Abstract
The integrity of a eukaryotic genome is often challenged by DNA double-strand breaks (DSBs). Even a single, unrepaired DSB can be a lethal event, or such unrepaired damage can result in chromosomal instability and loss of genetic information. Furthermore, defects in the pathways that respond to and repair DSBs can lead to the onset of several human pathologic disorders with pleiotropic clinical features, including age-related diseases and cancer. For decades, studies have focused on elucidating the enzymatic mechanisms involved in recognizing, signaling and repairing DSBs within eukaryotic cells. The majority of biochemical and genetic studies have used simple, DNA substrates, whereas only recently efforts have been geared towards understanding how the repair machinery deals with DSBs within chromatin fibers, the nucleoprotein complex that packages DNA within the eukaryotic nucleus. The aim of this review is to discuss our recent understanding of the relationship between chromatin structure and the repair of DSBs by homologous recombination. In particular, we discuss recent studies implicating specialized roles for several, distinct ATP-dependent chromatin remodeling enzymes in facilitating multiple steps within the homologous recombination process.
Keywords: chromatin remodeling, DNA repair, homologous recombination, INO80, RSC, SWI/SNF, SWR1
Cell viability and genomic stability are frequently threatened by chromosomal DNA double-strand breaks (DSBs) [1–3] (for recent reviews, see [4–9]). Indeed, this type of DNA damage is a natural consequence of basic cell metabolism. DSBs are induced by endogenous free oxygen radicals, collapsed replication forks or by the physical stress generated when dicentric or catenated chromosomes are pulled to opposite poles during mitosis [10]. Several cellular processes also employ DSBs as key steps for programmed genomic rearrangements, such as yeast mating type switching [11], variable (diversity) joining (V[D]J) recombination [12], class-switch recombination [13] and meiotic recombination [14]. In addition to these endogenous sources, DSBs are also produced exogenously when cells are exposed to DNA-damaging agents, such as ionizing radiation (IR), chemicals, chemotherapeutics that poison topoisomerase I or topoisomerase II [15], or UV light that creates alkyl adducts, pyrimidine dimers and crosslinks [16,17]. The failure or improper repair of DSBs can result in cell death or gross chromosomal changes, including deletions, translocations and fusions that promote genome instability and tumorigenesis. Consequently, cells have developed complex signaling networks that sense DSBs, arrest the cell cycle and activate repair pathways. Moreover, extensive DNA damage can activate overlapping signaling pathways that trigger apoptosis and prevent propagation of cells with highly unstable genomes [18].
Repair pathways: nonhomologous end-joining & homologous recombination
Eukaryotic cells have evolved two major mechanisms that repair chromosomal DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR). NHEJ is the predominant DSB repair mechanism in the G1 phase of the cell cycle, although NHEJ can take place throughout the cell cycle. In this pathway, the broken DNA ends are recognized and bound by the Ku70/Ku80 heterodimer, a complex which is conserved throughout the eukaryotes. In Saccharomyces cerevisiae, the Mre11/Rad50/Xrs2 (MRX) complex is also involved in the processing of DSBs for NHEJ; whereas in mammals NHEJ is associated with a complex of DNA-dependent protein kinase (DNA-PK) and the nuclease, artemis [19]. Both the MRX complex (in S. cerevisiae; Mre11–Rad50–Nbs1 [MRN] in mammals) and the DNA-PK are also important for keeping the two broken DNA ends together. Finally, the ligation of the broken DNA ends is accomplished by the Lig4/Lif1 complex in S. cerevisiae and Lig4/XRCC4 and XLF/Cernunnos factors in mammals [20–22]. Owing to nucleolytic processing of DNA ends to make them compatible for subsequent ligation, NHEJ can result in short deletions, and thus this process is typically error-prone [19].
Homologous recombination functions primarily in late S–G2 phase of the cell cycle, and relies on sequence homology from an undamaged sister chromatid or a homologous DNA sequence to use as a template for copying the missing information [23]. Therefore, in contrast to NHEJ, the HR pathway is essentially error-free. Genetic analyses in S. cerevisiae revealed that the proteins coded by the RAD52 epistasis group – RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11 and XRS2 – mediate HR [11]. The basic HR pathway is highly conserved throughout eukaryotes, as most of the known gene products involved in yeast HR have functional homologs in higher eukaryotes [24,25].
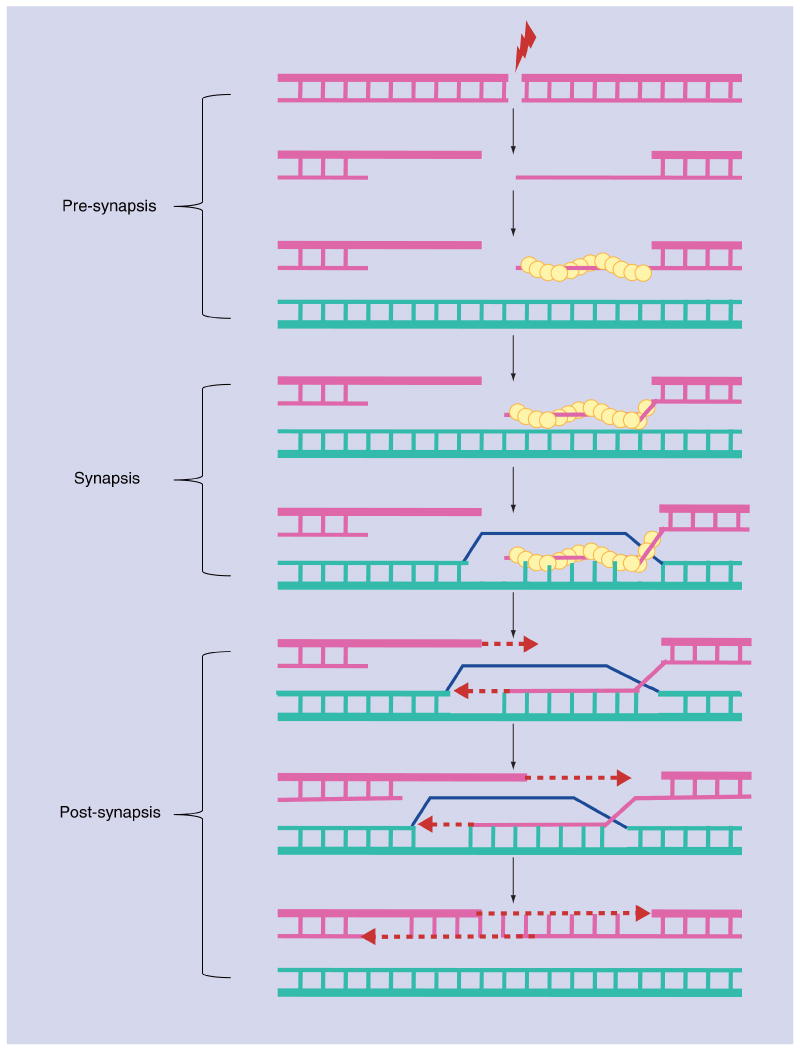
Much of what we know about the HR process is owing to in vivo studies that have exploited the HR event that governs mating type switching in S. cerevisiae [26]. Haploid budding yeast can exist as either a or α mating type, and the mating type is controlled by transcription factors encoded within the MAT locus on chromosome III. On the opposite arms of chromosome III, there also exist two donor loci, HMLα and HMRa, which contain the genes that determine α or a mating type, respectively. The HM loci are organized into transcriptionally silent heterochromatin, and thus only the genes at the MAT locus are expressed. In wild-type yeast cells, expression of the site-specific endonuclease, HO, initiates a mating type switching event by introducing a unique DSB at the MAT locus. This DSB is then repaired by HR, using one of the two HM loci as a homologous donor. The completion of this HR event leads to replacement of the information at MAT with HM sequences, but the silent donor locus remains unchanged. Strains that express the HO endonuclease from the galactose-dependent GAL10 promoter have provided a powerful tool to control DSB formation and to synchronize the HR process [27]. Furthermore, PCR, Southern blot and chromatin immunoprecipitation (ChIP) assays have been developed to identify HR intermediates and to follow the sequence of events during HR (Figure 1) [28].
