Abstract
Several attempts have been made at systematically mapping protein-protein interaction, or “interactome” networks. However, it remains difficult to assess the quality and coverage of existing datasets. We describe a framework that uses an empirically-based approach to rigorously dissect quality parameters of currently available human interactome maps. Our results indicate that high-throughput yeast two-hybrid (HT-Y2H) interactions for human are superior in precision to literature-curated interactions supported by only a single publication, suggesting that HT-Y2H is suitable to map a significant portion of the human interactome. We estimate that the human interactome contains ~130,000 binary interactions, most of which remain to be mapped. Similar to estimates of DNA sequence data quality and genome size early in the human genome project, estimates of protein interaction data quality and interactome size are critical to establish the magnitude of the task of comprehensive human interactome mapping and to illuminate a path towards this goal.
The protein-protein interactome of an organism is the network formed by all protein-protein interactions that can occur in a range of physiologically relevant protein concentrations. Mapping protein-protein interactions is crucial, albeit not sufficient, for unraveling the dynamic aspects of cellular networks, including when, where, and for what purpose protein interactions do occur in vivo1. Currently available human protein-protein interactome maps have been derived using (i) high-throughput yeast two-hybrid (HT-Y2H)2,3, (ii) HT co-affinity purification followed by mass spectrometry4, (iii) curation of published low-throughput experiments5–10, or (iv) computational predictions11,12. Despite a few attempts2,3,13,14, it remains difficult to accurately estimate the quality of these interactome maps and how far away we are from a complete map of the human interactome.
Differentiation between sets of protein pairs that can interact (biophysical interactions) and do interact (biological interactions) is only possible with reliable biophysical interactome maps. What proportion of currently available interactome maps represents true biophysical interactions and what proportion represents artifacts? Are the interactions provided by curated low-throughput experiments superior in quality to those obtained by HT strategies, as suggested previously15–17? Do the currently available interactome maps represent a significant or a negligible fraction of the human biophysical interactome? Here we provide insights that are crucial for developing a strategy for comprehensive interactome mapping, i.e., for estimating the size of the human interactome and thus an endpoint to the project, and for selecting suitable technologies, a realistic timeline and a funding model to achieve this goal.
Previous attempts to assess the quality of interactome maps for human13,14,18 or other species13,15,18–23 relied on measuring (i) the extent to which interacting proteins share other biological attributes, e.g., co-expression, or (ii) the extent to which different maps of the same interactome share common interactions. Both approaches suffer several inherent limitations. Methods that evaluate the quality of interactions with respect to mRNA co-expression22,23 are systematically biased against true biological interactions between proteins whose mRNAs are not necessarily correlated or are even anti-correlated in expression. Since available annotations for protein function and localization are far from comprehensive, lack of evidence for co-localization of a given pair of proteins does not imply that the interaction observed between these proteins is an artifact. Methods based on measuring the extent of overlap between two interactome maps13,20,21 require that the corresponding datasets be derived from identical or similar assays. Existing analyses have not always fulfilled this requirement13. Most existing methods for quality assessment do not distinguish between the multiple sources of false negatives and false positives associated with any interactome mapping strategy. For instance, those interactions missed by a single screen of an assay but identifiable after multiple screens must be distinguished from the interactions that would never be identified by that assay even after a saturating number of screens.
Here we developed a framework to estimate various quality parameters associated with currently used protein-protein interaction assays, namely screening completeness, assay sensitivity, sampling sensitivity and precision. We generated empirical data to rigorously dissect these quality parameters, without relying on correlation with other biological attributes. Combining these parameters provides an estimate of the size of the human binary biophysical interactome and projects a path towards the completion of its mapping.
RESULTS
An interaction mapping framework
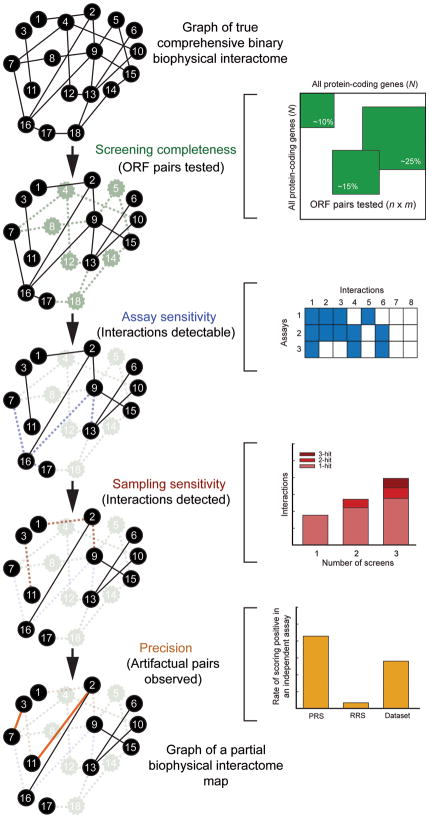
To accurately assess the quality of a given interactome map, we need to consider every possible source of false negatives (true interactions missing) and false positives (spurious pairs reported) associated with the assay used to generate the map. Our framework considers four parameters to estimate quality: “screening completeness”, “assay sensitivity”, “sampling sensitivity” and “precision” (Fig. 1).
