Abstract
The orientation of the second extracellular loop (ECL2) is divergent in G-protein coupled receptor (GPCR) structures determined. This discovery provoked the question, is the ECL2 conformation differentially regulated in the GPCRs that respond to diffusible ligands? We have determined the conformation of the ECL2 of the angiotensin II type 1 receptor by reporter-cysteine accessibility mapping in different receptor states (i.e. empty, agonist-bound and antagonist-bound). We introduced cysteines at each position of ECL2 of an N-terminal epitope-tagged receptor surrogate lacking all non-essential cysteines and then measured reaction of these with a cysteine-reactive biotin probe. The ability of biotinylated mutant receptors to react with a steptavidin-HRP-conjugated antibody was used as the basis for examining differences in accessibility. Two segments of ECL2 were accessible in the empty receptor, indicating an open conformation of ECL2. These segments were inaccessible in the ligand-bound states of the receptor. Using the accessibility constraint, we performed molecular dynamics simulation to predict ECL2 conformation in different states of the receptor. Analysis suggested that a lid conformation similar to that of ECL2 in rhodopsin was induced upon binding both agonist and antagonist, but exposing different accessible segments delimited by the highly conserved disulfide bond. Our study reveals the ability of ECL2 to interact with diffusing ligands and to adopt a ligand-specific lid conformation, thus, slowing down dissociation of ligands when bound. Distinct conformations induced by the bound agonist and the antagonist around the conserved disulfide bond suggest an important role for this disulfide bond in producing different functional states of the receptor.
Keywords: Cooperativity, Drug Action, G-Protein-coupled Receptors (GPCR), G-Proteins, 7-Helix Receptor, AT1R, Ang II, ECL2, Ligand-induced Conformational Change
Introduction
The G-protein-coupled receptors (GPCRs)2 have taken center stage in the study of cellular adaptation to environment, sensory perception, cell signaling, growth, and differentiation (1–3). Because of the wide ranging role of GPCRs in biology, understanding ligand-induced activation and inhibition mechanisms is critical for drug discovery endeavors. The unifying structural feature of GPCRs (Fig. 1) consists of seven transmembrane (TM) helices connected by short cytoplasmic and extracellular loops (ECL) (1–3). Sequence variability allows this largest and most diverse membrane receptor superfamily to respond to a wide variety of ligands and physical stimuli, including mechanical stretch and light. Small molecule ligands bind within the helical core of the receptor, whereas larger ligands (peptide and proteins) may bind to both TM helices and ECLs or only at ECLs. However, transmembrane signal transduction during the activation of GPCRs adhere to a common molecular mechanism (4). Binding of activating ligands to a GPCR induces rigid body movements of TM helices, which facilitates G-protein activation leading to cytoplasmic signal transduction. The crystallographic structures and accumulating structure-function studies reveal an unanticipated canonical role for ECL2 in ligand-induced activation of GPCRs (3–6).
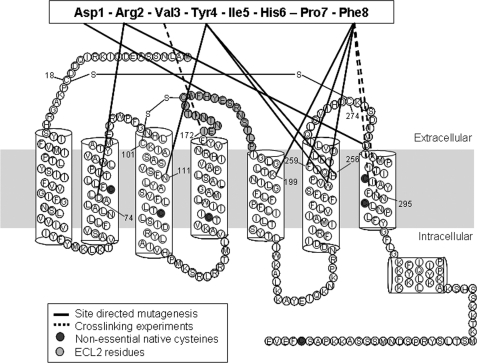
FIGURE 1.
Secondary structure model of a typical GPCR. The rat AT1a receptor shows the interactions of AT1R with Ang II previously mapped by site-directed mutagenesis (solid black lines) and cross-linking experiments (dashed lines) (20, 21, 24, 45, 55–58). ECL2 residues are highlighted in light gray. Native free cysteines are shown in dark filled circles, which were replaced with Ala in the HA-CYS−AT1R.
ECL2 in the prototypical GPCR, bovine rhodopsin, adopts a β-hairpin structure that projects into the site of covalently bound retinal as a stable lid directly interacting with the ligand (7, 8). Upon light activation ECL2 moves away (8, 9). Absence of a similar lid in the recently solved structures of GPCRs suggests that a path for diffusible ligands to the binding pocket may be a specialization that evolved in other GPCRs. The ECL2 in β2- and β1-adrenergic receptors has an α-helical conformation stabilized by an intraloop disulfide bond (10, 11). The ECL2 of A2A adenosine receptor with three disulfide bonds is unstructured (12). Despite these variations, all GPCR structures predict the ECL2 as a part of the ligand-binding pocket. ECL2 makes direct contacts with bound inverse agonist, 11-cis-retinal in rhodopsin (Ser186 to Ile189, Glu181, and Tyr191), the antagonist cyanopindolol in β1-adrenergic receptor (Thr203 and Phe201), the inverse agonist carazolol in β2-adrenergic receptor (Phe193), and the antagonist ZM241385 in A2A-adenosine receptor (Phe168 and Glu169) (7, 8, 10–12).
Positioned at the entrance to the binding cavity, ECL2 may regulate the entree of ligands in rhodopsin-like GPCRs (4, 6). In several human pathologies, autoantibodies directed against ECL2 region directly activate receptor signaling in different GPCRs (13–17). Nonetheless, spatio-temporal changes in ECL2 that occur upon ligand and autoantibody binding remain to be elucidated. In this study, we examine the conformation of ECL2 of the angiotensin II type 1 receptor (AT1R) specifically in empty and agonist- or antagonist-bound states. The AT1R is a 359-amino acid peptide hormone GPCR, which productively couples to Gq/11 and other intracellular signal transduction pathways, including the extracellular signal-regulated kinases (ERK1/2) in cells upon Ang II stimulation (18). Abnormal AT1R function causes hypertension, water-electrolyte imbalance, cardiac hypertrophy and heart failure. AT1R blockers (ARBs) are used to lower blood pressure and to prevent cardiac and vascular hypertrophy (18). Therefore, a better understanding of the mechanism of ligand interactions with AT1R is important to target future drug development efforts.
The predicted structure of ECL2 in AT1R resembles that of bovine rhodopsin (19). In AT1R, the ECL2 region is between residues Ile172 and Pro192, contains the highly conserved disulfide bond between Cys101 and Cys180, which connects ECL2 with TMIII (Fig. 1). As in rhodopsin, the residues located in ECL2 directly interact with the ligands, Ang II (20, 21) and losartan (21, 26). However, AT1R responds to diffusible ligands, similar to adrenergic and adenosine receptors, but lacks intraloop disulfide bond stabilizing ECL2. AT1R-directed autoantibodies bind to ECL2 and activate signaling (15, 17). Thus, ECL2 is a pivotal structural element in generating different conformational states of AT1R, but the role it plays in reversible binding of ligands is unknown. Here we show that ECL2 conformations are different in basal, Ang II-bound, and ARB-bound states by reporter-cysteine accessibility mapping (RCAM).