Figure 1. Biochemical steps of homologous recombination.
The presynapsis stage includes nucleolytic processing of the 5′-ends of DNA that flank a double-strand break to generate long 3′ single-stranded DNA tails, following assembly of the Rad51 presynaptic filament (yellow balls). The synapsis stage includes search for homology, followed by formation of paranemic and plectonemic joints. The post-synapsis stage involves extension of the joints, new DNA synthesis, formation of Holliday junctions, resolution of joints and ligation of broken DNA ends. The 5′–3′ orientation of DNA strands in the duplexes are marked by bold lines.
The first step of HR involves processing of the DSB so that the 5′-ends of the DNA duplex that flank the DSB are nucleolytically processed to generate long, 3′-single-stranded tails (Figure 1). This resection reaction requires a myriad of proteins, including the MRX complex, the Sgs1 helicase and several other nucleases, such as Sae2, Dna2 and Exo1[29–32]. The processed DSB ends are initially bound by the single strand DNA-binding protein, replication protein A (RPA), which is subsequently replaced by the key recombinase, Rad51. The Rad51 protein has a highly conserved structure among eukaryotes, and it can assemble onto single-stranded DNA (ssDNA) as well as double-stranded DNA (dsDNA) to form a right-handed helical filament in which approximately six protein monomers span approximately 18–19 bases (bp) of DNA per helical turn, with a pitch of approximately 10 nm [33,34]. Although Rad51 can bind and hydrolyze ATP, only the binding of ATP is required for the assembly of the presynaptic filament [35–37], and ATP hydrolysis promotes dissociation of Rad51 from DNA, resulting in high turnover and recycling of the recombinase [38–41]. Formation of the Rad51-presynaptic filament is facilitated by several ‘mediator’ proteins. Amongst them, Rad52 and Rad54 stimulate the catalytic activity of Rad51 through a direct interaction. In addition, other mediators include Rad55 and Rad57 in yeast [42], and at least five proteins in higher eukaryotes, including several Rad51 paralogs (Rad51B, Rad51C and Rad51D). Likewise, in higher eukaryotes, the BRCA1 and BRCA2 tumor suppressors interact and mediate the function of Rad51 in HR [43,44].
The primary role of the Rad51 nucleoprotein filament is to perform a search for a homologous duplex DNA and catalyze formation of a DNA joint molecule (Figure 1). A common source for this duplex DNA ‘donor’ is the undamaged sister chromatid, but homologous sequences on either the same or a different chromosome can be captured by the presynaptic filament. In fact, studies in both yeast and mammalian cells have suggested that this homology search process may occur on a genome-wide level [45–47]. After the initial synapsis of the Rad51 nucleoprotein filament with a duplex donor locus, an initial, transient joint molecule is formed, called a paranemic joint, whose stability depends on DNA–Rad51 interactions. In a paranemic joint, the 3′-end of the invading presynaptic filament is not engaged with the duplex donor. Subsequently, the paranemic joint is converted into a plectonemic joint, whose stability no longer depends on DNA–protein interactions, and the 3′-end of the invading presynaptic filament forms Watson–Crick bps with the complementary strand of the duplex donor. The characteristic structure of this stable DNA joint led to it being termed the displacement loop (D-loop) [43]. Conversion of the initial paranemic joint to the stable D-loop requires the Rad54 ATPase which appears to use the energy from ATP hydrolysis to alter DNA topology [48]. The 3′-end of the presynaptic filament within the D-loop serves as a primer for new DNA synthesis by DNA polymerase. HR intermediates are further extended by branch migration, changing the D-loop to a more prominently cruciform structure known as a Holliday junction. Finally, Holliday junctions are resolved by cleavage and ligation, or by dissociation of the extended strand from the donor and re-annealing to ssDNA on the opposite side of the DSB (termed synthesis-dependent strand annealing [SDSA]) [43].
Cellular response to DSBs
The cellular response to DSBs is mediated by several different classes of proteins, which are commonly known as sensors, mediators and effectors of checkpoint control pathways. The cellular response to DSBs is initiated by rapid recruitment of the highly conserved MRX (S. cerevisiae) or MRN (mammals) complex that contains Mre11, Rad50 and Xrs2 (MRX in S. cerevisiae) or Nbs1 (MRN in mammals). The MRN(X) complex has an integral role in the initial recognition of the DSB, cell-cycle checkpoint activation, signal amplification, as well as bridging the two ends of a DSB and facilitating DSB resection [49]. MRN(X) also activates the ataxia telengiectasia mutated ([ATM], Tel1 in yeast) kinase in vivo and in vitro, which is one of the key checkpoint cascade kinases. Once recruited to the DSB, MRN(X) and ATM can in turn activate another sensor kinase, ATM-related [ATR] Mec1 in yeast [50]. Long stretches of ssDNA generated by resection, as well as the binding of RPA, provide a critical signal in the damage repair signaling cascade. RPA recruitment at the DSB is necessary for recruitment of sensor kinases like ATR and ATR interacting partner ([ATRIP], Ddc2 in S. cerevisiae) [51]. The subclass of downstream substrates that act as either recruiters or activators of a plethora of additional complexes (‘effectors’) or scaffolds on which complexes can be assembled [52] are known as mediators. Finally, in mammals, checkpoint kinases (Chk1 and Chk2) work downstream of the signaling cascade and inactivate CDC25 phosphatases, culminating in cell-cycle arrest [53]. In contrast, arrest of cell-cycle progression through anaphase in S. cerevisiae takes place via inhibition of cohesin degradation, a step that is necessary for separation of sister chromatids [54].