Figure 1. Conceptual framework for interactome mapping.
The concepts of “screening completeness” (fraction of all pair-wise protein combinations tested), “assay sensitivity” (fraction of all biophysical interactions identifiable by a given assay), “sampling sensitivity” (fraction of all identifiable interactions that are detected in a single trial) and “precision” (fraction of pairs reported by a given assay that are true positives) can be estimated independently and combined to empirically estimate the size of binary interactomes. PRS: the set of positive reference set interactions; RRS: the random reference set. Solid black lines in a given network graph represent true biophysical interactions present in that network, dashed lines represent true biophysical interactions missing in that network, and solid colored lines represent biophysical artifactual pairs present in that network.
“Screening completeness” is the fraction of the total possible space of open reading frame (ORF) pairs that is tested to generate a given interactome map (Fig. 1a). Since currently available ORF resources3,24 only allow proteome-wide investigations of one protein isoform per gene, we ignore isoforms encoded by alternatively spliced transcripts here. For example, if we assume that the human genome consists of 22,500 protein-coding genes (N = 22,500 × 22,500/2 protein pairs), then the screening completeness of CCSB-HI12, a proteome-scale HT-Y2H effort that tested n = 7,000 × 7,000/2 human protein pairs, is n/N, or ~10%.
“Assay sensitivity” is the fraction of all biophysical interactions that can possibly be identified by an assay performed under a specific set of experimental conditions (Fig. 1b). For example, a given HT-Y2H assay may be unable to detect interactions involving specific types of membrane proteins or requiring post-translational modifications that do not occur in yeast cells.
“Sampling sensitivity” is the fraction of all identifiable interactions that are found in a single trial of an assay performed under a specific set of experimental conditions (Fig. 1c). When testing tens if not hundreds of millions of protein pairs in any space of pair-wise combinations, it might be necessary to sample that space multiple times to report all identifiable interactions.
Lastly, “precision” is the fraction of observed pairs in an interactome dataset that are true positives (Fig. 1d). False positive pairs reflect technical artifacts that erroneously score positive in a given assay performed under a specific set of experimental conditions. We distinguish between two types of artifactual pairs, “stochastic false positives”, which are observed in only one or a few trials of an assay and “systematic false positives”, which are observed in many or all trials.
Estimation of assay sensitivity
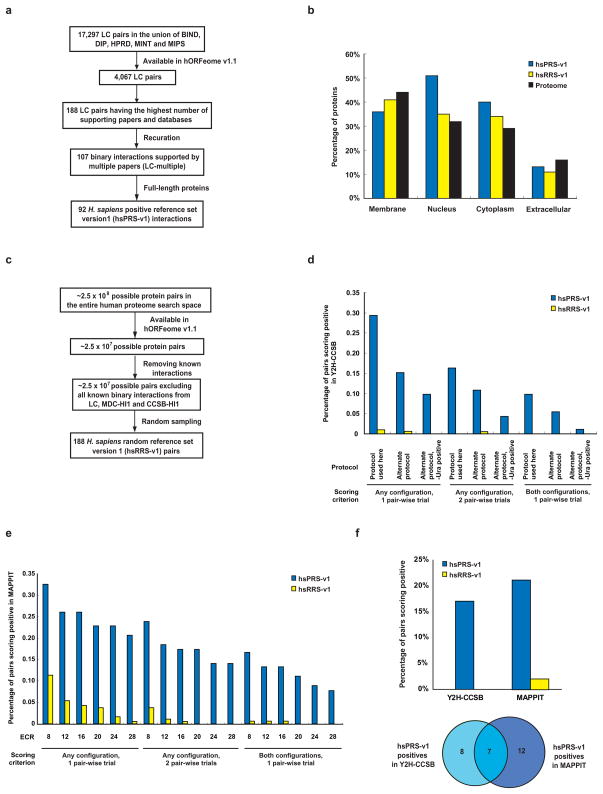
Estimation of the assay parameters described above requires reference sets of positive and negative interacting pairs. To compile a positive reference set of high-confidence human binary protein-protein interactions we started with interactions curated from the literature (“literature curated” or LC interactions) and from these, we chose 188 pairs present in our human ORFeome v1.1 clone collection24 that are supported by the greatest number of publications and curated by the highest number of databases. Systematic recuration of all publications thought to support these 188 protein pairs25 verified 107 direct binary interactions that involve human proteins and that are supported by multiple publications. Ninety-two of these interactions involve full-length proteins and constituted our Homo sapiens Positive Reference Set version 1 or “hsPRS-v1” (Fig. 2a and Supplementary Table 1 online). Proteins involved in the 92 hsPRS-v1 interactions exhibit broad cellular localization (Fig. 2b), suggesting that they are representative of the entire human proteome. It is impossible to generate a set of negative interacting pairs with absolute confidence. So we compiled a surrogate Random Reference Set (“hsRRS-v1”) of 188 protein pairs chosen randomly from the space of all ORFeome v1.1 pairs after excluding known interactions (Fig. 2c).
