Abstract
Although cell-to-cell variability has been recognized as an unavoidable consequence of stochasticity in gene expression, it may also serve a functional role for tuning physiological responses within a cell population. In the immune system, remarkably large variability in the expression of cytokine genes has been observed in homogeneous populations of lymphocytes, but the underlying molecular mechanisms are incompletely understood. Here, we study the interleukin-4 gene (il4) in T-helper lymphocytes, combining mathematical modeling with the experimental quantification of expression variability and critical parameters. We show that a stochastic rate-limiting step upstream of transcription initiation, but acting at the level of an individual allele, controls il4 expression. Only a fraction of cells reaches an active, transcription-competent state in the transient time window determined by antigen stimulation. We support this finding by experimental evidence of a previously unknown short-term memory that was predicted by the model to arise from the long lifetime of the active state. Our analysis shows how a stochastic mechanism acting at the chromatin level can be integrated with transcriptional regulation to quantitatively control cell-to-cell variability.
Keywords: cytokines, cytokine secretion assay, epigenetic regulation, gene expression, stochastic model
Introduction
Cross-population heterogeneity in gene expression has often been perceived as a mere by-product of microscopic fluctuations in molecular interactions that can impair the reliability of cellular processes and must be minimized through specific regulatory mechanisms (Elowitz et al, 2002; Blake et al, 2003; Fraser et al, 2004; Raser and O'Shea, 2004; Pedraza and van Oudenaarden, 2005). However, several examples have been described where such heterogeneity seems beneficial to generate phenotypic variability that is required for evolution and differentiation (Chang et al, 2008; Feinerman et al, 2008; Losick and Desplan, 2008). To understand the heterogeneity in multicellular organisms, one must consider that many tasks are carried out by cells acting together. Then, the population behavior must be tightly regulated, whereas cell-to-cell variability in gene expression might be used to tune the overall response (Hume, 2000).
Two principal modes of gene regulation have been proposed. In the graded mode, transcriptional regulators determine the level of gene expression by regulating the rate of transcription (Hazzalin and Mahadevan, 2002). In the binary mode, transcriptional regulators control the probability of a gene being transcribed, meaning that the gene is either in an ‘ON’ or an ‘OFF’ state (Walters et al, 1995; Hume, 2000; Biggar and Crabtree, 2001). To quantitatively control a population response in the binary mode, the mechanism of gene induction must be inherently stochastic, implying that, in the presence of activating transcription factors (TFs), a gene is transcribed only with a certain probability (Pirone and Elston, 2004; Raser and O'Shea, 2004). This probability determines the fraction of responding cells, and thereby the strength of the response. However, the molecular basis of such a binary mechanism has remained elusive. Binary gene expression may involve positive feedback and some form of ‘intrinsic’ (as yet not understood) heterogeneity between cells in a population (Becskei et al, 2001; Raj et al, 2006). Alternatively, stochasticity in the promoter activation due to slow or infrequent remodeling of the chromatin structure has been invoked (Pirone and Elston, 2004; Raser and O'Shea, 2004).
Several mammalian genes are regulated in a binary manner (Hume, 2000), of which cytokine genes of the immune system, such as IFN-β and several interleukins (ILs), belong to the best-studied examples (Bix and Locksley, 1998; Holländer et al, 1998; Riviere et al, 1998; Kelly and Locksley, 2000; Hu-Li et al, 2001; Calado et al, 2006; Apostolou and Thanos, 2008). For all these cytokines genes, one observes large variability and stochasticity of expression at the single-cell level. Rather than preventing tight control of immune responses, this variability may offer a unique means of regulation, by inducing stimuli controlling the fraction of cells expressing a given cytokine. To investigate how a stochastic mechanism of gene expression is used to control a physiologic response at the population level, we examined the regulation of the cytokine IL-4 in CD4+ T-helper type 2 (Th2) lymphocytes.
In response to antigenic stimulation, IL-4 is expressed transiently for several hours by Th2 cells and promotes an antibody-mediated immune reaction directed at infections with parasites (Zhu and Paul, 2008). The differentiation of naive Th cells into specialized IL-4-producing Th2 cells is accompanied by the long-term stable upregulation of the Th2-specific TF Gata-3 and a progressive increase in the accessibility of multiple regulatory sites across the ∼200 kb extended il4 locus, which depends on Gata-3 (Ouyang et al, 2000; Hofer et al, 2002; Ansel et al, 2006; Hegazy et al, 2010). To initiate il4 transcription, antigenic stimulation must activate the TF NFAT1 and also further reorganize the chromatin at the il4 locus (Guo et al, 2004; Cai et al, 2006). Therefore, IL-4 expression is regulated at multiple levels: progressive differentiation increases the accessibility of the locus, whereas antigenic stimulation induces the acute expression of the gene.