DNA double-strand break repair & chromatin
In eukaryotes, the recognition, processing and repair of DSBs must occur within the nucleoprotein complex of chromatin. How this complex process functions within the chromatin environment has been an area of intense research within the past 5 years [55]. Below we provide a general overview of chromatin structure, introduce three mechanisms for regulating chromatin dynamics, and then focus on the role of ATP-dependent chromatin remodeling enzymes in controlling discrete steps in the HR process.
The basic unit of chromatin is the nucleosome core particle, which consists of 147 bp of DNA wrapped in left-handed superhelical turns approximately 1.7-times around an octamer of histone proteins [56]. The histone octamer is composed of one tetramer of histones H3 and H4 that is flanked by two heterodimers of H2A and H2B. The histone proteins have a low molecular weight, are very basic in nature and highly conserved across all eukaryotes. They each harbor a structured, three-helix bundle called a histone fold motif, which mediates histone–histone and histone–DNA interactions. The structured histone fold domains are flanked by short, flexible N-terminal and C-terminal domains or tails, which protrude from the nucleosome core particle [56]. Although the histone tails are not required for assembly of either the octamer or a nucleosome, they are essential for regulation of many biological processes. Numerous post-translational modifications occur at different amino acid residues of the tails [57], regulating key biological processes. The tails are also important for both intramolecular and intermolecular folding of nucleosomal arrays in vitro to mediate different levels of compaction [58–60].
The primary structure of chromatin is a linear array of nucleosomes, forming the 10-nm fiber or ‘beads on a string’ structure as seen by electron microscopy [61]. This linear array is folded into a 3D secondary structure, called a 30-nm fiber, via internucleosomal interactions and stabilized by the association of linker histones, such as H1 or H5 [60,62]. The linker histones bind nucleosomes at the DNA entry/exit point, forming a particle called a chromatosome that stabilizes an additional 20 bp of DNA within the nucleosome. Chromatin fibers are also organized into enormous tertiary structures due to self-association of the 30 nm fibers into 100–400 nm thick filaments, called chromonema filaments, which are visualized by electron microscopy in interphase cells and detected by biophysical methods [58,63].
Eukaryotic chromosomes are organized into specialized domains that can regulate nuclear functions. Early cytological studies defined regions of the genome that underwent decondensation as the cells progressed from metaphase to interphase as euchromatin [64]. On the other hand, the regions that remained visibly condensed and deeply stained throughout the cell cycle were defined as heterochromatin. These latter regions are mainly found at centromeres and telomeres, and they play key roles in maintaining chromosome structure, stability and integrity. This type of heterochromatin is often referred to as constitutive heterochromatin. In some cases, the state of heterochromatin can change in response to cellular signals and revert back to that of euchromatin [65]. Reversible heterochromatin is termed facultative heterochromatin, and it may provide a means of controlling DNA metabolism, such as replication and transcription. Structural features that characterize heterochromatin include the presence of predominantly repetitive DNA sequences, low or absent gene density, late S-phase replication timing, regular nucleosome spacing, decreased accessibility to nucleases, loss of nuclease hypersensitive sites and hypoacetylation of histones [65–67]. These structural features distinguish heterochromatin from euchromatin and are conserved throughout eukaryotes. In addition, except for budding yeast, methylation of histone H3 at position K9 and its associated chromo-domain-containing protein, HP1, are also hallmarks of heterochromatin, conserved from fission yeast to mammals. Moreover, in vertebrates and plants, heterochromatin is supplemented with cytosine hypermethylation and associated proteins.
Chromatin dynamics: histone variants, histone modifications & chromatin remodeling enzymes
DNA double-strand breaks occur within both euchromatic and heterochromatic regions of the genome, and in both cases, the various steps of HR require that chromatin structure is disrupted, at least transiently. For instance, processing of the DNA ends to generate the 3′-ssDNA tails requires that the resection machinery navigate through nucleosomal DNA. Likewise, the genome-wide search for homology will involve scanning for Watson–Crick basepairing within duplex DNA that is wrapped around histone octamers and nucleosomal arrays that are folded into thick, condensed fibers. Extension of joint molecules requires that DNA synthesis progress through adjacent nucleosomes, and final resolution of the repair event will require restoration of chromatin structure, possibly accompanied by deposition of new nucleosomes. This myriad of events requires the chromatin structure to be dynamic in nature – capable of rapid unfolding, disassembly, assembly and refolding. Currently, we know of three distinct mechanisms that control the dynamics of chromatin structure:
Histone variants
Histone modifications
ATP-dependent chromatin remodeling enzymes.
Histone variants
In addition to the canonical histones H2A, H2B, H3 and H4, which are expressed during S phase and incorporated into chromatin at the replication fork, eukaryotes also employ a variety of histone variants that provide specialized structures and functions to the chromatin fiber. In contrast to the canonical histones, the histone variants are expressed throughout the cell cycle, and they are incorporated into chromatin independent of DNA replication [68]. Histone H2A has the largest number of variants, including H2A.X, H2A.Z, macroH2A and H2ABbd. Amongst these variants, H2A.X and H2A.Z have been well-characterized and linked to the repair of DNA DSBs and regulation of transcription [4,60]. In the case of DSB repair, H2A.Z facilitates the exonucleolytic resection of DSBs. Nucleosomal arrays that contain H2A.Z have an increased propensity to form 30-nm fibers compared with arrays that contain canonical H2A, suggesting that it may control fiber dynamics [69]. Interestingly, nucleosomes that harbor H2A.Z flank most RNA polymerase II promoters, and these nucleosomes tend to be inherently dynamic and easily evicted during transcriptional induction [70]. Thus, H2A.Z may contribute to both active and inactive chromatin states. This dynamic nature of H2A.Z provides one explanation for how this histone variant might facilitate DSB resection. Other well-studied histone variants are related to histone H3. For instance, Cse4/CENP-A, which is essential for centromere function, is found in yeast and mammalian centromeres, respectively [71,72], and H3.3 is a histone variant that replaces H3.1 during transcription [73].
Histone post-translational modifications
Each of the core histones are subject to a vast array of post-translational modifications, including lysine acetylation, methylation, sumoylation and ubiquitylation, serine/threonine phosphorylation, proline isomerization and arginine methylation [74]. Most of the well-characterized histone modifications occur within the extended N-terminal or C-terminal histone tail regions, but there are a growing number of post-translational modifications within the nucleosome core. Indeed, it may be that every solution-exposed histone residue may be subject to a modification event. Histone modifications control chromatin dynamics by two broad mechanisms. First, histone modifications can either create or eliminate binding sites for nonhistone proteins that influence the structure and function of the chromatin fiber [75]. For instance, methylation of histone H3 at lysine 9 promotes the binding of the HP1 protein, which facilitates formation of condensed heterochromatin structures. Second, histone modifications may directly impact either the stability of individual nucleosomes or influence the ability of chromatin fibers to fold into higher order structures. Although most of the histone modifications that occur on the N-terminal tail domains do not contribute to nucleosome stability, it is possible that some of the modifications within the core domain may directly impact nucleosome structure. To date, only one histone modification, acetylation of histone H4 at lysine 16, has been shown to directly impact chromatin structure. In this case, acetylation of this one lysine residue is sufficient to block formation of 30-nm fibers [76].
