Abstract
Voltage-gated potassium (Kv) channels, such as Kv1.2, are involved in the generation and propagation of action potentials. The Kv channel is a homotetramer, and each monomer is composed of a voltage-sensing domain (VSD) and a pore domain (PD). We analyzed the fluctuations of a model structure of Kv1.2 using elastic network models. The analysis suggested a network of coupled fluctuations of eight rigid structural units and seven hinges that may control the transition between the active and inactive states of the channel. For the most part, the network is composed of amino acids that are known to affect channel activity. The results suggested allosteric interactions and cooperativity between the subunits in the coupling between the motion of the VSD and the selectivity filter of the PD, in accordance with recent empirical data. There are no direct contacts between the VSDs of the four subunits, and the contacts between these and the PDs are loose, suggesting that the VSDs are capable of functioning independently. Indeed, they manifest many inherent fluctuations that are decoupled from the rest of the structure. In general, the analysis suggests that the two domains contribute to the channel function both individually and cooperatively.
Introduction
Voltage-gated potassium (Kv) channels aid in the repolarization phase of action potentials, and thus are important for proper communication between neurons and other excitable cells. These channels are located in the brain, ear, and heart. Mutations in Kv channels cause episodic ataxia syndromes, epilepsy, deafness, long QT syndrome, and other disorders (1).
Several x-ray structures of Kv channels are available. MacKinnon's group (2,3) solved the structure of rat Kv1.2 in its open state. Yarov-Yarovoy's group (4,5) created model structures that include the VSD loops that were missing in MacKinnon's structure (3). MacKinnon's group also solved the structure of a chimera protein Kv1.2 with the S3–S4 paddle from Kv2.1 (6) and the structure of KvAP, a homologous protein from archea (7). All of these structures have similar pore domains (PDs) but different voltage-sensing domains (VSDs). A detailed description of all the x-ray and modeled structures of Kv channels analyzed here is provided in the Supporting Material.
Kv channels are homotetramers, with each monomer containing two domains (VSD and PD; Fig. 1 b). The VSD includes helices S1–S4. The PD includes helices S5–S6, which are connected by a reentrant loop that is involved in ion selectivity. It is thought that voltage changes in the membrane affect the pore opening through four conserved arginine residues (R294, R297, R300, and R303) that are located in helix S4 (8–12). Mutations of arginine residues in the S4 helix allow the nonselective conductance of various cations, a phenomenon called “omega currents”. Mutating specific residues in the S4 helix causes a leak of protons (13). It was recently shown that omega currents are associated with four omega pores per K-channel (14).
Figure 1.
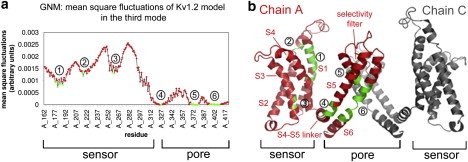
Identification of hinges in the third fluctuation mode. (a) MSF. The minima, corresponding to the hinges, are labeled in green and marked 1–6. (b) Two juxtaposed monomers of Kv1.2's model structure (5): chain A (red) and chain C (gray). Transmembrane helices S1–S6 are marked in red, and the six hinge regions (green) are marked by the gray encircled numbers. The figure was prepared using PyMol (71).
The VSD is also found in calcium (Cav) and sodium (Nav) channels (15). The same voltage sensor also controls a phosphatase activity in the ascidian VSD protein Ci-VSP (16–18). Kohout et al. (19) suggested that the protein works as a monomer and uses a multistep rearrangement for its activity, in similarity to the VSD of voltage-gated ion channels. Recently, a voltage-gated proton channel (VSOP or Hv) consisting of the VSD alone was discovered (20). This protein shares ∼30% sequence identity with the Kv1.2 VSD. They are also believed to share the same fold. Alabi et al. (21) transferred the S3–S4 paddle from Hv and Ci-VSP (one from each) into Kv2.1 and showed that the chimera proteins are functional. This suggests that the voltage-sensing mechanism is conserved between these remotely homologous proteins. The fact that the VSD preserves its function in various structural architectures supports the idea that it is an autonomous modular unit.
Three main models have been proposed for voltage sensing in Kv channels and the movement of the S4 helix in particular. The transporter model suggests that during activation, the S4 arginines move from a crevice with an intracellular solution to a crevice with an extracellular solution. The motion of the S4 helix involves rotation and tilting. Moreover, motions within the VSD change the shape of the crevices (22–24). In the helical screw model, the S4 helix changes its location in the plane of the membrane and rotates (25,26). It has been suggested that this motion is induced by a gating pore, or omega pore, that insulates the S4 helix from the membrane (27–29). In the paddle model, which was proposed based on the KvAP structure, S3 and S4 form a helical hairpin that moves across the membrane as a unit. This model suggests that the paddle is exposed to the lipid in both states (2,30).
In two previous studies, normal-mode analysis (NMA) of K-channels was used to investigate the global motions of the PD (31,32). Here, we extend those works with the goal of identifying communication between the voltage sensor and the pore. Searching for large conformational changes in such model structures by atomistic simulations is computationally demanding, and NMA using elastic network models (ENMs) provides an alternative. These coarse-grained models may not take the place of full-atom simulations, but analysis of the fluctuation modes often provides mechanistic insight into protein motion and its implications for function (33).
Here, we used the Gaussian network model (GNM) (34,35) and anisotropic network model (ANM) (36,37) to analyze the dynamic fluctuations around equilibrium of the Kv1.2 model structure. The combined analysis of GNM and ANM allowed the prediction of the hinge regions, the rigid structural elements that connect them, and the cooperative movements that may be involved in the channel's function.
