Abstract
We have developed a novel real-time PCR assay to identify and perform preliminary genotyping of mycobacteria in a manner tailored to our local service. Within a single thermocycler run, mycobacterial 16S rDNA and the Mycobacterium tuberculosis global lineage-defining RD750 polymorphism are targeted in separate reaction tubes, each of which includes both TaqMan and SYBR Green chemistries. The results of this 16S-RD assay differentiate M. tuberculosis complex (MTBC) from nontuberculous mycobacteria (NTM) and recognize whether or not MTBC isolates belong to the East African-Indian lineage, the single most frequently isolated global MTBC lineage in our service. If required, NTM amplicons may be sequenced to provide more specific identities. We report the technical performance of this assay on 88 mycobacteria-positive cultures and discuss its use in the initial management of mycobacterial infections. The 16S-RD assay correctly identified all 70 MTBC-positive cultures and 17 NTM-positive cultures while contemporaneously recognizing 26 MTBC isolates as within and 44 outside the East African-Indian lineage. In artificial samples, the combined assay also showed limited potential to detect mixed mycobacterial infections (MTBC/NTM) and tuberculosis infections involving more than one global MTBC lineage. The approach we have established can be readily tailored to targets of particular value for any mycobacterial diagnostic service, thereby optimizing the value of the results for local clinical and public health management of mycobacterial infections.
When a possible case of mycobacterial infection is detected, both the clinical and public health management depend critically on whether the causative agent is Mycobacterium tuberculosis or a nontuberculous mycobacterium (NTM). In the former case, treatment and prevention of further person-to-person transmission are urgent concerns while, in the latter, infections are predominantly sporadic, derive from environmental sources of infection, and often reflect pre-existing conditions in the patient.1,2,3 Although there are several promising direct molecular assays for M. tuberculosis,4,5,6,7,8,9 culture is the most sensitive means to detect specific mycobacterial infections and remains the mainstay of diagnostic services in well-resourced laboratories. However, when cultures are detected positive and the presence of mycobacteria confirmed by acid fast staining, it is not known whether the isolated mycobacteria belong to the Mycobacterium tuberculosis complex (MTBC) or the NTM group. Furthermore, when the possibility of recent transmission of tuberculosis (TB) arises, no information is available regarding the relatedness of the isolate to other contemporary isolates with potential epidemiological connections to the new case.
Cultures positive for mycobacteria are referred for further testing to address these points and, in the UK, this generally involves transmission to a reference laboratory where the caseload enables economies of scale in deploying established molecular identification and MTBC genotyping procedures. Thus, definitive recognition of infections as MTBC or NTM and the detection of potential epidemiological links may be delayed by days in the former case and weeks in the latter. In our service, where TB and NTM infections affect between 200 and 400 patients per annum, these delays may affect the management of a significant number of patients. Until identity and genotyping information is available, a precautionary response to a particular case in which mycobacteria have been isolated may lead to inappropriate isolation and treatment of patients for TB and unnecessary public health investigations.
To address these issues, we have developed a real-time PCR assay for analysis of mycobacteria-positive cultures. The assay targets the mycobacterial 16S rDNA and the global lineage-defining RD750 polymorphism in separate reactions within a single PCR run; it also combines TaqMan and SYBR Green technologies to simultaneously differentiate two products in each reaction tube. The 16S assay distinguishes between MTBC and NTM by combining newly designed universal mycobacterial 16S primers and a MTBC-specific 16S-targeted TaqMan probe. The RD assay enables differentiation of MTBC strains with intact (RD750+) and deleted RD750 (RD750−) regions by use of hemi-nested primers and an RD750+-specific TaqMan probe. SYBR Green chemistry is used in both assays to obtain a total PCR amplicon readout. The principles of the combined 16S-RD assay are illustrated in Figures 1 and 2.
Figure 1.
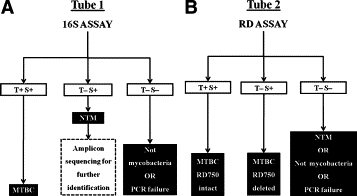
Schematic illustration of the combined 16S-RD assay. A: For the 16S assay (tube 1), TaqMan (T) signals show the presence or absence of MTBC-specific 16S rDNA sequence, whereas the SYBR Green (S) signals indicate the presence or absence of Mycobacterium genus-specific 16S rDNA sequence. B: For the RD assay (tube 2), T signals show the presence or absence of RD750, whereas the S signals act as further confirmation of MTBC-specific sequence and a positive assay control when the T signal is negative. T+ S− signals would be anomalous in both assays, indicating amplicons that do not interact with SYBR Green; no results were obtained in this category.