Figure 2. Assay sensitivity and background positive rate of binary interactome mapping assays.
(a) How positive reference set interactions were chosen from among the interactions available in the curated literature of low-throughput experimentally derived interactions (LC). (b) How random reference set pairs were chosen from among the possible pairs in our human ORFeome v1.1 clone collection24. (c) Distribution of cellular location of proteins making up the positive and random reference sets. (d) Assay sensitivity (fraction of hsPRS-v1 pairs scoring positive) and background positive rate (fraction of hsRRS-v1 pairs scoring positive) of the Y2H-CCSB assay based on varying experimental and scoring conditions, including the use of an alternate protocol (Supplementary Methods). We did not use the results of testing the hsRRS-v1 pairs here to estimate the false discovery rate of the Y2H-CCSB assay due to limited sample size. (e) Assay sensitivity and background positive rate of the MAPPIT assay upon varying experiment-to-control-ratio (ECR) scores (Supplementary Methods). (f) Upper panel: assay sensitivity and background positive rate of Y2H-CCSB and MAPPIT under the specific experimental conditions used (Supplementary Methods). For Y2H-CCSB, the fraction of hsPRS-v1 pairs scoring positive in at least one configuration and in both pair-wise mating experiments is depicted. This condition reflects the assay sensitivity of the specific experimental and scoring conditions of Y2H-CCSB used to generate CCSB-HI12. Lower panel: Venn diagram of hsPRS-v1 pairs scoring positive in the two assays. (g) Results of testing each hsPRS-v1 pair and each hsRRS-v1 pair using Y2H-CCSB and MAPPIT. Blue or yellow shaded squares represent protein pairs scored positive by a given assay.
PRS and RRS pairs can be used to experimentally calibrate conditions of an assay to achieve an optimal trade-off between the fraction of PRS and RRS pairs reporting positive26. We measured the fraction of hsPRS-v1/hsRRS-v1 pairs scoring positive across a range of experimental and scoring conditions of a stringent version of the Y2H system (Y2H-CCSB)2 and the mammalian protein-protein interaction trap assay (MAPPIT)27 (Supplementary Table 2 online and Fig. 2d,e).
The results observed with the hsPRS-v1 and hsRRS-v1 pairs for Y2H-CCSB confirm that the specific experimental conditions used in generating our first human interactome map, CCSB-HI12, reflected good assay design. We also derived suitable experimental conditions for the MAPPIT assay. Under these experimental conditions we estimated the assay sensitivity of Y2H-CCSB and MAPPIT to be 17% and 21% respectively (Fig. 2f and Supplementary Table 3 online). Using a larger, more recently updated set of ~1,500 LC interactions that are supported by multiple publications we estimated an assay sensitivity of 20% for Y2H-CCSB, consistent with our hsPRS-v1-based estimate. Y2H-CCSB and MAPPIT recovered partially overlapping sets of hsPRS-v1 interactions. Of the hsPRS-v1 pairs 29% (27/92) were reported by at least one assay, and of these, 26% (7/27) were detected by both assays (Figs. 2f,g). That 74% (20/27) of positive hits are specific to a single assay reflects the complementarities between the two assays.
We estimated the false positive rate (rate of scoring hsRRS-v1 pairs positive) of Y2H-CCSB and MAPPIT to be < 0.5% and 2% respectively (Fig. 2f and Supplementary Table 4 online). The results of testing hsRRS-v1 pairs by Y2H-CCSB do not permit a direct and reasonable estimate of the false discovery rate associated with the CCSB-HI1 dataset. The millions of pairs tested by Y2H-CCSB in the HT screen leading to the generation of CCSB-HI1 consist mostly of non-interacting pairs, so the number of non-interacting pairs tested in the HT-Y2H screen is orders of magnitude higher than the size of hsRRS-v1. Consequently, small changes in the hsRRS-v1-based estimate of the false positive rate of Y2H-CCSB can have a large effect on the resulting estimate of the false discovery rate of CCSB-HI1. Rather than using the Y2H-CCSB experiments on the hsRRS-v1 pairs, we instead used two alternate and independent approaches to estimate the false discovery rate of our Y2H-CCSB assay: (i) retesting Y2H-CCSB interactions in MAPPIT and (ii) modeling repeated screens of Y2H-CCSB (see below).