EXPERIMENTAL PROCEDURES
Cysteine-scanning Mutagenesis
The synthetic rat AT1R gene was cloned in pMT3 vector, N-terminally tagged with hemagglutinin (HA) epitope and used for mutagenesis as described previously (27, 28). HA-CYS−AT1R construct and single cysteine mutants were generated by PCR mutagenesis.
Western Blotting
COS1 cells cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum were transfected using FuGENE6 Transfection Reagent (Roche) according to the manufacturer's instructions. Transfected cells cultured for 48 h were harvested and lysed in Mammalian Protein Extraction Reagent (Pierce) containing protease (Sigma) and phosphatase inhibitors (Thermo Scientific). 50 μg of protein were run on 10% SDS-PAGE and blotted onto nitrocellulose membranes (Bio-Rad). The membrane was probed with mouse anti-HA monoclonal antibody (Roche) and HRP-conjugated anti-mouse IgG (GE Healthcare). The receptor expression was detected using ECL Plus Western blotting detection system (Amersham Biosciences).
Immunocytochemistry
24-h post-transfection, cells were plated onto poly-l-lysine-coated coverslips and serum-starved for 18 h before treatment. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, and blocked with 5% nonfat dry milk. Cells were incubated overnight with mouse anti-HA monoclonal antibody (1:1000), washed with PBS, and incubated with AlexaFluor 568 anti-mouse antibody (1:2000). Cells were then washed and mounted for confocal microscopy analysis.
Saturation Binding Assay
Assays were performed under equilibrium conditions, with [125I]-[Sar1,Ile8]Ang II (Dr. Robert Speth, University of Mississippi) concentrations ranging between 0.125 and 12 nm (specific activity: 2176 Ci/mmol) in total volume of 125 μl for 1 h as previously described (27, 28). Nonspecific binding was measured in the presence of 10−5 m 127I-[Sar1Ile8]Ang II (Bachem). The experiment was stopped by filtering the binding mixture through Whatman GF/C glass fiber filters, which were extensively washed with washing buffer. The bound ligand fraction was determined from the counts per minute remaining on the membrane. The binding kinetics was analyzed by program LigandR, which yields mean ± S.D. for Kd and Bmax values.
Analysis of MAPK Activity
48-h post-transfection, cells were serum-starved for 18 h and treated with 1 μm Ang II (Bachem) for 10 min. Cells were immunoblotted as described above and probed with phospho-ERK 42/44MAPK polyclonal antibody (1:1000) and HRP-conjugated anti-rabbit IgG (1:5000, GE Healthcare). Immunoreactivity was detected using ECL Plus. Same blots were then analyzed for total ERK 42/44MAPK reactivity. Results were used to normalize pERK 42/44MAPK measurements.
Intracellular [Ca2+] Measurement
Changes in cytoplasmic Ca2+ were measured using fluorescent calcium indicator Fura-2 (excitation maximum 340 and 380 nm; emission maximum 510 nm) (Invitrogen) (29). Cells were loaded with 1 μm Fura-2 acetoxymethyl ester at 37 °C following 48 h transfection with HA-AT1R or HA-CYS−AT1R. After 30 min of incubation with Fura-2 in BSS-Ca2+ buffer (140 mm NaCl, 5 mm KCl, 1.2 mm MgCl2, 5.5 mm glucose, 10 mm Hepes, 0.1% bovine serum albumin, and 2 mm CaCl2, pH 7.4), cells were washed to remove the extracellular dye. Calcium measurements were done in single cells using an inverted microscope (Zeiss Axiovert 135) connected to a CCD camera (Photon Technology International). The data were collected at 1.5-s intervals and analyzed using Image Master software (Photon Technology International). The release of intracellular Ca2+ in individual cells was measured after exposure to 1 μm Ang II in BSS by rapid solution exchange. Results are presented as average changes in the ratio of Fura-2 fluorescence upon excitation at 340 nm and 380 nm and plotted using SigmaPlot.
MTSEA-biotin Labeling
MTSEA-biotin (N-biotinylaminoethylmethanethiosulfonate, Toronto Research Chemicals) was dissolved in DMSO, and aliquots of 0.1 m stock solutions were thawed just prior to use. Experiments were repeated at least three times as described below. 48 h post-transfection, cells were harvested using non-enzymatic cell dissociation solution (Sigma), washed, and suspended in PBS. 500-μl aliquots of cells were incubated with 10 mm MTSEA-biotin at 25 °C for 30 min. The cells were diluted with cold PBS buffer, pelleted and resuspended in Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mm Tris (pH 7.4), 137 mm NaCl, 20% glycerol, protease, and phosphatase inhibitors). 750 μg of protein were incubated overnight with 4 μl of mouse anti-HA monoclonal antibody in lysis buffer. The receptors were immunoprecipitated with protein G-agarose beads (Millipore). Beads were washed four times with cold PBS, and proteins were extracted by heating at 60 °C in sample buffer for 10 min. Samples were immunoblotted and probed with Streptavidin-HRP (Amersham Biosciences). Total density of biotinylated mature monomeric receptor band was determined using KODAK 1D 3.6. To verify the receptor expression levels, the same blot was reprobed with anti-HA antibody as described above. The biotinylation levels of receptor proteins were normalized to expression levels. The same procedure was repeated following either 1 μm Ang II (Bachem) or 1 μm losartan (DuPont Merck Co, Wilmington, DE) treatment for 10 min.
Molecular Dynamics (MD) Simulations
MD simulations were performed using the MD software package NAMD (30) and the CHARMM 22. The model structure of AT1R (PDB ID: 2AC6) and AT1R with Ang II (PDB ID: 2AH3) was downloaded from the Protein Data Bank, and explicit hydrogens were added using the package VEGA ZZ 2.0.7 (31). This structure was subjected to 1000 steps of energy minimization. The energy-minimized system was heated from 0 to 310 °C in 31 °C intervals over the course of 20,000 steps. Simulations were carried out for 14 ns. The root-mean-square deviation of the backbone of AT1R with Ang II was stabilized after 7 ns. MD simulations were carried out using the molecular modeling software VEGA ZZ 2.0.7. We simulated ECL2 residues (Ile165-Pro192) with selected loop constraints, and other receptor regions were not simulated. The ECL2 simulations were carried out by imposing Cys101–Cys180 disulfide bond as a constraint in different states. The disulfide bond constraint was relaxed for 1 ns at the end of 14 ns of simulation. We used PyMOL (DeLano Scientific LLC) to visualize the output.
Losartan Docking
Blind docking was carried out using AutoDock4 Vina (32). The starting conformation of losartan was an energy minimized form. The receptor was held rigid during the docking process, while the ligand was allowed to be flexible. The grid box size was 38.0 Å - 20.8 Å - 26.4 Å in the x, y, and z dimensions, with the center of the grid corresponding to the central axis of the pore at Lys199 and His256 at the base of the selectivity filter. Exhaustiveness was set as default. We used PyMOL to visualize the output.