Figure 2.
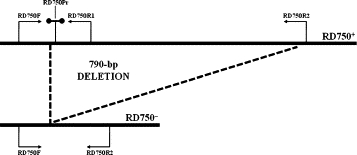
Diagrammatic representation of the RD assay targets.
RD750 was first described as a deletion in Rv1519 present in M. tuberculosis strain CH, a local isolate responsible for a large school-associated TB outbreak in Leicester in 2001.10 This large sequence polymorphism was subsequently designated RD750 and recognized as one of six phylogeographical lineage-defining deletions in a global collection of M. tuberculosis strains.11 It defines the East African-Indian (EA-I) lineage,11 the differentiation of which is also supported by genomic and multilocus sequencing data11,12 as well as spoligotyping. By the latter method, it is also designated as the Central Asian lineage.13 Intriguingly, the RD750 deletion has been associated with an immunosubversive phenotype14; it also occurs in approximately half of our local M. tuberculosis isolates (J. Malkin and H. Perera, unpublished data). The predominance of this lineage among Leicester M. tuberculosis isolates likely reflects our diverse ethnic population. In the 2001 census, 31% of Leicester city was reported to have an Asian ethnic background with the majority being Indian from either East Africa or Gujarat.15 In accordance with previous studies in the UK, our regional genotyping service reports a preponderance of the five-locus variable-number tandem repeat (VNTR) profile x2234/5 in this ethnic group.16 In this context, it will be appreciated that, in our service, early recognition of whether new M. tuberculosis isolates belong to the EA-I group through a single rapid assay offers the highest probability of obtaining strain information that might be discriminatory in a local context. For example, where two potentially linked cases yield RD750+and RD750− isolates, transmission can be excluded. Also, in the longer term, specific identification of M. tuberculosis lineages will facilitate detection of associations between clinical phenotypes and bacterial genotypes.17,18,19 This approach can be adapted to target any lineage-defining polymorphisms that are appropriate to a particular local service.
In the present study, we report evaluation of the 16S-RD assay on positive cultures. A by-product of the assay design (Figure 1A and 1B) is that is has the potential to detect mixed infections (MTBC + NTM or RD750+ + RD750−) in clinical specimens, a phenomenon previously recognized by molecular analysis.20 We therefore also report on the performance of our combined assay to detect mixed mycobacterial infections.
Materials and Methods
Bacterial Strains
Strains and DNA used in this study included clinical isolates from the Department of Clinical Microbiology, University Hospitals of Leicester (UHL), departmental stocks (DS), and research materials from Colorado State University (CSU). The following were used: RD750+ (CDC 155121 from CSU) and RD750− (CH10 from DS) strains of M. tuberculosis, M. smegmatis MC2155 (DS), Corynebacterium glutamicum ATCC 13032 (DS), Streptomyces fradiae T59235 (DS), Rhodococcus spp. (DS), Streptococcus pneumoniae D39 (DS), Staphylococcus aureus Newman (DS), Acinetobacter baumannii AYE (DS), Pseudomonas aeruginosa PA14 (DS), Escherichia coli K-12 (DS) Haemophilus influenzae (UHL), and Moraxella catarrhalis (UHL).
DNA Isolation
Mycobacterial growth indicator tube (MGIT) cultures were centrifuged at 755 × g for 22 minutes, and the resulting pellets were resuspended in 0.2 ml of sterile water. Colonies were scrapped off Löwenstein-Jensen (LJ) slopes and resuspended in 0.2 ml of sterile water. Culture suspensions from MGIT and LJ were boiled at 100°C for 30 minutes, centrifuged at 16,100 × g for 2 minutes, and the resulting supernatants used for PCR analyses.