ATP-dependent chromatin remodeling enzymes
The ATP-dependent chromatin remodeling enzymes typically consist of multi-subunit complexes that can use the energy from ATP hydrolysis to actively disrupt histone–DNA contacts. Recent studies indicate that ATP hydrolysis is used to ‘pump’ or translocate DNA over the histone octamer surface, changing nucleosome positioning [77]. This ATP-dependent reaction can also catalyze the eviction of histone octamers or promote the exchange of histone H2A–H2B dimers [78]. The hallmark of the ATP-dependent remodeling enzymes is the presence of an ATPase subunit that is related to the SWI/SNF2 subfamily of the Asp-Glu-Ala-Asp/His (DEAD/H) superfamily of nucleic acid-stimulated ATPases [79].
ATP-dependent remodeling enzymes from different organisms have been subdivided into groups based on sequence homology within the catalytic subunit and the presence of unique sequence motifs such as a bromodomain (Swi2/Snf2 family), SANT domain (ISWI family) and chromo-domain (Mi-2 Family) [80]. So far, six distinct remodeling complexes have been purified from S. cerevesiae: SWI/SNF [81], RSC [82], ISW1, ISW2 [83], INO80 [84] and SWR1 [85]. With the exception of the SWR1 enzyme, each of these enzymes can catalyze movements of histone octamers along DNA in cis [86–90]. The SWR1 enzyme is dedicated to the ATP-dependent incorporation of the histone variant, H2A.Z [85].
An exciting theme has emerged that histone-modifying enzymes and ATP-dependent chromatin remodeling enzymes do not necessarily function independently of each other in regulating basic chromatin-mediated processes like replication, transcription and DNA damage repair. Several lines of evidence indicate that modified histones facilitate recruitment of ATP-dependent chromatin remodeling enzymes to target sites [91]. For instance, the bromodomain within the Swi2 subunit of yeast SWI/SNF interacts with lysines acetylated by the Spt-Ada-Gcn5 acetyltransferase (SAGA) histone acetyltransferase complex, and this interaction stabilizes SWI/SNF at target genes, like HO [92]. In contrast, chromodomains, tudor domains and plant-homeodomain-finger domains bind distinct methylated lysine residues [93–96], and these interactions may help target enzymes like Mi-2 to target loci. On the other hand, ATP-dependent chromatin remodeling is also a prerequisite for recruitment of histone-modifying enzymes at certain target sites [97].
Chromatin dynamics that regulate HR
As discussed above, recombinational repair of a DSB requires extensive DNA ‘gymnastics’ – exonucleolytic processing, a genome-wide homology search, D-loop formation, joint extension and so on. Every one of these steps has the potential to be regulated by the organization of DNA into chromatin. Thus, it should come as no surprise that virtually every ATP-dependent chromatin remodeling enzyme and nearly every type of histone modification has been shown to be essential for, or be associated with, recombinational repair of a DSB. As a broad generalization, histone modifications appear to modulate the early recruitment of repair factors and checkpoint signaling molecules, whereas subsequent recruitment of ATP-dependent remodeling enzymes play key roles in facilitating the enzymatic steps of HR. However, remodeling enzymes also contribute to establishing and maintaining the cell-cycle checkpoint response.
Histone modifications in response to a DSB
The first chromatin-associated event that occurs in response to a DSB is the rapid phosphorylation of a serine residue (S129 in S. cerevisiae and S139 in the mammals) within the Ser-Gln-Glu (SQE) motif of the C-terminal domain of the histone variant, H2AX. In mammalian cells, H2AX is found within approximately 10% of nucleosomes, whereas in S. cerevisiae, H2AX represents the bulk H2A. The phosphorylated version of H2AX is typically referred to as γ-H2AX, and this phosphorylation event encompasses megabases of chromatin flanking a single DSB in mammalian cells [98], and approximately 50 kb in S. cerevisisae [99]. γ-H2AX is catalyzed by the ATM, ATR and DNA-PK kinases in mammalian cells, whereas the Mec1 and Tel1 checkpoint kinases perform this role in budding yeast [100]. The formation of γ-H2AX nuclear foci, which are easily detectable by immunofluorescence microscopy, has proven to be a useful experimental marker for DSB induction in mammalian cells. Although γ-H2AX is not essential for the initial recruitment of DSB response factors to a DSB, this phosphorylation event does stabilize the binding of checkpoint factors, and it is required for effective repair of DSBs by both the NHEJ and HR pathways [101–103].
Recent studies have suggested that γ-H2AX leads to the recruitment of proteins that ubiquitinate γ-H2AX, which in turn can recruit downstream players of the DNA damage response [104–106]. Another established role of γ-H2AX is the recruitment of different subunits of the cohesin complex in response to DSBs, specifically in S and G2 phases, promoting HR using the sister chromatid [107,108]. Importantly, the other major role of γ-H2AX is to provide binding sites for ATP-dependent chromatin remodeling (INO80 and SWR1) and histone modification enzymes (NuA4) at the site of a DSB [109–111]. In addition, recent studies show that dephosphorylation of γ-H2AX following DNA repair is necessary for efficient recovery from the DNA damage checkpoint [112]. These studies indicate that a three-protein complex, HTP-C (histone phosphatase H2A complex), containing the phosphoserine-specific phosphatase Pph3 (in S. cerevisiae) and protein phosphatase 2A (in mammalian cells), regulates the dephosphorylation of H2AX [113].
A second histone modification that plays a key role in the early recruitment of checkpoint factors to a DSB is the methylation of histone H3 lysine 79 by the Dot1 methyltransferase. In mammalian cells, H3K79me provides a binding site for 53BP1 which functions in checkpoint activation and rapidly co-localizes with γ-H2AX [114]. 53BP1 is related to yeast Rad9, and both 53BP1 and Rad9 bind to H3K79me in vitro [115]. In vivo, H3K79 is constitutively methylated, and it has been hypothesized that H3K79me is exposed following DSB formation, and that it subsequently recruits 53BP1 and promotes the checkpoint signal [114]. In mammals, trimethylation of H4K20 also appears to enhance the recruitment of 53BP1 within both pericentric heterochromatin and euchromatic locations [116,117].