RESULTS
Experimental Setup for Cys-scanning Mutagenesis on ECL2
The MTS (methylthiosulfonate) reagent-insensitive receptor construct, CYS−AT1R was used as the background to substitute reporter Cys residues in the ECL2. In the CYS−AT1R, native non-essential free Cys residues were substituted with Ala to prevent interference with the reporter Cys residues. Native disulfide bonds, Cys18–Cys274 and Cys101–Cys180, which are essential for receptor stability (33, 34) were not mutated (Fig. 1). Thus, the highly conserved (Cys101–Cys180) disulfide bond linking ECL2 with TMIII is preserved in all mutants.
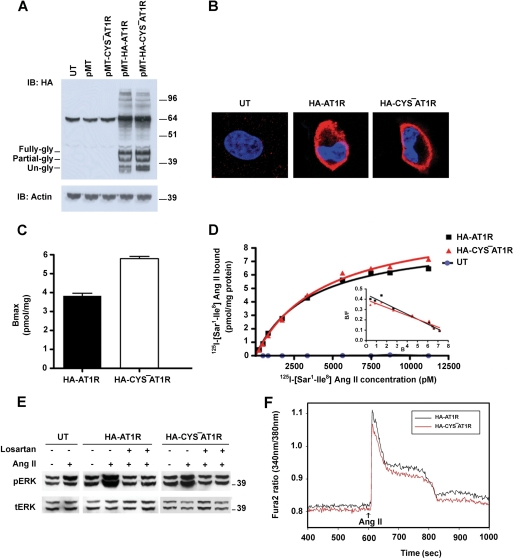
The properties of N-terminal HA-tagged AT1R and CYS−AT1R were similar when expressed in COS1 cells. Both receptors appear to be glycosylated (41.9 kDa). When analyzed by SDS-PAGE, unglycosylated, and glycosylated monomeric forms of the receptors as well as small amounts of higher molecular weight oligomers were present (Fig. 2A). HA-CYS−AT1R is expressed on the plasma membrane (Fig. 2B) with a Bmax 5.8 pmol/mg (Fig. 2C). The Kd value for 125I-[Sar1, Ile8]Ang II obtained by saturation binding analysis performed on intact cells was 4.4 nm (Fig. 2D). Ang II-induced signal transduction capacity measured by ERK1/2 activation suggested that HA-CYS−AT1R is comparable to HA-AT1R. The Ang II-induced ERK1/2 activation was inhibited by AT1R-selective antagonist, losartan (Fig. 2E). Ang II-induced mobilization of intracellular calcium, a measure of G-protein activation, was similar in HA-CYS−AT1R and HA-AT1R (Fig. 2F). Based on these similarities, we used the HA-CYS−AT1R as a template to create the ECL2 mutants used in this study.
FIGURE 2.
Comparison of HA-AT1R and HA-CYS−AT1R. A, Western blot analysis of HA-AT1R (lane 4) and HA-CYS−AT1R (lane 5) expression in COS1 cells. Cells which are untransfected (UT) (lane 1), transfected with pMT3 (lane 2) and CYS−AT1R without HA tag (lane 3) are shown as negative controls. Three differentially glycosylated monomeric bands of the receptor and higher molecular weight oligomeric forms of the receptor can be seen. B, localization of HA-AT1R (panel 2) and HA-CYS−AT1R (panel 3) on the plasma membrane of COS1 cells. Untransfected cells are shown as negative control (panel 1). Cells were labeled with DAPI (blue) for nucleus and with HA primary antibody and Alexa fluor 568 (red) secondary antibody for HA-tagged receptors. C, saturation curves of HA-AT1R and HA-CYS−AT1R determined using [125I]-[Sar1, Ile8]Ang II. The inset shows the corresponding Scatchard plot. D, cell surface density of HA-AT1R and HA-CYS−AT1R expressed as mean Bmax values derived from Scatchard plots. Error bars indicate the S.E., n = 3. E, ERK1/2 phosphorylation upon treatment with 1 μm Ang II on untransfected cells (lanes 1 and 2), HA-AT1R (lanes 3 and 4)-, and HA-CYS−AT1R (lanes 7 and 8)-transfected cells. ERK1/2 phosphorylation is inhibited by losartan treatment prior to Ang II treatment (lanes 5 and 9) and together with Ang II treatment (lanes 6 and 10). Total ERK levels are shown as loading control. F, measurement of intracellular Ca2+ mobilization upon 1 μm Ang II treatment in HA-AT1R- and HA-CYS−AT1R-transfected COS1 cells. The time point of Ang II treatment is indicated by an arrow.
Effect of Substitution of Reporter Cys in ECL2
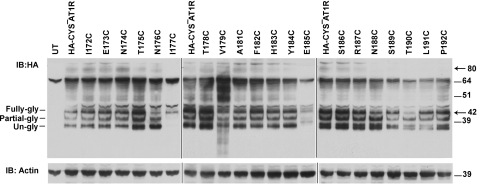
We constructed 20 single Cys substitution mutants in the lle172–Pro192 region in the HA-CYS−AT1R background. DNA sequencing confirmed that each native residue was substituted by a Cys. Each mutant expression plasmid transfected in COS1 cells (which lack endogenous AT1R) yielded ≈42-kDa mature form of the functional receptor protein (Fig. 3). Variable expression seen due to variability in transient transfection was not significant. The expression level of mutant I177C was highly reduced, and the V179C mutant receptor protein appeared to be degraded, indicating instability of these two mutant receptors. Substitution of Ser for Ile177 significantly reduced the expression; whereas Ala substitution was normal, suggesting that a hydrophobic residue is essential at position 177 for normal expression of the receptor (supplemental Fig. S1) Substitution of Ser and Ala for Val179 produced stable receptor protein, suggesting that Cys179 substitution causes instability. Presumably, Cys179 interferes with the Cys101–Cys180 disulfide bond formation. Cell surface receptor expression (Bmax and receptor number) of twenty Cys mutants measured in intact cells is shown in supplemental Table S1. All mutants bound the ligand with high affinity and specificity. The calculated cell surface receptor number shows that there are enough receptors on the cell surface to perform RCAM analysis in mutants with lower Bmax values. Binding of agonist Ang II elicited ERK1/2 activation response in all ECL2 mutants (supplemental Table S1), demonstrating the capacity of mutants to assume active conformation. Ang II-induced ERK1/2 activation capacity displayed by mutants E173C, T175C, E185C, S186C, S189C is reduced, however statistical analysis did not show significance. The antagonist losartan inhibited Ang II-dependent activation of mutants. Taken together, properties of ECL2 mutants suggest that the engineered Cys residues did not cause gross defect, which indicates that substituted Cys residue functions similar to that of the WT residue at each position.
FIGURE 3.