Oligonucleotide Primer and Probe Design
The oligonucleotide primers and TaqMan probes used are listed in Table 1. Full-length 16S ribosomal DNA sequences representing mycobacteria isolated from humans were retrieved from The Institute for Genomic Research (TIGR) or the Ribosomal Database Project II.22 The sequences were aligned using the ClustalX program with default settings, and alignments were processed with a custom built algorithm. MYCO16SF and MYCO16SR primers were designed targeting 16S rDNA region conserved in all of the retrieved mycobacterial sequences. A BLASTN search with these primers reveals that several other actinobacteria may give amplicons with these primers. However, with the possible exception of Nocardia spp., none of these are acid fast. The MTBC-specific 16S TaqMan probe (MYCO16SPr) was that previously reported but with a different fluorophore-quencher pair to allow combination with SYBR Green.23 The three RD750 primers were designed using Primer3 to give products of 126 and 193 bp with M. tuberculosis strains H37Rv and CH, respectively (J. Malkin, unpublished data). RD750 TaqMan probe (RD750Pr) was designed and synthesized by TIB MOLBIOL (Berlin, Germany). A BLASTN search demonstrated that none of the primers and TaqMan probes for both the 16S and RD assays shared significant similarity with other known nucleotide sequences or each other. Rv1519F and Rv1519R primers were from our previous study.10 All primers and MYCO16SPr were synthesized by MWG Biotech (Ebersberg, Germany).
Table 1.
Oligonucleotide Primers and TaqMan Probes for the Combined 16S-RD Assay
| PCR assay | Primer/Probe designation | Nucleotide sequence |
|---|---|---|
| 16S | MYCO16SF | 5′-GAAACTGGGTCTAATACCG-3′ |
| MYCO16SR | 5′-ATCTCAGTCCCAGTGTGG-3′ | |
| MYCO16SPr | 5′-ROX-TCCACCACAAGACATGCATCCCGTG-BHQ2-3′ | |
| RD | RD750F | 5′-CTTAAGCGTCCGCGTATCC-3′ |
| RD750R1 | 5′-GCCACAGCTGTACAGGTCAA-3′ | |
| RD750R2 | 5′-AACTTTCGGCGGTCAGTGTA-3′ | |
| RD750Pr | 5′-Cy5-CCGTTGGCGCAGAGCACTCC-BBQ-3′ | |
| Rv1519 | Rv1519F | 5′-GGTGGAGAGCGACGACATCAAG-3′ |
| Rv1519R | 5′-TGTAGCGACAGCATCGTCATCC-3′ |
ROX, 6-carboxy-X-rhodamine (excitation, 585 nm/detection, 610 nm); BHQ2, Black Hole Quencher 2 (absorbance 550–650 nm); Cy5, cyanine 5 (excitation 625 nm/detection 660 nm); BBQ, BlackBerry Quencher (absorbance 550–750 nm).
Combined 16S-RD Assay
Each DNA lysate was tested operator-blinded in duplicate for both the 16S and RD assays. For the 16S assay, PCRs were performed in 25-μl volumes containing 5 μl of DNA lysate, 250 nmol/L of each primer MYCO16SF and MYCO16SR, 150 nmol/L MYCO16SPr, and 1× ABsolute QPCR SYBR Green Mix (ABgene, Epsom, UK). For the RD assay, PCRs were performed in 25-μl volumes containing 5 μl of DNA lysate, 80 nmol/L of each primer RD750F, RD750R1, and RD750R2, 150 nmol/L RD750Pr, and 1× ABsolute QPCR SYBR Green Mix. PCRs were performed in a Rotor-Gene machine (Qiagen, Valencia, CA) as follows: 95°C for 15 minutes to activate Taq polymerase, followed by 40 cycles of 95°C for 15 seconds, 60°C for 60 seconds, 72°C for 20 seconds, and 84°C for 20 seconds. ROX and Cy5 signals were acquired at 60°C, whereas SYBR Green signals were acquired at 84°C. When testing the effect of combining TaqMan and SYBR Green on assay performance, TaqMan only reactions were performed using ABsolute QPCR Mix (ABgene), and all determinations were done in triplicate.
PCRs were repeated for all samples yielding no or anomalous signals with neat and diluted DNA lysates (1/10 and 1/100). Melt curves were examined for all assays. Mycobacterial 16S amplicons consistently yielded melt temperatures of 88.2 ± 0.2°C. 16S PCR products for all reactions yielding SYBR Green signals only and randomly selected reactions yielding both TaqMan and SYBR Green signals were subjected to gel analysis, gel purification using QIAquick Gel Extraction Kit (Qiagen), and DNA sequencing (MWG Biotech). 16S rDNA sequences were processed and mycobacterial species were identified using the Ribosomal Differentiation of Medical Microorganisms database.24
Rv1519 PCR
All samples were tested for the presence of intact or deleted Rv1519 by our established conventional PCR assay as described previously.10 Each DNA lysate was tested operator-blinded. PCR was repeated for all samples yielding no bands or multiple bands in 25-μl volumes with 5 μl of neat and diluted DNA lysates (1/10 and 1/100).
