Abstract
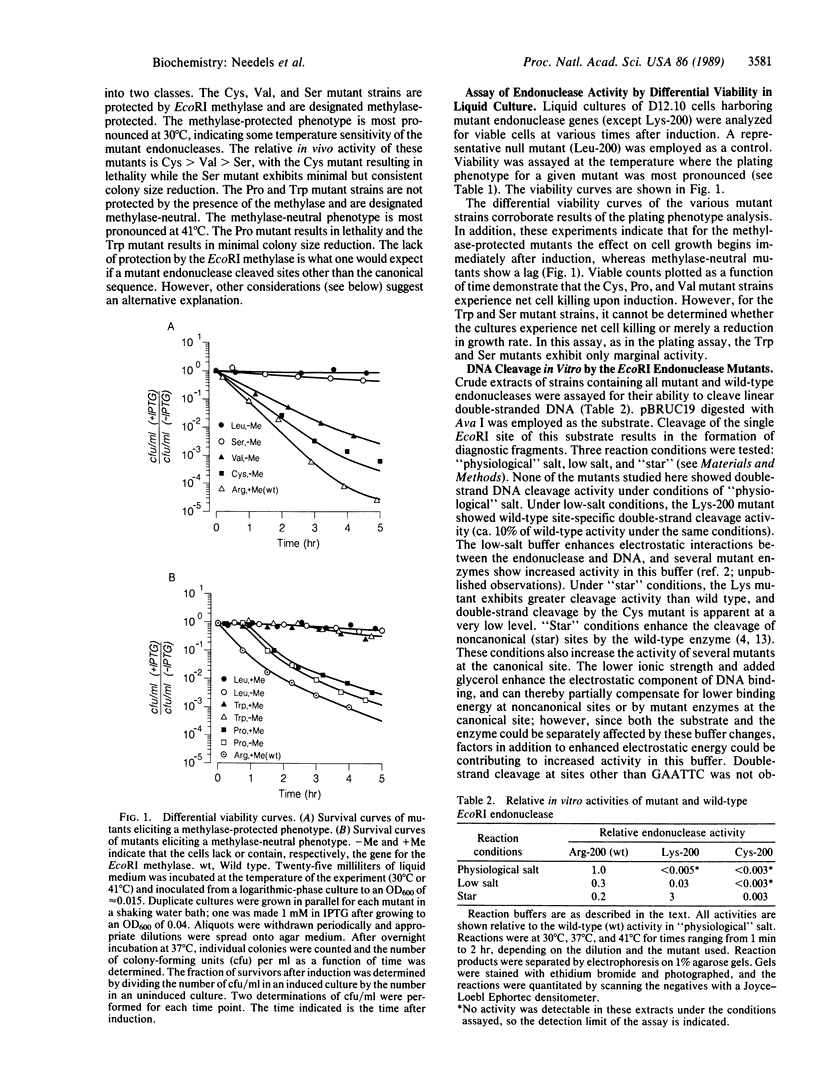
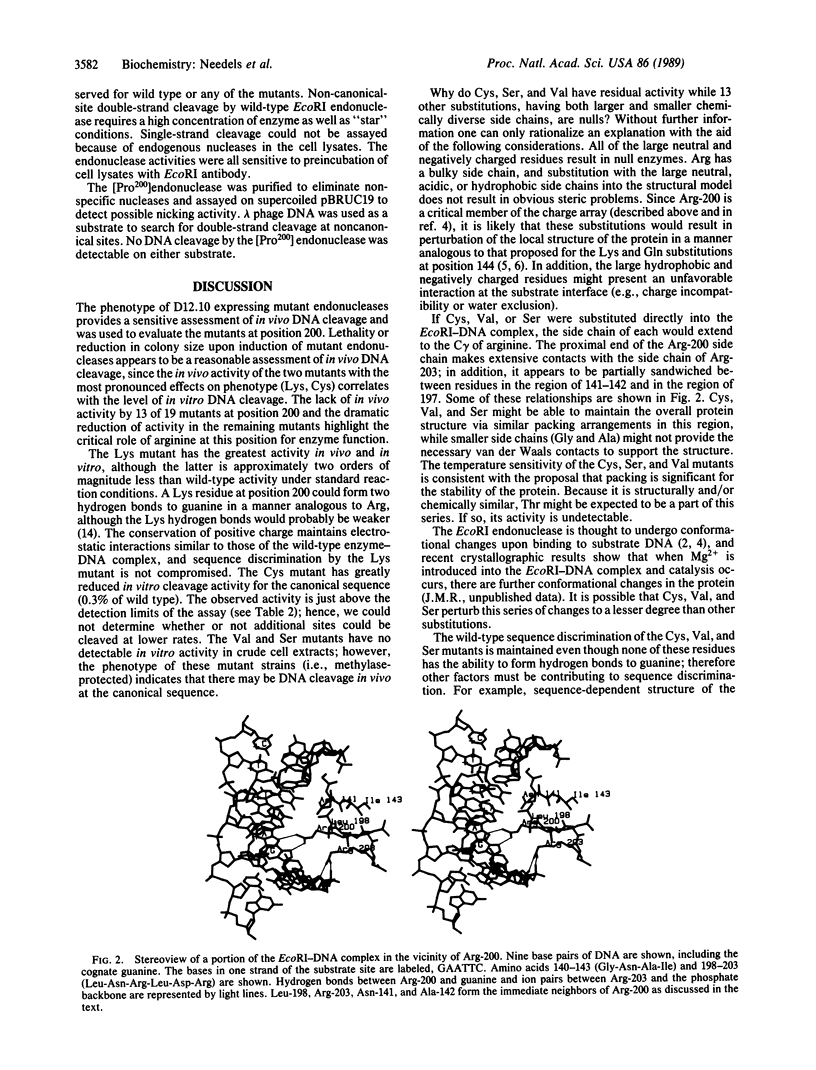
The arginine at position 200 of EcoRI endonuclease is thought to make two hydrogen bonds to the guanine of the sequence GAATTC and thus be an important determinant of sequence discrimination. Arg-200 was replaced by each of the other 19 naturally occurring amino acids, and the mutant endonucleases were assessed for activities in vivo and in vitro. The mutant endonuclease with lysine at position 200 exhibits the most in vivo activity of all the position 200 mutants, although the in vitro activity is less than 1/100th of wild-type activity. Five other mutants show more drastically reduced levels of in vivo activity (Cys, Pro, Val, Ser, and Trp). The Cys, Val, and Ser mutant enzymes appear to have in vivo activity which is specific for the wild-type canonical site despite the loss of hydrogen bonding potential at position 200. The Pro and Trp mutants retain in vivo activity which is independent of the presence of the EcoRI methylase. In crude cell lysates, only the Cys mutant shows a very low level of in vitro activity. None of the mutant enzymes show a preference for alternative sites in assays in vitro. The implications of these results are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Bass S., Sorrells V., Youderian P. Mutant Trp repressors with new DNA-binding specificities. Science. 1988 Oct 14;242(4876):240–245. doi: 10.1126/science.3140377. [DOI] [PubMed] [Google Scholar]
- Betlach M., Hershfield V., Chow L., Brown W., Goodman H., Boyer H. W. A restriction endonuclease analysis of the bacterial plasmid controlling the ecoRI restriction and modification of DNA. Fed Proc. 1976 Jul;35(9):2037–2043. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Ballard B. T., Stephenson F., Kohr W. J., Rodriguez H., Rosenberg J. M., Boyer H. W. Purification and characterization of the restriction endonuclease RsrI, an isoschizomer of EcoRI. Gene. 1988 Aug 15;68(1):43–51. doi: 10.1016/0378-1119(88)90597-5. [DOI] [PubMed] [Google Scholar]
