Abstract
ALR/Lt, a NOD-related mouse strain, was selected for resistance to alloxan free radical mediated diabetes (ALD). Despite extensive genomic identity with NOD (>70%), ALR mice display strong resistance to autoimmune Type 1 diabetes (T1D) due to an unusual elevation in both systemic antioxidant defenses as well as ROS production that extend to the beta cell level. Reciprocal backcross to NOD previously linked the ALR-derived T1D resistance to Chr. 3, 8, and 17 as well as to the ALR mt-Nd2a allele encoded by the mitochondrial genome (mtDNA). To determine whether any of the ALR-derived loci protecting against T1D also protected against ALD, 296 six-wk-old F2 mice from reciprocal outcrosses were alloxan treated, assessed for diabetes onset, and a genome wide scan (GWS) was conducted. GWS linked mt-Nd2 as well as three nuclear loci with alloxan-induced diabetes. A dominant ALR-derived ALD resistance locus on Chr. 8 co-localized with the ALR-derived T1D resistance locus identified in the previous backcross analysis. In contrast, whereas ALR contributed a novel TID resistance locus on Chr. 3 marked by Susp, a more proximal ALR-derived region marked by Il-2, contributed ALD susceptibility, not resistance. In addition, a locus was mapped on Chr. 2, where heterozygosity provided heightened susceptibility. Tests for alloxan sensitivity in ALR conplastic mice encoding the NOD mt-Nd2c allele and NOD mice congenic for the protective Chr. 8 locus supported our mapping results. Alloxan sensitivity was increased in ALR.mtNOD mice, whereas it was decreased by congenic introduction of ALR genome on Chr. 8 into NOD. These data demonstrate both similarities and differences in the genetic control of T1D versus ROS-induced diabetes.
Keywords: diabetes, mice, genetics, free radicals, autoimmunity
INTRODUCTION
Pancreatic beta cells are particularly sensitive to killing via reactive oxygen species (ROS), whether generated by the complex proinflammatory environment of an autoimmune cellular infiltrate (insulitis), or by the free radical generating toxin alloxan [1, 2]. This exquisite sensitivity has been proposed to play a role in the pathogenesis of T1D [3–12]. Production of ROS and down-regulation of antioxidant defenses characterized by a reduced glutathione [GSH] level and a progressive decline in the transcripts for catalase [CAS], superoxide dismutase [SOD], and thioredoxin [TRX] have been observed in apoptotic processes [13–15]. Hence, the ability to maintain redox potential in beta cells may counteract apoptotic signaling, protecting beta cells and inhibiting T1D [4, 5, 16, 17]. Indeed, GSH, SOD, and TRX have all been shown to inhibit apoptotic signaling by blocking the actions of apoptosis signal-regulating kinase [ASK1], AP-1, and NF-kB in islets [18–20]. Recombinant TRX has been shown to protect cells against apoptosis mediated through TNF and Fas pathways [21], and when overexpressed in beta cells, TRX protects against T1D in transgenic NOD mice [22]. The rapid rejection of islet grafts in diabetic NOD recipients has also been attributed in part to damage mediated by ROS, and treatment with ROS scavengers slowed allograft rejection [23, 24]. Likewise, increasing the stress response in NOD islets can prevent or reduce immune mediated damage including rejection of syngeneic islet grafts in recurrent autoimmune disease [22, 25, 26]. Therefore, identification of genetic elements that reduce cellular ROS production or increase anti-radical defenses could allow for better protection of islets both before and after diabetes onset. Studies with the ALR mouse have strongly supported this concept [3–6, 16, 17, 27, 28].
ALR mice were selected for resistance to alloxan-mediated diabetes [29]. Alloxan is a diabetogenic agent that destroys pancreatic beta cells by free radical damage [30, 31]. Alloxan, a glucose mimetic, is selectively transferred into beta cells where it decomposes to produce hydroxyl radicals. Inbred mouse strains are not equally susceptible to alloxan-induced diabetes. ALR/Lt represents an ICR-derived strain selected for resistance to alloxan at doses that are diabetogenic to related strains [29]. One of the most closely-related strains is NOD, sharing >70% genetic identity, including the diabetogenic MHC class II alleles. Previous studies between these two related strains have demonstrated an unusually strong ability of ALR islets to resist both cellular and cytokine-mediated beta cell killing that typifies T1D pathogenesis in NOD mice [5]. This resistance to autoimmune T1D development was correlated with the ALR strain’s ability to dissipate ROS systemically and at the beta cell level. Genetic analysis has demonstrated that resistance was, in part due to an interaction between nuclear and mitochondrial genes [3]. ALR-contributed loci on Chr. 3 and 8 (Idd22), as well as an MHC-linked locus on Chr. 17 (Idd16), were linked to T1D resistance in segregating populations following outcross of NOD and ALR [28]. The ALR-contributed linkage on Chr. 3 was correlated with both heightened superoxide dismutase 1 (SOD1) activity as well as suppression of superoxide generation in activated neutrophils and designated Susp [28]. In addition, reciprocal outcrosses between these two strains identified a single nucleotide polymorphism in mt-Nd2 encoded by the mitochondrial genome (mtDNA) unique to ALR [3].
While this prior analysis allowed for the association of loci with autoimmune diabetes, this analysis was not able to delineate the linkages that protected at the beta cell level from linkages that controlled dysfunction at the level of the immune system. Outcrosses between T1D-susceptible NOD mice with TID-resistant strains have identified well over 30 chromosomal loci exerting control of autoimmune diabetes development, there is little information as to what chromosomal loci distinguish mouse strains susceptible or resistant to chemically-induced diabetes, and whether there may be genetic overlap. Because beta cells in NOD and ALR islets are diametric opposites both in terms of susceptibility to spontaneous T1D development in vivo and the ability to withstand stress mediated by cytokines and dissipate ROS in vitro, we initiated a genetic analysis to identify loci controlling diabetogenic responsiveness to alloxan. We were particularly interested to determine which, if any, of these overlapped with the loci identified to confer resistance to spontaneous T1D.
METHODS
Mice
Breeding colonies of NOD/ShiLtJ and ALR/Lt were maintained at The Jackson Laboratory and the Children’s Hospital of Pittsburgh. Reciprocal F1 crosses were initiated (NOD females crossed to ALR males, F1 denoted as DR, and ALR females crossed to NOD males, F1 denoted as RD hereafter). From these reciprocal F1 mice, the four classes of F2 genotypes shown in Table 1 were produced. NOD/ShiLtJ.mtALR/LtJ/Mx mitochondrial conplastic mice (henceforth designated NOD.mtALR) and ALR/LtJ.mtNOD/ShiLtDvs (ALR.mtNOD) were produced and maintained as previously described [6]. The NOD.mtALR mice carried the ALR allele of mt-Nd2 that results in a L to M amino acid substitution at residue 276 of this complex I subunit. Alloxan sensitivity of these conplastic mice was tested at N12F2. To test whether the ALR locus on Chr. 8 contributing to T1D resistance (provisionally designated Idd22) also controlled alloxan responsiveness, NOD.ALR-(D8Mit205-D8Mit137)/Mx marker assisted congenic mice (henceforth designated as NOD.ALRc8) were created by crossing NOD/ShiLtJ to ALR/LtMx followed by 10 generations of successive backcrossing as previously described [6]. Homozygous congenic mice were then obtained by intercrossing N10 mice and testing progeny for alloxan resistance at F4.