Routine Testing of Clinical Mycobacterial Isolates at the Reference Laboratory
All clinical mycobacterial isolates were identified as part of the routine service by the Regional Centre for Mycobacteriology, Birmingham Heartlands Hospital, Birmingham, UK (reference laboratory) using the GenoType MTBC DNA strips for identification of MTBC and the GenoType Mycobacterium (consisting of two kits: Clinical Mycobacteria and Additional Species) for identification of NTM (Hain Lifescience, Nehren, Germany). Our 16S assay results were compared against these identification results as the gold standard. This laboratory also subjects all MTBC isolates to 15-locus VNTR-mycobacterial interspersed repetitive units genotyping. The five-locus VNTR analysis targets ETR A to ETR E as originally described by Frothingham and Meeker-O'Connell,25 and mycobacterial interspersed repetitive units analysis follows the 10-locus method of Supply et al.26
Simulated Mixed Infection Samples
For the mixed MTB-NTM infection tests, M. smegmatis was used to represent NTM. For the mixed MTBC strain studies, M. tuberculosis strains CDC 1551 (RD750+) and CH (RD750−) were used. Target DNA was diluted to 105, 104, and 103 16S rDNA copies/μl. These preparations were mixed in a checkerboard format. PCRs were performed as above except that only 2 μl of mixed template DNA was used per reaction. All PCR assays were performed in duplicate.
Results
Effect of Combining TaqMan and SYBR Green Chemistries in Single Reactions
We tested the effects of combining TaqMan and SYBR Green chemistries in a single-reaction tube with M. tuberculosis CDC 1551 and M. smegmatis DNA for the 16S assay and M. tuberculosis CDC 1551 and CH DNA for the RD assay with target amounts in the range of 103–107 gene copies, and illustrative signal tracings are shown in Figure 3A and 3B. Cycle threshold (Ct) values generated by TaqMan and SYBR Green for single and combined assays (six pairs of values for each chemistry) were compared by paired t tests. Although presence of the TaqMan probe had no measurable effect on SYBR Green Ct values, irrespective of the probe binding status of the reaction (P > 0.1), adding SYBR Green significantly increased the Ct for TaqMan signals (P < 0.05). Ct values were greater by approximately half a cycle and three cycles at high and at low target concentrations respectively (data not shown). The effect was more pronounced for the RD assay.
Figure 3.
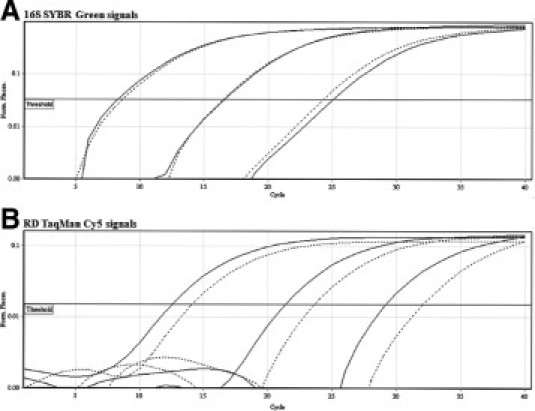
Effect of combining TaqMan and SYBR Green chemistries in single real-time PCRs. M. tuberculosis CDC 1551 target at 107 gene copies and two 100-fold serial dilutions were tested. Dotted lines represent reactions containing both TaqMan and SYBR Green, whereas solid lines represent reactions containing either SYBR Green (A) or TaqMan (B) alone. A shows 16S SYBR Green signals, and B shows RD TaqMan Cy5 signals.
Specificity of the 16S-RD Assay
In view of the potential of the mycobacterial 16S primers to produce amplicons from nonmycobacterial actinobacteria, DNA extracts from Corynebacterium glutamicum, Rhodococcus sp., and Streptomyces fradiae were studied. Although the first two organisms yielded positive 16S SYBR Green signals, TaqMan signals were absent and the RD assay was negative for both signals (16S: T− S+; RD: T− S−). There were no significant signals detected for all other organisms tested (see Materials and Methods for details).