Materials and Methods
Structure
We used an existing model structure of the rat Kv1.2 channel in an open conformation (5). The structure was built using ROSETTA (5) based on the incomplete x-ray structure of Long et al. (3) (PDB: 2a79). The channel is a homotetramer, and each monomer has 260 residues in the membrane. The monomer contains two transmembrane domains (the VSD and PD; Fig. 1 b) and an intracellular tetramerization domain (T1). The VSD is composed of four helical transmembranal segments: S1 (A162–Q193), S2 (D220–C244), S3 (N253–K277), and S4 (G284–S311). The PD is composed of two helical transmembranal segments (S5 (M325–Q350) and S6 (I385–T421)) connected by a reentrant loop (A351–T384). The VSD and PD are connected by a short helical segment (S4–S5 linker; K312–S324). In addition, each monomer has 161 residues and 78 residues in its N- and C-termini that were deleted from the structure. The analysis was performed on the transmembrane domains (the T1 domain was deleted). The calculations were also conducted for several other 3D structures (see the Supporting Material).
ENMs
We analyzed the Kv1.2 model structure using two different ENMs: GNM (34,35) and ANM (36,37). In these models, the protein structure, simplified into Cα atoms, is treated as an elastic network of residues connected by Hookean springs within a cutoff distance. Despite their inadequacy in describing nonlinear motions, ENMs have been shown to be capable of producing the near-native fluctuation behavior of residues in a given structure (38). GNM, being an isotropic and one-dimensional model, characterizes only the sizes (magnitudes) of the fluctuations. ANM, on the other hand, determines also the direction of the fluctuations. Thus, the fluctuations can be decomposed into N-1 and 3N-6 normal modes in GNM and ANM, respectively, and the real motion can be expressed as a linear combination of the fluctuations in these normal modes. The contribution of each motion is scaled with the inverse frequency of that mode. The slowest modes thus contribute most to the predicted fluctuations. Often, the slowest modes are related to motion that is associated with biological functions of the macromolecule (33,39). More details about the GNM and ANM models are provided in the Supporting Material.
Results
First, we describe the global motions of the channel suggested by the elastic modes. Next, we focus on the correlations between the fluctuations of specific elements and explore the coupling between the motions within and between the two domains, and between the subunits. We correlate the results with empirical data about the importance of amino acids based on evolutionary conservation (Fig. S1), and with clinical and mutagenesis data.
Hinges and rigid elements
GNM
We sorted the modes from the slowest to the fastest and looked at the distribution of their eigenvalues (eigenfrequencies). We used the first eight modes, which appeared to be sufficient to elucidate the motion of the channel. The addition of modes 9–12 to the analysis did not provide more insight, and the contribution of modes above 12 was significantly smaller (Fig. S2). The mean-square fluctuations (MSFs) and dynamic correlations of all 1040 modes were very similar to these obtained by averaging over the eight slowest modes (Fig. S3).
The three slowest modes of the tetramer make the greatest contribution to the overall motion (Fig. S2). The two slowest modes, sharing the same eigenvalue, are degenerate. In the first, the PDs of monomers A and C are mobile but the VSDs of these monomers are immobile, whereas the VSDs of monomers B and D are mobile but their PDs are immobile. In the second mode, a similar behavior was observed for the respective juxtaposed monomer pairs. The shape of the average of these two modes is the same as that of the third (Fig. 1 a), where all four monomers were observed to be involved in the cooperative motion of the structure. The third mode is the slowest individual mode that incorporates the motion of both the VSD and PD in the same monomer and in all the monomers. The shape of the average of the three slowest modes is similar to the shape of the third alone. According to this main mode (Fig. 1 a), each monomer is composed of seven rigid elements (Fig. 1 b, red, and Table S1) connected by six hinges (Fig. 1 b, green, and Table S2). Three of these hinges (M325–L328 between the VSD and PD, I402–V410 in the internal gate, and T373–D379 around the selectivity filter) were previously shown to be functional (40) (Fig. 1).
The next five modes (4–8) describe the motion of substructures within the domains, particularly in the VSD. Analysis of these modes revealed more hinges (Table S3), including a hinge in the S4 helix that was previously detected in experiments (41). The main hinges identified in the eight slowest modes are presented in Fig. 2.
Figure 2.
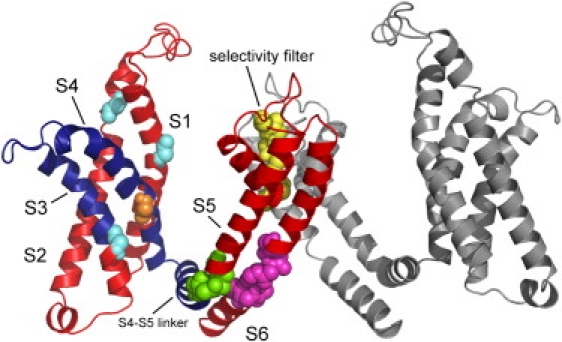
The main hinges as inferred from the eight slowest modes, presented on the 3D model structure of Kv1.2 (5). Only two chains are shown: chain A (left) in colors and chain C (right) in gray. The flexible segment identified in the three slowest modes, containing the S3 and S4 helices and the linker in chain A, is in dark blue. The main chain atoms of the amino acids that were found to serve as hinges are presented as spheres. The hinge in the selectivity filter (T373-D379) is in yellow. The hinge in the S6 helix (I402-V410) is in magenta. The hinge between the linker and the S5 helix (M325-L328) is in green. The hinges in the S1 helix (C181), S2 helix (S217), and S3 helix (V261) are in cyan. These hinges were identified in the first three modes. The hinge in the S4 helix (F302, orange) was identified in the next five modes (4–8). The network of hinges and rigid elements appears to couple between the VSD and PD. The figure was prepared using PyMol (71).