- Gresh N., Pullman B. A theoretical study of the interaction of guanine and cytosine with specific amino acid side chains. Biochim Biophys Acta. 1980 Jun 27;608(1):47–53. doi: 10.1016/0005-2787(80)90132-x. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L., Kurpiewski M., Lesser D., Grable J., Boyer H. W., Rosenberg J. M., Greene P. J. Coordinate ion pair formation between EcoRI endonuclease and DNA. J Biol Chem. 1983 Dec 10;258(23):14638–14646. [PubMed] [Google Scholar]
- Koudelka G. B., Harbury P., Harrison S. C., Ptashne M. DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4633–4637. doi: 10.1073/pnas.85.13.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn I., Stephenson F. H., Boyer H. W., Greene P. J. Positive-selection vectors utilizing lethality of the EcoRI endonuclease. Gene. 1986;42(3):253–263. doi: 10.1016/0378-1119(86)90229-5. [DOI] [PubMed] [Google Scholar]
- MOSELEY B. E., LASER H. REPAIR OF X-RAY IN MICROCOCCUS RADIODURANS. Proc R Soc Lond B Biol Sci. 1965 Apr 13;162:210–222. doi: 10.1098/rspb.1965.0035. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Hare D. Nuclear magnetic resonance and distance geometry studies of DNA structures in solution. Annu Rev Biophys Biophys Chem. 1987;16:423–454. doi: 10.1146/annurev.bb.16.060187.002231. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Richards J. H. Site-saturation studies of beta-lactamase: production and characterization of mutant beta-lactamases with all possible amino acid substitutions at residue 71. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1588–1592. doi: 10.1073/pnas.83.6.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky S. D., Love R., McClarin J. A., Rosenberg J. M., Boyer H. W., Greene P. J. Clustering of null mutations in the EcoRI endonuclease. Proteins. 1987;2(4):273–282. doi: 10.1002/prot.340020403. [DOI] [PubMed] [Google Scholar]
- Youderian P., Vershon A., Bouvier S., Sauer R. T., Susskind M. M. Changing the DNA-binding specificity of a repressor. Cell. 1983 Dec;35(3 Pt 2):777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]


