Abstract
Chromosomal translocations involving the MLL gene are associated with infant acute lymphoblastic and mixed lineage leukemia. There are a large number of translocation partners of MLL that share very little sequence or seemingly functional similarities, however, their translocations into MLL result in the pathogenesis of leukemia. To define the molecular reason why these translocations result in the pathogenesis of leukemia, we purified several of the commonly occurring MLL chimeras. We have identified a novel super elongation complex (SEC) associated with all chimeras purified. SEC includes ELL, P-TEFb, AFF4 and several other factors. AFF4 is required for SEC stability and proper transcription by poised RNA polymerase II in metazoans. Knockdown of AFF4 within SEC in leukemic cells shows reduction in MLL chimera target gene expression suggesting that AFF4/SEC could be a key regulator in the pathogenesis of leukemia through many of the MLL partners.
INTRODUCTION
Transcriptional control by RNA polymerase II (Pol II) is a multi-step process requiring the concerted action of multiple factors and contacts with the DNA template for the proper synthesis of nascent RNA (Conaway and Conaway, 1993; Kornberg, 2007; Shilatifard et al., 2003). For many years, it was considered that transcriptional regulation mainly occurs at the level of transcriptional initiation. However, we now know that transcriptional elongation and the factors regulating this process are also highly essential for the proper regulation of gene expression (Conaway et al., 2000; Reines et al., 1996; Shilatifard et al., 2003). The ELL gene on chromosome 19p13.1 was identified as one of the translocation partners of Mixed Lineage Leukemia (MLL) found in hematological malignancies (Thirman et al., 1994). Human ELL1 was demonstrated to be a Pol II elongation factor capable of increasing the catalytic rate of transcription elongation by reducing transient pausing by the enzyme (Shilatifard et al., 1996). The report of ELL1 being a Pol II elongation factor was the first biochemical and molecular characterization of any of the MLL partners in leukemia, and to date functionally, ELL1 is the best characterized of the MLL partners. In humans, there are three family members of ELL1, including ELL2 and ELL3 (Miller et al., 2000; Shilatifard et al., 2003; Shilatifard et al., 1997). In Drosophila, there is only one ELL family member, dELL, also capable of functioning as a Pol II elongation factor both in vitro and in vivo (Eissenberg et al., 2002; Gerber et al., 2001). Recent findings demonstrated that a large number of developmentally regulated genes contain Pol II poised at their promoters in the absence of detectable full-length transcripts suggesting that the regulation of transcription elongation is a key step in governing gene expression and development in eukaryotes (Boettiger and Levine, 2009; Muse et al., 2007; Zeitlinger et al., 2007). In support of the role for transcription elongation in the regulation of the activity of poised Pol II, ELL was demonstrated to be a major regulator of transcription by the poised Pol II at the heat shock loci (Ardehali et al., 2009; Gerber et al., 2001; Smith et al., 2008).
The human MLL gene on chromosome 11q23 undergoes frequent translocations with a variety of genes all resulting in the pathogenesis of hematological malignancies (Rowley, 1998; Tenney and Shilatifard, 2005). The MLL translocation-based leukemia involves a large number of fusion partners, many of which share little sequence or known functional similarity. Since ELL1 has been well characterized as a Pol II elongation factor, it has been postulated that perhaps other MLL partners may also function in the regulation of transcription elongation as well (Shilatifard et al., 2003). The most common translocation partners of MLL include AFF1, AF9, ENL, AF10 and ELL1 (Rowley, 1998; Tenney and Shilatifard, 2005). AF9, AF10 and ENL have been shown to interact directly with the histone methyltransferase Dot1, leading to the suggestion that Dot1-mediated methylation of H3K79 was central to leukemogenesis in patients with MLL translocations (Bitoun et al., 2007; Krivtsov et al., 2008; Mueller et al., 2007; Mueller et al., 2009; Okada et al., 2005; Zhang et al., 2006). However, at this time, there is little evidence, and no mechanistic understanding, for how H3K79 methylation by Dot1 could lead to gene activation. Furthermore, ELL1, one notable translocation partner of MLL, which is a focus of our studies and has a demonstrated role in transcription elongation, was not reported to be a part of these complexes (Mueller et al., 2007; Mueller et al., 2009).
To learn more about the molecular mechanisms of MLL-based chromosomal translocations in the pathogenesis of leukemia, we began our biochemical search for the identification of commonalities in the disparate MLL-fusions. Therefore, we generated several cell lines containing epitope tagged versions of some of the most common MLL fusion partners and purified these protein complexes. This report is the first biochemical characterization of any of the MLL-chimeras and the results have been extremely informative. Analysis of the purified MLL-chimera complexes by mass spectrometry resulted in the identification of the Pol II elongation factor ELL with the purified MLL-chimeras. We also found that all of the purified MLL-chimeras are associated with AFF4, itself a rare translocation partner of MLL. Further biochemical purification of the ELLs and the AFF4 complexes from nuclear extracts resulted in the identification of novel complexes containing the elongation factors ELL1, ELL2, ELL3, P-TEFb, with AFF4. These findings link two elongation factors, ELL and P-TEFb in one super elongation complex (SEC) with AFF4, which we now know is a shared subunit of the purified MLL-chimeras. We also demonstrated that SEC is capable of phosphorylating the C-terminal domain (CTD) of Pol II and that the stability of the complex requires the presence of AFF4, indicating that AFF4 is central for complex assembly.