We analyzed other structures, including a model of the closed state, and found that they share approximately the same MSF (see Supporting Material).
ANM
By comparing the slowest mode shapes (MSF) using GNM and ANM, we were able to identify the correspondence between the two elastic models, and thus to predict the direction of the motion controlled by the identified hinges (Table S4). The slowest ANM mode, as well as the second and third ANM modes, which are degenerate, describe hinges that are suggested by the three slowest GNM modes. The motion of the VSD dominates in this mode, and in comparison the PD appears to be immobile.
The fourth ANM mode, which shows an iris-like motion of the pore (Movie S1), corresponds to the fourth and fifth GNM modes (Table S4). The same iris-like motion was previously observed by NMA of various K-channels (31,32) and recently detected in single-molecule experiments (42). It was also shown for the MscL mechanosensitive channel in Escherichia coli (43). This iris-like motion is interesting for two reasons. First, it involves all four monomers and represents a coupled motion of the tetramer to open and close the channel. Second, it involves coupling between the PDs and VSDs. Thus, it may be associated with gating and the transmission of the voltage sensing to the gate. Indeed, a particularly high overlap of 0.49 was obtained between the direction of the fluctuations in this mode and the direction of the motion of the channel from the open to the closed states (5).
Correlation between the fluctuations
The hinges can mediate cooperative motions of the rigid elements, and the GNM analysis may reveal such motions.
Identification of motion in groups of slow modes
The average over the eight slowest modes describes the overall motion. The cooperative fluctuations within the domains, particularly in the VSD, were not apparent in the average mode. Thus, we divided these modes into two groups in accordance with the distribution of their eigenvalues: modes 1–3 and modes 4–8. In modes 1–3 (Fig. 3, a–c), the cooperative motion reflected the division of each subunit into two domains (VSD and PD), and the main hinge was detected between the linker and S5 helix. In these modes of motion, the VSD behaved as a rigid element. However, the PD was divided into further structural elements by the hinges at the internal gate (the S6 helix) and around the selectivity filter. The PD and VSD associate through these flexible regions. In the next five modes (4–8), substructural cooperative units also appear in the VSD and the extracellular mouth of the PD (Fig. 3, d–f).
Figure 3.
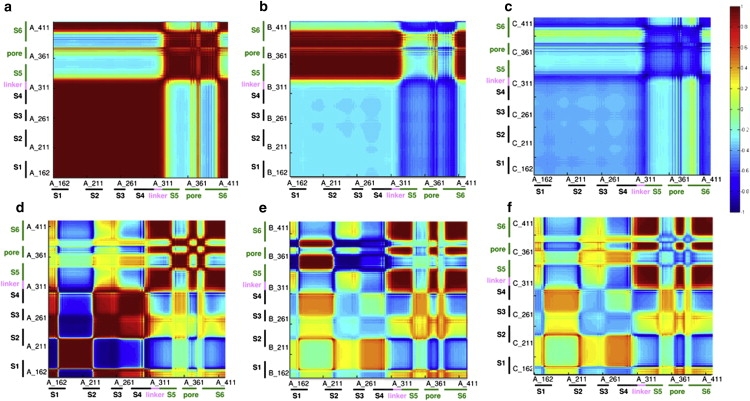
The cooperative motions between residue pairs in the tetramer. The axes mark the residue numbers in each chain. The magnitude of the positive and negative correlations between the fluctuations of the amino acids is color-coded using the red-to-blue scale on the right. (a and d) Interactions within chain A. (b and e) Interactions between residues in chain A and residues in its nearest neighbor (chain B). (c and f) Interactions between residues in chain A and residues in its juxtaposed neighbor (chain C). The different structural segments are marked on the axes. Panels a–c present averages over the three slowest modes, and panels d–f show averages over the next five modes (4–8). A positive correlation indicates a motion of the two residues with the same sense (e.g., in the same direction), whereas a negative correlation indicates a motion with the opposite sense (e.g., in the opposite direction).
First, we report the motions of the PD, which have already been identified in previous studies of potassium channels (31,32,42,44–48). Next, we describe the motions of the VSD. We then describe the hinges and correlated motions in each domain and between the domains, and show the independent motion of each domain. Last, we show the influence of the VSD on the PD and demonstrate the importance of the tetrameric form of the channel.
PD
The three slowest modes suggested two hinges in the PD, near the two gates of the channel (Fig. 1, marked as 5 and 6). The first (residues I402–V410) is near the internal gate of helix S6, and the second (T373–D379) is in the selectivity filter. The hinge in the S6 helix includes the conserved PVP motif (P405–P407). This hinge allows motion of the C-terminus of the helix, resulting in opening and closing of the internal gate. A ConSeq analysis (49) revealed that the helix, especially the PVP motif and the adjacent amino acids; Fig. S1), is conserved. The T373–D379 segment is also highly conserved.
The hinge in the selectivity filter is consistent with previously published GNM studies of PDs from various potassium channels (31,32). However, the inner helix of KcsA, a potassium channel lacking the VSD, was found to contain two hinges around G99 and L110 (31), instead of one in Kv1.2. These residues are equivalent to G398 and I409 in Kv1.2. In the most cooperative fluctuation modes of Kv1.2, the hinge in the inner helix (S6) appears in I402–V410. The second hinge, around G398, appears in the full tetramer in less cooperative modes (data not shown). The change between the locations of the hinges in the two channels may be due to limited motion of the internal gate in the Kv1.2 structure because of the connection to the large VSD. In support of this, GNM calculations of Kv1.2 in the absence of the VSD resulted in hinges similar to those observed in KcsA. Details concerning specific residues in the PD that are associated with various syndromes are available in the Supporting Material.