Table 1.
Incidence of alloxan induced diabetes in F2 crosses
| F1 Intercross | Total Incidence | Female Incidence | Male Incidence | Y Chr | mtDNA |
|---|---|---|---|---|---|
| DRxDR | 23.6% | 19.1%(9/47) | 28.6%(12/42) | R | D |
| RDxRD | 12.3% | 10.3%(3/29) | 13.6%(6/44) | D | R |
| DRxRD | 35.8% | 18.2%(4/22) | 44.4%(20/45) | D | D |
| RDxDR | 11.9% | 11.9%(5/42) | 12.0%(3/25) | R | R |
D = NOD; R = ALR. For cross directions the female breeders appear first.
All mice used were bred and maintained in the specific pathogen-free research animal facility at either The Jackson Laboratory, Bar Harbor, ME, or the Children’s Hospital of Pittsburgh, Pittsburgh, PA. Mice were allowed free access to acidified drinking water and NIH-31 diet (Purina Mills, Richmond, IN, containing 6% fat for NOD and both F1 and F2 progeny, but only 4% fat for ALR breeders. All procedures involving in the use of animals were approved by the Animal Care and Use Committees at both institutions.
Alloxan treatment
Alloxan was prepared in PBS immediately prior to injection and kept on ice during the procedure. F2 mice at 6 week of age were treated with alloxan at 52mg/kg body [3, 16]. For dose-dependent sensitivity test for conplastic and congenic male mice, alloxan was injected at different doses as indicated. Alloxan was administered via tail vein injection.
Plasma or Blood glucose measurement
For determination of plasma glucose, blood was drawn, without previous fasting, immediately prior to the injection and then one week after alloxan treatment. Mice that did not develop hyperglycemia by seven days were screened at 2 weeks after alloxan injection. Plasma glucose was determined using a Beckman Glucose Analyzer II (Beckman Instrument, Fullerton, CA). Alloxan-induced diabetes was diagnosed on non-fasting plasma glucose values ≥ 250mg/dl. Blood glucose in congenic and conplastic mice was measured using a glucometer (Freestyle Flash, Alameda, CA) before as well as 7 and 14 days after treatment with different alloxan doses.
DNA extraction and genome-wide scan
All F2 mice were sacrificed after two sequential blood glucose readings greater than 250 mg/dL or if diabetes did not occur, two weeks after alloxan administration. At time of sacrifice, a kidney was removed from each mouse and frozen immediately in liquid nitrogen, then kept at −80°C until DNA was extracted. DNA was purified from kidney using standard methods [27, 28]. Genome-wide scan was performed using a total of 152 markers (71 microsatellite and 81 SNP markers) that distinguish NOD and ALR (Supplemental Table 1). Microsatellite primer pairs were used as previously described [27, 28]. Fifty-eight of the SNPs were typed by KBioscience (Hoddesdon, UK). The other twenty-three SNPs were typed via Pyrosequencing as previously described [3, 6]. The primers for pyrosequencing (Supplemental Table 2) were designed using PSQ Assay Design (Biotage AB, Uppsala, Sweden) and Integrated DNA Technology software (Coralville, IA). Marker positions are based on Ensembl (http://www.ensembl.org/Mus_musculus/index.html) and NCBI (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Linkage analysis and statistics
The association of nuclear and mitochondrial genotypes with alloxan-induced diabetes incidence is determined by χ2 test using the JMP software (SAS institute, inc) using P<0.01 for suggestive and P<0.001 for significant linkages. QTL linkage analysis is performed with R/qtl (The Jackson Laboratory, Bar Harbor, ME. http://www.rqtl.org/) [32]. Thresholds for LOD scores are based on 1000 permutation tests [33]. In this intercross the values of LOD score thresholds are 2.2 and 3.2 for suggestive (P<0.63) and significant (P<0.05) levels, respectively, when sex is included as an additive covariate. Confidence intervals of QTLs are determined by using the posterior probability density method [34].
RESULTS
mt-Nd2c is linked with higher sensitivity to alloxan induced diabetes
We showed previously that alloxan at a dose of 52 mg/kg is diabetogenic to NOD but not ALR mice [3]. Further, (ALRxNOD)F1 hybrids are resistant to diabetes induced by this alloxan dose when ALR is the maternal parent, whereas (NODxALR)F1 mice are susceptible when NOD is the maternal parent [3]. Table 1 shows the effect of direction of cross (affecting both the mitochondrial genome and the male sex chromosome origin) on alloxan diabetes induction in the entire cohort of 296 F2 mice. As expected, males (n=156) were more sensitive to alloxan than females (n=140) (p=0.01). The highest diabetes induction (44.4%) was in F2 males inheriting both their Y Chromosome and their mitochondrial genome from NOD. However, neither the NOD nor the ALR Y Chromosome was linked to alloxan sensitivity (χ2=0.92, P=0.34). Regardless of Y Chromosome origin, F2 males inheriting their mtDNA from ALR exhibited lower diabetes incidence than those with the NOD mitochondrial genome (χ2=11.19, P=0.0008). All mice, females and males, inheriting the ALR mtDNA were significantly more protected against diabetes than those inheriting the NOD mitochondrial genome (χ2=12.025, P=0.0005). As noted above, the NOD and ALR mtDNA only differ at a single SNP in mt-Nd2, with NOD harboring a C (mt-Nd2c) and ALR an A (mt-Nd2a) at nucleotide 4738 [3]. This sequence variation leads to an amino acid substitution, leucine to methionine, respectively.
Genome-wide scan for alloxan resistance
Genome wide scan of alloxan-treated mice of both sexes (n=296) revealed suggestive evidence for linkages on Chr. 2 (LOD=2.2), 3 (LOD=2.6), and 8 (LOD=2.2). The peak of linkage and the confidence interval (CI) on each chromosome is: Chr. 2, rs3711897 at 87.4Mb, χ2=10.169, P=0.0062, 95% CI=64–145Mb; Chr. 3, D3Nds6 at 36.9 Mb, χ2=12.097, P=0.0024, 95% CI=20–50.7 Mb; Chr. 8, rs3699199 at 77.2Mb, χ2=9.992, P=0.0068, 95% CI=61–103 Mb.