During Th2 differentiation, the probability of a cell expressing IL-4 increases progressively (from ∼10 to ∼50%), and in the majority of IL-4-expressing cells only one of the two il4 alleles is active (Bix and Locksley, 1998; Riviere et al, 1998). The active allele is not imprinted but chosen randomly upon each stimulation, suggesting an underlying stochastic process (Hu-Li et al, 2001). IL-4-producing cells are enriched for il4 alleles with higher chromatin accessibility (Guo et al, 2004), reduced DNA methylation (Tykocinski et al, 2005) and different architecture of the extended locus (Cai et al, 2006). Although the regulation of IL-4 has been analyzed in considerable detail, the molecular basis for its probabilistic expression remains incompletely understood.
In this paper, we attempt to explain the dynamic and stochastic properties of IL-4 expression on the basis of the biochemical rates for chromatin rearrangement, transcription, and translation. To this end, we develop a mathematical model of IL-4 expression and quantify its parameters experimentally. This model leads to predictions on the time scales of il4 chromatin dynamics during acute stimulation and differentiation of Th2 cells. We verify these predictions experimentally and obtain a quantitative picture of how slow changes in chromatin accessibility during Th2-cell differentiation modulate the probability of chromatin opening required for transcription. Here, a stochastic mechanism at the single-cell level is used to tune the IL-4 response at the population level.
Results
IL-4 expression in Th2 cells is a transient, stochastic and cell-autonomous process
To quantitatively analyze IL-4 expression, we generated T-helper (Th) cells competent to express the il4 gene. Cell culture under appropriate conditions caused murine naive Th cells to differentiate into Th2 cells, which express IL-4 upon stimulation with PMA and ionomycin (P/I), mimicking encounter with cognate antigen (Openshaw et al, 1995). As IL-4 is a secreted protein, we stimulated the cells in the presence of the secretion inhibitor brefeldin A (BfA) to detect all cells that had expressed IL-4 throughout the time course of stimulation. In agreement with earlier reports (Bix and Locksley, 1998; Riviere et al, 1998; Hu-Li et al, 2001), we found that only a fraction of cells expressed IL-4 during a given stimulation, resulting in a bimodal distribution with distinct peaks of IL-4-producing and non-producing cells (Figure 1A, red line). We calculated the expression variability of IL-4, as measured by the normalized variance νP=(σP/<PROTEIN>)2. For the entire population, we measured a variability of νP=1.5, and for the fraction of IL-4-producing cells νP+=0.6 (Supplementary Figure S1). Mainly due to the pronounced bimodal expression pattern, the expression variability was considerably higher than in other reported mammalian genes (νP=0.01–0.1) (Sigal et al, 2006).
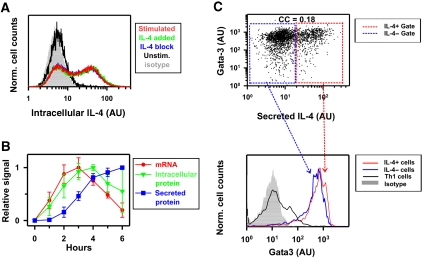
Figure 1.
IL-4 expression is a transient, probabilistic, and cell-autonomous process. (A) Two-week-differentiated Th2 cells from wild-type mice were stimulated for 4 h with PMA/ionomycin/brefeldin A alone (red), in the presence of IL-4-specific blocking antibodies (blue), or with the addition of recombinant IL-4 (green), and analyzed by flow cytometry. Background fluorescence was measured by staining with an isotype antibody (gray area). (B) IL-4 expression kinetics was measured during stimulation with PMA/ionomycin in 1, 2, and 3-week-differentiated wild-type Th2 cells. mRNA was analyzed by quantitative PCR (• red), intracellular IL-4 protein was quantified by flow cytometry (▾ green), and secreted IL-4 was measured by ELISA (▪ blue). All measurements were normalized to the maximal value of each stimulation. The mean and s.d. of 3 (mRNA), 6 (secreted protein), or 9 (intracellular protein) kinetics are shown. (C) One-week-differentiated Th2 cells were stimulated with anti-CD3/28 for 3.5 h. Secreted IL-4 was detected using IL-4 secretion assay (see Materials and methods), then cells were fixed and stained for Gata-3 and measured with flow cytometry. Top panel: Gata-3 expression and IL-4 secretion exhibit a correlation coefficient equal to 0.18. The presented experiment is representative of two independent repetitions using wild-type mice and two independent repetitions using heterozygous il4wt/il4gfp mice (Hu-Li et al, 2001). Bottom panel: IL-4-producing and non-producing cells were gated as shown in the top panel (blue and red insets). Gata-3 distributions of IL-4-producing (red curve) and non-producing (blue curve) subpopulations show similar expression profiles, distinguished from the isotype control (gray shadow) and from a control population polarized for one week in Th1 conditions (black line). Source data is available for this figure at www.nature.com/msb.
A bimodal expression pattern is the hallmark of the binary mode of gene induction and has previously been associated with positive feedback regulation (Becskei et al, 2001). As external IL-4 itself could potentially induce its own expression in a para- and/or autocrine manner through the IL-4 receptor/Stat6 pathway (Ansel et al, 2006; Schulz et al, 2009), we tested whether this positive feedback loop affects bimodal IL-4 expression. Stimulation in the presence of blocking anti-IL-4 antibodies, which interrupt autocrine signaling, did not change the IL-4 expression pattern in the population (Figure 1A, compare blue with red). Moreover, the addition of saturating amounts of IL-4 to reinforce the potential feedback loop did not alter IL-4 expression (Figure 1A). We conclude that IL-4 expression in Th2 cells is a cell-autonomous process independent of para- and autocrine positive feedback.