Expression of ECL2 single cysteine mutants. Expression analysis of HA-CYS−AT1R (lane 2) and HA-tagged single cysteine mutants (lanes 3–22) in transiently transfected COS1 cells. Untransfected cells served as negative control (lane 1). Actin expression levels are shown as loading control.
MTSEA-biotin Reaction with Reporter Cys
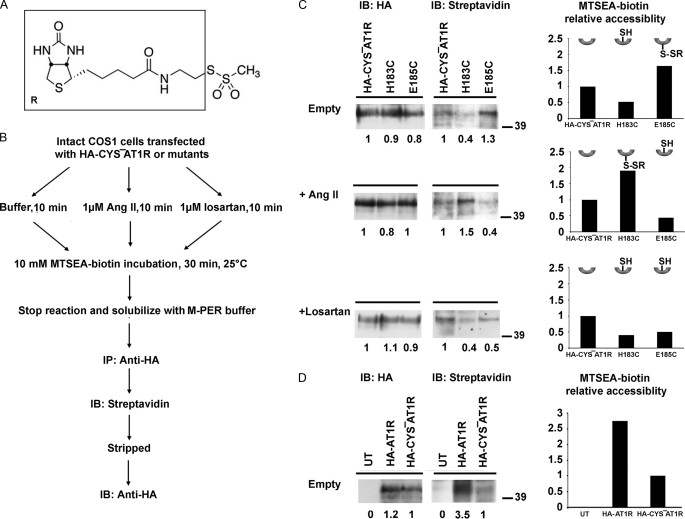
RCAM methodology previously used to determine activation-induced conformational changes in AT1R employed radioligand binding as a read-out (27, 28, 35–37). In this study we used MTSEA-biotin (N-biotinylaminoethylmethanethiosulfonate) (Fig. 4A); a thiol-labeling reagent that does not react with disulfide bonded or buried Cys residues, but rapidly reacts with water-exposed reactive cysteines (38). The resulting site-specific attachment of biotin to engineered Cys was detected by streptavidin-HRP. Fig. 4B charts the experimental strategy. The receptor topology was preserved during MTSEA-biotin reaction as closely as possible to the native state in the plasma membrane by using adherent cells rather than fractionated membranes. Notably, the MTSEA-biotin labeling was compared in different ligand-bound states concurrently (Fig. 4B) for each mutant and the HA-CYS−AT1R control, to minimize confounding factors, which might influence due to different cell surface expression levels. Differential labeling of a particular mutant with and without agonist/antagonist suggests that it is the binding of ligand that influences position/conformation of ECL2 and alters accessibility of the reporter.
FIGURE 4.
MTSEA-biotin accessibility of representative mutants. A, structure of MTSEA-biotin, the part of the molecule that modify with reporter Cys is shown in the box. B, schematic representation of experimental design for measuring MTSEA-biotin accessibility. C, immunoprecipitated receptors were probed for both HA (left) and streptavidin-HRP (right) to detect receptor expression and biotinylation levels, respectively. Please note that the same blot is used for probing with HA and streptavidin-HRP. The blots for representative mutants H183C and E185C are shown under three experimental conditions; in the absence of ligand (upper), in the presence of 1 μm Ang II (middle), and in the presence of 1 μm losartan (lower). The fully glycosylated monomeric receptor band at 41.9 kDa was used for determination of MTSEA-biotin accessibility. HA signal intensity and streptavidin-HRP signal intensity of each sample is compared with the HA-CYS−AT1R in the same gel as indicated by numbers below the bands. The corresponding plots show the MTSEA-biotin relative accessibility, which is the ratio of relative streptavidin-HRP signal to relative HA signal for particular sample. Relative MTSEA-biotin accessibility of each mutant is compared with the HA-CYS−AT1R in the same gel. The inset is the schematic representation of reporter cysteines which point up when inaccessible and point down when accessible, reacted with MTSEA-biotin (shown as -SR). D, MTSEA-biotin accessibility of HA-AT1R and HA-CYS−AT1R.
Immunoprecipitation of detergent-solubilized HA-tagged receptor after MTSEA-biotin treatment of representative samples is shown in Fig. 4C. Fully glycosylated monomeric receptor band was monitored for the mapping studies. As seen in supplemental Table S2, examination of monomeric band yields more consistent results with lower standard error compared with multimeric band. The overall accessibility pattern was same regardless of monomeric or multimeric bands were analyzed. Band intensities observed by streptavidin-HRP blot followed by HA blot estimated the level of biotinylation in each experiment. MTSEA-biotin modification of HA-AT1R is ≈2-fold more than HA-CYS−AT1R (Fig. 4D). This result is anticipated, because Miura and Karnik (27) demonstrated that native Cys76 in TMII of AT1R is MTSEA-sensitive. We used HA-CYS−AT1R as the control and the labeling of all other mutants was quantified in comparison to labeling of HA-CYS−AT1R in the same experiment. Fig. 4C shows changes in the accessibility of Cys reporters replacing His183 and Glu185. The H183C is not accessible in the absence of ligand, but is highly reactive in the Ang II-bound state. In contrast, Ang II binding renders E185C less accessible, although it was highly reactive in the absence of Ang II. In the presence of antagonist losartan, the accessibility of both Cys reporters is reduced. These results provided the first indication that the binding of agonists and antagonists induce distinct changes in the conformation or position of ECL2 in AT1R.
Of note, the significantly less MTSEA-biotin accessibility observed for H183C, which has a free cysteine compared with none in HA-CYS−AT1R (Fig. 4C, top panel) was real. In some other mutants, MTSEA-biotin accessibility lower than that of HA-CYS−AT1R was also observed (Fig. 5A). The basis for lower reactivity is not clear at this time. Theoretically, the MTSEA-biotin reaction for substituted cysteine should be greater than or at least equal to HA-CYS−AT1R. The discrepancy is not correlated with the expression of H183C or other mutants. Because Ang II binding increased the H183C reactivity (Fig. 4C, middle panel), the substituted cysteine at this position was acceptable as a sensor of local conformational change.
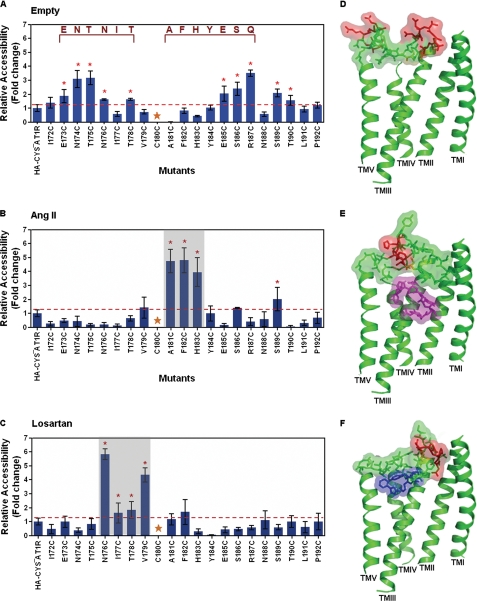
FIGURE 5.