The alloxan diabetes protective locus mapping to Chr. 8 overlaps with Idd22
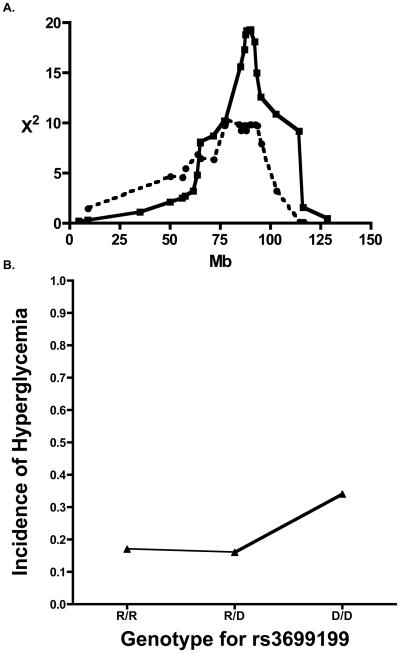
Previously, an ALR-contributed T1D-protective locus, Idd22, was mapped to Chr. 8 using a backcross of (NOD x ALR)F1 mice to NOD [28]. In the present study, genome-wide scan of 296 F2 individuals mapped an ALR-derived alloxan-resistance locus to Chr. 8. The peak linkage was at 77.2Mb (rs3699199) for alloxan-induced diabetes overlapping with the previously reported T1D resistance locus (Fig. 1A). Analysis of mice with each genotype clearly demonstrates that this ALR inherited alloxan resistance allele provides dominant protection as the incidence of diabetes in mice with only a single ALR allele was equal to that of mice with two copies of the ALR allele (16% and 17%, respectively, Fig. 1B) and significantly less that mice that were homozygous for the NOD allele (34%).
Fig. 1.
ALR-derived loci on Chromosome 8 provide resistance against both T1D and alloxan-induced diabetes. (1A) Linkage of ALR-derived protection against alloxan-induced diabetes on Chromosome 8 (dotted line, peak χ2 =9.992, P=0.0068) overlaps with previously reported and recently updated linkage against type 1 diabetes (solid line). (1B) Effect plot of phenotype by the single nucleotide polymorphism marker rs3699199. The ALR allele provides dominant protection against the development of diabetes as mice inheriting R/R and R/D have an equal incidence of hyperglycemia that is significantly reduced compared to mice that inherited both alleles of rs3699199 from NOD. Incidence of alloxan-induced Diabetes is on vertical axis while genotype is on the horizontal axis (D = NOD; R = ALR).
An unexpected alloxan susceptibility allele contributed by ALR was mapped on Chr. 3
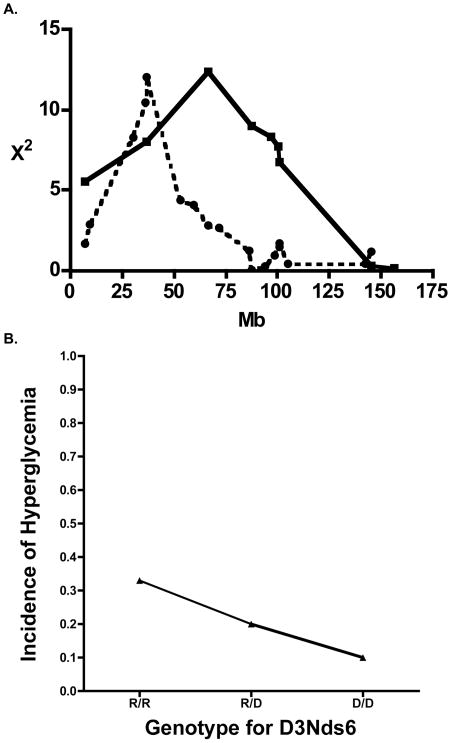
In the previously-published NOD backcross analysis of ALR-derived loci conferring protection to autoimmune T1D, a resistance locus was mapped on medial Chr. 3 (peak at D3Mit241, 33cM) [28]. This locus coincided with the mapping location of Susp, the suppressor of superoxide production by activated neutrophils. In the present F2 cross tested for alloxan resistance, suggestive evidence for a more proximal, non-overlapping locus on the same chromosome was mapped with the peak linkage at D3Nds6, marking the Il2/Idd3 locus 19.2 cM (χ2 =12.1, P=0.0024, Fig. 2A). Surprisingly, in this case, ALR was not the contributor of the resistance allele, but rather contributed an allele that increased sensitivity (Fig. 2B). The mode of inheritance of this ALR-derived susceptibility allele was co-dominant as witnessed by the gene dosage effect on incidence of alloxan-induced diabetes (Fig 2B).
Fig. 2.
Linkages on Chromosome 3 suggest T1D and alloxan-induced diabetes may be controlled by different genes on this chromosome. (2A) The linkage on Chr. 3 mapped in a F2 population subjected to alloxan administration (Dotted line, peak χ2 =12.1, P=0.0024) does not overlap with previously reported linkage mapped in backcross population for type 1 diabetes. (2B) Effect plot of phenotype by microsatellite marker D3Nds6 shows NOD allele contributes to resistance while ALR allele contributes to susceptibility. Incidence of alloxan-induced Diabetes is on y-axis while genotype is on the x-axis (D = NOD; R = ALR).
Heterozygosity to a Chr. 2 locus contributes to higher susceptibility to alloxan
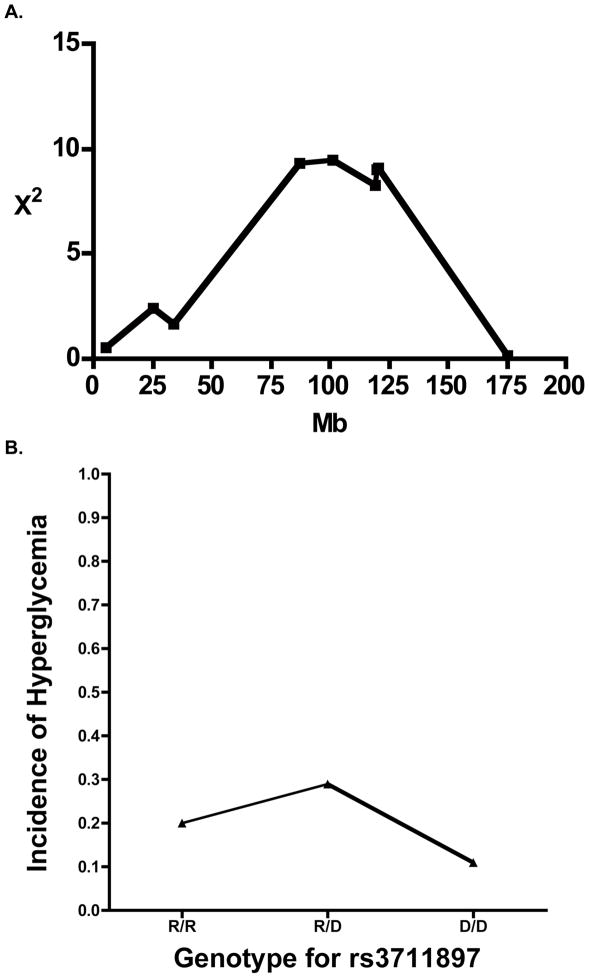
Suggestive evidence was obtained for a locus affecting alloxan sensitivity on Chr. 2 with peak significance linking to the single nucleotide polymorphism marker rs3711897 at 87.03Mb (χ2 of 10.2 and P=0.0062, Fig. 3A). This is more proximal than the Idd13 complex marked by the B2m-Il1 loci. Interestingly, heterozygosity was linked to higher susceptibility to induced diabetes when compared to homozygosity for either parental allele (Fig. 3B).