To analyze the kinetics of IL-4 induction, we measured mRNA, intracellular and secreted protein over 6 h in the presence of the P/I stimulus. IL-4 mRNA levels reached a peak after ∼3 h and dropped back to a low level within the next 3 h (Figure 1B). Intracellular protein followed with a delay, reaching its maximum approximately at hour 4 (Figure 1B). IL-4 accumulation rate in the extracellular medium attained its maximum at the same time (Figure 1B; Supplementary Figure S2). Thus, the P/I stimulus induces rapid and transient IL-4 expression in a Th2 cell population.
To investigate whether the bimodality in IL-4 expression reflects the existence of distinct subpopulations with different competence for IL-4 expression, we measured the Th2 lineage-specifying TF Gata-3 whose expression is the hallmark of Th2 cells. We found uniformly elevated Gata-3 expression in both IL-4-producing and non-producing cells, which was significantly higher than Gata-3 levels in Th1 cells used as negative control (Figure 1C). Interestingly, there was a weak positive correlation between IL-4 production and Gata-3 expression that, however, was not predictive of whether a cell expresses IL-4 (correlation coefficient (CC)=0.18). These results show that IL-4 expression is heterogeneous in a population of uniformly differentiated, Gata-3+ Th2 cells.
Random fluctuations in mRNA and protein levels do not contribute substantially to the observed variability in IL-4 expression
To study this phenomenon in detail, we used Th2 cells from heterozygous mice expressing IL-4 from one allele and GFP under the control of the il4 promoter from the other allele (Hu-Li et al, 2001). When monitored separately, the two il4 alleles are expressed in an uncorrelated manner (CC⩽0.07, Figure 2A). Therefore, we analyzed models for single-allele regulation to investigate the origins of IL-4 expression variability. Such allele-intrinsic variability can be due to the randomness in the reactions governing production and degradation of mRNA and protein (Elowitz et al, 2002; Ozbudak et al, 2002; Raser and O'Shea, 2004; Thattai and van Oudenaarden, 2001). In a simple model where transcription is activated by stimulation-induced TFs and the resulting mRNA is translated into protein with a constant rate (Figure 2B), the expression variability is given by
![]()
at steady state (Thattai and van Oudenaarden, 2001; Paulsson, 2004), where <PROTEIN> and <mRNA> are the mean number of molecules in a cell, and τR and τP represent mRNA and protein lifetimes, respectively (see Supplementary Mathematical Appendix 1). To estimate the expression variability arising from transcription and translation, we measured these parameters experimentally. From the expression kinetics (Figure 1B), we found the lifetimes of IL-4 mRNA and intracellular protein to be ∼90 min (τR) and ∼45 min (τP), respectively (Supplementary Figure S3), which is in agreement with an earlier report (Yarovinsky et al, 2006). Absolute quantification of secreted IL-4 by ELISA yielded a mean number of protein molecules per cell (<PROTEIN>) of ∼5 × 105 (see Materials and methods). With RT–PCR, we measured ∼100-fold higher mRNA concentrations for IL-4 than for the house-keeping gene HPRT, whose mRNA level can be assumed to lie in the order of, at least, a dozen copies. With a realistic transcription rate in the order of 1/s (Alberts et al, 2002), we estimated the mean number of mRNA molecules (<mRNA>) to lie in the range of ∼103 per IL-4-expressing cell. The expression variability arising from transcription and translation (equation (1)) is then obtained νT<0.01. As the variability of the measured data, νP∼1.5 (cf. Figure 1A), is several orders of magnitude larger, we concluded that the random birth–death turnover of mRNA and protein does not contribute significantly to the observed variations.
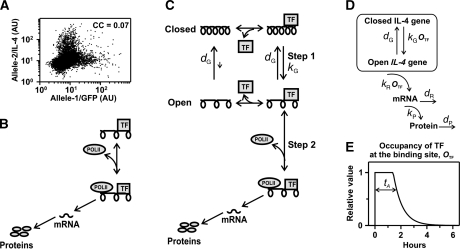
Figure 2.