Investigation of Positive Cultures
A total of 88 cultures from different patients deemed probable mycobacteria-positive following Ziehl-Neelsen (ZN) acid fast staining obtained from our routine service between January and November 2007 were analyzed. All cultures except for one were confirmed to be positive for mycobacteria by the reference laboratory. This MGIT lysate gave no signal in either the 16S (T− S−) or RD assay (T− S−), consistent with the reference laboratory report of “no acid-fast bacilli seen.”
Of the 88 positive cultures, 42 were MGIT and 46 were LJ cultures. The combined 16S-RD assay results for analysis of these cultures are displayed in Table 2. The 16S assay and the RD assay identified 68 and 70 MTBC-positive cultures, respectively. Two MGIT lysates did not give an MTBC signal with the 16S TaqMan probe but were positive for 16S SYBR Green (T− S+) and for both RD TaqMan and SYBR Green signals (T+ S+). Direct sequencing of these two 16S amplicons yielded indeterminate results. Since both the RD750 primers and TaqMan probe used in this assay are MTBC specific, we concluded that the “false-negative” 16S TaqMan results associated with these two MGIT lysates probably may have been due to cross-reactive nonmycobacterial DNA present within these lysates. Reference laboratory results subsequently confirmed that the RD assay had correctly identified all 70 MTBC-positive cultures within the set of 88 cultures investigated. Seventeen 16S-RD results indicated NTM-positive cultures (16S T− S+ and RD T− S−); these again were subsequently confirmed by the reference laboratory.
Table 2.
Combined 16S-RD Assay Applied to 88 Consecutive Mycobacteria-Positive Cultures
| RD assay | ||||
|---|---|---|---|---|
| T+ S+ | T− S+ | T− S− | Total | |
| 16S assay | ||||
| T+ S+ | 42 | 26 | 0 | 68 |
| T− S+ | 2 | 0 | 17 | 19 |
| T− S− | 0 | 0 | 1 | 1 |
| Total | 44 | 26 | 18 | 88 |
T, TaqMan signal; S, SYBR Green signal. See Fig. 1 for further explanation.
16S Amplicon Sequencing and Identification of NTM
Comparisons of sequences of the 16S amplicons from the 17 probable NTM-positive cultures with those in the Ribosomal Differentiation of Medical Microorganisms database, yielded definitive results for 15 cultures (8 M. avium complex, 2 M. gordonae, 2 M. kansasii, 1 M. abscessus, 1 M. peregrinum, and 1 M. malmoense), which were also found to be concordant with those of the reference laboratory. 16S amplicons from two NTM-positive MGIT cultures failed to yield DNA sequence data; the two cultures were eventually identified as M. gordonae by the reference laboratory.
M. tuberculosis RD750 and VNTR Genotyping
Initial results indicated that 44 of the MTBC strains were RD750+ (RD T+ S+) and 26 were RD750− (RD T− S+). These 70 lysates were subsequently analyzed by our established Rv1519 PCR assay, and their VNTR profiles were received from the reference laboratory. Although the majority of our RD750− isolates genotyped as VNTR 42234/5, a more extensive study has revealed that all can be encompassed within the profile x2234/5 (R.J. Smith, unpublished data), and we have used this latter profile to recognize concordance or otherwise with our real-time RD assay. For 62 lysates, the results of all three genotyping assays (RD, Rv1519 PCR, and VNTR) were concordant. In eight cases, no amplicons were obtained with the Rv1519 PCR, but RD and VNTR results were concordant. Occasional Rv1519 PCR failures are expected given the relatively large 1- to 3-kb amplicon sought and the crude DNA preparation used in this assay.