Fig. 3.
A linkage on Chromosome 2 is unique to alloxan-induced diabetes. (3A) Negative heterosis is demonstrated by the linkage on Chr. 2. The peak for this linkage was at the single nucleotide polymorphic marker rs3711897. (3B) Effect plot of phenotype by microsatellite marker rs3711879 (02–087029735-M) shows heterozygosity on this locus contributes to a higher susceptibility than both NOD and ALR allele. Incidence of alloxan-induced Diabetes is on vertical axis while genotype is on the horizontal axis (D = NOD; R = ALR).
Use of conplastic mice confirms a role for mt-Nd2 in susceptibility/resistance to alloxan
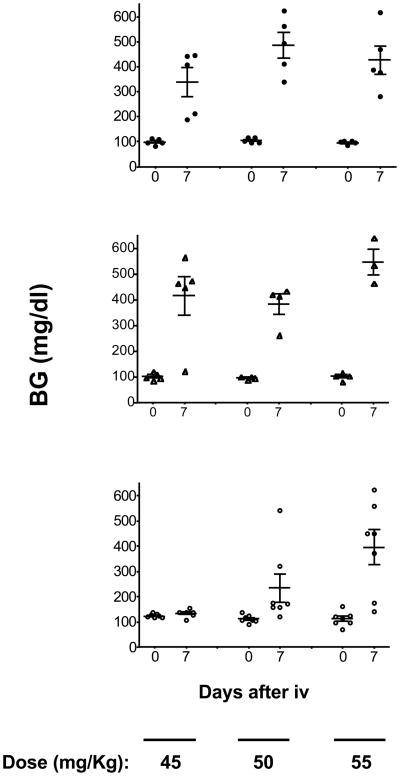
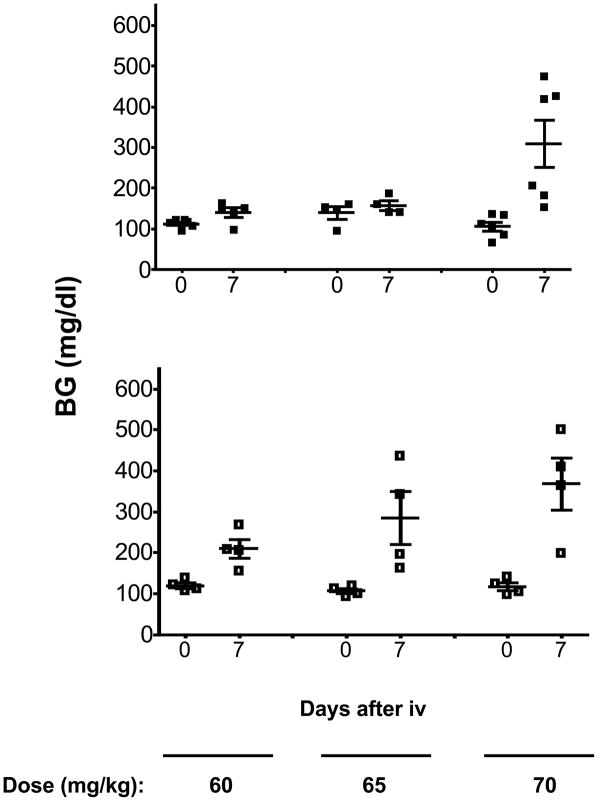
Six-wk-old NOD and NOD.mtALR males were challenged with increasing doses of alloxan starting with 35mg/kg body weight (Fig. 4 upper and middle panels). The two strains responded equally at each dose used. Hence, in the absence of ALR-derived nuclear-encoded genes, the unique mtNd2a allele of ALR does not confer detectable alloxan resistance. However, as shown in Fig. 5, replacement of the ALR mtDNA with that from NOD reduced resistance to alloxan. When 6-wk-old ALR.mtNOD males were compared to standard ALR males over alloxan doses of 60, 65, and 70 mg/kg, both stocks were resistant to the 60 mg/kg dose. However, diabetes was induced in 2 out of 4 ALR.mtNOD conplastic mice at a dose of 65 mg/kg body weight, while none of 5 control standard ALR/Lt males responded to this dosage (Fig. 5). Thus, the mt-Nd2a contributes to free radical resistance in the presence of nuclear loci from ALR.
Fig. 4.
NOD.ALRc8 mice have increased resistance against alloxan-induced diabetes. Six-week-old male NOD (upper panel), NOD.mtALR (middle panel) and NOD.ALRc8 (lower panel) mice were challenged with different dosage of alloxan intravenously. Blood glucose reported in mg/dL was measured before and 7 days after alloxan injection.
Fig. 5.
ALR.mtNOD mice exhibited increased susceptibility to alloxan-induced diabetes. Six-wk-old male ALR (upper panel) and ALR.mtNOD (lower panel) mice were challenged with different doses of alloxan intravenously. Blood glucose was measured before and 7 days after alloxan injection.
NOD.ALRc8 congenic mice exhibit alloxan resistance
To confirm the ALR-derived alloxan resistance linkage on Chr. 8, we assessed the degree of alloxan resistance in males of our NOD.ALRc8 congenic stock (spanning the markers D8Mit205-D8Mit137). We compared alloxan sensitivity of 6-wk-old congenic males versus standard NOD males to doses of alloxan between 45–55 mg/kg body weight (Fig. 4). Diabetes was induced at a dose of 45 mg/kg body weight and higher in standard NOD males, whereas NOD.ALRc8 males were completely resistant to the 45 mg/kg dose. However, in contrast to ALR males, which are resistant to doses in excess of 65 mg/kg (Fig. 4), NOD.ALRc8 males were susceptible to a dose of 50mg/kg body weight. Hence, the Chr.8 linkage only accounts for one component of a genetically more complex ALR strain resistance to alloxan.
DISCUSSION
While inbred strains differ in their susceptibility to chemically mediated diabetes [35], the majority of the studies designed to better understand the mechanisms of streptozotocin or alloxan-induced diabetes have done so using single gene knockouts [36–53]. Missing from these knockout studies, demonstrating either increased sensitivity or resistance to chemically-mediated diabetes, have been direct comparisons to spontaneous T1D. The onset of T1D is controlled by the interplay of multiple loci resulting in aberrant immune responses as well as requirements at the beta cell level. The goal of this study was to use comparative genetics to identify linkages involved in the control of both T1D and chemically-induced diabetes. We obtained suggestive evidence for linkage of 4 loci contributing to alloxan-induced diabetes susceptibility in F2 mice. Among these two, mt-Nd2a and Idd22, are involved in both models.