MTSEA-biotin accessibility maps of ECL2 single cysteine mutants. The MTSEA-biotin relative accessibility of mutants are expressed as mean ± S.E., n = 3. The red line shown on the graph designates the significance cutoff (S.E. of HA-CYS−AT1R accessibility, n = 9) that determines the accessibility of mutants. Mutants with significantly higher accessibility compared with HA-CYS−AT1R are indicated with an red asterisk. The conserved Cys180 residue is marked by gold star. A, MTSEA-biotin accessibility map of ECL2 mutants in the absence of ligand. The autoantibody binding epitope sequences are indicated in dark red. B, MTSEA-biotin accessibility map of ECL2 mutants in the presence of AngII. C, MTSEA-biotin accessibility map of ECL2 in the presence of losartan. D, molecular dynamics simulation of ECL2 in the absence of ligand. TMI, TMII, TMIII, TMVI, and TMV are shown in the model. The ECL2 residues and side chains are shown in a stick and surface model. The accessible residues are shown in red. The disulfide-bonded cysteines Cys101 (TMIII) and Cys180 (ECL2) are shaded in yellow. E, molecular dynamics simulation of ECL2 in the presence of Ang II. Ang II is shown as a magenta stick. The accessible residues are shown in red. F, molecular dynamics simulation of ECL2 in the presence of losartan. Losartan is shown as blue stick. The accessible residues are shown in red.
ECL2 Conformation Constrained by the Cys101–Cys180 Disulfide Bond
The accessibility map of Cys reporters in ECL2 without adding any ligand is shown in Fig. 5A. The mean ± S.E. observed for HA-CYS−AT1R in nine independent experiments represents the streptavidin-HRP signal for the receptor that lacks free Cys residues. Significantly higher streptavidin-HRP signal (mean ± S.E.) was observed for reporter Cys residues in two distinct regions of ECL2. The accessible N-terminal (Glu173, Asn174, Thr175, Asn176, and Thr178) and the C-terminal (Glu185, Ser186, Arg187, Ser189, and Thr190) regions were separated by a region of low MTSEA-biotin reactivity. The inaccessible Ile177-Tyr184 segment contains at its center the disulfide bond linking ECL2 to TMIII. In addition, reporter Cys residues at Ile172, Ile177, Asn188, Leu191, and Pro192 were also inaccessible. All of the mutants in which MTSEA-biotin labeling did not increase were expressed on the cell surface (supplemental Table S1) and efficiently immunoprecipitated as shown by the HA blot, suggesting that the absence of signal was due to inaccessibility of the particular Cys reporter, and not due to the lack of cell surface expression. Unlabeled receptors with reduced cell surface expression levels (I177C, P192C) were still expressed at sufficient quantity to yield accessibility signal, as seen upon ligand exposure (see below).
Induced Change in Accessibility of ECL2 upon Ligand Binding
Surprisingly, Ang II treatment prior to MTSEA-biotin reaction significantly reduced the overall accessibility of ECL2 (Fig. 5B). Residues found to be highly reactive in the absence of ligand did not react in the presence of Ang II, which suggests two possibilities: either Ang II masked these residues directly or the ECL2 undergoes an Ang II-dependent conformational change. Decreased accessibility of consecutive residues of ECL2 in two segments rather than individual residues supports the idea that Ang II induces conformational rearrangement of ECL2. These results do not support the notion that ECL2 acts as a reversible gate that opens upon agonist activation, as proposed previously (5). Rather, ECL2 may adopt different conformations in both AngII- and losartan-bound states, which argues against the idea that ECL2 opens to allow uptake or release of ligands. Ang II binding increased the accessibility of Ala181, Phe182, and His183, which are immediately C-terminal to the disulfide-bonded Cys180. Thus Ang II changes local conformation involving the disulfide bond.
Binding of losartan before the MTSEA-biotin labeling (Fig. 5C) also decreased the overall reactivity of ECL2. These accessibility changes are similar to that observed in rhodopsin (7, 9) and dopamine receptor (39). Increase in the reactivity of residues, Asn176-Val179, indicates that losartan binding induced flexibility immediately N-terminal to the disulfide bond. Thus, conformational changes induced near the disulfide bond upon binding the agonist are different from binding the antagonist.
Models shown in Fig. 5, D–F are based on molecular dynamic simulation of ECL2 conformation incorporating the accessibility constraints. In the absence of ligand, ECL2 conformation is open toward the extracellular space (Fig. 5D). The Val179 to Tyr184 region harboring the conserved disulfide bond (Cys101 and Cys180 are shaded in yellow) is deeper and more restricted than the N-terminal and C-terminal regions of the loop (accessible residues are shown in red). The access to ligand-binding pocket is wider near TMV-TMVII. The model also depicts ECL2 forming a tightly packed lid against bound ligands, Ang II (magenta in Fig. 5E) and losartan (blue in Fig. 5F). Varying positions of the disulfide bond and the neighboring accessible residues (red) in Ang II- and losartan-bound conformations of ECL2 indicates flexibility of the region that is normally thought to be constrained because of the disulfide bond.
Hydrogen-bonding Networks Involving Ligand-associated ECL2
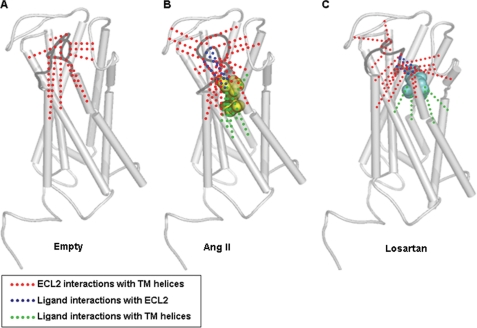
The overall decrease of ECL2-accessibility in the Ang II-bound state indicates that ECL2 conformation changes upon receptor activation. We used inaccessible residues as constraint to perform molecular dynamic simulation to gain insight regarding the nature of ECL2 conformational rearrangement. The hydrogen bonding of the inaccessible residues with a 3.2 Å cutoff are displayed in the empty, Ang II-bound, and losartan-bound states of the AT1R (Fig. 6, supplemental Fig. S3 for more details). In the empty state, ten inaccessible residues are involved in extensive interaction with TM domain and ECL3 (red in Fig. 6A). In the Ang II-bound state, sixteen residues inaccessible to MTSEA-biotin increase intramolecular contacts (red in Fig. 6B, supplemental Fig. 3B) of ECL2 with six TM helices and extracellular segments, N-terminal tail, ECL1 and ECL3. The intramolecular hydrogen-bonding network in the presence of losartan (Fig. 6C, supplemental Fig. 3C) is even more extensive (red) with distinctly different contacts in TM domain.
FIGURE 6.