Potential to Detect Mixed Infections
To examine the capacity of our combined assay to detect mixed infections involving either a combination of MTBC and NTM or MTBC strains with intact and deleted RD750, we studied its performance using mixtures of purified DNA as detailed in Table 3. We note that it is only possible to reliably recognize the target detected solely by SYBR Green chemistry (NTM or RD750− in the 16S and RD assays, respectively) when the copy number detected by the SYBR Green signal significantly exceeds that detected by the TaqMan probe in the same sample. Only in this situation can the higher SYBR Green signal be confidently interpreted to show the presence of the second TaqMan-negative target. This noted, it is apparent that the combined assay will not detect mixed infections when the SYBR Green signals are approximately equal to the TaqMan signals. The presence of NTM species in mixed MTBC-NTM samples did not affect the performance of the RD assay (data not shown). We emphasize that where signals suggestive of a mixed infection are obtained, further analyses such as cloning and sequencing in the case of the 16S assay and gel electrophoresis in the case of the RD assay could be applied to obtain further confirmation.
Table 3.
Potential of the Combined 16S-RD Assay to Detect Mixed Infections
|
M. smegmatis (16S SYBR Green signal) |
M. tuberculosis CH (RD SYBR Green signal) |
|||||
|---|---|---|---|---|---|---|
| 105* | 104 | 103 | 105 | 104 | 103 | |
| M. tuberculosis CDC 1551 signals† | ||||||
| 105 | BOTH‡ | CDC | CDC | BOTH | CDC | CDC |
| 104 | BOTH | BOTH§ | CDC | BOTH | BOTH | CDC |
| 103 | MS | BOTH | CDC | BOTH | BOTH | BOTH |
Figures denote number of target DNA copies added.
TaqMan signals from both 16S and RD assays.
BOTH, both targets detected; CDC, only M. tuberculosis CDC 1551 (RD750+) detected; MS, only M. smegmatis detected. No M. tuberculosis CH (RD750−) only signals were obtained.
Borderline M. smegmatis signal.
Discussion
We have developed a molecular assay capable of distinguishing MTBC and NTM when applied to positive LJ and MGIT cultures; further identification of NTM species is achieved by 16S rDNA sequencing. When positive for MTBC, the assay also determines whether the MTBC strain belongs to the EA-I lineage. Since roughly half of the MTBC isolates obtained by our service belong to this lineage, the result provides an opportunity for preliminary epidemiological discrimination. As far as we are aware, the simultaneous use of TaqMan and SYBR Green chemistries within a single tube, as deployed in our assay, has not been reported previously.
The combination of the two signal generating chemistries had little consequence for the performance of the assay on positive mycobacterial cultures. Inclusion of TaqMan probes had no detectable effect on SYBR Green results. In contrast, inclusion of SYBR Green led to an increase in TaqMan Ct values, particularly at low target concentrations. This might reflect the impact of SYBR Green binding to the same region as the TaqMan probes and resultant lower probe cleavage rates. Although this had little consequence for the present study, it could lead to decreased assay sensitivity in clinical samples.
As expected, the combined assay gave 16S amplicons with some actinobacteria but was universally negative with the other bacterial targets tested. Since the assay is designed for use in conjunction with ZN-positive samples this should not lead to ambiguous results. Although Nocardia spp. are weakly ZN positive and can be isolated with both MGIT and LJ media, they will also stain Gram-positive. Thus, when applied to positive cultures, which are routinely ZN stained, additional consideration of Gram stain results and the results of conventional cultures should rule out any ambiguity in interpretation.
The 16S-RD assay correctly identified all 70 MTBC-positive cultures. Two MTBC-positive MGIT cultures failed to give TaqMan signals in the 16S assay. These results derived from MGIT culture lysates in which the possible presence of nonmycobacterial signals may have been a confounding factor. Interestingly the 16S melt temperature values obtained for these samples were more than 1°C lower than those obtained from confirmed mycobacterial amplicons and sequencing revealed amplicons with no clear identity (data not shown). It seems that occasionally the 16S assay produces anomalous amplicons that may be recognized by their low melt temperature values. MTBC DNA in these samples can be detected by the parallel RD assay. Thus, at the present time, there appear to be limitations to the negative predictive value of the 16S assay alone. We currently advocate use of the combined 16S-RD assay where our available data indicate all negative RD results are true negatives for MTBC.
Of the remaining 18 positive cultures analyzed, the combined assay correctly identified the presence of NTM sequence in all 17 reference laboratory-confirmed cases. Subsequent sequencing and Ribosomal Differentiation of Medical Microorganisms analysis produced NTM identities concordant with the reference laboratory in 15 cases. There were two primary sequencing failures, both from cultures yielding M. gordonae. A final MGIT culture graded positive failed to yield any PCR signals and was subsequently determined to be negative for mycobacteria by the reference laboratory. This occurs in up to 5% of the MGIT cultures graded positive for acid-fast bacilli by our service. Application of the 16S-RD assay to such samples graded acid-fast bacilli-positive therefore has the potential to recognize such true negatives before forwarding for analysis by the reference laboratory. Since a preliminary report indicating the likely isolation of mycobacteria is routinely issued to clinicians when cultures are forwarded to the reference laboratory, use of our assay to eliminate these preliminary false-positive results warrants further analysis.