The motion of the internal gate in helix S6 (P407–T421) is positively correlated with the motion of the selectivity filter in each monomer in the three slowest modes (Fig. 3 a) as well as in the next five modes (Fig. 3 d and Fig. S4). The positive correlation indicates that the two rigid elements may move in the same direction. This correlation is consistent with experimental studies that showed a correlation between the two gates in the KcsA and Shaker channels (44,47). The correlations between the motions of different parts of the PD (Fig. 3) were found to be identical to the correlations in the absence of the VSD (data not shown), which demonstrates the independence of this domain. In this respect, it is noteworthy that in the presence of the VSD, the gate and selectivity filter mediate the cooperativity between this domain and the PD.
Motion of the VSD
The most important motions of the VSD are presented here; other motions are presented in the Supporting Material.
Hinge between the domains
Analysis of the three slowest GNM modes suggested that the VSD and PD are connected by a main hinge between the linker and the S5 helix, around residues M325–L328 (Fig. 1, marked as 4). Analysis of the corresponding ANM mode also revealed the same region as a hinge (Fig. 4). Adjacent to this hinge region, S324 is in close proximity to V408 and I409 in the S6 helix, where they appear to interact via hydrogen bonds. An equivalent position to E327 in Kv7.1 is associated with two syndromes (see Supporting Material).
Figure 4.
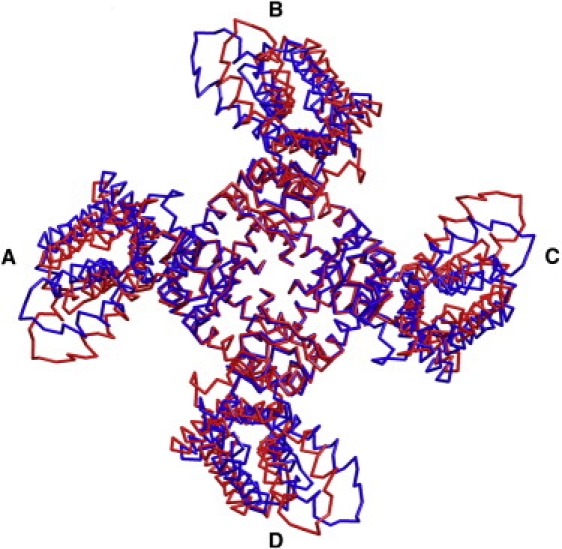
The first ANM mode of fluctuations. The two deformed tetrameric structures, approximately reflecting the motion end-points, are colored blue and red, with the four subunits marked A–D. The original model structure used for the analysis is not presented. The positions of the VSDs (periphery) differ between the two conformations, whereas the PD (center) is, in essence, immobile. The picture was prepared using PyMol (71).
Another important observation emerged from the analysis of the same three modes (and the corresponding ANM modes): The S3–S4 paddle (A262–V301) and linker (S311–S324) move as a rigid unit. This motion appears to be responsible for the mechanical coupling between the VSD and the PD. There are no hinges in the S3–S4 loop, the S4 helix, and the linker (Table S2). These regions appear to form a rigid structural element that may transmit the motion from the voltage sensor to the pore.
S4 helix
Fig. 5 shows the average fluctuations of the three slowest GNM modes, and the fluctuations of the fourth and fifth modes. The S4 helix is a rigid segment in the lowest modes, but a hinge appears in the vicinity of residue F302 in the next two modes. The hinge is located in the middle of the S4 helix, between R300 and R303, which are highly conserved (Fig. S1) and involved in voltage sensing (8–12). The importance of this hinge has been demonstrated experimentally (41,50,51).
Figure 5.
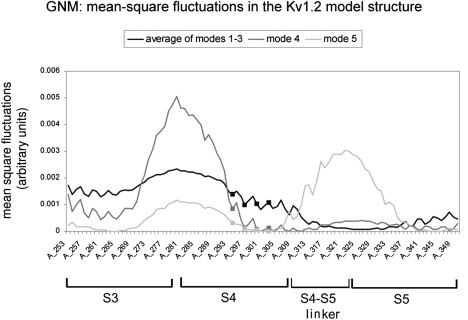
MSF of the Kv1.2 model structure in the five slowest modes. The averaged MSFs of modes 1–3 are in black (identical to Fig. 1a), and the fluctuations of the fourth and fifth modes are in dark and light gray, respectively. The hinge in the S4 helix appears only in the fourth and higher modes, in residue F302. Arginine residues (294, 297, 300, and 303) are marked in squares. Only fluctuations of the S3–S5 helices are presented here, even though the calculation was done for the entire structure. A minimum within the S4 helix appears only from the fourth mode.
Independent motion of the VSD
GNM calculations showed that the dynamic correlations in the isolated VSD were similar to the correlations observed in the VSDs of the whole channel (Fig. S5; see Supporting Material for more details). This suggests that the VSD may sense the changes in membrane voltage regardless of its coupling to the PD and the other monomers of the channel. ANM calculations of the four VSDs in the absence of the PDs showed independent motion for each VSD. This again implies that the VSDs operate independently, and is consistent with recent experiments in Hv, which is comprised of a VSD only (52,53). The experiments showed that even though it is expressed as a dimer, the monomer is still functional as a proton channel.
Correlations and cooperative behavior
Voltage sensing is thought to be an independent process for each monomer, and gating is a cooperative motion of the tetrameric form (54–56). To investigate how the cooperativity is reflected by the fluctuations, we compared the GNM MSF of the first three modes of the monomer and the first eight modes of the tetramer (Fig. 6). These appear to approximate the overall behavior in both cases, according to the eigenvalues. The shapes of the average modes are very similar. Overall, there are seven hinges in the first three modes of the monomer, compared with six hinges in the first three modes of the tetramer and nine in the next five modes (Fig. 6, Table S2, and Table S3). The hinges in the VSD and PD are identical except for the hinge near R300 of the S4 helix, which appears in a more cooperative mode in the isolated monomer. Then again, the hinge between the domains disappeared in the fluctuations of the isolated monomer. The motion of the PD is less restricted in the isolated monomer, and thus the C-terminus of the S6 helix appears to be more mobile than in the tetramer.