To determine the ALR-derived loci that provide enhanced protection at the beta cell level, outcrosses of ALR to NOD were performed and the progeny tested for the development of hyperglycemia after administration of alloxan. NOD mice spontaneously develop autoimmune T cell-mediated destruction of pancreatic islets and this may interfere with the destruction of islets induced by free radicals released by alloxan, since hyperglycemia is the common phenotype for both types of destructions. We tried to avoid potential autoimmune-mediated damage by treating F2 mice at six weeks of age and ended the experiment by eight weeks of age. Although insulitis may occur as early as four weeks of age in NOD mice, in our facility the earliest clinical diabetes is observed at ten weeks of age in female NOD and later in males of this strain. The fact that the F2 male population showed a higher incidence of diabetes after alloxan treatment suggests that diabetogenesis was via direct alloxan cytotoxicity and not autoimmunity.
F1 hybrids between NOD and ALR were protected against alloxan-induced diabetes only when the maternal parent was ALR [3], suggesting that maternal factors contribute to protection against not only autoimmune diabetes but also alloxan induced free radical mediated diabetes. We crossed NOD to ALR and created F2 individuals in all 4 potential maternal/paternal directions (Table 1). Phenotypic analysis of these four populations clearly demonstrated that diabetes incidence in mice encoding NOD mt-Nd2c allele was increased compared to mice encoding mt-Nd2a (Table 1). This finding was consistent with previously observed NOD maternal contribution to alloxan-induced diabetes in reciprocal F1 crosses of NOD to ALR [3]. Furthermore, F2 crosses clarified that the origination of the maternal contribution observed in F1 hybrids was from mt-Nd2c and not the result of an X-linked locus or imprinting. Recently, we have demonstrated that mitochondrial ROS production is decreased in ALR compared to ALR.mtNOD and increased in NOD compared to NOD.mtALR [6, 54]. Therefore, mt-Nd2c is associated with alloxan susceptibility and elevated mitochondrial ROS production, while mt-Nd2a is reciprocally associated with reduced mitochondria ROS and protection from alloxan. Likely, the mt-Nd2c allele decreases the threshold to free radical damage by increasing intrinsic ROS. The decreased threshold increases susceptibility to alloxan as well as the damaging ROS produced during the pathogenesis of T1D.
To confirm the linkage, we tested newly created reciprocal conplastic mouse strains [6] for alloxan sensitivity. In ALR.mtNOD conplastic mice, an increased sensitivity to alloxan-induced hyperglycemia was observed when compared to parental ALR mice (Fig. 5). The ALR.mtNOD mice maintained a higher threshold for alloxan sensitivity when compared to NOD and NOD.mtALR showing that mt-Nd2c is not the sole locus providing alloxan susceptibility. In the conplastic mouse strain NOD.mtALR, susceptibility was not altered compared with parental NOD mice (Fig. 4). This reinforces the concept that nuclear genes play an important role in the alloxan resistance measured in parental ALR mice.
The linkage on Chr. 8, where the ALR allele provides dominant resistance against alloxan-induced diabetes, overlaps with a previously mapped ALR-derived locus (Idd22) that protects against autoimmune diabetes in a backcross cohort [28]. Given the elevated free radical scavenge capability in ALR mice, it is likely that the protection against both types of diabetes provided by ALR gene or genes on Chr. 8 may work at pancreatic beta cell level rather than in immune system. The protective role of ALR allele on Chr. 8 is further illustrated by the fact that congenically introducing the alloxan and T1D-resistance locus from ALR into NOD background results in increased resistance of NOD.ALRc8 mice to alloxan (Fig. 4).
The common effect of beta cell damage in both autoimmune and alloxan-induced diabetes again suggests that this protection may be due to elevated ROS dissipation, as previously observed in ALR mice [4, 16, 17]. Given the role of oxidative stress in the pathogenesis in both types of beta cell damage, a plausible candidate gene in this region would be Prdx2, located at 87.6MB. This gene product belongs a ubiquitous family of antioxidant enzymes, the peroxiredoxins. Q-RT-PCR analysis did not reveal any increase in Prdx2 expression comparing ALR with NOD. Furthermore, at this time no connections have been made between peroxiredoxins and alloxan resistance. In contrast, inactivation of NF-kB has been shown to protect against alloxan-induced diabetes [20, 55–58] and has been proposed to protect islets during T1D [4, 59]. A candidate in this region is Kruppel Like Factor 2 (Klf2). Klf2, which is elevated 4 fold in ALR islets and 3.6 fold in NOD-Idd22 islets compared to NOD islets [our unpublished data], is capable of both upregulating antioxidant gene expression [60] as well as inhibiting NF-κB activation [61]. NF-κB plays a key role in the cytokine-induced beta cell death [3]. Indeed, ALR islets are cytokine resistant and showed defective nuclear translocation of NF-κB P65 subunit after cytokine treatment, correlating with reduced kinetics of IκB degradation and suppressed iNOS induction [3].
Free radical mediated alloxan-induced diabetes can be prevented by overexpression of the antioxidant enzyme SOD1 [36]. ALR mice have a constitutive and systemic increase in SOD1 activity that maps in trans to the Suppressor of Superoxide Production (Susp) on Chr. 3 [27]. This Susp locus co-localized with an ALR-derived locus that exerted dominant protection against spontaneous T1D. Both phenotypes had a peak for linkage on Chr. 3 with D3Mit241 (33.0 cM; 66.68 Mbp). Therefore, we were surprised to have identified an ALR derived allele on Chr. 3 that increased susceptibility to alloxan-induced diabetes. This locus is approximately 30 million bases proximal [peak for linkage with D3Nds6 (19.2 cM; 37.02 Mbp)] to the T1D protective Suppressor of Superoxide Production, Susp, locus [27, 28], suggesting different genes controlling sensitivity to alloxan and resistance to T1D on this chromosome (Fig. 2A). The newly mapped ALR-derived alloxan-sensitive locus overlaps with Idd3 [62], Il2 the candidate gene for Idd3 which impairs regulatory T cell function resulting in autoimmunity [63]. Given the nature of alloxan-induced diabetes and the fact that ALR and NOD have identical cDNA sequences [28], IL-2 is not likely the candidate gene. A gene proximal to this peak, Slc2a2, encodes GLUT2, the glucose transporter on liver, kidney, and beta cell surfaces. Alloxan is transported by GLUT2 into beta cells [31]. We did not detect differences in the cDNA sequence of Slc2a2 comparing NOD with ALR, nor were we able to measure any differences in GLUT2 in islets by quantitative Real-time PCR or western blot comparing NOD with ALR (Data not shown). Therefore, Slc2a2 is also not a likely candidate.
Despite a very high degree of homology comparing ALR to NOD throughout Chr. 17, our previous genetic study of the autoimmune diabetes backcross population linked ALR-derived T1D resistance to Chr. 17, suggesting an immune system involvement, because the peak of linkage mapped very close to the Major Histocompatibility Complex [28, 64–66]. In the current F2 cross for sensitivity to alloxan, it is not surprising that this Chr. 17 linkage associated with immune responsiveness did not appear because of the differences between toxin-mediated beta cell destruction versus immune-mediate beta cell killing. Likewise, a linkage unique to alloxan-induced diabetes was mapped on Chr. 2 where mice heterozygous for this locus had a higher susceptibility than either parental type. Heterosis is not unusual in sensitivity to chemicals, possibly by altering the function of a metabolic enzyme. This was previously demonstrated in a report of increased sensitivity to thiazolidinedione induced hepatosteatosis in a F1 cross between two genetically diverse inbred strains, NON/Lt and NZO/Lt [67].