Of 70 MTBC signals obtained, the RD assay assigned 44 isolates to RD750 intact and 26 to RD750 deleted. In each case where the conventional Rv1519 PCR assay produced an Rv1519-related amplicon, the RD assay results were confirmed. As expected, all of the RD750− strains fell into the VNTR x2234/5 group.
Our results demonstrate that within a total processing time of 4 hours the combined 16S-RD assay reliably identifies mycobacteria-positive cultures, differentiates between MTBC and NTM, and assigns the MTBC signals to within or outside the EA-I lineage. In our series of 88 positive cultures, this rapid assay gave primary results that were concordant with subsequent reference laboratory analysis in every case.
We also explored the potential of our combined assay to detect mixed infections. The results show that our assay has potential to detect mixed infections due to MTBC strains belonging to different lineages and mixed MTBC/NTM infections. This potential is limited to samples in which the SYBR Green signals generated by our assay indicate a significant excess of copy numbers over those attributable to the TaqMan probe in the same reaction. In this way, our study has demonstrated both the value and limitation of using a combination of TaqMan and SYBR Green signals in real-time assays.
Turning to the current clinical utility of the 16S-RD assay, early confirmation of MTBC or NTM infection is of established value and allows for appropriate clinical and public health management. Assignment of MTBC-positive cultures to within or outside a particular global lineage allows for recognition or exclusion of links between contemporary isolates and could therefore facilitate appropriate responses. Evaluation of the practical utility of such preliminary information is currently in progress.
Our assay could be adapted to target other lineage-defining RDs appropriate to different geographic locations. Indeed, one might even envisage applying an individually selected RD target analyses to different samples depending on the specific epidemiological question to be resolved. This assay can therefore be adapted to complement the local, routine mycobacterial service in specified populations.
We conclude that we have established a rapid molecular assay that confirms the presence of mycobacteria, differentiates between MTBC and NTM, and assigns MTBC signals to within or outside the EA-I lineage. The assay is reliable when applied to positive LJ and MGIT cultures, has potential to detect mixed infections, and is being further developed for direct application to clinical samples.
Acknowledgements
We thank the Department of Clinical Microbiology, University Hospitals of Leicester for all mycobacteria-positive cultures.
Footnotes
Supported by the Henry-Smith Charity and the UK Medical Research Council. M. tuberculosis CDC 1551 genomic DNA was provided by Colorado State University (part of National Institutes of Health, National Institute of Allergy and Infectious Diseases contract HHSN266200400091C, entitled “Tuberculosis Vaccine Testing and Research Materials”).
E.S.G.C. and J.M. contributed equally to this study.