Several other histone modifications have been implicated in the DSB response. Additional residues in the C-terminal domain of H2A, S122 [118] and T126 [119] have been reported to be essential for survival of S. cerevisiae after different types of DNA damage [120]. However, the precise roles of these residues have yet to be determined. Furthermore, phosphorylation of S14 (in mammals) or S10 (in S. cerevisiae) at the N-terminal tail of H2B has also been implicated in the DSB response, and immunofluorescence studies have shown that H2B phosphorylation co-localizes with γ-H2AX foci in response to DNA DSBs [121]. In addition, ubiquitination of H2B lysine 123 by the Rad–BreI complex appears to be necessary for activation of the DNA-damage checkpoint in budding yeast [122]. And finally, acetylation of conserved lysine residues in the N-terminal domains of H3 and H4 is important for repair of DSBs by both NHEJ and HR, and these modifications are mediated by a number of histone acetyltransferases (HATs), such as NuA4, Gcn5 and Hat1 [123–125]. The levels of acetylation of H3 lysines (K9, 14, 18, 23 and 27) and H4 lysines (K5, 8, 12 and 16) increase within nucleosomes very close to the DSB, and decrease as the DSB is repaired. In mammalian embryonic stem cells, the Tip60 HAT complex is recruited to DSBs, which induces acetylation of H4 and facilitates HR [126]. How histone acetylation promotes DSB repair is not yet clear.
Distinct roles for ATP-dependent chromatin remodeling enzymes
Chromatin immunoprecipitation studies have exploited the galactose-inducible HO endonuclease to monitor the recruitment of ATP-dependent remodeling enzymes to the single HO-induced DSB at the budding yeast MAT locus. These studies have demonstrated that five different remodeling enzymes, SWI/SNF, RSC, INO80, SWR1 and Rad54, are rapidly recruited to a HO DSB (Table 1). Although many of these enzymes show similar biochemical activities, yeast genetic studies have demonstrated that each of these enzymes plays distinct roles in the HR process (Figure 2).
Table 1. Chromatin remodeling enzymes and double-strand break repair.
| Study | Chromatin remodeling enzymes | Time of recruitment at a DSB | Proposed functions in pathway of homologous recombinational repair | Ref. |
|---|---|---|---|---|
| Chai et al. (2005), Liang et al. (2007), Shim et al. (2007), Kent et al. (2007), Shim et al. (2005) | RSC | 20 min | Induced nucleosome mobility immediately after DSB formation | [127–131] |
| Recruitment of Ku70/80 and MRX at a DSB | ||||
| Recruitment of cohesins | ||||
| Enhancement of final ligation step | ||||
| Sinha et al. (2009) | SWI/SNF | 40 min | Homlogy search and joint formation on heterochromatic donor | [142] |
| Mazin et al. (2003), Bugreev et al. (2006), Wolner et al. (2005), Bugreev et al. (2007), Sinha et al. (2008), Sugawara et al. (2003) | Rad54 | 40 min | Stabilization of Rad51-presynaptic filament | [139,140,143–146] |
| Conversion of paranemic to plectonemic joints | ||||
| Branch migration of joint intermediates | ||||
| Dissociation of Rad51 from presynaptic filament | ||||
| Papamichos-Chronakis et al. (2006), Tsukuda et al. (2005), Tsukuda et al. (2009), van Attikum et al. (2007 | INO80 | 1 h | Processing of a DSB to generate ssDNA tail | [133–135,147] |
| Recruitment of Ku70/80 at a DSB | ||||
| Cell-cycle checkpoint adaptation | ||||
| Nucleosome loss from donor chromatin after synapsis/joint formation | ||||
| van Attikum et al. (2007), Mizuguchi et al. (2004) | SWR1 | 1 h | Recruitment of Ku70/80 at a DSB | [85,147] |
| Deposition of Htz1 within cellular chromatin | ||||
DSB: Double strand break; ssDNA: single-stranded DNA.
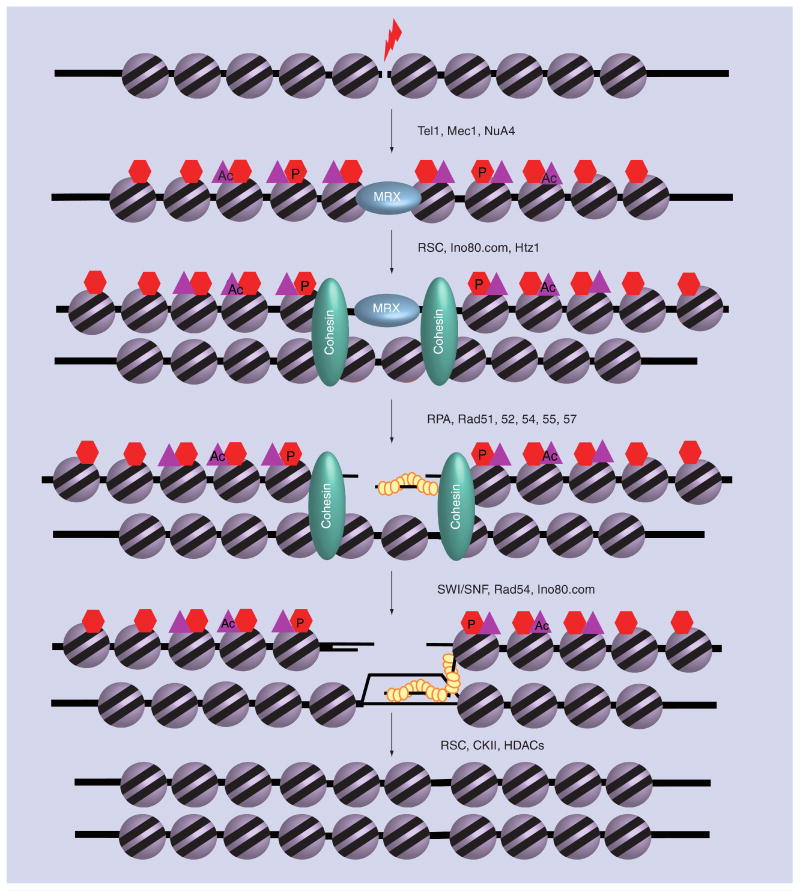
Figure 2. ATP-remodeling enzymes are implicated in different steps of the recombinational repair process of a chromosomal DNA DSB.
Following the rapid phosphorylation of H2AX, the RSC remodeling enzyme is recruited to DSB chromatin. RSC is then believed to mobilize a few nucleosomes flanking the break. RSC action helps to load subunits of the cohesin complex, Ku70/80 and MRX. INO80 is recruited by γ-H2AX and facilitates MRX-dependent nucleolytic processing of the DSB. INO80 and SWR1 also facilitate Ku70/80 recruitment. Next, Rad51 assembles onto ssDNA tails to generate functional presynaptic filament (yellow balls). ATP-dependent remodeling activity of SWI/SNF, Rad54 and INO80 are involved in formation of stable recombination intermediates during the strand invasion event by remodeling the donor nucleosomes. Finally, RSC is required for the final ligation of the newly synthesized DNA strand, and CKII and HDACs are believed to function in restoration of chromatin structure following homologous recombination.