Predicted hydrogen-bonding network formed by inaccessible residues of ECL2. The ECL2 is highlighted in dark gray. A, intramolecular interactions of ECL2 with the TM helices (red) in the absence of ligand. B, intramolecular interactions of ECL2 with the TM helices (red) in the presence of Ang II. Ang II is shown in yellow spheres. Two intermolecular hydrogen bonding network between Ang II and ECL2 (blue) and between Ang II and TM helices (green) are also shown. C, intramolecular interactions of ECL2 with the TM helices (red) in the presence of losartan. Losartan is shown in cyan spheres. Two intermolecular hydrogen-bonding network between losartan and ECL2 (blue) and between losartan and TM helices (green) are also shown.
Two intermolecular hydrogen-bonding networks, the first (green) between Ang II and TM-helices and the second (blue) between Ang II and ECL2 (Fig. 6B, supplemental Fig. 3B) highlights the major difference between basal and active states of AT1R. The rearrangement of hydrogen bonding network in the inhibited receptor state upon losartan binding is shown in Fig. 6C. The intermolecular hydrogen-bonding network between TM domain and losartan (green) is less extensive than that between losartan and ECL2 (blue) (see supplemental Fig. 3C for details).
Together, molecular dynamic simulation studies predict a flaccid ECL2 conformation in the empty state. In the ligand-bound state extensive hydrogen bonding of ECL2 with the ligand, TM domain, ECL1, ECL3, and N-terminal tail stabilizes a compact lid-like structure. These hydrogen-bonding networks may govern the high affinity of AT1R toward Ang II and losartan. Leaning of Ang II-bound ECL2 toward a set of residues and losartan-bound ECL2 toward a different set appears to contribute respectively to receptor activation and inhibition.
DISCUSSION
Results presented here shed new light on ligand-specific structural dynamics of ECL2 in AT1R. In bovine rhodopsin, the EC-domain forming a compact lid interacts with the inverse agonist form of the chromophore (5–9). A less compact lid conformation is maintained in opsin with an aqueous channel opening near the extracellular ends of TMV-TMVII and ECL3 (8). Similar lid is not found in subsequently solved GPCR structures. Although, tethered to TMIII the ECL2 in these GPCRs is stabilized outside the ligand pocket by additional disulfide bonds (10–12). This observation led to the speculation that open ECL2 conformation plays a specific role, namely capturing diffusible ligands and provide an aqueous channel to facilitate the ligand entry and exit from the binding pocket in milliseconds (5, 10–12). Whether ligands modulate conformation of ECL2 is not known. In many peptide hormone receptors including AT1R, ECL2 directly interacts with agonists and is also target of autoantibodies. Hence, the propensity of ECL2 in AT1R to assume ligand-specific conformation described here is important. The ECL2 sequence of AT1R and rhodopsin share 11 of 21 residues (supplemental Fig. S4), and the predicted structure of ECL2 is ideally positioned to form multiple interactions with the bound ligand and other segments of the receptor (19).
Autoantibody epitope mapping studies in human AT1R independently support the open conformation of the ECL2 in the empty receptor (15–17). Circulating autoantibodies recognize the readily accessible Ile172–Thr178 and Glu185–Thr190 segments. For instance, agonistic autoantibodies from preeclampsia patients bind to the epitope AFHYESQ (17), and from patients with malignant hypertension and refractory, vascular allograft rejection bind to epitopes ENTNIT and AFHYESQ in ECL2 (15, 16). It is interesting to note that the antigenic epitopes (dark red in Fig. 5A) are interrupted by the Val179–Tyr184 region adjoining the disulfide bond that was found inaccessible by RCAM. Autoantibody binding to ECL2 epitopes may exert strain on the disulfide bond to activate the AT1R directly or may open the entrance channel for circulating Ang II. Losartan binding, which masks epitopes, is known to protect patients harboring autoantibodies. In many other GPCRs, the autoantibodies directed toward ECL2 may also modulate conformation of the EC-domain (13, 14). In a more general sense, conformational dynamics of ECL2 may regulate the fundamental process of autoantibody-mediated GPCR activation and thus initiate the pathophysiological process.
At least in some GPCRs, the proposed function of the ECL2 is to regulate inactive receptor conformation (6, 40). Mutagenesis directed to ECL2 in our study, as well as in a previous study (50), did not find activating mutations. ECL2 possibly serves an essential function in protein folding and maintaining the folded state of the AT1R as indicated by I177C and V179C mutations. Reducing agents that break the Cys101–Cys180 disulfide linkage with TMIII do inactivate folded AT1R (33, 34). We speculate that the inaccessible residues of ECL2 may actually regulate low basal activity of ligand free receptor. Out of ten inaccessible residues, seven are similar to the residues in Ang II (supplemental Fig. S5), which can form an Ang II-analogous pharmacophore hidden within ECL2. Whether the predicted Ang II-analogous pharmacophore in ECL2 works like an extremely weak agonist or antagonist will need further study. When primary sequence is scrambled in Ang II, an inhibitory pharmacophore is generated as shown earlier (41–44).
The open conformation of ECL2 in AT1R may facilitate capturing the ligands. Photoaffinity cross-linking of Val3 and Phe8 side chains in Ang II with Ile172 in ECL2 and Phe293/Asn294 in TMVII, supports the ECL2 folding hypothesis (45). Site-directed mutagenesis studies further support the idea because Val3 and Asp1 side chains in Ang II specifically interact with Ile172 and His183 in ECL2 (20, 21) and remaining Ang II residues directly interact with TM helices (Fig. 1). Molecular model of AT1R predicted that ligands bind in the TM domain ≈25 Å deeper from the membrane border (19). The model suggests that the proximal part of ECL2 dips into the binding pocket reaching the bound Ang II (19). Direct interaction of Ang II possibly induces folding of ECL2 to a lid conformation (Fig. 5E), which is different when compared with the movement of ECL2 away from the activating ligand, all-trans-retinal in light-activated rhodopsin (9). In other GPCRs the conformation of ECL2 in the presence of agonist is not yet known. Thus, the ligand-induced folding of ECL2 requires considerable rearrangement in the hydrogen bonding networks (Fig. 6 and supplemental Fig. S3) in AT1R. Similar conformational changes are likely in other GPCRs, where a direct role of ECL2 in ligand specificity is documented (46–48).
A rhodopsin-like lid formed by ECL2 in the losartan-bound state is quite similar to the proposed arrangement of ECL2 in the antagonist-bound state of D2 dopamine receptor (39). Mutagenesis, photoaffinity cross-linking, and modeling studies (21–26, 49) show that losartan competitively occupies the Ang II binding site. Molecular modeling studies show that the only possibility for both ECL2 and TM domain to interact with losartan would be through folding of ECL2 as shown in Fig. 5F, forming a lid over bound losartan.
