Abstract
Motivation: Significance analysis of microarrays (SAM) is a widely used permutation-based approach to identifying differentially expressed genes in microarray datasets. While SAM is freely available as an Excel plug-in and as an R-package, analyses are often limited for large datasets due to very high memory requirements.
Summary: We have developed a parallelized version of the SAM algorithm called ParaSAM to overcome the memory limitations. This high performance multithreaded application provides the scientific community with an easy and manageable client-server Windows application with graphical user interface and does not require programming experience to run. The parallel nature of the application comes from the use of web services to perform the permutations. Our results indicate that ParaSAM is not only faster than the serial version, but also can analyze extremely large datasets that cannot be performed using existing implementations.
Availability:A web version open to the public is available at http://bioanalysis.genomics.mcg.edu/parasam. For local installations, both the windows and web implementations of ParaSAM are available for free at http://www.amdcc.org/bioinformatics/software/parasam.aspx
Contact: rmcindoe@mail.mcg.edu
Supplementary information: Supplementary Data is available at Bioinformatics online.
1 INTRODUCTION
Shrinkage-based approaches to testing for differential expression in microarray experiments have proven to best identify the genes of scientific interest (Kerr, 2009). The computational demand of analyzing these data continues to increase due to the reduced cost of microarrays and increases in the number of replicates being measured. The computational burden is especially evident with shrinkage-based tests where permutations are often used to assess statistical significance and the false discovery rate (FDR) (Benjamini and Hochberg, 1995).
The significance analysis of microarrays (SAM) algorithm is a popular method for differential expression analyses using a shrinkage-based-test and permutations for the FDR (Tusher et al., 2001). The popularity of SAM is enhanced by its free availability as an Excel plug-in or R package. SAM is a computationally demanding algorithm and analysis of larger datasets is challenging due to the high memory requirements. Creating a parallel version of SAM could address problems often encountered with larger experiments. With a parallel algorithm, the computational workload is divided among multiple CPUs and the main memory of all participating computers is utilized to avoid caching operations to the disk which will significantly decrease algorithm execution time. The most time and memory intensive portion of the SAM algorithm is permuting the columns to determine the null distribution. For large datasets, users of the serial version of SAM will decrease the number of permutations used in analyses to allow the algorithm to complete without error. This results in a poorly characterized null distribution. To overcome this limitation, we developed a high performance parallelized version of the SAM algorithm called ParaSAM. This version divides the permutations across multiple compute nodes, allowing ParaSAM to perform a large number of permutations (e.g. 1000).
2 IMPLEMENTATION
We designed ParaSAM with an easy and manageable client-server application model that can be easily deployed in most research laboratories, using the same model design we implemented in ParaKMeans (Kraj et al., 2008). A web service (ParallelSAM) performs the permutations, allowing for parallel computation. The main Application Programming Interface (API) is a .NET dynamic link library that connects to and uses the ParallelSAM web service(s). The API is used by the user interfaces to orchestrate the activities of the compute nodes and provides the methods to load the data, send the data to the nodes and orchestrate the asynchronous multithreaded connections to the ParallelSAM web services (Supplementary Fig. S1).
We have developed both a stand-alone windows GUI that can be installed on any Windows machine and an AJAX-based (Asynchronous JavaScript and XML) web GUI. The stand-alone GUI also provides easy file management, compute node management, program options and a results window for saving the data. The one-class, two-class unpaired and multiclass response types are implemented in both versions.
ParaSAM is easily installed using the built-in Windows Installer (MSI files). The ParallelSAM web service is installed on each machine that will be used as a compute node and the GUI is installed on the computer to be directly used by the end user. The web-based GUI requires IIS to be functional. Because most laboratory personnel use Windows-based computers, we felt we would reach a larger number of people by writing the software to run in the Windows operating system.
This parallel implementation of the SAM algorithm does not require expensive hardware and the number of compute nodes can be as small as one. In fact, any number of inexpensive desktop computers connected by a network can be used. The permutation partitioning scheme is not restricted and is entirely dependent on the number of compute nodes participating in the algorithm. In ParaSAM, we have utilized the memory efficiently using C# and have achieved almost a 2-fold speed-up even on a single machine.
3 PERFORMANCE AND TESTING
3.1 Configuration of test system used to assess ParaSAM
The performance of ParaSAM was evaluated using one master computer with between 1 to 7 compute nodes. For comparisons, we installed R 2.8.0 and the samr package on the master computer. The master computer and the seven compute nodes were all identical machines: Dell Poweredge 2650 with Dual 3.06 GHz/512K Cache Xeon Processors and 8.0 GB DDR 266 Mhz RAM.
3.2 Increased capacity of handling larger datasets
We analyzed an experimentally derived microarray dataset (NCBI Gene Expression Omnibus; GSE9006: Gene expression in PBMCs from children with diabetes) (Kaizer et al., 2007). Combined data from both chips (HG-U133A and HG-U133B) were used for a 44 760 gene dataset. The data from only the HG-U133A chip was used for the 22 283 genes dataset. The entire dataset contains 117 arrays with three groups: Healthy, 24 arrays; Type-1 Diabetes, 81 arrays; and Type-2 Diabetes, 12 arrays. The arrays were further subdivided into smaller datasets as shown in Supplementary Table S1. A multiclass SAM analysis was performed using the samr-package and ParaSAM with 1 to 7 nodes.