Ac: Acetylation; CKII: Casein kinase II; DSB: Double-strand break; HDAC: Histone deacetylase; P: Phosphorylation; RPA: Replication protein A.
RSC plays multiple roles during HR
Three different ATP-dependent remodeling enzymes, RSC, INO80 and SWR1, appear to regulate early chromatin transitions that regulate the efficient processing of the DSB. Amongst them, three functional roles have been attributed to the remodeling activities of the RSC complex alone. RSC is rapidly recruited to the DSB [127], where it stimulates recruitment of the Ku70/Ku80 and MRX complexes, and facilitates H2AX phosphorylation [128]. RSC is required for changes in nuclease accessibility within chromatin adjacent to the DSB, suggesting that RSC may mobilize a few nucleosomes flanking the break to enhance recruitment of repair machinery to DSB chromatin [129,130]. In addition to these early steps, RSC is also required for proper loading of cohesins during HR [129,131], and RSC is required for the final ligation of the DSB [127].
Roles for INO80 & SWR1 during early steps in HR
The INO80 and SWR1 remodeling enzymes are also recruited to chromatin surrounding an HO-induced DSB within an hour of DSB induction [109,111]. Recruitment of these complexes requires H2AX phosphorylation, and the shared Arp4 subunit interacts directly with γ-H2AX in vitro [110]. Interaction with γ-H2AX may also involve the INO80 subunit, Nhp10 [111]. SWR1 appears to function in concert with the NuA4 acetyltransferase to promote the exchange of γ-H2AX for the Htz1 (H2A.Z) histone variant [132]. Both INO80 and SWR1 are required for efficient processing of the DSB, as mutations affecting the Arp8 subunit of INO80, or loss of the Htz1 histone variant (the product of SWR1 action) lead to significantly decreased kinetics of ssDNA formation [109]. The decreased kinetics lead to a delay in the activation of the cell-cycle checkpoint, but they do not significantly alter the kinetics of HR. Interestingly, both INO80 and SWR1 are required for the optimal recruitment of the Ku70/Ku80 complex to the DSB, but only the SWR1 enzyme is required for full levels of NHEJ. Thus, there is a surprisingly large commitment of remodeling enzymes to the recruitment of Ku70/Ku80 to a DSB (RSC, INO80 and SWR1). How these enzymes facilitate Ku70/80 recruitment is not clear, though a simple model would posit that Ku70/80 binding to DSB ends is exquisitely sensitive to neighboring nucleosomes, more so than other proteins like MRX. The INO80 enzyme is also required for cell-cycle checkpoint adaptation, a process that allows cells to turn off the checkpoint and enter mitosis when a DSB persists. This role for INO80 is linked to the SWR1-dependent incorporation of Htz1 [133].
Two reports have also suggested that the INO80 enzyme may facilitate removal of nucleosomes during the exonucleolytic processing of DSBs [134,135]. Likewise, ChIP analyses by the Gasser group demonstrated an approximately twofold loss of histone signal during DSB resection. However, significant controversy does surround the fate of nucleosomes during this early step of HR. Early ChIP studies from Shroff and Haber indicated that histone H2B was not lost from DSB proximal chromatin, even at timepoints where Rad51 was bound to at least 2 kb of ssDNA at the DSB [99]. Furthermore, in a recent study from the Tyler group, careful normalization of ChIP signals to input DNA samples suggested that histones may not be significantly lost from the DSB region during resection [136]. We have also observed variable histone loss at DSBs during resection [Manisha Sinha & Craig L Peterson, University of Massachusetts Medical School, MA, USA. Unpublished data]. Why have these different results been found? If nucleosomes are retained on the ssDNA that results from resection, such histone–DNA complexes may exhibit altered formaldehyde cross-linking efficiency. Cross-linking of histones to ssDNA may be less efficient, leading to the incorrect conclusion that histones are lost during resection, or cross-linking may be highly favored, and if this is the case then histone loss may be grossly underestimated. Variability in cross-linking efficiency among different laboratories may have led to the current discrepancies. Notably, early biochemical studies have demonstrated that nucleosome-like particles can form efficiently on ssDNA [137,138]. Even in cases where ChIP studies indicate histone loss, the change in histone density is usually only two- to four-fold, whereas histone ChIP signals are reduced at least five- to ten-fold when nucleosomes are displaced during transcriptional activation. Thus, by all accounts, the ssDNA that is adjacent to the DSB is unlikely to be histone free.
Why do we care about the fate of nucleosomes at processed DSBs? First, if nucleosomes remain on ssDNA, then such complexes will need to be ‘remodeled’ during later stages of HR when Holliday junctions are resolved and DNA strands are exchanged. The presence of histones on ssDNA may also impact the efficiency of the checkpoint response via inhibition of RPA binding, imparting perhaps another role for remodeling enzymes. The presence of nucleosomes on ssDNA would also require a change in how we view the Rad51 presynaptic filament. Typical views propose a continuous Rad51–ssDNA complex, whereas the presence of histones would suggest some type of discontinous structure. Unfortunately, there are no easy methods to detect and map the formation or position of histone–ssDNA complexes in vivo. Typical nucleases, like DNase or micrococcal nuclease (MNase), that are often used for chromatin mapping, will efficiently digest histone–ssDNA complexes [137,138]. Thus, a major technical challenge awaits the field, and resolution of these issues may await biochemical studies where the fate of histones can be directly followed during a resection reaction.
SWI/SNF facilitates the homology search within heterochromatin
Following DSB processing, the search for a homologous DNA donor would seem to be a likely step for involvement of chromatin remodeling enzymes. However, the INO80, SWR1, RSC and Rad54 remodeling enzymes are not required for capture of the homologous duplex in vivo, even when the donor is assembled into condensed heterochromatin [133,139,140; Manisha Sinha and Craig L Peterson, University of Massachusetts Medical School, MA, USA. Unpublished data]. Recently, we successfully reconstituted the homology search process in vitro with recombinant nucleosomes and purified factors [141]. Surprisingly, the Rad51 nucleoprotein filament appears to be sufficient in vitro to search and capture homology, even when the homologous sequences are located on the surface of a positioned nucleosome [141].