Figure 6.
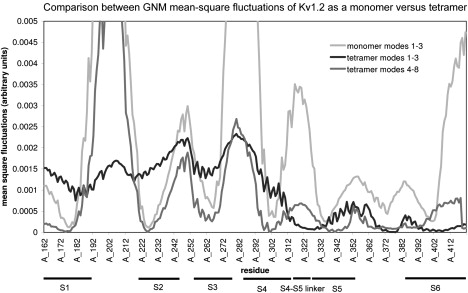
GNM MSF of the tetramer and the monomer. The MSF of the three slowest modes of the monomer are presented in light gray. The MSF of the three slowest modes of the tetramer are in black (identical to Fig. 1a) and the next five modes are in dark gray. The locations of the S1–S6 helices are indicated. Most of the hinges identified in the tetramer also appear in the monomer. The main differences are the higher mobility of helix S6 and the shift in the location of the hinge between the domains.
The dynamic correlation between the selectivity filter and the VSD within each monomer disappeared in the absence of intermonomer contacts (Fig. S6, a and b). Moreover, the correlation between the motion of the selectivity filter and the S6 helix diminished (Fig. S6, a and b). The cooperativity between the inner gate (S6 helix) and the VSD also weakened in this case.
To identify the exact interface that caused the changes in motion mentioned above, we deleted specific contacts and repeated the calculation. Excluding only the contacts at the interface between the near-neighbor monomers led to the appearance of most of the hinges as in the full tetramer, but in less cooperative modes. However, the hinge between the domains disappeared in the slowest modes (as well as in the isolated monomer). Excluding the interface between the S4 and S5 helices of each neighboring monomers caused changes in the motions of the S1–S2 and S3–S4 loops in the three slowest modes. The deletion also led to the disappearance of the dynamic correlation between the selectivity filter and the VSD (Fig. S7). These changes may indicate that the coupling between the PD and VSD involves interactions via this interface. The dynamic correlations between residues of the VSD in each monomer were the same even in the absence of contacts between S4 and S5 of the neighbor monomer. However, the loss of this interface affected the dynamic correlations between the VSD and PD and inside the PD. The voltage-sensing motion was unaffected, but the coupling between the two domains and the coupling inside the PD were different (Fig. S7).
A complementary ANM analysis was conducted to search for cooperativity with respect to the direction of the motions. In all of the slowest ANM modes of the isolated monomer, the selectivity filter moved together with the extracellular regions of the PD as a rigid element. A rotated motion of the S1–S2 and S3–S4 loops was also observed in one of the cooperative modes. This motion was similar to the opening of a pore in an iris-like motion of the whole Kv1.2 tetramer. Here the inner parts of the VSD helices, closer to the intracellular part, were less mobile and did not show a counterclockwise motion.
Influence of the sensor on the pore
Correlations between the motion of the gate and the VSD
The analysis of the first three modes suggested that the C-terminal region of the S6 helix (P407–T421), the internal gate, is positively correlated with the whole VSD and selectivity filter in each monomer (Fig. 3 a). Yet, these regions were negatively correlated with the extracellular regions of the PD (V339–S371 and D379–C394). Thus, the internal gate, the selectivity filter, and the VSD behave as elements of the same dynamic unit. The internal gate is known to be susceptible to depolarization (57,58). Thus, the observed dynamic correlation between the gate and the VSD may be considered to be functional. Indeed, such a functional correlation was found in the human Kv1.5 homolog. The mutation R401N (R297 in rat Kv1.2; helix S4) shifted the voltage dependence of activation to negative potentials, whereas the P511G (P405) mutation (helix S6) had the opposite effect (59).
In the second group of modes, where the hinge in the S4 helix is visible, the sense of the correlation between the S6 helix and the S3–S4 paddle became negative (Fig. 3 d and Fig. S4). The coupling between the near-neighbor and juxtaposed monomers was negative in the eight slowest modes (Fig. 3, b, c, e, and f). Still, the positively correlated fluctuations between these two regions were lost in the absence of the intermonomer interactions (Fig. S6 b and Fig. S9). This implies that the negative intramonomer coupling of these two regions is intrinsic to the monomer, but the positive intramonomer coupling requires the interactions between monomers. Thus, the most cooperative modes could be ascribed to the gating process, and the next modes could be associated with the voltage sensing.
A recent sequence analysis detected a group of evolutionarily correlated residues with several positions known to be important for voltage sensing and gating functions (Table S5) (60). Some of these positions are located in the paddle (T264, A275, and K277) and internal gate (H418 and E420). The fact that amino acids in the internal gate are both evolutionarily (60) and dynamically (this work) correlated with amino acids in the S3–S4 paddle helps to reinforce the functional connection between these regions.
This was not the only correlation between the motion of the VSD and the PD. Strong positive intermonomer correlated fluctuations were detected in the three slowest modes between the S3–S4 paddle and the S5 helix (the S3–S4 helices of monomer A, and the S5 helix of monomer B; Fig. 3 b). This correlation was negative in the next-slowest modes within the monomer and between the monomers (Fig. 3, d–f). The negative correlation in the fluctuations of the S3–S4 paddle and S5 helix remained in the absence of intermonomer contacts (Fig. S6 b).