Precision of existing human interactome datasets
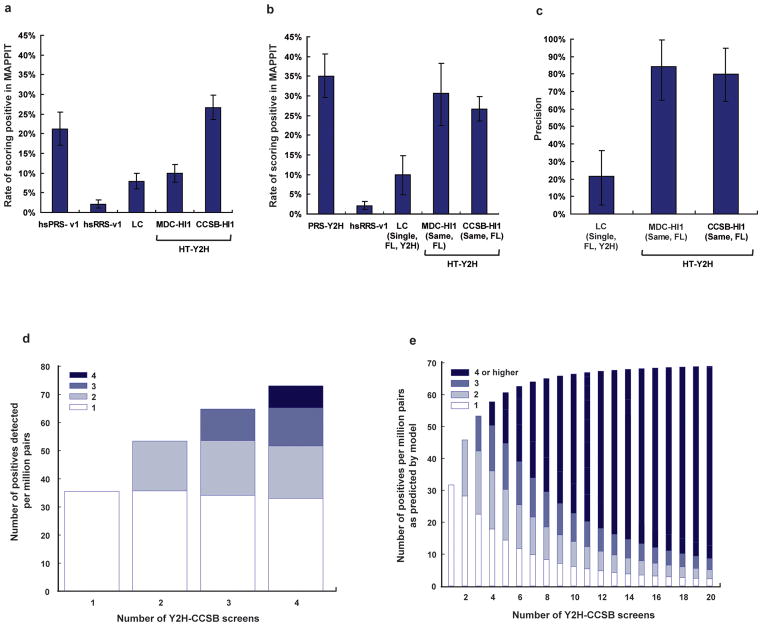
We estimated the precision of the two existing HT-Y2H human interactome datasets, CCSB-HI12 and MDC-HI13, and a low-throughput LC human interactome dataset2 by measuring the extent to which a subset of 188 positive pairs chosen randomly from each dataset (Supplementary Table 1) retested in MAPPIT. To do so, we first benchmarked the performance of a dataset in MAPPIT experiments against (i) the false positive rate of MAPPIT and (ii) the false negative rate of MAPPIT. We estimated these benchmarks by evaluating the fraction of hsRRS-v1 and hsPRS-v1 pairs reporting positive by MAPPIT, respectively. The results with the hsRRS-v1 pairs provided an estimate of MAPPIT’s false positive rate that is sufficiently resolved for estimating false discovery rates of the various interactome datasets, since the size of the hsRRS-v1 is similar to the size of each of the three different interactome datasets tested. Relative to the proportion of hsPRS-v1 and hsRRS-v1 pairs scoring positive (21% and 2%, respectively), the fractions of pairs that scored positive in the three datasets were: LC, 8%; MDC-HI1, 10%; and CCSB-HI1, 27% (Fig. 3a and Supplementary Table 4).
Figure 3. Precision and sampling sensitivity in interactome datasets.
(a) Comparison of interactome datasets by comparing the rate of observing a positive by MAPPIT given a positive in the dataset. (b) Interactome datasets were further compared after removing various biases by considering interactions originally derived using full-length (FL) proteins and using Y2H assays. (c) Precision of each tested dataset computed by accounting for the rate of detecting hsRRS-v1 pairs and Y2H-supported hsPRS-v1 pairs by MAPPIT in b. Error bars represent estimated standard deviation of the mean based on a Monte Carlo simulation of scores observed in a given assay. (d and e) Sampling sensitivity and Y2H-CCSB repeat screens. Bars filled with white represent protein pairs uncovered in only one screen and progressively dark shades of blue represent protein pairs reported in increasing number of multiple screens. (d) Data observed in Y2H-CCSB repeat screens indicating the total number of positive pairs reported after one, two, three or four screens. (e) Predicted saturation curve of the number of uncovered interactions against the number of screens for Y2H-CCSB after modeling the data in d and assuming a single isoform per gene in the respective tested spaces.
This analysis needs to be adjusted for potential dataset biases. First, we minimized the effect due to differences between the sequences of the clones originally used to report the interactions and sequences of the full-length clones used here. We considered only pairs for which the proteins originally used were described as full-length (FL) or, whenever identifiable, pairs for which the isoforms originally used were the same (“Same”) as the ones used here. Second, since the CCSB-HI1 and MDC-HI1 datasets were each described in a single publication, we compared them to the subset of LC interactions also supported by a single publication (“Single”), which represents most currently available literature-curated interaction information25. Including interactions supported by multiple publications in the LC dataset would be circular since our hsPRS-v1 benchmark was derived from LC interactions supported by multiple publications. Lastly, to account for the moderate bias of MAPPIT in detecting Y2H-supported (“Y2H”) interactions, we considered the subset of hsPRS-v1 and LC interaction pairs supported by at least one Y2H experiment in the corresponding curated publications (Supplementary Data 1 and Supplementary Table 5 online). Based on these consolidated datasets, 34% of Y2H-supported hsPRS-v1 pairs (PRS-Y2H) and 2% of hsRRS-v1 pairs scored positive. Relative to this, the fractions of pairs that scored positive in the three subsets of protein pairs were: LC (Single, FL, Y2H), 10%; MDC-HI1 (Same, FL), 31%; and CCSB-HI1 (Same, FL), 27% (Fig. 3b). Thus, the two HT-Y2H datasets performed comparably to the PRS-Y2H pairs in MAPPIT, while the literature-curated interactions supported by a single publication performed poorly. Given the fraction of PRS-Y2H pairs and hsRRS-v1 pairs scoring positive by MAPPIT, the precision of each of the three datasets can be computed as: LC (Single, FL, Y2H), 25%; MDC-HI1 (Same, FL), 83%; and CCSB-HI1 (Same, FL), 79% (Fig. 3c and Supplementary Table 3).