Summary
Using F2 crosses of NOD and the closely related ALR mouse strains, we analyzed genetic controls of T1D and alloxan-induced free radical mediated diabetes. Suggestive evidence for 4 loci contributing to alloxan-induced diabetes susceptibility in F2 mice were mapped. Among the three nuclear linkages, the ALR contribution on Chr. 8 to alloxan resistance was confirmed by congenic stock construction and co-localized to a region of the chromosome previously associated with resistance to spontaneous T1D. In addition, an ALR stock conplastic for the NOD allele of mt-Nd2, which was previously linked to T1D susceptibility, showed increased susceptibility to alloxan-induced diabetes. In conclusion, both the mitochondrial and nuclear genomes are important in the genetic control of these two types of diabetes and subsets of genes that provide resistance to free radical mediated damage also confer resistance to spontaneous T1D.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance provided by Pamela Stanley for F2 animal breeding, Aaron Gusdon and Vadim Pliner for typing and breeding of conplastic and congenic mice. This project is supported by grants from the Juvenile Diabetes Research Foundation (C.E.M.), and the National Institutes of Health DK27722 and DK36175 (E.H.L.) and AI56374 and DK74656 (C.E.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takasu N, Asawa T, Komiya I, Nagasawa Y, Yamada T. Alloxan-induced DNA strand breaks in pancreatic islets. Evidence for H2O2 as an intermediate. J Biol Chem. 1991;266:2112–2114. [PubMed] [Google Scholar]
- 2.Takasu N, Komiya I, Asawa T, Nagasawa Y, Yamada T. Streptozocin- and alloxan-induced H2O2 generation and DNA fragmentation in pancreatic islets. H2O2 as mediator for DNA fragmentation. Diabetes. 1991;40:1141–1145. doi: 10.2337/diab.40.9.1141. [DOI] [PubMed] [Google Scholar]
- 3.Mathews CE, Leiter EH, Spirina O, Bykhovskaya Y, Gusdon AM, Ringquist S, Fischel-Ghodsian N. mt-Nd2 Allele of the ALR/Lt mouse confers resistance against both chemically induced and autoimmune diabetes. Diabetologia. 2005;48:261–267. doi: 10.1007/s00125-004-1644-8. [DOI] [PubMed] [Google Scholar]
- 4.Mathews CE, Suarez-Pinzon WL, Baust JJ, Strynadka K, Leiter EH, Rabinovitch A. Mechanisms underlying resistance of pancreatic islets from ALR/Lt mice to cytokine-induced destruction. J Immunol. 2005;175:1248–1256. doi: 10.4049/jimmunol.175.2.1248. [DOI] [PubMed] [Google Scholar]
- 5.Mathews CE, Graser RT, Savinov A, Serreze DV, Leiter EH. Unusual resistance of ALR/Lt mouse beta cells to autoimmune destruction: role for beta cell-expressed resistance determinants. Proc Natl Acad Sci USA. 2001;98:235–240. doi: 10.1073/pnas.98.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusdon AM, Votyakova TV, Reynolds IJ, Mathews CE. Nuclear and mitochondrial interaction involving mt-Nd2 leads to increased mitochondrial reactive oxygen species production. J Biol Chem. 2007;282:5171–5179. doi: 10.1074/jbc.M609367200. [DOI] [PubMed] [Google Scholar]
- 7.Gale EA. Molecular mechanisms of beta-cell destruction in IDDM: the role of nicotinamide. Hormone Research. 1996;45:39–43. [PubMed] [Google Scholar]
- 8.Horio F, Fukuda M, Katoh H, Petruzzelli M, Yano N, Rittershaus C, Bonnerweir S, Hattori M. Reactive oxygen intermediates in autoimmune islet cell destruction of the nod mouse induced by peritoneal exudate cells (rich in macrophages) but not t cells. Diabetologia. 1994;37:22–31. doi: 10.1007/BF00428773. [DOI] [PubMed] [Google Scholar]
- 9.Nerup J, Mandrup-Poulsen T, Molvig J, Helquist S, Wogensen L, Egeberg J. Mechanisms of panbreatic β cell destruction in type 1 diabetes. Diabetes Care. 1988;11:16–23. [PubMed] [Google Scholar]
- 10.Suarez-Pinzon WL, Mabley JG, Strynadka K, Power RF, Szabo C, Rabinovitch A. An inhibitor of inducible nitric oxide synthase and scavenger of peroxynitrite prevents diabetes development in NOD mice. J Autoimmun. 2001;16:449–455. doi: 10.1006/jaut.2001.0507. [DOI] [PubMed] [Google Scholar]
- 11.Suarez-Pinzon WL, Strynadka K, Rabinovitch A. Destruction of rat pancreatic islet beta-cells by cytokines involves the production of cytotoxic aldehydes. Endocrinology. 1996;137:5290–5296. doi: 10.1210/endo.137.12.8940348. [DOI] [PubMed] [Google Scholar]
- 12.Tabatabaie T, Waldon AM, Jacob JM, Floyd RA, Kotake Y. COX-2 inhibition prevents insulin-dependent diabetes in low-dose streptozotocin-treated mice. Biochem Biophys Res Commun. 2000;273:699–704. doi: 10.1006/bbrc.2000.2959. [DOI] [PubMed] [Google Scholar]
- 13.Briehl MM, Baker AF. Modulation of the anti-oxidant defence as a factor in apoptosis. CellDeath Differ. 1996;3:63–70. [PubMed] [Google Scholar]
- 14.Briehl MM, Cotgreave IA, Powis G. Downregulation of antioxidant defence during glucocorticoid-mediated apoptosis. Cell Death Differ. 1995;2:41–46. [PubMed] [Google Scholar]
- 15.Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunology today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 16.Mathews CE, Leiter EH. Resistance of ALR/Lt islets to free radical-mediated diabetogenic stress is inherited as a dominant trait. Diabetes. 1999;48:2189–2196. doi: 10.2337/diabetes.48.11.2189. [DOI] [PubMed] [Google Scholar]
- 17.Mathews CE, Leiter EH. Constitutive differences in antioxidant defense status distinguish alloxan-resistant and alloxan-susceptible mice. Free radical biology & medicine. 1999;27:449–455. doi: 10.1016/s0891-5849(99)00084-2. [DOI] [PubMed] [Google Scholar]
- 18.Lizard G, Gueldry S, Sordet O, Monier S, Athias A, Miguet C, Bessede G, Lemaire S, Solary E, Gambert P. Glutathione is implied in the control of 7-ketocholesterol-induced apoptosis, which is associated with radical oxygen species production. FASEB J. 1998;12:1654–1663. doi: 10.1096/fasebj.12.15.1651. [DOI] [PubMed] [Google Scholar]