Several mutants in the S4, S5, and S6 helices were shown to change the voltage sensitivity (51,61,62). The specific mutations in the S5 and S6 helices are located at the interface between the VSD (helix S4) and PD of the near-neighbor monomers. The mutations perturbed the equilibrium between the activated and resting states of the sensor, in favor of the resting state (61). A reorientation of the N-terminal region of the S4 helix toward residues in the neighboring PD was shown for Kv1.2 (62). Pathak et al. (51) revealed a cooperative gating motion by monitoring the motion of helix S4 in mutated monomers using an environmentally sensitive fluorophore. Mutations of residues V369, I372, and S376 in the Shaker's S4 helix to I, L, and T, respectively, caused the activation and channel opening to occur over different voltage ranges. Other studies that mutated one monomer and compared its function with that of a WT channel and a channel in which all monomers are mutated led to the notion that the gating motion of the S4 helix is cooperative (54,63). The necessity of the intermonomer connectivity for the dynamic correlations between the S3–S4 paddle with the S5 and S6 helices is consistent with the latter experimental studies.
Motion of the S1 helix
Several residues in the C-terminus of the S1 helix have been shown to be important. For example, T184 is thought to bind E350 of the PD of an adjacent monomer (64). A statistical coupling analysis of 360 Kv channel sequences showed a coevolved interface between S1 and the pore helix near the extracellular surface (65). Futhermore, cross-linking was shown for the corresponding residues of T184 and I361 in Shaker. Our results suggest that the N-terminus of the S1 helix is also important. Specifically, it correlated with the extracellular mouth of the PD, the N-terminus of the S2 helix, the S2–S3 loop, the S3–S4 paddle, and the internal part of the S6 helix (Fig. S8). The sense of these correlations changed depending on whether they were located in the same or different monomers (Fig. 3, d–f). These suggest an interpretation of the correlated substitutions of residues I18 and D20 in the KvAP's S1 helix with the gate and paddle (60); the C-terminus of the S1 helix may be involved in gating. A similar analysis of the full Kv1.2 structure, including the intracellular tetramerization domain (T1), revealed similar correlations (data not shown).
Discussion
The dynamics of biomolecular structures is complex and involves various motions, both local and global, that occur over a wide range of timescales. Here, within the approximations of the ENM, to reflect the intrinsic dynamics of the channel structure, we searched for cooperative modes of motion. Based on their eigenfrequencies, the slow fluctuation modes clustered into two groups: modes 1–3 and modes 4–8. Thus, the analysis reflects the dynamics on two different timescales.
We found that the two groups of a few slow modes were sufficient to reveal interesting correlations in the dynamic fluctuations between the domains and between the monomers. The first group, representing fluctuations on the longest timescale, i.e., the most cooperative motion, included the three slowest modes. An analysis of this group revealed that the hinges that are sufficient to describe the gating motions and some of the internal motions of the VSD. Moreover, the fluctuations in these modes presented correlations between the two domains and within the PD (but not the VSD, which moves only as a rigid body). The second group, comprising the next five modes, also revealed a hinge in the S4 helix. Further, the correlated fluctuations in these modes displayed more rigid elements in the VSD and extracellular mouth of the PD.
Overall, the results are consistent with the notion that movement of the charged arginines of the VSD propagates to the outer helix of the PD, thereby opening the pore (66). The network of seven main hinges (Fig. 2) suggests how coupling between the two domains is possible: The hinge in the internal gate of the S6 helix is located near the hinge in the linker. The linker, on the other hand, forms a rigid unit with the S3–S4 paddle (residues A262–S324; Fig. 2). Thus, a motion of the paddle in the membrane may pull the N-terminus of helix S5, which is very close to the hinge in helix S6. Changes in the packing around the gate hinge may then influence the motion of the internal gate. This motion, which was identified in the first group of slow modes, leads us to suggest that these modes are responsible for the gating motion, i.e., motion toward the opening of the gate in the PD. Yet, the identification of a hinge in the S4 helix in the next group of modes may contribute to defining these modes as activation motion, i.e., motion that is associated with the transduction of voltage sensing into conformational changes, ultimately leading to gating.
The motion of the S4 helix, which was observed here, is thought to be responsible for voltage sensing. The appearance of the hinge near the helix center in the isolated VSD, the monomer, and the tetramer implies that each voltage sensor has the capacity to function independently. This suggestion is supported by studies of chimeras of Kv2.1 with the paddle from Hv and Ci-VSP (21), and concatenated heterotetramer Shaker channels in which the four gating charges on helix S4 were neutralized in three of the four subunits (67). However, some of the motions observed here involve cooperativity in the dynamics of the internal part of the S1 helix and the selectivity filter, suggesting that these regions, which are remote from each other in two different structural domains, are allosterically coupled.
The analysis revealed a correlation between the fluctuations of the selectivity filter, located in the external region of the channel, and the internal parts of the PD and VSD. The correlation was observed in both groups of the slowest modes as well as in the average over the eight slowest modes. This indicates a functional connection between the fluctuations of the outer gate (selectivity filter) and the fluctuations of the internal parts of the structure. The selectivity filter and internal gate were also found to be dynamically and evolutionarily correlated with residues in the internal part of helix S1 (60). Because these correlations were detected using two complementary computational approaches, it would be interesting to also examine them experimentally.
The motions described here are consistent with previously proposed gating mechanisms, in that the S3 and S4 helices appeared as a rigid structural unit (40). They are also consistent with data showing that the paddle moves as a unit in response to changes in membrane voltage when tarantula toxins bind the paddle (21). Monte Carlo and molecular-dynamics simulations predicted a rotation of the S4 helix relative to the S3 helix during the conformational change (5,68), in contradiction to the paddle model. This suggestion is supported by results from luminescence resonance energy transfer (69) and cystein cross-linking (70) experiments in Shaker. We did not detect such a motion. In this context, it is important to note that the NMA used here suffers from some simplifications in its underlying model and theory (see Supporting Material), such as the use of harmonic approximation and nonspecific interactions. Thus, any motion that is dominated by nonlinear dynamics would not be clearly observed with this approach. More importantly, the predictions rely on the model structure; any phenomena that involve significantly different structures would not be observed.