References
- 1.Horsburgh CR., Jr Epidemiology of mycobacterial diseases in AIDS. Res Microbiol. 1992;143:372–377. doi: 10.1016/0923-2508(92)90048-s. [DOI] [PubMed] [Google Scholar]
- 2.Management of opportunist mycobacterial infections: Joint Tuberculosis Committee Guidelines 1999. Subcommittee of the Joint Tuberculosis Committee of the Br Thoracic Society. Thorax. 2000;55:210–218. doi: 10.1136/thorax.55.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 4.Miller N, Cleary T, Kraus G, Young AK, Spruill G, Hnatyszyn HJ. Rapid and specific detection of Mycobacterium tuberculosis from acid-fast bacillus smear-positive respiratory specimens and BacT/ALERT MP culture bottles by using fluorogenic probes and real-time PCR. J Clin Microbiol. 2002;40:4143–4147. doi: 10.1128/JCM.40.11.4143-4147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broccolo F, Scarpellini P, Locatelli G, Zingale A, Brambilla AM, Cichero P, Sechi LA, Lazzarin A, Lusso P, Malnati MS. Rapid diagnosis of mycobacterial infections and quantitation of Mycobacterium tuberculosis load by two real-time calibrated PCR assays. J Clin Microbiol. 2003;41:4565–4572. doi: 10.1128/JCM.41.10.4565-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shrestha NK, Tuohy MJ, Hall GS, Reischl U, Gordon SM, Procop GW. Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J Clin Microbiol. 2003;41:5121–5126. doi: 10.1128/JCM.41.11.5121-5126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurabachew M, Enger O, Sandaa RA, Skuce R, Bjorvatn B. A multiplex polymerase chain reaction assay for genus-, group- and species-specific detection of mycobacteria. Diagn Microbiol Infect Dis. 2004;49:99–104. doi: 10.1016/j.diagmicrobio.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Papaparaskevas J, Houhoula DP, Siatelis A, Tsakris A. Molecular-beacon-based real-time PCR for detection and quantification of Mycobacterium tuberculosis DNA in clinical samples. J Clin Microbiol. 2008;46:3177–3178. doi: 10.1128/JCM.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu P, Zhong H, Luo Y, Mei J, Gao Q. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47:845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajakumar K, Shafi J, Smith RJ, Stabler RA, Andrew PW, Modha D, Bryant G, Monk P, Hinds J, Butcher PD, Barer MR. Use of genome level-informed PCR as a new investigational approach for analysis of outbreak-associated Mycobacterium tuberculosis isolates. J Clin Microbiol. 2004;42:1890–1896. doi: 10.1128/JCM.42.5.1890-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:2658–2671. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 14.Newton SM, Smith RJ, Wilkinson KA, Nicol MP, Garton NJ, Staples KJ, Stewart GR, Wain JR, Martineau AR, Fandrich S, Smallie T, Foxwell B, Al-Obaidi A, Shafi J, Rajakumar K, Kampmann B, Andrew PW, Ziegler-Heitbrock L, Barer MR, Wilkinson RJ. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc Natl Acad Sci USA. 2006;103:15594–15598. doi: 10.1073/pnas.0604283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Diversity of Leicester: A Demographic Profile. Edited by Leicester City Council; 2008. pp. 1–28. [Google Scholar]
- 16.Menendez MC, Buxton RS, Evans JT, Gascoyne-Binzi D, Barlow RE, Hinds J, Hawkey PM, Colston MJ. Genome analysis shows a common evolutionary origin for the dominant strains of Mycobacterium tuberculosis in a UK South Asian community. Tuberculosis. 2007;87:426–436. doi: 10.1016/j.tube.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NT, Thuong NT, Stepniewska K, Huyen MN, Bang ND, Loc TH, Gagneux S, van Soolingen D, Kremer K, van der Sande M, Small P, Anh PT, Chinh NT, Quy HT, Duyen NT, Tho DQ, Hieu NT, Torok E, Hien TT, Dung NH, Nhu NT, Duy PM, van Vinh Chau N, Farrar J. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:1–9. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, Jackson-Sillah DJ, Fox A, Deriemer K, Gagneux S, Borgdorff MW, McAdam KP, Corrah T, Small PM, Adegbola RA. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thwaites G, Caws M, Chau TT, D'Sa A, Lan NT, Huyen MN, Gagneux S, Anh PT, Tho DQ, Torok E, Nhu NT, Duyen NT, Duy PM, Richenberg J, Simmons C, Hien TT, Farrar J. Relationship between Mycobacterium tuberculosis genotype and the clinical phenotype of pulmonary and meningeal tuberculosis. J Clin Microbiol. 2008;46:1363–1368. doi: 10.1128/JCM.02180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, van Helden PD. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR, Jr, Venter JC, Fraser CM. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson RJ, DesJardin LE, Islam N, Gibson BM, Kanost RA, Wilkinson KA, Poelman D, Eisenach KD, Toossi Z. An increase in expression of a Mycobacterium tuberculosis mycolyl transferase gene (fbpB) occurs early after infection of human monocytes. Mol Microbiol. 2001;39:813–821. doi: 10.1046/j.1365-2958.2001.02280.x. [DOI] [PubMed] [Google Scholar]
- 24.Harmsen D, Rothganger J, Frosch M, Albert J. RIDOM: Ribosomal Differentiation of Medical Micro-organisms Database. Nucleic Acids Res. 2002;30:416–417. doi: 10.1093/nar/30.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frothingham R, Meeker-O'Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144(Pt 5):1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 26.Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39:3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