Sampling sensitivity and stochastic false discovery rate
To estimate sampling sensitivity and the number of screens required to achieve saturation, we repeated four Y2H-CCSB screens (“repeat screens”) in a defined search space of 1,822 DB-Xs (or “baits”; representing 1,744 unique genes) against 1,796 AD-Ys (or “preys”; representing 1,752 unique genes), representing approximately three million pair-wise combinations (Supplementary Table 6 online). We developed a probabilistic model that considered the search space of three million protein pairs to be a mixture of true biophysical interactions and non-interacting pairs. Using a Bayesian approach, our model estimated (i) the fraction of all identifiable true biophysical interactions found in one, two, or a saturating number of screens and (ii) the fraction of non-interacting pairs erroneously detected in a screen. In short, our approach estimated distributions of values of the above parameters that fit the experimental results observed in the repeat screens.
Out of the three million pair-wise combinations tested, the four Y2H-CCSB repeat screens together reported 240 protein-protein interactions (Supplementary Tables 7 and 8 online). Of these interactions 49% appeared in multiple screens. The total number of new interactions identified after successive screens showed an increasing trend, indicating that more interactions would be found with additional screens (Fig. 3d). Based on our model, we estimated that the sampling sensitivity per screen is 45% and that after a saturating number of screens, Y2H-CCSB can identify 71 interactions per million pairs tested (Fig. 3e). Approximately six screens are needed to reach 90% saturation. Importantly, the number of single hits (interactions found in only one out of several screens) decreases while the contribution of multiple hits dominates after multiple screens. Adjusting for these repeat screens being done in only one Y2H configuration (bait-prey vs. prey-bait), we estimated that upon testing both configurations, the sampling sensitivity per screen is 53% and that after a saturating number of screens, Y2H-CCSB can identify 118 interactions per million pairs tested (Supplementary Table 3).
Our model estimated that approximately eight out of every million non-interacting pairs tested falsely report positive in Y2H-CCSB. Consequently, our model estimated a stochastic false discovery rate of 12%, meaning that 12% of the interactions reported in a single Y2H-CCSB screen correspond to stochastic false positives. Since the MAPPIT experiments (Fig. 3c) evaluate the union of systematic and stochastic false positives in a given dataset and estimated an overall false discovery rate of 21% for CCSB-HI1, we deduce a systematic false discovery rate of 14% (Supplementary Methods online).
The MAPPIT experiments show that existing human HT-Y2H maps have high precision. However, the fraction of CCSB-HI1 and MDC-HI1 interactions common to both maps is small although statistically significant, only 6% and 2%, respectively (P = 10−18, Supplementary Data 2 and Supplementary Fig. 1 online). Our results indicate that low sampling sensitivity and differences in assay sensitivity likely account for the small overlap.
Estimation of the size of the human interactome
We estimated four important parameters associated with the quality of human binary interactome maps (Fig. 1). For the Y2H-CCSB assay evaluated here, we computed the screening completeness of the repeat screens as ~1%; the hsPRS-v1 experiment estimated an assay-sensitivity of ~17% (Fig. 2f); the model of the repeat screens estimated a sampling sensitivity of ~53% (Fig. 3e); and the MAPPIT experiment estimated a precision of ~79% (Fig. 3c). We also estimated the variation of these estimates associated with sampling (Supplementary Table 3). Integrating these parameters, we predict that the entire human interactome, excluding splice variant complexity, contains approximately 74,000–200,000 binary biophysical interactions (Table 1).
Table 1. Sizing the human interactome.
| Average number of interactions detected per screen | Screening completeness of the repeat screen search space | Assay sensitivity | Estimated sampling sensitivity per repeat screen | Precision | Systematic false discovery rate | Stochastic false discovery rate | Size of the human interactome |
|---|---|---|---|---|---|---|---|
| Repeat screen experiment | Ensembl version 44.36f | hsPRS-v1 experiment | Repeat screen experiment | MAPPIT experiment | MAPPIT and repeat screen experiments | Repeat screen experiment | Combining all parameters |
| 199 | 1.2% | 17% ± 3.8% | 53.1% ± 10% | 79.4% ± 15.9% | 13.6% ± 14.5% | 11.7% ± 6.1% | 130,111 ± 32,618 (73,548–199,688) |
Estimation of the parameters of the binary interactome mapping framework. Average values ± 1 s.d. (standard deviation) of each parameter based on Monte Carlo simulation (n = 10,000 runs) are indicated after accounting for testing protein pairs in both bait-prey and prey-bait configurations.
The range of interactome size reported corresponds to the 95% confidence interval of the size predicted.