Many Pol II elongation factors associate with transcribing Pol II and relocalize to heat shock loci upon stress. We find that both the human and Drosophila homologues of AFF4 associate with the elongating form of Pol II on chromatin and relocalize to heat shock loci upon stress. Furthermore, a reduction in the AFF4 levels by RNAi results in a failure of proper heat shock gene expression. Our biochemical studies indicate that AFF4 is a central factor required for the assembly and integrity of the ELL/P-TEFb/MLL partner elongation complex. Therefore, we anticipate that the translocation of MLL to its partners will result in the relocalization of AFF4 to the MLL target genes such as HOXA9 and HOXA10. Indeed, in support of this hypothesis, AFF4 is associated with the MLL target genes, HOXA9 and HOXA10 in the leukemia cell line MV-4–11, and the knockdown of AFF4 in these cells results in reduction of HOXA9 and HOXA10 expression. Overall, our study identifies AFF4 as a key component of the RNA polymerase II elongation complex and a shared subunit of many of the MLL-chimeras. This observation points to AFF4 and the transcriptional elongation complexes as a possible target for the treatment of hematological malignancies caused by some common MLL translocations found in infant leukemias.
RESULTS and DISCUSSION
In order to begin to understand how the misregulation of gene expression is caused by MLL-fusion proteins, we expressed some of the most common MLL fusion proteins, MLL-ELL1, MLL-ENL, MLL-AFF1 and MLL-AF9 in 293 cells with a Flag epitope tag under an inducible promoter, each integrated at the same site within the genome. Expression and purification of the MLL-N-terminal region most frequently found in MLL-fusion proteins, resulted in the isolation of Menin, which is known to associate with the N-terminus of MLL and LEDGF, an interactor of Menin (Supplemental Table 1, Yokoyama and Cleary, 2008). Following the biochemical isolation of MLL-ELL1, MLL-AFF1, MLL-AF9 and MLL-ENL (Figure 1A), these purified complexes were subjected to Multidimensional Protein Identification Technology (MudPIT) to carry out proteomic analyses for each complex. While the MLL-AF9 and MLL-ENL identified a few of the proteins previously described as ENL associated proteins, including AFF1, AFF4, and Dot1(Mueller et al., 2007; Mueller et al., 2009), the MLL-ELL1 and MLL-AFF1 chimera complexes included AFF4, but notably not Dot1 (Supplemental Table S1). In fact, AFF4, which itself is a rare translocation partner of MLL, is a shared subunit for all of the purified MLL-chimeras (Supplementary Table 1).
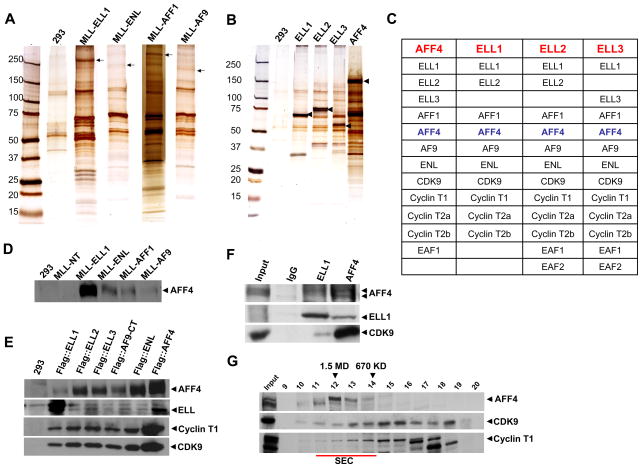
Figure 1. AFF4 is a shared subunit of several of the MLL-chimeras and associates with known RNA polymerase II elongation factors.
(A) Clonal cell lines expressing flag-tagged MLL-ELL1, MLL-ENL, MLL-AFF1 and MLL-AF9 were generated in 293 cells and the resulting protein complexes were purified using the FLAG-affinity purification method and analyzed by SDS-PAGE, silver staining and mass spectrometry. Arrows indicate the position of the Flag-tagged proteins. (B-C) Purification of Pol II elongation factors ELL1, ELL2, ELL3 and AFF4. Clonal cell lines expressing flag-tagged ELL1–3 were generated in 293 cells and the resulting protein complexes were purified and analyzed as in (A). Arrows in (B) indicate the position of the Flag-tagged subunit. (C) All three human ELL paralogs were identified in the Flag-AFF4 purification. ELL1 and its paralogs ELL2 and ELL3 were separately purified and demonstrated a similar set of associated proteins as found in the AFF4 purification. The Flag-ELL1 construct corresponded to the portion of ELL1 in MLL-ELL1 chimeras and lacks the N-terminal Eaf1/Eaf2 interaction domain, explaining the failure to identify these two proteins in this purification. AFF4 was found in all of the Flag-ELLs purifications indicating that it is a component of a novel RNA polymerase II elongation complex. (D–F) Confirmation of an interaction of AFF4 with the MLL-chimeras and components of the P-TEFb elongation factor by Flag and/or endogenous immunoprecipitations. Arrowheads show the position of the protein probed by Western analysis. (D) Flag immunoprecipitations of MLL-chimeras demonstrate an association of AFF4 with all chimeras, but not with a Flag-tagged MLL-N-terminal domain common to all chimeras. (E) Western blot analyses of ELL1, ELL2, ELL3, AF9, ENL and AFF4 immunoprecipitations confirm the observed interactions of Cyclin T1 and CDK9 with these factors. (F) Immunoprecipitations from 293 cells show the endogenous association of P-TEFb with AFF4 and ELL1. (G) Size exclusion chromatography of HeLa nuclear extracts demonstrated that AFF4-containing complexes elute at ~ 1.5 MDa (fractions 11–14, underlined in red) and contain a small portion of the CDK9 and CycT1-containing (P-TEFb) complexes from the nuclear extracts. Fractions corresponding to 1.5 MDa and 670 kDa are indicated with arrowheads. The 1.5 MDa complex is referred to as the super elongation complex (SEC).