- 19.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho E, Medeiros DM, Bray TM. Glutathione (GSH) inhibits activation of NF-kB, an oxidative stress response element in the pancreas and protects against alloxan-induced diabetes. FASEB J. 1998;12:A815. [Google Scholar]
- 21.Matsuda M, Masutani H, Nakamura H, Miyajima S, Yamauchi A, Yonehara S, Uchida A, Irimajiri K, Horiuchi A, Yodoi J. Protective activity of adult T cell leukemia-derived factor (ADF) against tumor necrosis factor-dependent cytotoxicity on U937 cells. J Immunol. 1991;147:3837–3841. [PubMed] [Google Scholar]
- 22.Hotta M, Tashiro F, Ikegami H, Niwa H, Ogihara T, Yodoi J, Miyazaki J. Pancreatic beta cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and streptozotocin-induced diabetes. J Exp Med. 1998;188:1445–1451. doi: 10.1084/jem.188.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomikos I, Wang Y, Lafferty K. Involvement of O2 radicals in autoimmune diabetes. Immunol Cell Biol. 1989;67:85–87. doi: 10.1038/icb.1989.12. [DOI] [PubMed] [Google Scholar]
- 24.Nomikos IN, Prowse SJ, Carotenuto P, Lafferty KJ. Combined treatment with nicotinamide and desferrioxamine prevents islet allograft destruction in NOD mice. Diabetes. 1986;35:1302–1304. doi: 10.2337/diab.35.11.1302. [DOI] [PubMed] [Google Scholar]
- 25.Bertera S, Crawford ML, Alexander AM, Papworth GD, Watkins SC, Robbins PD, Trucco M. Gene transfer of manganese superoxide dismutase extends islet graft function in a mouse model of autoimmune diabetes. Diabetes. 2003;52:387–393. doi: 10.2337/diabetes.52.2.387. [DOI] [PubMed] [Google Scholar]
- 26.Tiedge M, Lortz S, Munday R, Lenzen S. Complementary action of antioxidant enzymes in the protection of bioengineered insulin producing rinm5f cells against the toxicity of reactive oxygen species. Diabetes. 1998;47:1578–1585. doi: 10.2337/diabetes.47.10.1578. [DOI] [PubMed] [Google Scholar]
- 27.Mathews CE, Dunn BD, Hannigan MO, Huang CK, Leiter EH. Genetic control of neutrophil superoxide production in diabetes-resistant ALR/Lt mice. Free radical biology & medicine. 2002;32:744–751. doi: 10.1016/s0891-5849(02)00747-5. [DOI] [PubMed] [Google Scholar]
- 28.Mathews CE, Graser RT, Bagley RJ, Caldwell JW, Li R, Churchill GA, Serreze DV, Leiter EH. Genetic analysis of resistance to Type-1 Diabetes in ALR/Lt mice, a NOD-related strain with defenses against autoimmune-mediated diabetogenic stress. Immunogenetics. 2003;55:491–496. doi: 10.1007/s00251-003-0603-8. [DOI] [PubMed] [Google Scholar]
- 29.Ino T, Kawamoto Y, Sato K, Nishikawa K, Yamada A, Ishibashi K, Sekiguchi F. Selection of mouse strains showing high and low incidences of alloxan-induced diabetes. Jikken Dobutsu. 1991;40:61–67. doi: 10.1538/expanim1978.40.1_61. [DOI] [PubMed] [Google Scholar]
- 30.Elsner M, Tiedge M, Guldbakke B, Munday R, Lenzen S. Importance of the GLUT2 glucose transporter for pancreatic beta cell toxicity of alloxan. Diabetologia. 2002;45:1542–1549. doi: 10.1007/s00125-002-0955-x. [DOI] [PubMed] [Google Scholar]
- 31.Szkudelski T. The mechanism of alloxan and streptozotocin action in B cells of the rat pancreas. Physiol Res. 2001;50:537–546. [PubMed] [Google Scholar]
- 32.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics (Oxford, England) 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 33.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiter EH, Gerling IC, Flynn JC. Spontaneous insulin dependent diabetes mellitus (IDDM) in Nonobese Diabetic (NOD) mice: comparisons with experimentally-induced IDDM. In: McNeill JH, editor. Experimental Models of Diabetes. Boca Raton: CRC Press; 1999. pp. 257–295. [Google Scholar]
- 36.Kubisch HM, Wang J, Bray TM, Phillips JP. Targeted overexpression of Cu/Zn superoxide dismutase protects pancreatic beta-cells against oxidative stress. Diabetes. 1997;46:1563–1566. doi: 10.2337/diabetes.46.10.1563. [DOI] [PubMed] [Google Scholar]
- 37.Cardinal JW, Allan DJ, Cameron DP. Poly(ADP-ribose)polymerase activation determines strain sensitivity to streptozotocin-induced beta cell death in inbred mice. J Mol Endocrinol. 1999;22:65–70. doi: 10.1677/jme.0.0220065. [DOI] [PubMed] [Google Scholar]
- 38.Flodstrom M, Tyrberg B, Eizirik DL, Sandler S. Reduced sensitivity of inducible nitric oxide synthase-deficient mice to multiple low-dose streptozotocin-induced diabetes. Diabetes. 1999;48:706–713. doi: 10.2337/diabetes.48.4.706. [DOI] [PubMed] [Google Scholar]
- 39.Masutani M, Suzuki H, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, Ochiya T, Sugimura T. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pieper AA, Brat DJ, Krug DK, Watkins CC, Gupta A, Blackshaw S, Verma A, Wang ZQ, Snyder SH. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardinal JW, Margison GP, Mynett KJ, Yates AP, Cameron DP, Elder RH. Increased susceptibility to streptozotocin-induced beta-cell apoptosis and delayed autoimmune diabetes in alkylpurine-DNA-N-glycosylase-deficient mice. Mol Cell Biol. 2001;21:5605–5613. doi: 10.1128/MCB.21.16.5605-5613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosokawa M, Dolci W, Thorens B. Differential sensitivity of GLUT1- and GLUT2-expressing beta cells to streptozotocin. Biochem Biophys Res Commun. 2001;289:1114–1117. doi: 10.1006/bbrc.2001.6145. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Hansotia T, Yusta B, Ris F, Halban PA, Drucker DJ. Glucagon-like peptide-1 receptor signaling modulates beta cell apoptosis. J Biol Chem. 2003;278:471–478. doi: 10.1074/jbc.M209423200. [DOI] [PubMed] [Google Scholar]
- 44.Babaya N, Ikegami H, Fujisawa T, Nojima K, Itoi-Babaya M, Inoue K, Ohno T, Shibata M, Ogihara T. Susceptibility to streptozotocin-induced diabetes is mapped to mouse chromosome 11. Biochem Biophys Res Commun. 2005;328:158–164. doi: 10.1016/j.bbrc.2004.12.149. [DOI] [PubMed] [Google Scholar]