Mathematical models of stochastic IL-4 expression. (A) Three-day-differentiated Th2 cells from heterozygous il4wt/il4gfp mice, carrying a knocked-in GFP in one il4 allele, were stimulated for 6 h with PMA/ionomycin/brefeldin A and analyzed by flow cytometry to detect GFP (Allele-1/GFP) and intracellular IL-4 wild type (Allele-2/IL-4). The presented experiment is representative of three independent repetitions. In all repetitions, the correlation coefficient (CC) was ⩽0.07. (B) Model of gene expression. The presence of stimulation-dependent transcription factors (TFs) enables the il4 gene to recruit mRNA polymerase-II (POLII) for transcription. Before degradation, mRNA molecules are repeatedly translated into protein, which are secreted as cytokine signal. (C) The model in (B) was extended by addition of a chromatin-opening step: Upon binding, stimulation-dependent TFs open the il4 chromatin structure (with rate kG) and subsequently recruit POLII, which leads to mRNA and protein synthesis. Chromatin opening is reversed with rate dG. (D) Schematic representation of the model describing IL-4 expression as a two-step process (for the relation between TF and OTF, see Supplementary Mathematical Appendix 2). (E) Kinetics of TF occupancy at binding site (OTF) in the simulations: an initial phase of maximal occupancy (t<tA, OTF (t)=1) is followed by an exponential decay with decay time tA/2 (see Materials and methods: Stochastic model of IL-4 expression upon stimulation). Source data is available for this figure at www.nature.com/msb.
Bimodal IL-4 expression originates from stochastic chromatin opening
As a simple model of transcription and translation (Figure 2B) cannot account for the observed expression variability, we extended the model by including a stimulation-dependent chromatin opening step (Figure 2C), which is known to occur during IL-4 expression (Avni et al, 2002; Guo et al, 2004). For simplicity, we lumped the multiple (and as yet incompletely understood) molecular events that contribute to this opening (reviewed in Ansel et al, 2006) into a single transition that renders the chromatin structure permissive for transcription. As all processes that may be involved in chromatin opening (histone modification, chromatin remodeling, and long-range intrachromosomal interactions) are reversible, we also accounted for the recondensation of the chromatin. Chromatin opening (Figure 2C, Step 1) as well as the recruitment of the general transcription machinery (Figure 2C, Step 2) are dependent on antigenic stimulation and require the binding of stimulation-induced TFs. Assuming that binding and release of TF at regulatory sites are in fast equilibrium (see Supplementary Mathematical Appendix 2), we translated the model in Figure 2C into a mathematical description shown schematically in Figure 2D. The likelihood of chromatin opening (with rate kG) and the transcription rate (kR) are controlled by the relative occupancy (OTF) of TF at a binding site, whereas the closing of the chromatin structure proceeds with a basal rate dG. Thus, chromatin opening that allows transcription is an active, stimulation-dependent process.
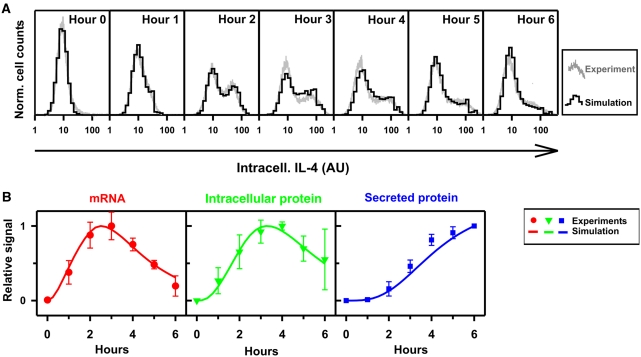
To investigate whether the extended model can account for the bimodal expression pattern observed for IL-4, we compared stochastic simulations of IL-4 expression with experimental data. Having estimated the rates of transcription, translation, and degradation from experimental measurements (see earlier section), three parameters remained unidentified: the rates of chromatin opening (kG) and closing (dG) and the dynamics of TF-binding site occupancy (OTF (t)). The stimulation-dependent TF NFAT1 that induces chromatin opening and transcription of the il4 gene (Rao et al, 1997; Agarwal et al, 2000; Avni et al, 2002; Hogan et al, 2003) is activated rapidly within a few minutes of stimulation and completely deactivated after 3 h of continuous stimulation (Loh et al, 1996). Therefore, the time course assumed for OTF consists of an initial phase of constant maximal occupancy followed by an exponential decay (Figure 2E). We simulated the expression kinetics of IL-4 with the extended model, considering stochastic transitions between the gene states (open and closed) and taking the duration of TF occupancy tA as a random variable with average value <tA> (see Materials and methods: Stochastic model of IL-4 expression upon stimulation). As only low variability arises from IL-4 transcription and translation (see above), these processes were modeled deterministically. By fitting kG, dG, and <tA> with a simulated annealing algorithm (Supplementary Figure S4), we found that the model accurately reproduces the distribution of IL-4 expression within the population over the entire time course of a stimulation (Figure 3A) and, at the same time, accounts for the kinetics of IL-4 mRNA, intracellular and secreted protein (Figure 3B). We estimated a gene-opening rate of kG=0.23/h, implying that ∼23% of the cells were activated in 1 h of maximal TF-binding site occupancy. Therefore, in the proposed extended model, slow and stochastic chromatin opening (Step 1) is the limiting step for the induction of IL-4 and the main source of the bimodal IL-4 expression.
Figure 3.