Given the unexpected observation of finding this largely uncharacterized protein associating with the MLL chimeras, we therefore generated a cell line expressing Flag epitope-tagged AFF4 and identified associated proteins (Figure 1B-C). Surprisingly, all three ELL proteins were found in the isolated complexes. In turn, expressing and isolating Flag-tagged ELL1, ELL2 and ELL3 revealed AFF4 associating with each ELL (Figure 1B-C). Furthermore, the ELL and AFF4-containing complexes also consist of additional MLL partners, AFF1, ENL, and AF9 (Figure 1C). Another subunit of the AFF4 – ELL1 complex is the component of the Pol II C-terminal domain (CTD) kinase, P-TEFb, consisting of Cdk9 with cyclin T1, T2a or T2b (Figure 1C). We also detected the previously identified ELL-associated factors EAFs (Simone et al., 2003; Simone et al., 2001) in the AFF4, ELL2 and ELL3 complexes (Figure 1C). Since the purification of the ELL1 complexes were performed with ELL1 lacking the first 50 amino acids (missing in the MLL-ELL1 chimera and required for interactions with EAFs), we did not detect any of the EAFs in the ELL1 purifications. Given the fact that EAFs enhance the in vitro transcription elongation properties of ELLs (Kong et al., 2005), it is interesting that we also observe these factors with the AFF4-containing complexes.
The observed interactions between ELL1–3, AFF4, and the components of P-TEFb were also confirmed by Flag and endogenous co-immunoprecipitations (Figure 1D-F). In different preparations and with different tagged subunits, the relative amounts of some subunits in the isolated complexes can vary (e.g. see ELL1 and AFF4 levels in Figure 1D-E). Therefore, it is important to tag and purify multiple subunits to get a clear picture of the complexes in vivo. Indeed, previous interpretations that Dot1, AFF1 and AFF4 exist in a single complex were primarily based on these proteins co-purifying with a single subunit, ENL (Mueller et al., 2007; Mueller et al., 2009). We also find that ENL associates with AFF1, AFF4 and Dot1, but importantly, we find that Dot1 is not associated with AFF1, AFF4 or the ELL complexes indicating that ENL is part of at least two distinct complexes (Supplemental Table 1 and data not shown).
To further characterize the AFF4-containing complexes, we analyzed nuclear extracts by their application to size exclusion chromatography, followed by SDS-PAGE and Western analysis of AFF4 and the components of the P-TEFb elongation complex (Figure 1G). These studies clearly indicate that a small portion of the P-TEFb co-purifies with the AFF4 large complex at about 1.5 MDa (Figure 1G, fraction 11–13), which we call the super elongation complex (SEC) due to the presence of multiple Pol II elongation factors. Overall, these studies reveal that many of the MLL partners found in leukemia, which have very little sequence or seemingly functional similarities, are found in large macromolecular complexes associated with the Pol II elongation factors ELL and P-TEFb.
P-TEFb is a CTD kinase involved in the regulation of transcription elongation by Pol II and can exist in both active and inactive forms (Peterlin and Price, 2006). To determine whether the purified ELL and AFF4-containing complexes contain active P-TEFb, we tested the kinase activity of these purified complexes towards the GST-Pol II C-terminal domain fusion protein (GST-CTD). The ELL2, ELL3, and AFF4 complexes were assayed in the presence and/or absence of ATP and the GST-CTD (Figure 2A). The resulting products were subjected to SDS-PAGE followed by Western analysis with antibodies specific to Pol II CTD either phosphorylated on serine 2 (pS2) or serine 5 (pS5) (Figure 2A). We also tested the autophosphorylation of CDK9 and the possible phosphorylation of AFF4 by P-TEFb (Figure 2A). From these studies, it appears that the purified ELL and AFF4-containing complexes are active as a Pol II CTD kinase. Our studies also suggest that AFF4, which sequence alignment demonstrates similarly bears repeated SP motifs (Figure 2B), is also phosphorylated by P-TEFb (Figure 2A), a phenomenon also observed previously for AFF1 (Bitoun et al., 2007). Similar CTD kinase activities are found in ELL1 and MLL-fusion protein complexes (Supplemental Figure S1).
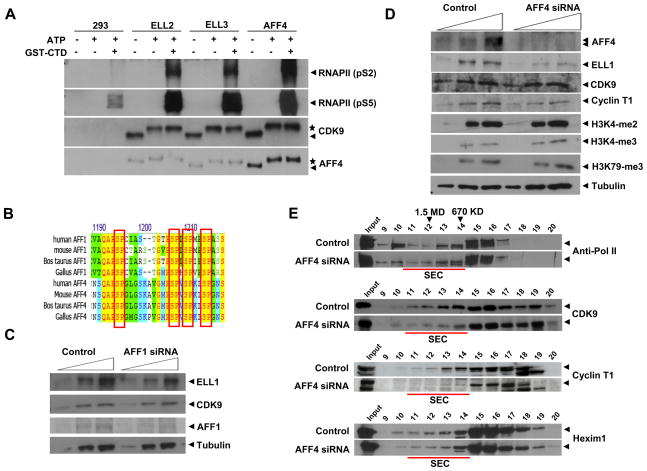
Figure 2. AFF4 is required for the assembly and the enzymatic activity of SEC containing ELLs, P-TEFb and MLL partners.
