tsutsumi R., Higashi H., Hatakeyama M. (2004) Cancer Sci. 95, 442–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaturvedi P., Sharma S., Reddy E. P. (1997) Mol. Cell. Biol. 17, 3295–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higashi H., Tsutsumi R., Muto S., Sugiyama T., Azuma T., Asaka M., Hatakeyama M. (2002) Science 295, 683–686 [DOI] [PubMed] [Google Scholar]
- 37.Bode J. G., Schweigart J., Kehrmann J., Ehlting C., Schaper F., Heinrich P. C., Häussinger D. (2003) J. Immunol. 171, 257–266 [DOI] [PubMed] [Google Scholar]
- 38.Ando T., Kusugami K., Ohsuga M., Ina K., Shinoda M., Konagaya T., Sakai T., Imada A., Kasuga N., Nada T., Ichiyama S., Blaser M. J. (1998) Infect. Immun. 66, 4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bronte-Tinkew D. M., Terebiznik M., Franco A., Ang M., Ahn D., Mimuro H., Sasakawa C., Ropeleski M. J., Peek R. M., Jr., Jones N. L. (2009) Cancer Res. 69, 632–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto T., Taga T., Akira S. (1994) Cell 76, 253–262 [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee M., Hönemann D., Lentzsch S., Bommert K., Sers C., Herrmann P., Mathas S., Dörken B., Bargou R. C. (2002) Blood 100, 3311–3318 [DOI] [PubMed] [Google Scholar]
- 42.Vogel W., Lammers R., Huang J., Ullrich A. (1993) Science 259, 1611–1614 [DOI] [PubMed] [Google Scholar]
- 43.Symes A., Stahl N., Reeves S. A., Farruggella T., Servidei T., Gearan T., Yancopoulos G., Fink J. S. (1997) Curr. Biol. 7, 697–700 [DOI] [PubMed] [Google Scholar]
- 44.Schaper F., Gendo C., Eck M., Schmitz J., Grimm C., Anhuf D., Kerr I. M., Heinrich P. C. (1998) Biochem. J. 335, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta T. K., Talbot E. S., Scherle P. A., Ivashkiv L. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11107–11112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung J., Uchida E., Grammer T. C., Blenis J. (1997) Mol. Cell. Biol. 17, 6508–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain N., Zhang T., Fong S. L., Lim C. P., Cao X. (1998) Oncogene 17, 3157–3167 [DOI] [PubMed] [Google Scholar]
- 48.Darnell J. E., Jr. (1997) Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 49.Sasse J., Hemmann U., Schwartz C., Schniertshauer U., Heesel B., Landgraf C., Schneider-Mergener J., Heinrich P. C., Horn F. (1997) Mol. Cell. Biol. 17, 4677–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horvath C. M., Wen Z., Darnell J. E., Jr. (1995) Genes Dev. 9, 984–994 [DOI] [PubMed] [Google Scholar]
- 51.Schindler C., Darnell J. E., Jr. (1995) Annu. Rev. Biochem. 64, 621–651 [DOI] [PubMed] [Google Scholar]
- 52.Ihle J. N. (1996) Cell 84, 331–334 [DOI] [PubMed] [Google Scholar]
- 53.O'Shea J. J. (1997) Immunity 7, 1–11 [DOI] [PubMed] [Google Scholar]
- 54.Dang C. V. (1999) Mol. Cell. Biol. 19, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiuchi N., Nakajima K., Ichiba M., Fukada T., Narimatsu M., Mizuno K., Hibi M., Hirano T. (1999) J. Exp. Med. 189, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray R., Miller D. M. (1991) Mol. Cell. Biol. 11, 2154–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sano S., Itami S., Takeda K., Tarutani M., Yamaguchi Y., Miura H., Yoshikawa K., Akira S., Takeda J. (1999) EMBO J. 18, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atherton J. C., Blaser M. J. (2009) J. Clin. Invest. 119, 2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang E. E., Chapeau E., Hagihara K., Feng G. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16064–16069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grossman R. M., Krueger J., Yourish D., Granelli-Piperno A., Murphy D. P., May L. T., Kupper T. S., Sehgal P. B., Gottlieb A. B. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6367–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsumoto K., Hashimoto K., Yoshikawa K., Nakamura T. (1991) Exp. Cell Res. 196, 114–120 [DOI] [PubMed] [Google Scholar]
- 62.Kang J., Blaser M. J. (2006) Nat. Rev. Microbiol. 4, 826–836 [DOI] [PubMed] [Google Scholar]
- 63.Blaser M. J., Kirschner D. (2007) Nature 449, 843–849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.