To summarize, in this work we utilized a simple computational model to explore the functional motions of the Kv1.2 channel. In general, our analysis of the fluctuations revealed the existence of independent and cooperative motions. These are reminiscent of modularity and hierarchical organization of structural units that appear to mediate the channel's function. Within this framework, the differences in the hinges and dynamic couplings observed between the monomer and the tetramer yield a dynamic fingerprint for Kv channels as reflected by the fluctuations. The voltage-sensing motion occurs independently within each monomer, whereas gating is a cooperative motion of all four monomers.
Supporting Material
Figures S1–S10, tables S1–S4, movies S1a and S1b, and the multiple sequence alignment are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00214-6.
Supporting Material
Acknowledgments
We thank Ugur Emekli for help with the HingeProt web server, and Guy Nimrod for helpful discussions. We also thank Diane Papazian, Vladimir Yarov-Yarovoy, and Roderick MacKinnon for providing the coordinates of their Kv structure models. Author contributions: Adva Yeheskel conducted the simulations, analyzed the results, and produced the first draft of the manuscript. Turkan Haliloglu and Nir Ben-Tal guided her jointly.
This study was supported by grants 222/04 and 611/07 from the Israel Science Foundation to N.B.-T. T.H. received partial support from the Turkish Academy of Sciences and EU-FP6-ACC-2004-SSA-2 contract No. 517991, and the Betil Fund. N.B.-T. and T.H. received a grant from NATO (No. LST.CLG.980180).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License(http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Sanguinetti M.C., Spector P.S. Potassium channelopathies. Neuropharmacology. 1997;36:755–762. doi: 10.1016/s0028-3908(97)00029-4. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y., Lee A., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 3.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 4.Pathak M.M., Yarov-Yarovoy V., Isacoff E.Y. Closing in on the resting state of the Shaker K(+) channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 5.Yarov-Yarovoy V., Baker D., Catterall W.A. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc. Natl. Acad. Sci. USA. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long S.B., Tao X., MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 7.Lee S.Y., Lee A., MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal S.K., MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 9.Ahern C.A., Horn R. Specificity of charge-carrying residues in the voltage sensor of potassium channels. J. Gen. Physiol. 2004;123:205–216. doi: 10.1085/jgp.200308993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker O.S., Larsson H.P., Isacoff E.Y. Three transmembrane conformations and sequence-dependent displacement of the S4 domain in Shaker K+ channel gating. Neuron. 1998;20:1283–1294. doi: 10.1016/s0896-6273(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 11.Seoh S.A., Sigg D., Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 12.Wang M.H., Yusaf S.P., Sivaprasadarao A. Effect of cysteine substitutions on the topology of the S4 segment of the Shaker potassium channel: implications for molecular models of gating. J. Physiol. 1999;521:315–326. doi: 10.1111/j.1469-7793.1999.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starace D.M., Stefani E., Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 14.Tombola F., Pathak M.M., Isacoff E.Y. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- 15.Yang N., Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 16.Murata Y., Iwasaki H., Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 17.Hossain M.I., Iwasaki H., Okamura Y. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J. Biol. Chem. 2008;283:18248–18259. doi: 10.1074/jbc.M706184200. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki H., Murata Y., Okamura Y. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 2008;105:7970–7975. doi: 10.1073/pnas.0803936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohout S.C., Ulbrich M.H., Isacoff E.Y. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 20.Ramsey I.S., Moran M.M., Clapham D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alabi A.A., Bahamonde M.I., Swartz K.J. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chanda B., Asamoah O.K., Bezanilla F. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 23.Starace D.M., Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 24.Bezanilla F. Voltage sensor movements. J. Gen. Physiol. 2002;120:465–473. doi: 10.1085/jgp.20028660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durell S.R., Shrivastava I.H., Guy H.R. Models of the structure and voltage-gating mechanism of the Shaker K+ channel. Biophys. J. 2004;87:2116–2130. doi: 10.1529/biophysj.104.040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahern C.A., Horn R. Stirring up controversy with a voltage sensor paddle. Trends Neurosci. 2004;27:303–307. doi: 10.1016/j.tins.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Yang N., George A.L., Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 28.Yang N., George A.L., Horn R. Probing the outer vestibule of a sodium channel voltage sensor. Biophys. J. 1997;73:2260–2268. doi: 10.1016/S0006-3495(97)78258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein S.A., Price L.A., Pausch M.H. ORK1, a potassium-selective leak channel with two pore domains cloned from Drosophila melanogaster by expression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93:13256–13261. doi: 10.1073/pnas.93.23.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Ruta V., MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 31.Haliloglu T., Ben-Tal N. Cooperative transition between open and closed conformations in potassium channels. PLOS Comput. Biol. 2008;4:e1000164. doi: 10.1371/journal.pcbi.1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrivastava I.H., Bahar I. Common mechanism of pore opening shared by five different potassium channels. Biophys. J. 2006;90:3929–3940. doi: 10.1529/biophysj.105.080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahar I., Rader A.J. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haliloglu T., Bahar I., Erman B. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997;79:3090–3093. [Google Scholar]