Ligand-specific folding of ECL2 appears to induce specific perturbation around the disulfide bond, which may play a critical role in the inhibition and activation of the receptor. The N-terminal residues in Ang II interact with ECL2 and the C terminus with the receptor's inter-helical crevice (Fig. 1), leading Ang II to fold into a C shape, a conformation predicted in many studies (41, 43, 51, 52). We propose that reorganization of the hydrogen-bond network with residues on ECL2 is then coupled to movements of TM helices through the Cys101-Cys180 bond. The ECL2 length (27 ± 13) is conserved in the GPCR family (4), but the frequency of activating mutations targeting this region is lower than TM helices. What is the significance of ligand-specific conformation of ECL2 in the receptor activation process? One possibility is that in the ligand bound state ECL2 interacts with multiple TM helices, which enables ECL2 to integrate conformational cues originating at multiple contacts between ligand and the receptor. The disulfide bond in ECL2 conserved in >90% of GPCRs may play a general role in efficiently coupling ligand binding to GPCR activation or inhibition. In all five main human GPCR families (rhodopsin, glutamate, secretin, adhesion, and frizzled/taste2) mutations targeting ECLs impair ligand-dependent activation, also suggests a conserved function of the EC-domain throughout the GPCR superfamily (4). Thus, the dynamic conformational changes in the ECL2 may be critical for the activation mechanism of the receptor.
Current ideas about how an EC-lid may limit the access of ligands to cognate GPCRs are based on extrapolation from elegant dynamic and structural studies on bovine rhodopsin (7–9). Our analysis allows us to reach a different conclusion, that a lid conformation is induced in AT1R upon binding of both agonists and antagonists. Transient formation of an EC-lid may be very important in other GPCRs for blocking rapid exit of the ligand from the pocket. Their ligands bind to the pocket rapidly and leave the binding pocket at a much slower rate. For example, the dissociation rate (k−a) is five times larger than the association rate (ka) of both Ang II and losartan for AT1R (53, 54). A transiently formed lid can prevent the ligands from leaving the pocket rapidly, thus sustaining the ligand-specific conformation longer. In the agonist-bound state, this would allow the receptor to amplify the signal by prolonged G-protein activation. Unlike rhodopsin, the activation in most other GPCRs may require calling to order different dynamic parts of the receptor to achieve a sharp transition to active state. In this way, the lid may help to ensure that the strain energy gain by agonist and/or antagonist binding is efficiently transformed into helix rearrangement. In other words, the lid allows the ligand to flip the GPCR switch more easily and perhaps faster, as suggested by Bourne and Meng (5).
Supplementary Material
Acknowledgments
We thank J. Boros and S. Miura for constructing the CYS−AT1R, R. Desnoyer, A. Bhatnagar, A. Shukla, S. Mazzei, J Drazba for experimental assistance, and S. V. Naga Prasad for fruitful discussions and advice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL57470 (to S. S. K.). This work was also supported by an assistantship from the Department of BGES, Cleveland State University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- GPCR
- G-protein-coupled receptor
- ECL2
- extracellular loop 2
- RCAM
- reporter cysteine accessibility mapping
- AngII
- angiotensin II
- AT1R
- angiotensin II type 1 receptor
- MTSEA-biotin
- N-biotinylaminoethyl methanethiosulfonate
- HRP
- horseradish peroxidase
- TM
- transmembrane
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- MD
- molecular dynamics
- MAPK
- mitogen-activated protein kinase.
REFERENCES
- 1.Ji T. H., Grossmann M., Ji I. (1998) J. Biol. Chem. 273, 17299–17302 [DOI] [PubMed] [Google Scholar]
- 2.Lagerström M. C., Schiöth H. B. (2008) Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum D. M., Rasmussen S. G., Kobilka B. K. (2009) Nature 459, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karnik S. S., Gogonea C., Patil S., Saad Y., Takezako T. (2003) Trends Endocrinol. Metab. 14, 431–437 [DOI] [PubMed] [Google Scholar]
- 5.Bourne H. R., Meng E. C. (2000) Science 289, 733–734 [DOI] [PubMed] [Google Scholar]
- 6.Massotte D., Kieffer B. L. (2005) Nat. Struct. Mol. Biol. 12, 287–288 [DOI] [PubMed] [Google Scholar]
- 7.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 8.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 9.Ahuja S., Hornak V., Yan E. C., Syrett N., Goncalves J. A., Hirshfeld A., Ziliox M., Sakmar T. P., Sheves M., Reeves P. J., Smith S. O., Eilers M. (2009) Nat. Struct. Mol. Biol. 16, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G. W., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebesgue D., Wallukat G., Mijares A., Granier C., Argibay J., Hoebeke J. (1998) Eur. J. Pharmacol. 348, 123–133 [DOI] [PubMed] [Google Scholar]
- 14.Wang W., Guo G., Tang J., Li J., Zhao R., Hjalmarson A., Fu L. X. (2000) Chin. Med. J. 113, 867–871 [PubMed] [Google Scholar]
- 15.Liao Y. H., Wei Y. M., Wang M., Wang Z. H., Yuan H. T., Cheng L. X. (2002) Hypertens. Res. 25, 641–646 [DOI] [PubMed] [Google Scholar]
- 16.Dragun D., Müller D. N., Bräsen J. H., Fritsche L., Nieminen-Kelhä M., Dechend R., Kintscher U., Rudolph B., Hoebeke J., Eckert D., Mazak I., Plehm R., Schönemann C., Unger T., Budde K., Neumayer H. H., Luft F. C., Wallukat G. (2005) N. Engl. J. Med. 352, 558–569 [DOI] [PubMed] [Google Scholar]
- 17.Zhou C. C., Zhang Y., Irani R. A., Zhang H., Mi T., Popek E. J., Hicks M. J., Ramin S. M., Kellems R. E., Xia Y. (2008) Nat. Med. 14, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunyady L., Catt K. J. (2006) Mol. Endocrinol. 20, 953–970 [DOI] [PubMed] [Google Scholar]