(A) Pol II C-terminal domain (CTD) Kinase assays with the ELL2, ELL3 and AFF4-containing complexes were performed with GST-Pol II C-terminal domain fusion protein (GST-CTD). ELL2, ELL3, or AFF4 complexes were assayed in the presence of ATP and/or the GST-CTD and subjected to Western blot analyses with antibodies specific to Pol II CTD phosphoserine 2 (pS2) and phosphoserine 5 (pS5), CDK9 and AFF4. Consistent with previous observations, serine 2 and serine 5 of Pol II CTD are good substrates for the P-TEFb complexes in vitro. CDK9, itself, is also known to be autophosphorylated, resulting in a shift in gel migration in SDS- PAGE (indicated by asterisk, while an arrow indicates the faster migrating unphosphorylated form). AFF4 shows a similar gel mobility shift as CDK9, also indicated by an asterisk, suggesting that it is a substrate for P-TEFb as well. See Figure S1 for additional kinase assays. (B) Sequence alignment of a potential site for multiple phosphorylation of AFF4-related proteins bearing SP motifs favored by P-TEFb. (C–D) AFF4, and not AFF1, is required for stability of the SEC containing ELL1, P-TEFb and MLL-partners in HeLa cells. Western blot analysis of ELL1, CDK9 and Cyclin T1 was performed in the presence and absence of AFF1 or AFF4. Nuclear extracts from the siRNA-mediated for AFF4 or AFF1 were analyzed by SDS-PAGE and Western blot analysis. Arrows indicate increasing protein loads. Bulk protein levels of ELL1 are reduced in AFF4, but not AFF1 knockdown in these cells. Bulk protein levels of P-TEFb were not affected by AFF1 or AFF4 RNAi. Global H3K4 and H3K79 methylation levels were not affected by AFF4 knockdown. Tubulin serves as a loading control. (E) Gel filtration analyses of nuclear extracts from control and AFF4-directed siRNA treated cells. Larger P-TEFb-containing complexes, SEC (fractions 10–14 also seen in Figure 1G and indicated by underlining in red) are reduced in AFF4 knockdown cells, indicating that the presence of AFF4 is required for the assembly of SEC.
To determine which of the components of the AFF4 complex are required for complex stability and association with P-TEFb kinase, we reduced the levels of several components of the complex using RNAi (Figure 2C). We observed that the reduction of the AFF4 homologue AFF1 does not alter ELL1 and P-TEFb stability in these cells (Figure 2C, Supplementary Figure S2). However, the loss of AFF4 results in the instability of ELL1 with no significant effect on the stability of the P-TEFb components (Figure 2D). Our studies so far indicate that the AFF4-containing complex associates with a small portion of the P-TEFb components either when the AFF4 complex is purified (Figure 1B-F), or when nuclear extracts were analyzed by size exclusion chromatography (Figure 1G). We, therefore, tested for the association of P-TEFb with the AFF4/ELL complex in the absence of AFF4 (Figure 2E). Nuclear extracts from cells treated with AFF4 RNAi were subjected to size exclusion chromatography and the fractions were analyzed by SDS-PAGE, followed by Western analyses using antibodies specific to Pol II, CDK9, Cyclin T1 and Hexim 1 (Figure 2E). This biochemical analysis demonstrated that reduction in AFF4 levels result in the loss of association of Cdk9 and Cyclin T1 with the large AFF4-containing complex (Figure 2E fractions 11–13), but not the Hexim1-containing P-TEFb complexes (Figure 2E fractions 15–17, see Peterlin and Price, 2006 for a review of known P-TEFb complexes). Together, these results demonstrate that AFF4 is a central component of the P-TEFb/ELL complexes, which we call SEC.
The first in vivo characterization of ELL as a transcription elongation factor was in Drosophila, where there is only one ELL-like protein, dELL (Eissenberg et al., 2002; Gerber et al., 2001). Indeed, the first hint of a connection between P-TEFb was the RNAi-mediated knockdown of Cdk9 and the loss of dELL from chromatin (Eissenberg et al., 2007). Additionally, although it took years to identify the affected genes, dELL and the sole Drosophila homologue of AFF4 (see Supplemental Figure S3) were part of a small set of genes isolated in a screen for Ras signaling components (Eissenberg et al., 2002; Neufeld et al., 1998; Su et al., 2001; Tang et al., 2001; Wittwer et al., 2001). Based on our new findings, we were interested to extend these intriguing links between ELL, P-TEFb and AFF4 in Drosophila.
Since many Drosophila Pol II elongation factors have been shown to associate with elongating Pol II on chromatin and relocalize to heat shock loci upon stress (Ardehali et al., 2009; Eissenberg et al., 2002; Gerber et al., 2001; Gerber et al., 2005y; Gerber et al., 2005b; Smith et al., 2008; Tenney et al., 2006), we generated polyclonal sera to dAFF4 and performed colocalization studies of dAFF4 with dELL and the elongating form of Pol II (Figure 3). dELL and the elongating form of Pol II colocalize extensively with dAFF4 (Figure 3A-B), not seen with preimmune sera (data not shown). The Hsp70 loci in Drosophila have been used as a model system for studying transcription elongation. The Hsp70 loci contain poised polymerase, which upon heat shock is phosphorylated at serine 2 in the CTD repeats by P-TEFb, allowing productive transcription (Lis et al., 2000). We assayed the presence of dAFF4 at Hsp70 after heat shock and observe that indeed dAFF4 colocalizes with dELL and the elongating form of Pol II on polytene chromosomes at major heat shock loci, including the Hsp70 genes at 87A and 87C (Figure 3C-D, Supplemental Figure S4). Chromatin immunoprecipitation of dAFF4 shows that it becomes associated with Hsp70 upon heat shock, and is present throughout the transcribed unit (Figure 3E), similar to what was previously observed for P-TEFb (Boehm et al., 2003).