- 35.Bahar I., Atilgan A.R., Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold. Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 36.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emekli U., Schneidman-Duhovny D., Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 38.Cui Q., Bahar I. Chapman & Hall/CRC; Boca Raton, FL: 2006. Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. [Google Scholar]
- 39.Bahar I., Lezon T.R., Bakan A., Shrivastava I.H. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 2009;110:1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tombola F., Pathak M.M., Isacoff E.Y. How does voltage open an ion channel? Annu. Rev. Cell Dev. Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 41.Cuello L.G., Cortes D.M., Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu H., Iwamoto M., Oiki S. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell. 2008;132:67–78. doi: 10.1016/j.cell.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 43.Valadié H., Lacapcre J.J., Etchebest C. Dynamical properties of the MscL of Escherichia coli: a normal mode analysis. J. Mol. Biol. 2003;332:657–674. doi: 10.1016/s0022-2836(03)00851-9. [DOI] [PubMed] [Google Scholar]
- 44.Blunck R., Cordero-Morales J.F., Bezanilla F. Detection of the opening of the bundle crossing in KcsA with fluorescence lifetime spectroscopy reveals the existence of two gates for ion conduction. J. Gen. Physiol. 2006;128:569–581. doi: 10.1085/jgp.200609638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franqueza L., Lin M., Sanguinetti M.C. Long QT syndrome-associated mutations in the S4-S5 linker of KvLQT1 potassium channels modify gating and interaction with minK subunits. J. Biol. Chem. 1999;274:21063–21070. doi: 10.1074/jbc.274.30.21063. [DOI] [PubMed] [Google Scholar]
- 46.Miloshevsky G.V., Jordan P.C. Open-state conformation of the KcsA K+ channel: Monte Carlo normal mode following simulations. Structure. 2007;15:1654–1662. doi: 10.1016/j.str.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 47.Yifrach O., MacKinnon R. Energetics of pore opening in a voltage-gated K(+) channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 48.Tranebjaerg L., Bathen J., Bitner-Glindzicz M. Jervell and Lange-Nielsen syndrome: a Norwegian perspective. Am. J. Med. Genet. 1999;89:137–146. [PubMed] [Google Scholar]
- 49.Berezin C., Glaser F., Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 50.Ledwell J.L., Aldrich R.W. Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J. Gen. Physiol. 1999;113:389–414. doi: 10.1085/jgp.113.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pathak M., Kurtz L., Isacoff E. The cooperative voltage sensor motion that gates a potassium channel. J. Gen. Physiol. 2005;125:57–69. doi: 10.1085/jgp.200409197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch H.P., Kurokawa T., Larsson H.P. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 2008;105:9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tombola F., Ulbrich M.H., Isacoff E.Y. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58:546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoppa N.E., Sigworth F.J. Activation of Shaker potassium channels. III. An activation gating model for wild-type and V2 mutant channels. J. Gen. Physiol. 1998;111:313–342. doi: 10.1085/jgp.111.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tytgat J., Hess P. Evidence for cooperative interactions in potassium channel gating. Nature. 1992;359:420–423. doi: 10.1038/359420a0. [DOI] [PubMed] [Google Scholar]
- 56.Zandany N., Ovadia M., Yifrach O. Direct analysis of cooperativity in multisubunit allosteric proteins. Proc. Natl. Acad. Sci. USA. 2008;105:11697–11702. doi: 10.1073/pnas.0804104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi K.L., Mossman C., Yellen G. The internal quaternary ammonium receptor site of Shaker potassium channels. Neuron. 1993;10:533–541. doi: 10.1016/0896-6273(93)90340-w. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y., Holmgren M., Yellen G. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 1997;19:175–184. doi: 10.1016/s0896-6273(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 59.Labro A.J., Raes A.L., Snyders D.J. Coupling of voltage sensing to channel opening reflects intrasubunit interactions in Kv channels. J. Gen. Physiol. 2005;125:71–80. doi: 10.1085/jgp.200409194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleishman S.J., Yifrach O., Ben-Tal N. An evolutionarily conserved network of amino acids mediates gating in voltage-dependent potassium channels. J. Mol. Biol. 2004;340:307–318. doi: 10.1016/j.jmb.2004.04.064. [DOI] [PubMed] [Google Scholar]
- 61.Soler-Llavina G.J., Chang T.H., Swartz K.J. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Lewis A., Jogini V., Roux B. Atomic constraints between the voltage sensor and the pore domain in a voltage-gated K+ channel of known structure. J. Gen. Physiol. 2008;131:549–561. doi: 10.1085/jgp.200809962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J. Gen. Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKeown L., Burnham M.P., Jones O.T. Identification of an evolutionarily-conserved extracellular threonine residue critical for surface expression and its potential coupling of adjacent voltage-sensing and gating domains in voltage-gated potassium channels. J. Biol. Chem. 2008;283:30421–30432. doi: 10.1074/jbc.M708921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S.Y., Banerjee A., MacKinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K(+) channels. PLoS Biol. 2009;7:e47. doi: 10.1371/journal.pbio.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banerjee A., MacKinnon R. Inferred motions of the S3a helix during voltage-dependent K+ channel gating. J. Mol. Biol. 2008;381:569–580. doi: 10.1016/j.jmb.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagnon D.G., Bezanilla F. A single charged voltage sensor is capable of gating the Shaker K+ channel. J. Gen. Physiol. 2009;133:467–483. doi: 10.1085/jgp.200810082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bjelkmar P., Niemelä P.S., Lindahl E. Conformational changes and slow dynamics through microsecond polarized atomistic molecular simulation of an integral Kv1.2 ion channel. PLOS Comput. Biol. 2009;5:e1000289. doi: 10.1371/journal.pcbi.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Posson D.J., Selvin P.R. Extent of voltage sensor movement during gating of Shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Broomand A., Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 71.DeLano W.L. DeLano Scientific; Palo Alto, CA: 2002. PyMol Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