Stochastic model of IL-4 expression can account for the experimental measurements. (A) Two-week-differentiated wild-type Th2 cells were stimulated with PMA/ionomycin and intracellular IL-4 was quantified by flow cytometry at the indicated time points (gray lines). Using the model shown in Figure 2D, the distributions of the IL-4 expression level were simulated (black histograms). For the simulation, model parameters kR, dR, kP, and dP were estimated from experimental measurements, whereas the stochastic parameters kG, dG, and tA were estimated using a best-fitting procedure (see Table I and Materials and methods: Kinetics rate estimations). Log-normally distributed background noise for the auto-fluorescence was inferred from the unstimulated control (hour 0). One representative experiment out of three is shown. (B) The kinetics of mRNA, intracellular and secreted protein were simulated (lines) and compared with the measurements shown in Figure 1B (•, ▾, ▪). Source data is available for this figure at www.nature.com/msb.
The long lifetime of the open gene state predicts short-term memory for IL-4 expression
Our fitting of the model to the data estimated the lifetime of the open gene state (dG−1) to be longer than 0.7 days (where values >0.7 are practically equally possible). As this lifetime is much longer than the mean duration of the TF-binding site occupancy during antigenic stimulation (<tA>=1.3 h), acute IL-4 expression is terminated by the cessation of TF activity (terminating Step 2; Figure 2C), rather than through the closing of the chromatin (Step 1). However, because of the long lifetime of the open chromatin state, the model predicts that the cells with an open locus will re-express IL-4 with higher probability upon a second stimulation. Specifically, the fraction of cells carrying an open gene (ON) at time t after the initial response decays as 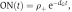 , where ρ+ is the fraction of IL-4-producing cells during the first stimulation (t=0). The increased fraction of IL-4-expressing cells in a second stimulation, ρ+II, is given by (see Materials and methods: Stochastic model of IL-4 induction upon second stimulation):
, where ρ+ is the fraction of IL-4-producing cells during the first stimulation (t=0). The increased fraction of IL-4-expressing cells in a second stimulation, ρ+II, is given by (see Materials and methods: Stochastic model of IL-4 induction upon second stimulation):
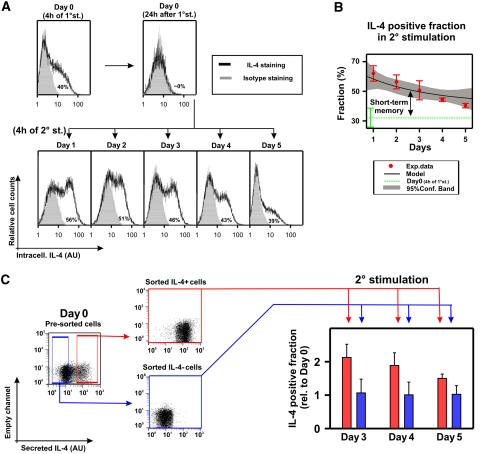
We measured the fraction of cells producing IL-4 upon a secondary response at different time points after the first stimulation (Figure 4A). As predicted by our analysis, we observed an elevated fraction of activated cells in the second stimulation. This fraction decreased progressively as the resting phase increased (Figure 4B, red dots), resulting in a short-term memory for il4 induction at the population level. This short-term memory was not caused by IL-4 protein that had remained inside the cells from the first response because protein was cleared completely from the cells within 24 h (Figure 4A, panel ‘Day 0 (24 h after 1° stim.)’). Fitting the data with the analytical solution (equation (2)) showed that the data can be accounted for by a lifetime of the open gene state of dG−1=2.9 days. This estimate is consistent with the previously published observation that 10 days after stimulation, the cells have no memory of the earlier stimulation (Hu-Li et al, 2001).
Figure 4.
IL-4-producing Th2 cells exhibited the short-term memory predicted by the model. (A) One-week-differentiated il4wt/il4gfp Th2 cells were stimulated with anti-CD3/28 in the presence of costimulatory signals (see Materials and methods). To assess their opening probability, an aliquot of unstimulated cells was instead stimulated with PMA/ionomycin/BfA and stained for intracellular IL-4 (Day 0). To return to the resting state, the cells were transferred onto a plate without anti-CD3 and rested overnight. At 1, 2, 3, 4, or 5 days after stimulation, a fraction of cells were stimulated with PMA/ionomycin/BfA and stained. To control that no IL-4 protein from the initial stimulation remained, the cells were stained 24 h after stimulation. Background fluorescence was measured by staining with an isotype antibody (gray area), and rescaled to the lower peak to quantify the presented percentages of IL-4-producing cells (for further details see Supplementary Figure S1). One representative experiment out of three is shown. (B) The decay of the IL-4+ fraction upon the second stimulation (red •, averages and s..d. from three independent experiments) was fitted to the exponential regression model 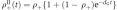 presented in equation (2) (SigmaPlot, Corr.Coef.=0.82, P-value<0.05). The best-fit curve (black solid line; chromatin closing rate dG=0.015/h and chromatin-opening probability upon stimulation ρ+=0.4) and 95% confidence bands (gray shadow) are presented. The IL-4+ fraction upon 4 h of first stimulation were 0.32±0.08 (Day 0, dotted green line), in agreement with the inferred value for the basal probability of chromatin-opening ρ+. The IL-4+ fraction upon the second stimulation showed a transient increase with respect to the first stimulation, with average decay time of 2.8 days (short-term memory). (C) One-week-differentiated il4wt/il4gfp Th2 cells were stimulated for 3.5 h with CD3/CD28-specific antibodies, and IL-4-positive and negative cells were purified using the IL-4 secretion assay, followed by flow-cytometric sorting (Supplementary Figure S6). IL-4 positive and negative cells were stimulated for an additional 20 h and rested (see Materials and methods). After 3–5 days in resting conditions, aliquots of sorted populations were restimulated for 4 h with PMA/ionomycin/BfA and the fractions of producing cells were determined by IL-4 intracellular staining followed by flow cytometry. Mean and s.d. of the values, relative to the producing fraction of the unsorted cells at Day 0, from five independent experiments are presented (for more detail see Supplementary Figure S9). By using t-test, no significant differences were found between the sorted IL-4 negative cells at Days 3, 4, or 5 (blue bars) and the unsorted cells at Day 0 (P-value >0.5). On contrary, for each time point, the sorted IL-4 positive (red bars) cells differed significantly (P-value<0.05) from the positive cells at different days, the negative cells at different days, and the unsorted cells at Day 0. Source data is available for this figure at www.nature.com/msb.