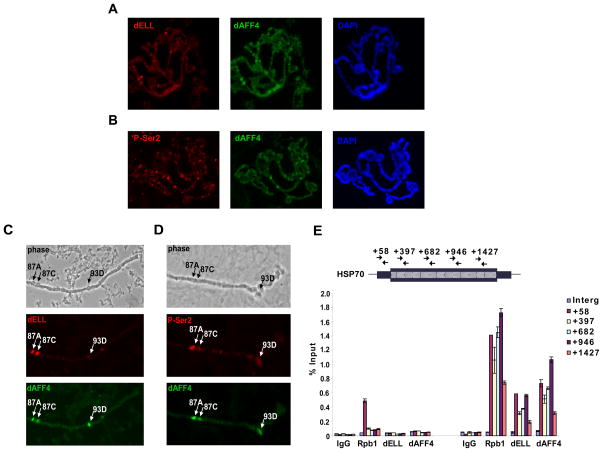
Figure 3. The Drosophila ortholog of AFF4 colocalizes with ELL and the elongating form of Pol II on Drosophila polytene chromosomes.
(A and B) Polytene chromosome preparations from 3rd instar larval salivary glands were probed with antibodies to dELL (A, red) or the H5 monoclonal antibody recognizing the Ser-2 phosphorylated (P-Ser2), elongating form of Pol II (B, red). Both antibodies show substantial colocalization with dAFF4 (A and B, green). Chromosomes were counterstained with DAPI (A and B, blue). (C and D) Polytene chromosomes were prepared from heat shocked 3rd instar larvae and stained as in (A and B). Phase contrast images show positions of the 87A, 87C and 93D major heat shock loci. dAFF4 is recruited along with dELL at these loci after heat shock, associated with the P-Ser2 form of RNA Pol II. See Figure S4 for additional images. (E) Chromatin immunoprecipitation of dAFF4 and RNA Pol II large subunit (Rpb1) at Hsp70 before and after 10 minutes of heat shock at 37° C. While Rpb1 is present at Hsp70 prior to heat shock, dAFF4 can only be detected at Hsp70 after heat shock, where it is found throughout the transcription unit along with Pol II. Hsp70 primers have been previously described (Boehm et al., 2003). Error bars represent standard deviations.
We next asked if AFF4 was similarly recruited to the human HSP70 gene. Using chromatin immunoprecipitation (ChIP), AFF4 levels were measured across the human HSP70 gene before and after heat shock in HeLa cells (Figure 4A-B, Supplemental Figure S5). Upon heat shock, AFF4 is found at the HSP70 promoter and throughout the transcribed region along with RNA Pol II. Interestingly, ELL2 and ELL3 are also recruited to the 5′ end of HSP70, but are not significantly enriched as far into the body of the gene as AFF4. This could reflect different sensitivities of the antibodies employed, or conceivably to differential usage of the elongation factors in this complex at distinct steps of transcription elongation. ELL1 antibodies did not work in any of our ChIP assays, which we believe reflects the relative higher quality of ELL2 and ELL3 antibodies, and not the differential usage of these factors on chromatin.
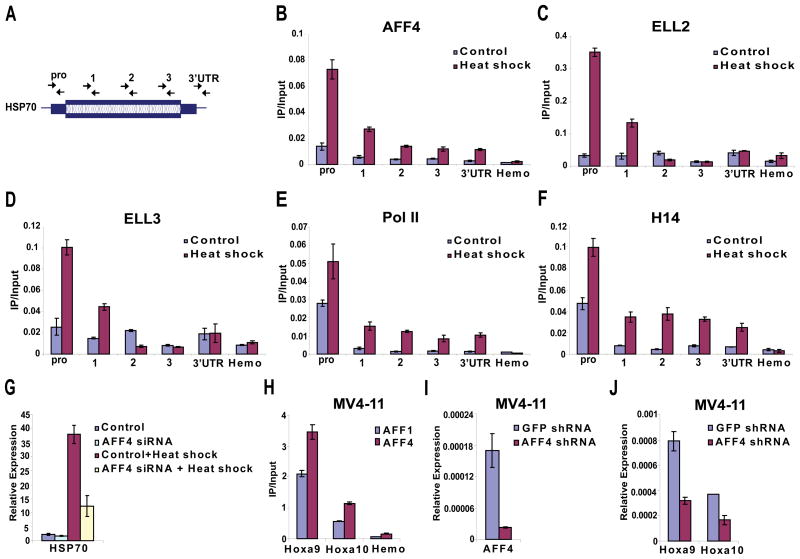
Figure 4. AFF4 is required for HSP70 induction and is recruited to MLL chimera target genes in leukemic cells.
(A–G) HeLa cells were heat shocked by incubation at 42° C for 2 hours. Non-heat shocked and heat-shocked cells were used in chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assays with AFF4, ELL2, ELL3, general Pol II and the H14 monoclonal antibody recognizing Serine 5 phosphorylated form of Pol II (B–F). (A) Position/primer pairs used for QPCR along the HSP70 gene are indicated. (B–F) AFF4 is recruited to the HSP70 gene after heat shock along with ELLs and RNA polymerase II. (G) Knockdown of AFF4 in HeLa cells by RNAi inhibits HSP70 induction. Control and AFF4 siRNA-treated cells were heat shocked as in (A) and HSP70 mRNA levels were assessed by quantitative RT-PCR and normalized to GAPDH mRNA levels. Non-heat shock control and AFF4 siRNA-treated cells are shown for comparison. (H-J) Recruitment of AFF4 to genes induced by the MLL-AFF1 chimera in MV4–11 cells. (H) Antibodies to the C-terminal domain of AFF1 found in the MLL chimera and antibodies raised against the N-terminal domain of AFF4 were used in ChIP-qPCR assays at HOXA9 and HOXA10 loci, known targets of the MLL-chimera found in human leukemia. As expected AFF1, shows recruitment to HOXA9 and HOXA10 in the leukemic MV4–11 cells. AFF4 is also recruited to HOXA9 and HOXA10, consistent with its co-purification with the MLL-AFF1 chimera in Figure 1A and Supplementary Table 1. Similar findings were observed with sera from two immunized rabbits (data not shown). The beta globin gene (Hemo), which is not expressed in HeLa or MV4–11 cells, is used as a negative control in (B–F and H). Note that experiments in Figure 4B and 4H were done with crude 4 week serum or affinity purified from 10 week serum, respectively and were normalized with different amounts of input DNA. See Figure S6 for additional ChIP experiments. (I) Knockdown of AFF4 in the leukemic MV4–11 cells by retroviral introduction of a shRNA targeting AFF4. (J) Reduction of HOXA9 and HOXA10 expression in MV4–11 cells after AFF4 knockdown. GFP shRNA is used as a non-targeting control shRNA. Expression levels were measured by quantitative RT-PCR and normalized to 18S rRNA. Error bars represent standard deviations.