- 19.Gogonea C., Karnik S. S. (2006) J. Mol. Mod. 12, 1610–2940 [Google Scholar]
- 20.Feng Y. H., Noda K., Saad Y., Liu X. P., Husain A., Karnik S. S. (1995) J. Biol. Chem. 270, 12846–12850 [DOI] [PubMed] [Google Scholar]
- 21.Boucard A. A., Wilkes B. C., Laporte S. A., Escher E., Guillemette G., Leduc R. (2000) Biochemistry 39, 9662–9670 [DOI] [PubMed] [Google Scholar]
- 22.Yamano Y., Ohyama K., Chaki S., Guo D. F., Inagami T. (1992) Biochem. Biophys. Res. Commun. 187, 1426–1431 [DOI] [PubMed] [Google Scholar]
- 23.Noda K., Saad Y., Kinoshita A., Boyle T. P., Graham R. M., Husain A., Karnik S. S. (1995) J. Biol. Chem. 270, 2284–2289 [DOI] [PubMed] [Google Scholar]
- 24.Noda K., Saad Y., Karnik S. S. (1995) J. Biol. Chem. 270, 28511–28514 [DOI] [PubMed] [Google Scholar]
- 25.Schambye H. T., Hjorth S. A., Bergsma D. J., Sathe G., Schwartz T. W. (1994) Proc. Natl. Acad. Sci. 91, 7046–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji H., Leung M., Zhang Y., Catt K. J., Sandberg K. (1994) J. Biol. Chem. 269, 16533–16536 [PubMed] [Google Scholar]
- 27.Miura S. I., Karnik S. S. (2002) J. Biol. Chem. 27, 24299–24305 [DOI] [PubMed] [Google Scholar]
- 28.Miura S., Zhang J., Boros J., Karnik S. S. (2003) J. Biol. Chem. 278, 3720–3725 [DOI] [PubMed] [Google Scholar]
- 29.Lee C., Hwang S. A., Jang S. H., Chung H. S., Bhat M. B., Karnik S. S. (2007) FEBS Lett. 581, 2517–2522 [DOI] [PubMed] [Google Scholar]
- 30.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedretti A., Villa L., Vistoli G. (2004) J. Comput. Aided Mol. Des. 18, 167–173 [DOI] [PubMed] [Google Scholar]
- 32.Trott O., Olson A. J. (2010) J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohyama K., Yamano Y., Sano T., Nakagomi Y., Hamakubo T., Morishima I., Inagami T. (1995) Regul. Pept. 57, 141–147 [DOI] [PubMed] [Google Scholar]
- 34.Feng Y. H., Saad Y., Karnik S. S. (2000) FEBS Lett. 484, 133–138 [DOI] [PubMed] [Google Scholar]
- 35.Boucard A. A., Roy M., Beaulieu M. E., Lavigne P., Escher E., Guillemette G., Leduc R. (2003) J. Biol. Chem. 278, 36628–36636 [DOI] [PubMed] [Google Scholar]
- 36.Martin S. S., Holleran B. J., Escher E., Guillemette G., Leduc R. (2007) Mol. Pharmacol. 72, 182–190 [DOI] [PubMed] [Google Scholar]
- 37.Yasuda N., Miura S., Akazawa H., Tanaka T., Qin Y., Kiya Y., Imaizumi S., Fujino M., Ito K., Zou Y., Fukuhara S., Kunimoto S., Fukuzaki K., Sato T., Ge J., Mochizuki N., Nakaya H., Saku K., Komuro I. (2008) EMBO Rep. 9, 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akabas M. H., Stauffer D. A., Xu M., Karlin A. (1992) Science 258, 307–310 [DOI] [PubMed] [Google Scholar]
- 39.Shi L., Javitch J. A. (2004) Proc. Natl. Acad. Sci. 101, 440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klco J. M., Wiegand C. B., Narzinski K., Baranski T. J. (2005) Nat. Struct. Mol. Biol. 12, 320–326 [DOI] [PubMed] [Google Scholar]
- 41.Marshall G. R., Bosshard H. E., Vine W. H., Glickson J. D., Needleman P. (1974) in Recent Advances in Renal Physiology and Pharmacology (Wesson L. G., Fanelli G. M. eds) pp. 215–256, University Park Press, Baltimore [Google Scholar]
- 42.Bumpus F. M., Khosla M. C. (1977) in Hypertension: Physiology and Treatment (Genest J., Koiw E., Kuchel O. eds) pp. 183–201, McGraw-Hill, New York [Google Scholar]
- 43.Samanen J., Regoli D. (1994) in Angiotensin II Receptors: Medicinal Chemistry (Rufolo R. ed) pp. 11–97, Vol. 2, CRC Press, Boca Raton, FL [Google Scholar]
- 44.Miura S., Feng Y. H., Husein A., Karnik S. S. (1999) J. Biol. Chem. 274, 7103–7110 [DOI] [PubMed] [Google Scholar]
- 45.Pérodin J., Deraët M., Auger-Messier M., Boucard A. A., Rihakova L., Beaulieu M. E., Lavigne P., Parent J. L., Guillemette G., Leduc R., Escher E. (2002) Biochemistry 41, 14348–14356 [DOI] [PubMed] [Google Scholar]
- 46.Zhao M. M., Hwa J., Perez D. M. (1996) Mol. Pharm. 50, 1118–1126 [PubMed] [Google Scholar]
- 47.Duprez L., Parma J., Costagliola S., Hermans J., Van Sande J., Dumont J. E., Vassart G. (1997) FEBS Lett. 409, 469–474 [DOI] [PubMed] [Google Scholar]
- 48.Banères J. L., Mesnier D., Martin A., Joubert L., Dumuis A., Bockaert J. (2005) J. Biol. Chem. 280, 20253–20260 [DOI] [PubMed] [Google Scholar]
- 49.Nouet S., Dodey P. R., Bondoux M. R., Pruneau D., Luccarini J. M., Groblewski T., Larguier R., Lombard C., Marie J., Renaut P. P., Leclerc G., Bonnafous J. C. (1999) J. Med. Chem. 42, 4572–4583 [DOI] [PubMed] [Google Scholar]
- 50.Parnot C., Bardin S., Miserey-Lenkei S., Guedin D., Corvol P., Clauser E. (2000) Proc. Natl. Acad. Sci. 97, 7615–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia K. C., Ronco P. M., Verroust P. J., Brünger A. T., Amzel L. M. (1992) Science 257, 502–507 [DOI] [PubMed] [Google Scholar]
- 52.Nikiforovich G. V., Kao J. L., Plucinska K., Zhang W. J., Marshall G. R. (1994) Biochemistry 33, 3591–3598 [DOI] [PubMed] [Google Scholar]
- 53.Timmermans P. B., Wong P. C., Chiu A. T., Herblin W. F., Benfield P., Carini D. J., Lee R. J., Wexler R. R., Saye J. A., Smith R. D. (1993) Pharmacol. Rev. 45, 205–251 [PubMed] [Google Scholar]
- 54.Ojima M., Inada Y., Shibouta Y., Wada T., Sanada T., Kubo K., Nishikawa K. (1997) Eur. J. Pharmacol. 319, 137–146 [DOI] [PubMed] [Google Scholar]
- 55.Yamano Y., Ohyama K., Kikyo M., Sano T., Nakagomi Y., Inoue Y., Nakamura N., Morishima I., Guo D. F., Hamakubo T. (1995) J. Biol. Chem. 270, 14024–14030 [DOI] [PubMed] [Google Scholar]
- 56.Feng Y. H., Miura S., Husain A., Karnik S. S. (1998) Biochemistry 37, 15791–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laporte S. A., Boucard A. A., Servant G., Guillemette G., Leduc R., Escher E. (1999) Mol. Endocrinol. 13, 578–586 [DOI] [PubMed] [Google Scholar]
- 58.Clément M., Martin S. S., Beaulieu M. E., Chamberland C., Lavigne P., Leduc R., Guillemette G., Escher E. (2005) J. Biol. Chem. 280, 27121–27129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.