Interacting protein pairs and shared functional annotation
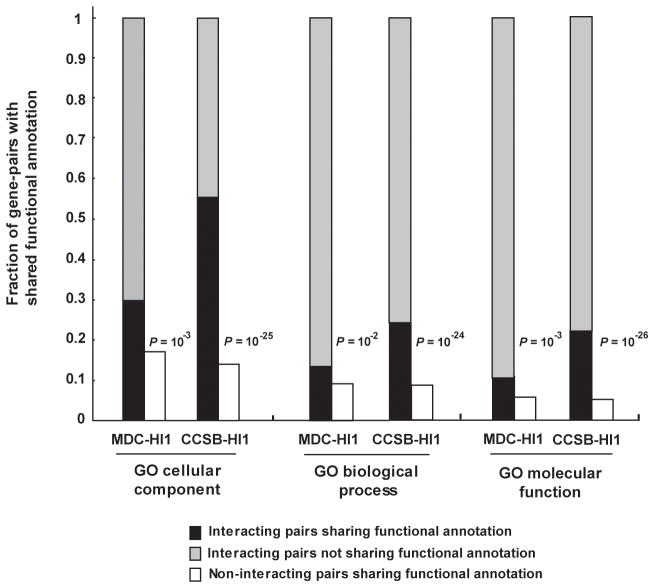
A statistically significant fraction (P < 10−3) but not all of the interacting protein pairs in CCSB-HI1 and MDC-HI1 showed correlation for shared functional annotations compared to random expectation (Fig. 4). Given the high technical quality of these datasets demonstrated here, interacting pairs that do not correlate with known functional annotations could be: (i) promising candidates for biological discovery, particularly true biological interactions that involve proteins currently lacking adequate functional annotations; or (ii) true biophysical interactions that do not occur physiologically. We call this second class “pseudo-interactions” by analogy to pseudo-genes. Pseudo-interactions could correspond to ancient biological interactions that have evolved to lose physiological relevance and provide interesting insights into the evolution of the interactome.
Figure 4. Correlation of interacting pairs for shared functional annotation.
Correlation of interacting pairs in CCSB-HI1 and MDC-HI1 interactome maps for specific shared Gene Ontology functional annotations. P-values indicate the probability of observing such a correlation by chance (compare black bars to white bars) computed using Fisher’s exact test. Analysis was performed on MDC-HI1 and CCSB-HI1 interactions reported using full-length ORFs.
DISCUSSION
Several previous studies have estimated the precision of existing maps or the size of interactomes13–15,18,20–23,28. Our empirical framework addresses limitations of these studies (detailed discussion in Supplementary Data 3 online). Methods that rely on correlation with other biological attributes to estimate precision of interactome maps (i) often use as benchmark, LC interaction datasets, which are sociologically biased, (ii) assume that knowledge of biological attributes such as Gene Ontology functional annotation is complete and unbiased, and (iii) are inherently constrained by pre-existing paradigms regarding the expectation for interacting protein pairs to share biological attributes. Approaches based on analyzing the extent of overlap between interactome maps13,20,21 suffer specific limitations in their implementations such as comparing maps that were not derived using the same assay, or using LC datasets as a reference set, which may not be appropriate given a potentially higher false positive rate than previously anticipated (Fig. 3c)25. Earlier studies also failed to consider one or more of the parameters that influence interactome map quality, i.e., completeness, systematic false discovery rate, stochastic false discovery rate, assay sensitivity and sampling sensitivity, which could in turn significantly affect estimates of interactome size. All these limitations together may have led to overestimated false discovery rates for HT-Y2H human interactome maps.
Our framework overcomes these limitations by (i) considering every possible source of false negatives and false positives, (ii) using a high-quality reference set requiring interactions to be supported by multiple publications and to pass additional recuration, (iii) assessing false discovery rates directly using information from independent protein-protein interaction assays and (iv) comparing overlaps between four homogeneously-derived repeat screens to assess the sampling sensitivity and stochastic false discovery rate of Y2H-CCSB. Close attention to these parameters will be vital to design the strategy, e.g., number of screens and types of assays to use, for future interactome mapping projects.
The hsPRS-v1 and hsRRS-v1 provide hundreds of experimentally testable clone pairs of positive and random reference sets for binary protein-protein interactions. Previous assays typically relied on testing one or a few positive control pairs and a few or no random control protein pairs. Though our reference sets are a first version and will be improved, they mark a substantial effort towards the standardized calibration of binary interaction mapping assays, an objective that has not been previously achieved systematically.
Although LC datasets are commonly perceived to be of better quality than datasets generated with HT technologies15–17, the results of our MAPPIT experiments indicate that stringent implementations of HT-Y2H assays produce interaction datasets with technical quality at least as good if not superior to low-throughput LC interactions (Fig. 3c). These results substantiate previous computational analyses of human29 and yeast30 interactome maps. Large-scale curation of the primary literature is challenging and may have higher error rates than previously anticipated25. HT interactome mapping strategies have several advantages over low-throughput strategies: (i) since defined search spaces are used, information about positives (pairs observed to interact) as well as negatives (pairs not observed to interact) is available; (ii) experiments are standardized therefore well-controlled, comparable and scalable; (iii) cost-efficient strategies can be developed; and (iv) HT strategies are less sociologically biased than low-throughput experiments.
Implementation of our framework can be improved in various ways. The statistical power of the analyses can be increased by testing more PRS interactions, by repeatedly screening larger search spaces, or by using additional independent assays for measuring precision. Our current implementation does not consider multiple splice isoforms per gene, so we most likely underestimated the interactome size. Additional modifications to the framework will be required to thoroughly analyze non-binary co-complex membership maps such as those generated by HT co-affinity purification followed by mass spectrometry4. While more refined estimates can be made with future enhancements, the central concepts and overall approach are in place for design and comprehensive evaluation of any interactome mapping assay. Recently, our group has developed an interaction toolkit consisting of four independent assays to evaluate the quality of any protein interaction dataset26. Ongoing technological advancements related to assay automation and cost reduction will enable testing of expanded versions of the PRS and thousands (rather than hundreds) of Y2H, LC and other interactions using these assays.
