Abstract
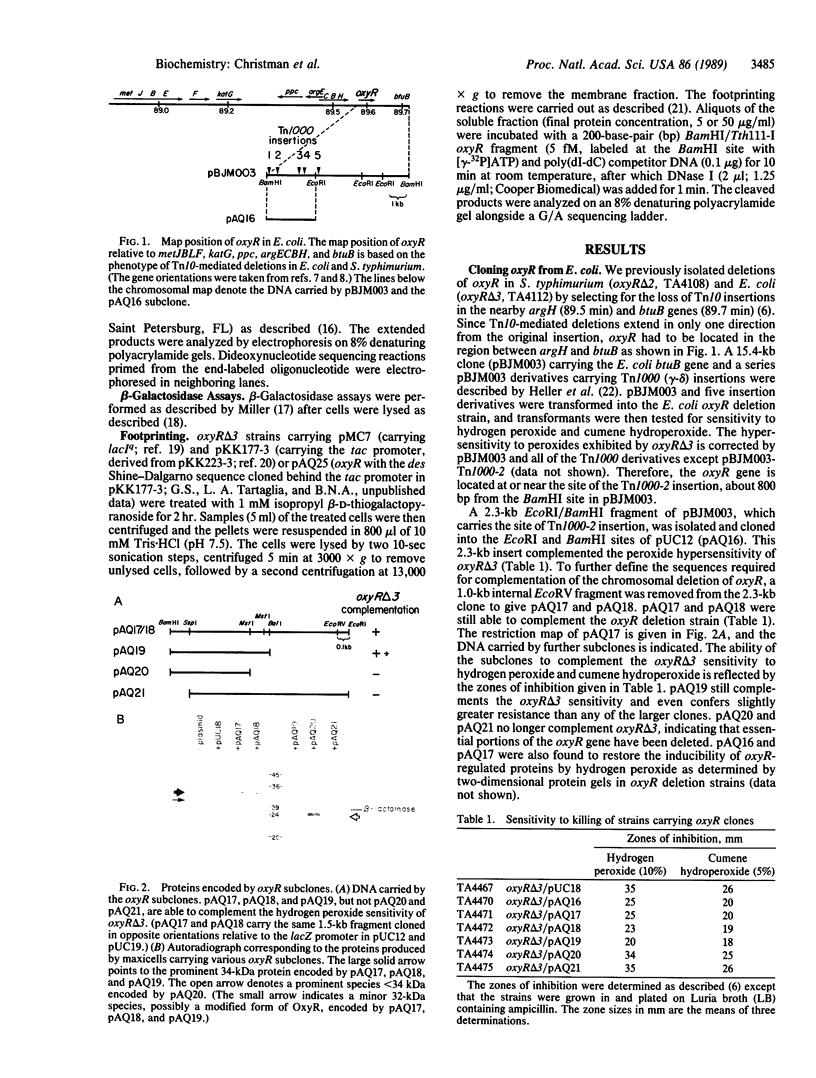
The oxyR gene is required for the induction of a regulon of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium. The E. coli oxyR gene has been cloned and sequenced, revealing an open reading frame (305 amino acids) that encodes a 34.4-kDa protein, which is produced in maxicells carrying the oxyR clone. The OxyR protein shows homology to a family of positive regulatory proteins including LysR in E. coli and NodD in Rhizobium. Like them, oxyR appears to be negatively autoregulated: an oxyR::lacZ gene fusion produced 5-fold higher levels of beta-galactosidase activity in oxyR null mutants compared to oxyR+ controls, and extracts from an OxyR-overproducing strain were able to protect regions (-27 to +21) of the oxyR promoter from DNase I digestion. DNA sequence analysis of the oxyR2 mutation, which causes overexpression of oxyR-regulated proteins in the absence of oxidative stress, showed that the oxyR2 phenotype is due to a missense mutation (C.G to T.A transition) that changes alanine to valine at amino acid position 234 of OxyR.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P. DNA sequence for a low-level promoter of the lac repressor gene and an 'up' promoter mutation. Nature. 1978 Aug 24;274(5673):762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985 Jul;41(3):753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J. Homeo boxes in the study of development. Science. 1987 Jun 5;236(4806):1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Heller K., Mann B. J., Kadner R. J. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985 Mar;161(3):896–903. doi: 10.1128/jb.161.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N., Nicolas M. H., Cole S. T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986 Dec 20;5(13):3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Gojobori T., Aota S., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1986;14 (Suppl):r151–r197. doi: 10.1093/nar/14.suppl.r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Morgan R. W., Christman M. F., Jacobson F. S., Storz G., Ames B. N. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski J., Jagura-Burdzy G., Kredich N. M. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1987 May 5;262(13):5999–6005. [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plamann L. S., Stauffer G. V. Nucleotide sequence of the Salmonella typhimurium metR gene and the metR-metE control region. J Bacteriol. 1987 Sep;169(9):3932–3937. doi: 10.1128/jb.169.9.3932-3937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam S. L., Koch A. L. Complications in the simplest cellular enzyme assay: lysis of Escherichia coli for the assay of beta-galactosidase. Anal Biochem. 1975 Feb;63(2):350–360. doi: 10.1016/0003-2697(75)90357-7. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Stragier P., Richaud F., Borne F., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J Mol Biol. 1983 Aug 5;168(2):307–320. doi: 10.1016/s0022-2836(83)80020-5. [DOI] [PubMed] [Google Scholar]
- Triggs-Raine B. L., Doble B. W., Mulvey M. R., Sorby P. A., Loewen P. C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988 Sep;170(9):4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986 Feb 15;261(5):2441–2450. [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]