Although capture of homology within euchromatin regions of the genome may not require remodeling enzymes, the SWI/SNF remodeling enzyme plays an essential role when synapsis occurs within condensed, heterochromatic loci. During HR, SWI/SNF is recruited to the DSB, and it subsequently associates with the heterochromatic donor locus during joint formation [127]. In the absence of the SWI2 ATPase or the Snf5 subunit, the invading MATa presynaptic filament is unable to locate and synapse with the heterochromatic HMLα donor locus. Paradoxically, yeast mutants that lack a functional SWI/SNF complex are not very sensitive to DSB-inducing agents, questioning its general requirement for DSB repair by HR. Moreover, our biochemical studies indicate that the chromatin remodeling activity of SWI/SNF does not stimulate a Rad51-mediated homology search on a nucleosomal donor [141]. Thus, SWI/SNF may only facilitate HR when donor sequences are assembled into heterochromatin. Recently, we have reconstituted a heterochromatic chromatin donor using recombinant nucleosomes and purified Sir2, Sir3 and Sir4 proteins [142]. Strikingly, assembly of the donor into heterochromatin eliminates the ability of Rad51 to mediate a successful homology search. However, addition of the SWI/SNF remodeling enzyme restores formation of a joint molecule, and synapsis is associated with eviction of the Sir3 protein. In contrast, the INO80, SWR1, RSC and Rad54 remodeling enzymes are unable to substitute for SWI/SNF, and they do not promote synapsis with a heterochromatic donor [142]. Thus, both in vivo and in vitro studies indicate that the SWI/SNF remodeling enzyme may specifically regulate HR when the donor locus is located with condensed heterochromatin.
Roles for Rad54 & INO80 during late stages of HR
Following the formation of the initial DNA joint, two distinct chromatin transitions have been described that require two different ATP-dependent remodeling enzymes. Following the initial synapsis of the nucleoprotein filament and the donor, conversion to a joint that can be extended by DNA polymerases requires the ATPase activity of the Rad54 enzyme, and Rad54 activity is associated with disruption of a single nucleosome at the joint [139]. Thus, during HR between MATa and HMLα, Rad54 remodels chromatin without large-scale rearrangement of nucleosomes. In addition, joint formation leads to the loss of at least two nucleosomes from chromatin surrounding the initial joint, and these nucleosome loss events require the Arp8 subunit of the INO80 remodeling enzyme. In this latter case, nucleosome loss that is catalyzed by INO80 may regulate branch migration [135].
Future perspective
The role of chromatin in the regulation of the cellular response and repair of chromosomal DSBs has only begun to be fully appreciated. Research in the last decade has identified an ever growing number of chromatin modifications and chromatin remodeling enzymes that have emerged as key regulators of recombinational repair. Great emphasis has fallen on the distinct functions of ATP-dependent chromatin remodeling enzymes and their key roles in maintaining genomic integrity. With the powerful combination of yeast genetics and in vitro HR reactions with chromatin substrates, new insights into the mechanics and regulation of HR should be realized.
Executive summary.
Successful completion of double strand break (DSB) repair by homologous recombination requires quite a bit of DNA gymnastics that provide regulatory possibilities for chromatin structure.
Although it is generally assumed that nucleosomes are displaced from DSB chromatin during exonucleolytic processing, most data indicate that processed single-stranded DNA (ssDNA) tails remain associated with significant levels of histones that may impact checkpoint responses and subsequent steps.
Many of the chromatin remodeling enzymes and histone modifying enzymes that were initially characterized for their roles in gene transcription are now known to play roles in recombinational repair of DSBs.
Histone modifications that are associated with DSB repair, including γ-H2AX, H3 K79 methylation and H4 K20 methylation, facilitate repair by enhancing the binding of repair and/or checkpoint factors to DSB chromatin.
The Rad51–ssDNA presynaptic filament is sufficient to search for and capture DNA sequence homology, even when the homology is present on the nucleosomal surface.
When the donor sequences are embedded within heterochromatin, such as Sir-containing heterochromatin of budding yeast, a successful homology search and joint capture requires the SWI/SNF chromatin remodeling enzyme.
The RSC remodeling enzyme remodels nucleosomes adjacent to the DSB, facilitating nonhomologous end-joining (NHEJ) and homologous recombination (HR) by stimulating recruitment of Ku70/Ku80, MRX and cohesins.
The INO80 remodeling enzyme facilitates exonucleolytic resection, promotes Ku70/80 recruitment and displaces nucleosomes during a successful strand invasion event. INO80 also contributes to the cell-cycle checkpoint response.
The SWR1 remodeling enzyme catalyzes the deposition of the yeast H2A.Z histone variant (also called Htz1), which stimulates exonucleolytic processing.
The Rad54 ATPase (also a chromatin remodeling enzyme) stimulates formation of the Rad51 presynaptic filament, promotes formation of a stable plectonemic joint molecule and displaces Rad51 from recombination intermediates. Rad54 may also disrupt nucleosomes during stable joint formation.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.O'Driscoll M, Jeggo PA. The role of double-strand break repair – insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 4.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 5.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol Histol. 2006;37:261–269. doi: 10.1007/s10735-006-9047-4. [DOI] [PubMed] [Google Scholar]
- 7.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagiannis TC, El-Osta A. Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia. 2007;21:195–200. doi: 10.1038/sj.leu.2404478. [DOI] [PubMed] [Google Scholar]
- 10.Acilan C, Potter DM, Saunders WS. DNA repair pathways involved in anaphase bridge formation. Genes Chromosomes Cancer. 2007;46:522–531. doi: 10.1002/gcc.20425. [DOI] [PubMed] [Google Scholar]
- 11.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhuri J, Basu U, Zarrin A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 14.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 15.Degrassi F, Fiore M, Palitti F. Chromosomal aberrations and genomic instability induced by topoisomerase-targeted antitumour drugs. Curr Med Chem Anticancer Agents. 2004;4:317–325. doi: 10.2174/1568011043352920. [DOI] [PubMed] [Google Scholar]
- 16.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, γ-H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci USA. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 19.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 20.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 21.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 22.van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 23.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 24.Sonoda E, Takata M, Yamashita YM, Morrison C, Takeda S. Homologous DNA recombination in vertebrate cells. Proc Natl Acad Sci USA. 2001;98:8388–8394. doi: 10.1073/pnas.111006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 26.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 27.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes A, Haber JE. Physical monitoring of HO-induced homologous recombination. Methods Mol Biol. 1999;113:403–415. doi: 10.1385/1-59259-675-4:403. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raynard S, Niu H, Sung P. DNA double-strand break processing: the beginning of the end. Genes Dev. 2008;22:2903–2907. doi: 10.1101/gad.1742408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 34.Conway AB, Lynch TW, Zhang Y, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 35.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 37.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J Biol Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 40.Ristic D, Modesti M, van der Heijden T, et al. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 44.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 45.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haber JE, Leung WY, Borts RH, Lichten M. The frequency of meiotic recombination in yeast is independent of the number and position of homologous donor sequences: implications for chromosome pairing. Proc Natl Acad Sci USA. 1991;88:1120–1124. doi: 10.1073/pnas.88.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 49.Williams RS, Williams JS, Tainer JA. Mre11–Rad50–Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 50.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 51.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 52.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Buolamwini JK. Cell cycle molecular targets in novel anticancer drug discovery. Curr Pharm Des. 2000;6:379–392. doi: 10.2174/1381612003400948. [DOI] [PubMed] [Google Scholar]
- 54.Heideker J, Lis ET, Romesberg FE. Phosphatases, DNA damage checkpoints and checkpoint deactivation. Cell Cycle. 2007;6:3058–3064. doi: 10.4161/cc.6.24.5100. [DOI] [PubMed] [Google Scholar]
- 55.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 56.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 57.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol Chem. 2008;389:353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 58.Fletcher TM, Hansen JC. The nucleosomal array: structure/function relationships. Crit Rev Eukaryot Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]; ▪▪ A must read for anyone interested in chromatin structure and function.