The effect of AFF4 recruitment to HSP70 was assessed by siRNA-mediated knockdown of AFF4. Knockdown of AFF4 leads to a defective heat shock response, showing reduced induction of HSP70 compared to control siRNA-treated cells (Figure 4G). While HSP70 is used as a model gene for studying transcription elongation, we recognize that other factors, not known to directly stimulate transcription elongation, also travel with the polymerase, such as components of the exosome (Andrulis et al., 2002). However, based upon the proven in vitro stimulation of transcription elongation by ELL1, the requirement for AFF4 in the stability of the P-TEFb-AFF4-ELL complex, the association of AFF4 with HSP70 upon heat shock and its requirement for full expression of HSP70, we propose that AFF4 is a central component of the SEC complex.
To begin to investigate the role of AFF4 as a common component of complexes formed by MLL chimeras, we assessed the recruitment of AFF4 to HOXA9 and HOXA10 loci in the MV4–11 cell line from a patient with a MLL-AFF1 translocation. As with many MLL translocations, HOXA9 and HOXA10 are up-regulated in these cells. Indeed, chromatin immunoprecipitation with antibodies corresponding to the C-terminal portion of AFF1, which is contained in this MLL chimera, shows recruitment to HOXA9 in the MV4–11 cells (Figure 4H), as well as in another MLL-AFF1 fusion cell line SEM, but not in an unrelated leukemia cell line, REH (Supplemental Figure S6). Interestingly, AFF4 is also recruited to HOXA9 and HOXA10 in the MV4–11 cells, despite the fact that it is the related AFF1 gene that is involved in the MLL translocation in these cells. The antibody to AFF4 was raised against an amino-terminal portion not found in MLL chimeras, ruling out cross-reaction with the related AFF1 protein that is part of the MLL chimera. To assess the functional significance of AFF4 recruitment to MLL-AFF1 target genes, we performed lentiviral delivery of shRNA to the MV4–11 cells. Significant reductions of Hoxa9 and Hoxa10 are observed upon knockdown of AFF4 in these cells (Figure 4I-J). Together, these findings lend support to our hypothesis that AFF4, a very rare translocation partner of MLL is nonetheless a component of many MLL-fusion protein complexes and participates in leukemogenesis. This finding is supported by our initial biochemical purifications of the MLL-chimeras (Figure 1A) and now with the analysis of the MLL-target genes from cell lines generated from a MLL-translocation patient.
Previous studies have provided evidence for links among different MLL translocation partners. ENL, AF9 and AF10 have been linked to the histone methyltransferase Dot1; and it was suggested that a common mechanism of MLL-translocation-based leukemia was through H3K79 methylation by Dot1 (Bitoun et al., 2007; Krivtsov et al., 2008; Mueller et al., 2007; Mueller et al., 2009; Okada et al., 2005). However, the most common translocation partner of MLL is AFF1, which our present studies show does not associate with Dot1. Other studies suggesting a physical interaction between Dot1 and AFF1 were based on the isolation of these two proteins in ENL immunoprecipitates and through building a network of 2-hybrid and other interactions. However, we have determined the proteins associated with some of the commonly occurring MLL chimeras found in infant leukemias. MLL-AFF1 does not physically associate with Dot1, so a role for Dot1 at genes upregulated in MLL-AFF1 leukemias may be subsequent to gene activation by this MLL chimera. We have recently shown that Dot1-mediated H3K79 methylation is linked to cell cycle control in yeast (Schulze et al., 2009) and methylation by Dot1 could also have some role in transcriptional enhancement in leukemogenesis. In contrast, MLL-AFF1 co-purified the SEC complex containing ELL1 and P-TEFb, two proven transcription elongation factors in vitro and in vivo, each with demonstrated abilities to activate transcription through transcription elongation. It will be interesting and essential to further determine the mechanisms and regulation of the SEC complex in transcriptional control in normal development and leukemogenesis. Both AFF1 and AFF4 copurify with the ELL proteins and another AFF protein, AFF3, which is also a rare translocation partner with MLL (von Bergh et al., 2002). The related AFF2 gene (FMR2) is silenced in a form of mental retardation (Knight et al., 1993), thus implicating all members of this family in human diseases (Bitoun and Davies, 2009). P-TEFb itself is involved in a number of malignancies and developmental diseases (Romano and Giordano, 2008), and it will be intriguing to determine which of these processes involve SEC or SEC-like complexes.