presented in equation (2) (SigmaPlot, Corr.Coef.=0.82, P-value<0.05). The best-fit curve (black solid line; chromatin closing rate dG=0.015/h and chromatin-opening probability upon stimulation ρ+=0.4) and 95% confidence bands (gray shadow) are presented. The IL-4+ fraction upon 4 h of first stimulation were 0.32±0.08 (Day 0, dotted green line), in agreement with the inferred value for the basal probability of chromatin-opening ρ+. The IL-4+ fraction upon the second stimulation showed a transient increase with respect to the first stimulation, with average decay time of 2.8 days (short-term memory). (C) One-week-differentiated il4wt/il4gfp Th2 cells were stimulated for 3.5 h with CD3/CD28-specific antibodies, and IL-4-positive and negative cells were purified using the IL-4 secretion assay, followed by flow-cytometric sorting (Supplementary Figure S6). IL-4 positive and negative cells were stimulated for an additional 20 h and rested (see Materials and methods). After 3–5 days in resting conditions, aliquots of sorted populations were restimulated for 4 h with PMA/ionomycin/BfA and the fractions of producing cells were determined by IL-4 intracellular staining followed by flow cytometry. Mean and s.d. of the values, relative to the producing fraction of the unsorted cells at Day 0, from five independent experiments are presented (for more detail see Supplementary Figure S9). By using t-test, no significant differences were found between the sorted IL-4 negative cells at Days 3, 4, or 5 (blue bars) and the unsorted cells at Day 0 (P-value >0.5). On contrary, for each time point, the sorted IL-4 positive (red bars) cells differed significantly (P-value<0.05) from the positive cells at different days, the negative cells at different days, and the unsorted cells at Day 0. Source data is available for this figure at www.nature.com/msb.
The chromatin memory time of the chromatin state found for IL-4 expression, 2.9 days, is one order of magnitude larger than the characteristic time of intracellular IL-4 turnover (∼2 h; Supplementary Mathematical Appendix 1) that provides a basal ‘memory’ for gene expression (Sigal et al, 2006). As changes of epigenetic states of a gene have previously been associated with cell division because of the chromatin disintegrated during DNA replication (Richter et al, 1999; Rando and Verstrepen, 2007), we asked whether the loss of IL-4 memory is linked to cell proliferation. Cells were labeled with the cell-proliferation marker DDAO at Day 0, and stimulated again at Day 3 and at Day 5. No correlation between IL-4 expression and cell proliferation was found at both time points (CC<0.05, see Supplementary Figure S5). Therefore, the loss of IL-4 memory seems to be a stochastic event independent of the cell cycle.
Next, we investigated whether the memory for IL-4 expression indeed resided in the IL-4 positive fraction of the first stimulation, as predicted by the model. During the first stimulation, we sorted the live cells into an IL-4-producing (positive) and a non-producing (negative) fraction, using the IL-4 secretion assay followed by flow cytometric sorting (Figure 4C; Supplementary Figure S6). The fractions were cultured separately and cells were restimulated after different resting periods during which they had ceased to express IL-4 (Figure 4C, left panels; Supplementary Figure S7). The fraction of IL-4-producing cells was quantified at Days 3–5 after cell sorting, to avoid any interference of the preceding secretion assay with intracellular staining of IL-4 (Supplementary Figure S8). As expected from the model, the probability of IL-4 re-expression in sorted IL-4 positive cells was consistently larger than in the sorted IL-4 negative cells and decreased progressively until almost reaching the values of the original unsorted population at Day 0 (Figure 4C, right panel). By contrast, the sorted IL-4 negative cells exhibited a constant induction probability that is undistinguishable from the unsorted population (Supplementary Figure S9). A similar behavior was observed when measuring IL-4 protein in the supernatant at Days 1 and 2 after the initial stimulation in that IL-4 concentrations were higher in the sorted IL-4 positive population compared with the negative population (Supplementary Figure S10). These findings substantiate the results of the Gata-3 analysis (cf. Figure 1C) that the IL-4-expressing and non-expressing cells arise stochastically from a uniform cell population.