Similar to estimates of the number of protein-encoding genes in the human genome, ~14,000–300,000 in the early 1990s31, empirical sizing of the interactome is critical to establish the complexity of the network and to estimate how far we are from a complete human interactome map. Assuming one splice isoform per gene, we predict that the size of the human interactome is ~130,000 interactions. This confirms two previous estimates of human interactome size, which ranged from 150,000–370,000 interactions2,13. Out of ~23,000 currently reported human interactions (a combination of ~17,000 LC interactions and ~6,000 HT-Y2H interactions), our measurements indicate that ~10,000 (~42%) are true binary physical interactions (Supplementary Data 4 online). Thus, the fraction of interactions identified so far represents ~8% of the full interactome.
With 22,500 protein-coding genes, nearly 250 million protein pairs need to be tested individually, clearly requiring unbiased, systematic and cost-effective HT approaches. Interactome mapping is gradual: six screens are necessary to reach 90% saturation with Y2H-CCSB. No single assay offers complete assay sensitivity. The fraction of protein-protein interactions detectable by the specific version of HT-Y2H used here (“Y2H-CCSB”) is ~17%. Combining different versions of the Y2H system and using increased expression of both hybrid proteins can increase this proportion to ~40% (data not shown and Braun et al.26). Still, comprehensive mapping of the interactome will require the development of additional HT versions of MAPPIT and other assays26.
The potential impact on biology of a complete and reliable biophysical protein interaction map cannot be overestimated. Our results offer a quantitative roadmap in this direction, uncovering both the magnitude of the task ahead as well as the potential roadblocks.
METHODS
The Y2H-CCSB experiments were performed as described2 with minor changes. MAPPIT experiments were performed essentially as described32. Mathematical modeling of the repeat screens was performed using a Bayesian approach. All parameters observed from either the experimental data or from the mixture model were used as inputs into a Monte Carlo simulation to calculate the corresponding magnitudes of corresponding numbers reported in the text. Detailed descriptions of all datasets and methods can be found in Supplementary Methods online.
Supplementary Material
Supplementary Figure 1. Overlap and screening completeness of existing human binary interactome maps
Supplementary Table 2. Comparison of the features of the two different technologies, Y2H-CCSB and MAPPIT
Supplementary Table 3. Estimate of various parameters using Monte Carlo simulations based on experimental data and the mixture model of repeat screens
Supplementary Table 5. Calculation of conditional dependence between Y2H-CCSB and MAPPIT
Supplementary Data 1. Consideration of the correlation between pairs scoring positive in Y2H and in MAPPIT assays in the analysis of MAPPIT experiments
Supplementary Data 2. Examination of screening completeness of search spaces and overlap between CCSB-HI1 and MDC-HI1 datasets
Supplementary Data 3. Limitations of previous approaches for estimating data quality and interactome size
Supplementary Data 4. Current status of available human interactome maps
Supplementary Table 1. List of interactions in various datasets used in pair wise test experiments using MAPPIT and Y2H-CCSB assays
Supplementary Table 4. Scores for the Y2H-CCSB and MAPPIT experiments on the hsPRS-v1 and hsRRS-v1 to compute assay sensitivity and background positive rate and scores on subsets of the LC, MDC-HI1 and CCSB-HI1 interaction datasets
Supplementary Table 6. Identity of the ORFs making up the Y2H-CCSB repeat screens
Supplementary Table 7. Interactions found in the Y2H-CCSB repeat screens
Supplementary Table 8. Interactions found in the Y2H-CCSB repeat screens reported according to MIMIX specifications
Supplementary Table 9. Identity of the ORFs making up the MDC-HI1 search space (Space II)
Acknowledgments
We thank members of CCSB and the Vidal, Barabasi, Wanker and Tavernier laboratories and S. Sahasrabuddhe, R. Bell, R. Chettier and C. Wiggins for helpful discussions, E. Smith for help with generating Figure 1, and Agencourt Biosciences for sequencing assistance. This work was supported by National Institutes of Health grants 2R01HG001715 (M.V. and F.P.R.) and 5P50HG004233 (M.V. and F.P.R.) from the National Human Genome Research Institute, 5U54CA112952 (J. Nevins, Duke University; M.V. subcontract) and 5U01CA105423 (S.H. Orkin, DFCI; M.V. project) from the National Cancer Institute, an Ellison Foundation grant (M.V.), a W.M. Keck Foundation grant (M.V.), Dana-Farber Cancer Institute ISR funds (M.V.), National Institutes of Health training grant postdoctoral fellowship T32CA09361 (K.V.), National Institutes of Health grants IH U01 A1070499-01 and U56 CA113004 and National Science Foundation grant ITR DMR-0926737 IIS-0513650 (A.-L.B.), Bundesministerium für Bildung und Forschung grants NGFN2, KB-P04T01, KB-P04T03, 01GR0471 and Deutsche Forschungsgemeinschaft grants SFB 577 and SFB618 (E.E.W. and U.S.), grant GOA12051401 from the University of Ghent and the “Fonds Wetenschappelijk Onderzoek – Vlanderen” (FWO-V) G.0031.06 (J.T.), and a research grant from the National Cancer Institute of Canada awarded (C.B.). I.L. is a postdoctoral fellow with the FWO-V. M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium).