- 59.Carruthers LM, Hansen JC. The core histone N-termini function independently of linker histones during chromatin condensation. J Biol Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 60.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding – wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 61.Woodcock CL. Chromatin architecture. Curr Opin Struct Biol. 2006;16:213–220. doi: 10.1016/j.sbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Woodcock CL, Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- 63.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 64.Passarge E. Emil Heitz and the concept of heterochromatin: longitudinal chromosome differentiation was recognized fifty years ago. Am J Hum Genet. 1979;31:106–115. [PMC free article] [PubMed] [Google Scholar]
- 65.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 66.Henikoff S. Heterochromatin function in complex genomes. Biochim Biophys Acta. 2000;1470:O1–O8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- 67.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 68.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 69.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 70.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 71.Basrai MA, Hieter P. Is there a unique form of chromatin at the Saccharomyces cerevisiae centromeres? Bioessays. 1995;17:669–672. doi: 10.1002/bies.950170802. [DOI] [PubMed] [Google Scholar]
- 72.Durand-Dubief M, Ekwall K. Heterochromatin tells CENP-A where to go. Bioessays. 2008;30:526–529. doi: 10.1002/bies.20763. [DOI] [PubMed] [Google Scholar]
- 73.Orsi GA, Couble P, Loppin B. Epigenetic and replacement roles of histone variant H3.3 in reproduction and development. Int J Dev Biol. 2009;53:231–243. doi: 10.1387/ijdb.082653go. [DOI] [PubMed] [Google Scholar]
- 74.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 75.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 76.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]; ▪ First demonstration that a specific histone modification can alter chromatin folding dynamics.
- 77.Zhang Y, Smith CL, Saha A, et al. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyer LA, Logie C, Bonte E, et al. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 81.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 82.Cairns BR, Lorch Y, Li Y, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 83.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 85.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]; ▪ Key paper that described novel histone exchange activity of the SWR1 enzyme.
- 86.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 88.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 89.Jin J, Cai Y, Yao T, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 90.Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 91.Hassan AH, Prochasson P, Neely KE, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 92.Chandy M, Gutierrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nielsen PR, Nietlispach D, Mott HR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 94.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 95.Shi X, Kachirskaia I, Walter KL, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J, Daniel J, Espejo A, et al. Tudor, MBT and chromodomains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fry CJ, Peterson CL. Chromatin remodeling enzymes: who's on first? Curr Biol. 2001;11:R185–R197. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- 98.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 99.Shroff R, Arbel-Eden A, Pilch D, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 101.Karagiannis TC, Harikrishnan KN, El-Osta A. Disparity of histone deacetylase inhibition on repair of radiation-induced DNA damage on euchromatin and constitutive heterochromatin compartments. Oncogene. 2007;26:3963–3971. doi: 10.1038/sj.onc.1210174. [DOI] [PubMed] [Google Scholar]
- 102.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 103.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huen MS, Grant R, Manke I, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mailand N, Bekker-Jensen S, Faustrup H, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 106.Kolas NK, Chapman JR, Nakada S, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 108.Unal E, Arbel-Eden A, Sattler U, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 109.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 110.Downs JA, Allard S, Jobin-Robitaille O, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 111.Morrison AJ, Highland J, Krogan NJ, et al. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 112.Keogh MC, Kim JA, Downey M, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 113.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 114.Huyen Y, Zgheib O, Ditullio RA, Jr, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 115.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 116.Lachner M, Sengupta R, Schotta G, Jenuwein T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harb Symp Quant Biol. 2004;69:209–218. doi: 10.1101/sqb.2004.69.209. [DOI] [PubMed] [Google Scholar]
- 117.Botuyan MV, Lee J, Ward IM, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harvey AC, Jackson SP, Downs JA. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics. 2005;170:543–553. doi: 10.1534/genetics.104.038570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wyatt HR, Liaw H, Green GR, Lustig AJ. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics. 2003;164:47–64. doi: 10.1093/genetics/164.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Moore JD, Yazgan O, Ataian Y, Krebs JE. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics. 2007;176:15–25. doi: 10.1534/genetics.106.063792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med. 2004;199:1671–1677. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 123.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bird AW, Yu DY, Pray-Grant MG, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 125.Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 127.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ One of the first studies to demonstrate recruitment of multiple chromatin remodeling enzymes to a HO-induced double-strand break (DSB). They also use yeast genetics to define functional roles for RSC and SWI/SNF.
- 128.Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell's response to DNA damage. Curr Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates RSC-dependent changes in chromatin structure adjacent to a DSB. Changes in chromatin structure are independent of DSB processing.
- 130.Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 131.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]; ▪▪ Landmark study, showing a functional link between histone acetylation and histone exchange at DSBs. Study indicated role for SWR1-like complexes in downregulation of γ-H2AX.
- 133.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsukuda T, Lo YC, Krishna S, Sterk R, Osley MA, Nickoloff JA. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst) 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 136.Chen CC, Carson JJ, Feser J, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Interesting study demonstrating requirement of new histone deposition in checkpoint inactivation.
- 137.Palter KB, Foe VE, Alberts BM. Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell. 1979;18:451–467. doi: 10.1016/0092-8674(79)90064-3. [DOI] [PubMed] [Google Scholar]
- 138.Palter KB, Alberts BM. The use of DNA-cellulose for analyzing histone-DNA interactions. Discovery of nucleosome-like histone binding to single-stranded DNA. J Biol Chem. 1979;254:11160–11169. [PubMed] [Google Scholar]
- 139.Wolner B, Peterson CL. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J Biol Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- 140.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54 and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 141.Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell. 2008;30:803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Biochemical studies indicate that a successful homology search on chromatin fibers does not require ATP-dependent chromatin remodeling or histone modifications.
- 142.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138(6):1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein: Stabilization of the Rad51 nucleoprotein filament. J Biol Chem. 2003;278(16):14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 144.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442(7102):590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 145.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol. 2007;14(8):746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 146.Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell. 2008;30(6):803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26(18):4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]