We then asked whether the memory for IL-4 expression was a cellular property affecting both il4 alleles or was a property of the expressed il4 allele, using cells heterozygous for an il4 wild-type allele and an il4 allele with GFP knock-in (Hu-Li et al, 2001). We found that the probability to express the GFP knock-in allele was similar in the sorted IL-4-positive and negative cell populations (Supplementary Figure S11). Moreover, IL-4-positive cells had no proliferative advantage, excluding selective proliferation as the basis of short-term memory (Supplementary Figure S12). Finally, the elevated expression of Gata-3 in both IL-4-producing and non-producing cells (Figure 1C) remained similar in the sorted IL-4 positive and negative Th2 cells (Supplementary Figure S13). These findings imply that short-term memory is predominantly a property of the expressed il4 allele that may be due to the opening of chromatin structure upon antigenic stimulation.
Taken together, our results show that the cells retain a short-term memory for IL-4 expression for several days after a given stimulation, resulting in an increased IL-4 expression probability. Our data are consistent with a model in which this increase is carried by the Th2 cells that remain in the open chromatin state reached during the first stimulation. These cells can readily start il4 gene transcription when the stimulus-dependent TFs are activated again in a second stimulation.
Antigen-dependent TFs affect both the fraction and the expression level of IL-4 producers
In the model, the stimulation-dependent TFs control IL-4 induction by promoting both the chromatin opening (Step 1) and recruitment of the general transcriptional machinery (Step 2). In our mathematical analysis (see ‘Mathematical appendix 3’ in Supplementary information), we find that both the duration (tA) and the strength (OTFMax) of the TF-binding site occupancy contribute to the fraction, ρ+, of IL-4 positive cells
and to the average level of IL-4 expression per positive cell
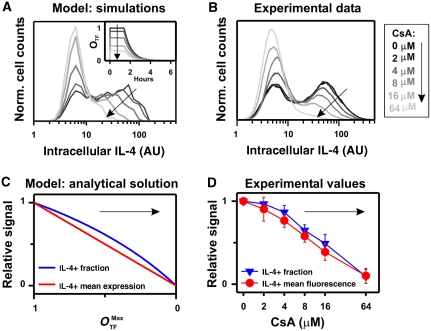
Consequently, a reduction of the TF activity should decrease the fraction and the expression level of IL-4 (Figure 5A). To test this prediction experimentally, we titrated the activity of the antigen-dependent TF NFAT1 during stimulation by adding increasing amounts of cyclosporine A (CsA), a pharmacologic inhibitor of NFAT activation, and assessed IL-4 expression by intracellular staining. Both the fraction of IL-4-producing cells and the mean fluorescence intensity (MFI) of the IL-4-producing cells were decreased by CsA in a dose-dependent manner (Figure 5B), and the decrease in both quantities was similar to the theoretical prediction (equations (3) and (4) and Figure 5C and D).
Figure 5.
Inhibition of the stimulation-induced transcription factor NFAT1 results in a decreased fraction and expression level of IL-4-producing cells, as predicted by the model. (A) Simulated distribution of IL-4 expression after 3 h of stimulation (parameter values in Table I, except kG=0.5/h) for different values of the initial transcription factor (TF) occupancy at the binding site, OTF (0)=OTFMax, as indicated in the inside panel. (B) Two-week-differentiated wild-type Th2 cells were stimulated with PMA/ionomycin/BfA in the presence of the indicated amounts of the NFAT1-inhibitor CsA and intracellular IL-4 expression was assessed. (C) The fraction and production in the IL-4-positive cells (IL-4+) from the model approximation (equations (3) and (4)) for different values of OTFMax are presented, relative to the value OTFMax=1. (D) The fraction and the mean fluorescence intensity for the IL-4-producing cells were determined in three independent repetitions of the experiment in Figure 5B. Mean and s.d., relative to the value in the absence of CsA (darkest curve in Figure 5B), are presented. Source data is available for this figure at www.nature.com/msb.
Thus, the antigen-dependent signaling has a dual role in regulating both gene opening of the il4 locus and the transcription initiation of the il4 gene.