- 45.Georgia S, Bhushan A. p27 Regulates the transition of beta-cells from quiescence to proliferation. Diabetes. 2006;55:2950–2956. doi: 10.2337/db06-0249. [DOI] [PubMed] [Google Scholar]
- 46.Stalvey MS, Muller C, Schatz DA, Wasserfall CH, Campbell-Thompson ML, Theriaque DW, Flotte TR, Atkinson MA. Cystic fibrosis transmembrane conductance regulator deficiency exacerbates islet cell dysfunction after beta-cell injury. Diabetes. 2006;55:1939–1945. doi: 10.2337/db05-1647. [DOI] [PubMed] [Google Scholar]
- 47.Stiles BL, Kuralwalla-Martinez C, Guo W, Gregorian C, Wang Y, Tian J, Magnuson MA, Wu H. Selective deletion of Pten in pancreatic beta cells leads to increased islet mass and resistance to STZ-induced diabetes. Mol Cell Biol. 2006;26:2772–2781. doi: 10.1128/MCB.26.7.2772-2781.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burns N, Gold B. The effect of 3-methyladenine DNA glycosylase-mediated DNA repair on the induction of toxicity and diabetes by the beta-cell toxicant streptozotocin. Toxicol Sci. 2007;95:391–400. doi: 10.1093/toxsci/kfl164. [DOI] [PubMed] [Google Scholar]
- 49.Callewaert HI, Gysemans CA, Ladriere L, D’Hertog W, Hagenbrock J, Overbergh L, Eizirik DL, Mathieu C. Deletion of STAT-1 pancreatic islets protects against streptozotocin-induced diabetes and early graft failure but not against late rejection. Diabetes. 2007;56:2169–2173. doi: 10.2337/db07-0052. [DOI] [PubMed] [Google Scholar]
- 50.Conarello SL, Jiang G, Mu J, Li Z, Woods J, Zycband E, Ronan J, Liu F, Roy RS, Zhu L, Charron MJ, Zhang BB. Glucagon receptor knockout mice are resistant to diet-induced obesity and streptozotocin-mediated beta cell loss and hyperglycaemia. Diabetologia. 2007;50:142–150. doi: 10.1007/s00125-006-0481-3. [DOI] [PubMed] [Google Scholar]
- 51.Fazio EN, Pin CL. Mist1-null mice are resistant to streptozotocin-induced beta cell damage. Biochem Biophys Res Commun. 2007;353:823–828. doi: 10.1016/j.bbrc.2006.12.110. [DOI] [PubMed] [Google Scholar]
- 52.Mori H, Shichita T, Yu Q, Yoshida R, Hashimoto M, Okamoto F, Torisu T, Nakaya M, Kobayashi T, Takaesu G, Yoshimura A. Suppression of SOCS3 expression in the pancreatic beta-cell leads to resistance to type 1 diabetes. Biochem Biophys Res Commun. 2007;359:952–958. doi: 10.1016/j.bbrc.2007.05.198. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Zhang L, Chou A, Allaby T, Belanger G, Radziuk J, Jasmin BJ, Miki T, Seino S, Renaud JM. KATP channel-deficient pancreatic beta-cells are streptozotocin resistant because of lower GLUT2 activity. Am J Physiol Endocrinol Metab. 2008;294:E326–335. doi: 10.1152/ajpendo.00296.2007. [DOI] [PubMed] [Google Scholar]
- 54.Gusdon AM, Votyakova TV, Mathews CE. mt-Nd2a Suppresses Reactive Oxygen Species Production by Mitochondrial Complexes I and III*. J Biol Chem. 2008;283:10690–10697. doi: 10.1074/jbc.M708801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho E, Chen G, Bray TM. Supplementation of N-acetylcysteine inhibits NFkappaB activation and protects against alloxan-induced diabetes in CD-1 mice. Faseb J. 1999;13:1845–1854. [PubMed] [Google Scholar]
- 56.Ho E, Chen G, Bray TM. Alpha-phenyl-tert-butylnitrone (PBN) inhibits NFkappaB activation offering protection against chemically induced diabetes. Free radical biology & medicine. 2000;28:604–614. doi: 10.1016/s0891-5849(99)00271-3. [DOI] [PubMed] [Google Scholar]
- 57.Ho E, Quan N, Tsai YH, Lai W, Bray TM. Dietary zinc supplementation inhibits NFkappaB activation and protects against chemically induced diabetes in CD1 mice. Experimental biology and medicine (Maywood, NJ. 2001;226:103–111. doi: 10.1177/153537020122600207. [DOI] [PubMed] [Google Scholar]
- 58.Quan N, Ho E, La W, Tsai YH, Bray T. Administration of NF-kappaB decoy inhibits pancreatic activation of NF-kappaB and prevents diabetogenesis by alloxan in mice. Faseb J. 2001;15:1616–1618. doi: 10.1096/fj.00-0855fje. [DOI] [PubMed] [Google Scholar]
- 59.Wang CY, She JX. SUMO4 and its role in type 1 diabetes pathogenesis. Diabetes/metabolism research and reviews. 2008;24:93–102. doi: 10.1002/dmrr.797. [DOI] [PubMed] [Google Scholar]
- 60.Ali F, Hamdulay SS, Kinderlerer AR, Boyle JJ, Lidington EA, Yamaguchi T, Soares MP, Haskard DO, Randi AM, Mason JC. Statin-mediated cytoprotection of human vascular endothelial cells: a role for Kruppel-like factor 2-dependent induction of heme oxygenase-1. J Thromb Haemost. 2007;5:2537–2546. doi: 10.1111/j.1538-7836.2007.02787.x. [DOI] [PubMed] [Google Scholar]
- 61.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JR, Hearne CM, Knight AM, Love JM, McAleer MA, Prins JB, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 63.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Graser RT, Mathews CE, Leiter EH, Serreze DV. MHC characterization of ALR and ALS mice: respective similarities to the NOD and NON strains. Immunogenetics. 1999;49:722–726. doi: 10.1007/s002510050673. [DOI] [PubMed] [Google Scholar]
- 65.Mathews CE, Graser RT, Serreze DV, Leiter EH. Reevaluation of the major histocompatibility complex genes of the NOD-progenitor CTS/Shi strain. Diabetes. 2000;49:131–134. doi: 10.2337/diabetes.49.1.131. [DOI] [PubMed] [Google Scholar]
- 66.Pomerleau DP, Bagley RJ, Serreze DV, Mathews CE, Leiter EH. Major histocompatibility complex-linked diabetes susceptibility in NOD/Lt mice: subcongenic analysis localizes a component of Idd16 at the H2-D end of the diabetogenic H2(g7) complex. Diabetes. 2005;54:1603–1606. doi: 10.2337/diabetes.54.5.1603. [DOI] [PubMed] [Google Scholar]
- 67.Pan HJ, Lin Y, Chen YE, Vance DE, Leiter EH. Adverse hepatic and cardiac responses to rosiglitazone in a new mouse model of type 2 diabetes: relation to dysregulated phosphatidylcholine metabolism. Vascul Pharmacol. 2006;45:65–71. doi: 10.1016/j.vph.2005.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.