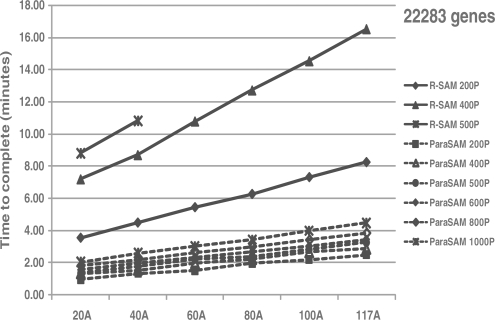
ParaSAM successfully analyzed datasets with 44 760 genes and 117 arrays using 1000 permutations, whereas R (samr) could only perform 200 permutations on this dataset (Table 1 and Supplementary Table S2). When evaluating smaller datasets (22 283 genes, 20 arrays), samr can only perform 500 permutations. While execution time for both ParaSAM and samr increased with the number of arrays analyzed, ParaSAM scaled significantly better than samr (Fig. 1).
Table 1.
Comparison of time (minutes) taken to complete the SAM analyses by R and ParaSAM (7 nodes) on two datasets (22 283 genes and 44 760 genes)
| R-SAM |
ParaSAM |
|||||
|---|---|---|---|---|---|---|
| Permutations | 20 arrays | 60 arrays | 117 arrays | 20 arrays | 60 arrays | 117 arrays |
| 22 283 genes | ||||||
| 200 | 3.57±0.08 | 5.46±0.06 | 8.26±0.25 | 0.96±0.01 | 1.54±0.03 | 2.50±0.06 |
| 400 | 7.21±0.22 | 10.81±0.13 | 16.54±0.34 | 1.35±0.03 | 1.98±0.03 | 2.89±0.06 |
| 1000 | PF | PF | PF | 2.04±0.03 | 3.02±0.06 | 4.48±0.11 |
| 44 760 genes | ||||||
| 200 | 7.60±0.19 | 12.97±0.50 | 20.76±0.38 | 2.29±0.05 | 3.37±0.11 | 5.45±0.15 |
| 400 | 14.22±0.33 | PF | PF | 2.71±0.07 | 4.30±0.17 | 6.23±0.10 |
| 1000 | PF | PF | PF | 4.35±0.09 | 6.32±0.09 | 10.32±0.31 |
PF, program failed. All times in minutes where N=12 runs.
Fig. 1.
Execution time of ParaSAM and ‘samr’ with increasing size of datasets and permutations. Each series indicates the program and number of permutations used.
3.3 Significantly increased speedup
The speed-up of ParaSAM was evaluated using a small dataset (22 283 genes, 40 arrays, 500 permutations) and compared with samr. We found that ParaSAM is 1.83X, 3.22X, 4.49X, 5.25X, 5.65X, 6.11X, 6.07X faster than samr using 1 to 7 nodes, respectively (Supplementary Figures S2 and S3). Interestingly, increases in speed start to plateau after five nodes are used (200 permutations/node).
3.4 ParaSAM produces the same results as SAM
We validated ParaSAM with samr using one-group, two-group and multigroup datasets using 1000 permutations on datasets with 3000 genes for 12 arrays (One class), 14 arrays (Two class) or 25 arrays (Multiclass). The results indicate that ParaSAM is not statistically different than R-SAM for the One class (P = 0.9055), Two class (P = 0.6721) or Multiclass (P = 0.8852) analyses (Supplementary Tables S3–S8 and Figures S4–S6).
4 CONCLUSIONS
As the number of publically available microarray experiments increases, the ability to analyze extremely large datasets across multiple experiments becomes critical. The ability to conduct such analyses depends on algorithms that are fast and memory efficient. ParaSAM is designed to provide the general scientific community with an easy and manageable client-server Windows application. ParaSAM is faster than existing implementations and is able to analyze much larger datasets. This software is freely available and can help the scientific community with accurate and effective microarray data analysis.
Funding: National Institute of Diabetes Digestive and Kidney Diseases (Grant U24DK076169) to R.A.M.
Conflicts of Interest: none declared.
REFERENCES
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Series B Methodol. 1995;57:289–300. [Google Scholar]
- Kaizer EC, et al. Gene expression in peripheral blood mononuclear cells from children with diabetes. J. Clin. Endocrinol. Metab. 2007;92:3705–3711. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- Kerr KF. Comments on the analysis of unbalanced microarray data. Bioinformatics. 2009;25:2035–2041. doi: 10.1093/bioinformatics/btp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraj P, et al. ParaKMeans: implementation of a parallelized K-means algorithm suitable for general laboratory use. BMC Bioinformatics. 2008;9:200. doi: 10.1186/1471-2105-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]