We originally proposed that ELL1, which we had identified as an activity from rat liver extracts that stimulated transcription elongation in vitro, and is homologous to the ELL gene in humans involved in chromosomal translocations in leukemia, is linked to transcription elongation by Pol II to leukemogenesis (Shilatifard et al., 1996). Recently, other groups have continued to propose a role for MLL-translocation partners with enhancing transcription elongation of the target genes of MLL chimeras (Bitoun et al., 2007; Krivtsov et al., 2008; Mueller et al., 2007; Mueller et al., 2009). These recent studies have focused primarily on a proposed Dot1-AFF1 interaction, which we now believe was the result of the isolation of ENL. Our evidence indicates that ENL participates in two distinct complexes, one with Dot1 and one within the SEC. An important area for future investigation is to define the relative contributions of these two types of complexes to leukemogenesis. This will require extensive conventional biochemical studies in hematopoietic cells bearing the relevant MLL chimeras.
In this study, we have demonstrated the following: 1) Many of the MLL fusion partners appear to interact with each other within a large macromolecular complex named SEC. 2) These large macromolecular complexes containing the MLL fusion partners also consist of several of the known Pol II elongation factors including ELL1, ELL2, ELL3 and the components of the P-TEFb kinase, Cdk9 and cyclin T1, T2a and T2b. 3) AFF4, which itself is a rare translocation partner of MLL, is a common factor associated with the purified MLL chimeras and Pol II elongation complexes. 4) AFF4 is required for the proper assembly and activity of large elongation complexes and association with P-TEFb. 5) AFF4 is a core component of the Pol II elongation complex containing ELLs and P-TEFb, associates with the elongating Pol II and relocalizes to heat shock puff sites upon stress, and is required for proper heat shock gene expression by Pol II. 6) We have demonstrated that AFF4 relocalizes to the MLL target genes HOXA9 and HOXA10 in cells bearing an MLL-AFF1 fusion, where it is required for the expression of these genes.
Collectively, the results of this study identify AFF4 as a component of the Pol II elongation complexes consisting of ELLs, P-TEFb and several of the common MLL fusion partners. These findings could prove critical for understanding the etiology of MLL-translocation based leukemias and for identifying additional targets for the treatment of the hematological malignancies resulting from these translocations, as well as for understanding fundamental aspects of transcription elongation control in development.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies recognizing tubulin and Flag tag were obtained from Sigma, H3K4me2, H3K79me3, CDK9, pS2 and pS5 (Phospho Pol II CTD), Cyclin T1, Hexim1 were obtained from Abcam, H5 and H14 (Phospho CTD) from Covance. H3K4me3 and general Pol II antibodies were previously described (Wang et al. 2009). ELL1 mouse monoclonal (recognizing an epitope within the first 100 amino acids (aa)), full length ELL2, full length ELL3, AFF1(aa 764–931), AFF4(aa 73–237), and Drosophila AFF4 (aa 1427–1673 of the longest isoform), human AFF1 and AFF4 were expressed as His-tag fusion proteins in PET-16b, purified on NTA-agarose according to Qiagen’s protocol and sent to Pocono Rabbit Farm and Labs for immunization into rabbits, except for dAFF4 which was injected into guinea pigs. Drosophila Rpb1 antibody was raised in rabbits against the synthetic peptide ERLMKKVFTDDVIKEMTDSG(C) conjugated via cysteine to KLH. dELL antibody and polytene chromosome immunostaining were previously described (Smith et al. 2008).
Expression Plasmids and Cell Lines
MLL-ELL1 cDNA was previously described (DiMartino et al. 2000). MLL-AF9, MLL-AFF1 and MLL-ENL cDNAs were a gift from Dr. Jay Hess (University of Michigan). AFF4 cDNA was obtained from Open Biosystems. ELL1, ELL2 and ELL3 cDNAs were previously described (Johnstone et al., 2001). Flag-tagged cDNAs were cloned into pCDNA5/FRT-TO vector (Invitrogen) modified with an N-terminal flag tag. The plasmids were then transfected into 293 Flp-in-TRex cells and selected by hygromycin. The single clones were picked and cultured up to 3 liters. The 293 Flp-in-TRex cells were grown in suspension with CD 293 medium (Invitrogen) as described by the manufacturer. MV4–11 cell line was a gift from Dr. Mike Thirman (U. Chicago, IL) and the SEM and REH cell lines were obtained from the ATCC. All three leukemia cell lines were grown according to the ATCC’s recommendations.
Flag purification and MudPIT analysis
Nuclear extracts were prepared and subjected to anti-Flag agarose immunoaffinity chromatography. Trichloroacetic acid-precipitated protein mixtures from purifications were digested with endoproteinase Lys-C and trypsin (Roche) and subjected to MudPIT analysis as previously described (Wu et al., 2008).
Immunoprecipitations and kinase assays
Approximately 107 cells for each assay were collected, washed with phosphate-buffered saline once, and lysed in high-salt lysis buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 mM dithiothreitol) containing proteinase inhibitors (Sigma). After incubation at 4°C for 30 min, the lysate was cleared twice by centrifugation at 4°C. The balance buffer (20 mM HEPES [pH 7.4], 1 mM MgCl2, 10 mM KCl) was added to the resulting supernatant to make the final NaCl concentration 300 mM. The lysate was then mixed with antibodies and protein A beads or with anti-Flag agarose (Sigma). After incubation at 4°C for 4 h, the beads were spun down and washed three times with wash buffer (10 mM HEPES [pH 7.4], 1 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100) before eluting by boiling in SDS gel sample buffer. Kinase assays done as previously described (Bitouin et al. 2007).