Footnotes
AUTHOR CONTRIBUTIONS
K.V., J.F.R., A. Vazquez, U.S., I.L., J.T., E.W., A.L.B. and M.V. conceived the project. K.V., J.F.R., A. Vazquez, U.S. and I.L. coordinated the experiments and data analyses. J.F.R., U.S., T.H.K., M.Z., X.X., K.H., F.G., J.M.S., P.B., H.Y., S.C., C.S., E.D., J. Timm, K.R. and C.B. performed the Y2H experiments. J.F.R, T.H.K and C.S. performed the HT ORF cloning for MAPPIT experiments. I.L. and A.S.d.S. performed the MAPPIT experiments. K.V., A. Vazquez, T.H., K.I.G., M.A.Y, A. Vinayagam, N.S., N.K., C.L., M.L. and F.P.R. performed the computational and statistical analyses. M.E.C., A.S., H.B., J.F.R and K.V. performed the LC interaction recuration analyses. D.S., A.D. and R.R.M. provided laboratory support. K.V., J.F.R., A. Vazquez, U.S., I.L., M.E.C., D.E.H., J.T., E.W., A.L.B. and M.V. wrote the manuscript. D.E.H, J.T., E.W., A.L.B and M.V. co-directed the project.
References
- 1.Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 2.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 3.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 4.Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peri S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanzoni A, et al. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 7.Bader GD, et al. BIND–The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mewes HW, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futschik ME, Chaurasia G, Herzel H. Comparison of human protein-protein interaction maps. Bioinformatics. 2007;23:605–611. doi: 10.1093/bioinformatics/btl683. [DOI] [PubMed] [Google Scholar]
- 15.von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 16.Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi TK, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 18.Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics. 2005;6:100. doi: 10.1186/1471-2105-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Jedynak BM, Bader JS. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput Biol. 2007;3:e214. doi: 10.1371/journal.pcbi.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Haeseleer P, Church GM. Estimating and improving protein interaction error rates. Proc. IEEE Comput. Syst. Bioinform. Conf; 2004. pp. 216–223. [DOI] [PubMed] [Google Scholar]
- 21.Grigoriev A. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 2003;31:4157–4161. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein-protein interaction data? J Mol Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 24.Rual JF, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusick ME, et al. Literature-curated protein interaction datasets. Nat Meth. doi: 10.1038/nmeth.1284. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Meth. doi: 10.1038/nmeth.1281. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyckerman S, et al. Design and application of a cytokine-receptor-based interaction trap. Nat Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 28.Stumpf MP, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F, Schlicker A, Assenov Y, Lengauer T, Albrecht M. Computational analysis of human protein interaction networks. Proteomics. 2007;7:2541–2552. doi: 10.1002/pmic.200600924. [DOI] [PubMed] [Google Scholar]
- 30.Collins SR, et al. Towards a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nat Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- 32.Lemmens I, Lievens S, Eyckerman S, Tavernier J. Reverse MAPPIT detects disruptors of protein-protein interactions in human cells. Nat Protoc. 2006;1:92–97. doi: 10.1038/nprot.2006.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Overlap and screening completeness of existing human binary interactome maps
Supplementary Table 2. Comparison of the features of the two different technologies, Y2H-CCSB and MAPPIT
Supplementary Table 3. Estimate of various parameters using Monte Carlo simulations based on experimental data and the mixture model of repeat screens
Supplementary Table 5. Calculation of conditional dependence between Y2H-CCSB and MAPPIT
Supplementary Data 1. Consideration of the correlation between pairs scoring positive in Y2H and in MAPPIT assays in the analysis of MAPPIT experiments
Supplementary Data 2. Examination of screening completeness of search spaces and overlap between CCSB-HI1 and MDC-HI1 datasets
Supplementary Data 3. Limitations of previous approaches for estimating data quality and interactome size
Supplementary Data 4. Current status of available human interactome maps
Supplementary Table 1. List of interactions in various datasets used in pair wise test experiments using MAPPIT and Y2H-CCSB assays
Supplementary Table 4. Scores for the Y2H-CCSB and MAPPIT experiments on the hsPRS-v1 and hsRRS-v1 to compute assay sensitivity and background positive rate and scores on subsets of the LC, MDC-HI1 and CCSB-HI1 interaction datasets
Supplementary Table 6. Identity of the ORFs making up the Y2H-CCSB repeat screens
Supplementary Table 7. Interactions found in the Y2H-CCSB repeat screens
Supplementary Table 8. Interactions found in the Y2H-CCSB repeat screens reported according to MIMIX specifications
Supplementary Table 9. Identity of the ORFs making up the MDC-HI1 search space (Space II)