RNAi, RT-PCR and Chromatin Immunoprecipitation
AFF1 and AFF4 SMARTpools from Dharmacon were used for all siRNA experiments. RNA was extracted with RNeasy from Qiagen and RNA levels were measured with Qiagen SYBR green 1-step RT-PCR reagent. Nuclear extract was prepared as described above and subjected to Western blotting or Superose 6 size exclusion analysis on the SMART system. ChIP and QPCR for human cells was done as previously described (Wang et al. 2009). HeLa cell heat shock experiments were performed by incubating sub-confluent cells at 42° C for 2 hours. Drosophila S2 cells (D-mel2 cells, from Invitrogen) were heat shocked by adding prewarmed media to cells and incubating at 37° C for 10 minutes before fixation and chromatin immunoprecipitation. Hsp70 primers were previously described (Boehm et al., 2003). Intergenic primers to a non-transcribed region of chromosome 2L are, 5 prime to 3′, TCGTGAAATGTTTGCTACTCGAATA and AATTGCATCGCAACACAATGAG. Enrichment was measured using a standard curve of input DNA.
AFF4 shRNA construct was purchased from Open Biosystems (TRCN0000015823) and contains 10 mismatches with the related AFF1 gene over the 23 nucleotide hairpin region. Lentiviral particle preparation and infection was done as previously described. Briefly, 293T cells were plated at 60% confluency in 150 mm dishes in DMEM supplemented with 10% FBS. After incubation for 14 hours (37 °C, 5% CO2), 293T cells were co-transfected with 8μg of AFF4 shRNA or GFP control shRNA, 2.5 μg of packaging plasmids containing gag, tat and rev genes and 2.5 μg of VSV-G expressing plasmid using FuGENE 6 (Roche). The media was replaced with fresh DMEM supplemented with 10% FBS after 16 hours of transfection. The lentiviral supernatants were collected 48 to 72 hours after the transfection, filtered through 0.22 μm filters and concentrated at 18K rpm for 2 hours. For lentiviral infection of MV4–11 leukemia cells, 1×105 cells were seeded in RPMI 1640 media. Polybrene (Sigma) was added at a final concentration of 8μg/ml. After adding 50 μl lentiviral particles (MOI ~10), spin transduction was performed at 2000 RPM for 120 minutes at 32° C. 6 hours after infection, the media was replaced with 100 μl of RPMI 1640 media supplemented with 10% FBS and 5 ng/ml recombinant human granulocyte M-CSF (GM-CSF) (Prospect Protein Specialist). Cells were incubated at 37°C for four days before RNA extraction and RT-PCR as described for the siRNA experiments.
Supplementary Material
Acknowledgments
We thank Dr. Michael Thirman (U. Chicago) for providing the leukemic MV4–11 translocation cell line and Dr. Linheng Li for lentiviral packaging plasmids. We thank Tari Parmely and the Stowers Institute Tissue Culture Facility for assistance with growing cell lines. We are also grateful to Laura Shilatifard for editorial assistance. This work was performed to fulfill, in part, requirements for CQL’s PhD thesis research as a student registered with the Open University. The study was supported in part by a grant from National Institute of Health R01CA089455-10 to ASH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Davies KE. The robotic mouse: understanding the role of AF4, a cofactor of transcriptional elongation and chromatin remodelling, in purkinje cell function. Cerebellum. 2009;8:175–183. doi: 10.1007/s12311-009-0101-0. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics. 2007;277:101–114. doi: 10.1007/s00438-006-0164-2. [DOI] [PubMed] [Google Scholar]
- Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. Embo J. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Tenney K, Conaway JW, Conaway RC, Eissenberg JC, Shilatifard A. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J Biol Chem. 2005a;280:4017–4020. doi: 10.1074/jbc.C400487200. [DOI] [PubMed] [Google Scholar]
- Gerber MA, Shilatifard A, Eissenberg JC. Mutational analysis of an RNA polymerase II elongation factor in Drosophila melanogaster. Mol Cell Biol. 2005b;25:7803–7811. doi: 10.1128/MCB.25.17.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Gerber M, Landewe T, Tollefson A, Wold WS, Shilatifard A. Functional analysis of the leukemia protein ELL: evidence for a role in the regulation of cell growth and survival. Mol Cell Biol. 2001;21:1672–1681. doi: 10.1128/MCB.21.5.1672-1681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Flannery AV, Hirst MC, Campbell L, Christodoulou Z, Phelps SR, Pointon J, Middleton-Price HR, Barnicoat A, Pembrey ME, et al. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci U S A. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, Tang AH, Rubin GM. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics. 1998;148:277–286. doi: 10.1093/genetics/148.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, Conaway JW, Conaway RC. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci U S A. 1997;94:3639–3643. doi: 10.1073/pnas.94.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355–2362. doi: 10.1182/blood-2002-06-1664. [DOI] [PubMed] [Google Scholar]
- Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood. 2001;98:201–209. doi: 10.1182/blood.v98.1.201. [DOI] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su MA, Wisotzkey RG, Newfeld SJ. A screen for modifiers of decapentaplegic mutant phenotypes identifies lilliputian, the only member of the Fragile-X/Burkitt’s Lymphoma family of transcription factors in Drosophila melanogaster. Genetics. 2001;157:717–725. doi: 10.1093/genetics/157.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Rubin GM, Muller HA. Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian. Development. 2001;128:801–813. doi: 10.1242/dev.128.5.801. [DOI] [PubMed] [Google Scholar]
- Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergh AR, Beverloo HB, Rombout P, van Wering ER, van Weel MH, Beverstock GC, Kluin PM, Slater RM, Schuuring E. LAF4, an AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2002;35:92–96. doi: 10.1002/gcc.10091. [DOI] [PubMed] [Google Scholar]
- Wittwer F, van der Straten A, Keleman K, Dickson BJ, Hafen E. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development. 2001;128:791–800. doi: 10.1242/dev.128.5.791. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.