Progressive Th2 cell differentiation selectively controls the chromatin-opening rate
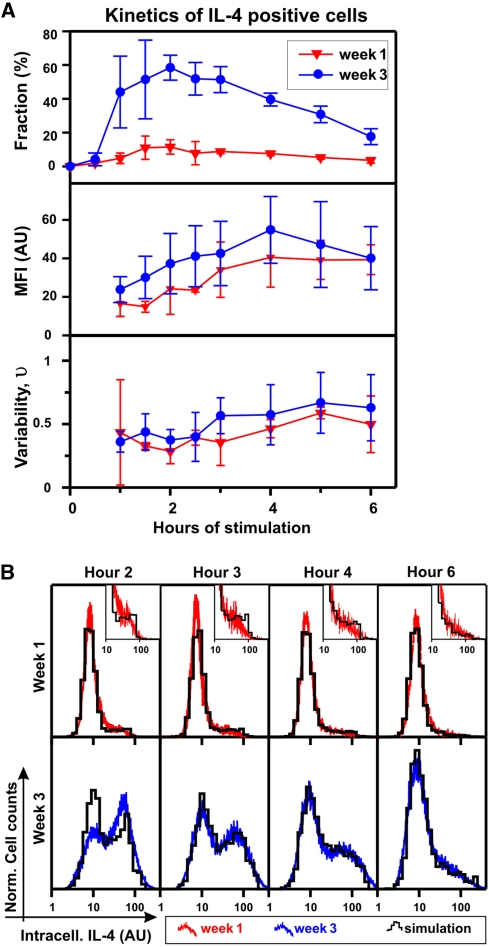
When stimulated repeatedly in the presence of IL-4, Th cells mature progressively towards the terminally differentiated Th2 cell phenotype. This Th2 cell differentiation process is accompanied by chromatin rearrangement at the il4 locus (Guo et al, 2004; Ansel et al, 2006). In the model, this differentiation will affect only the gene-opening probability kG, which is modulated by the structural properties of the chromatin. According to equations (3) and (4), an increased opening probability kG will result in an elevated fraction of IL-4-producing cells, whereas their expression level should remain rather constant. To test this prediction, we compared the fraction and expression level (MFI and variability) of the IL-4-producing cells in 1-week-differentiated and 3-week-differentiated Th2 cells over the time course of a response (6 h). We found that the IL-4 positive fraction increased 4–5-fold within 2 weeks of differentiation (Figure 6A). By contrast, the mean expression level in the positive fraction increased only by 16%, whereas the normalized variance remained unchanged (Figure 6A). The slight increase in the mean expression level can be explained by a higher fraction of cells expressing IL-4 from two alleles (which is expected to result in an increase of 18%; Supplementary Figure S14).
Figure 6.
Progressive changes in IL-4 expression during Th2 differentiation can be accounted for by a single parameter, the chromatin-opening rate kG. (A) One-week- and 3-week-differentiated wild-type Th2 cells were restimulated with PMA/ionomycin and IL-4 expression was assessed by intracellular staining at different time points during the response. By comparison with the isotype staining, IL-4 positive cells were identified (Supplementary Figure S1). The fraction, mean fluorescence intensity (MFI), and variability (νP) within the IL-4 positive cells were determined at indicated time points. Mean and s.d. of three independent experiments are shown. (B) The measured IL-4 distributions (red-top and blue-bottom lines for 1-week- and 3-week-diffferentiated Th2 cells, respectively) are compared with model simulations (black histograms) at indicated time points. The chromatin-opening rate kG was fitted to the measured distributions (Supplementary Figure S15), whereas the remaining parameters were fixed to the values in Table I. One representative experiment out of three is shown. Source data is available for this figure at www.nature.com/msb.
To validate our model, we tested whether the measured IL-4 expression kinetics at different stages of Th2 cell differentiation can be reproduced by adapting only the chromatin-opening rate kG. Indeed, we could quantitatively account for the measured distributions of IL-4-producing cells over the entire time course of the stimulation by increasing the chromatin-opening rate kG from 0.042/h for 1-week-differentiated cells to 0.31/h for 3-week-differentiated cells (Figure 6B; Supplementary Figure S15). Consistent with these numbers, the estimated value of kG for 2-week-differentiated Th2 cells (Figure 3A) was intermediate (0.23/h). In summary, we find that the changes in IL-4 variability during Th2 cell differentiation can be quantitatively accounted for by the progressive increase in a single-model parameter, namely the rate of chromatin opening upon antigenic stimulation. This implies that progressive Th2 differentiation facilitates the access of the transcription machinery to the il4 gene.
Discussion
In this study, we have investigated the expression dynamics of an inducible mammalian gene known to be highly regulated at the epigenetic level (Ansel et al, 2006). Our experimental data are consistent with a mathematical model of a two-step process for IL-4 induction that entails stimulation-induced chromatin rearrangement followed by the initiation of transcription. We present evidence for a rate-limiting chromatin-opening step that controls the probability of IL-4 expression and is itself regulated by the differentiation state of the cell. The experimentally inferred rate of stimulation-induced chromatin opening is remarkably low (in the order of 0.1/h), implying that, during the limited time window of TF activity induced by antigenic stimulation, only a fraction of cells manage to rearrange one or both il4 alleles. As a consequence, stochastic heterogeneity is observed in a Th2 cell population with regard to IL-4 expression. The subsequent reversal of il4 gene activity occurs on two very different time scales: after the withdrawal of the antigen stimulus, transcription is rapidly and efficiently shut down (within tens of minutes), whereas the increased competence to express the gene is reversed much more slowly over several days. We provide experimental evidence for a previously unknown short-term memory for IL-4 induction that was predicted by the model and may reside in the open chromatin conformation retained at the end of the stimulation.
What is the source of the bimodal expression pattern of IL-4 and of its high expression variability? In principle, the observed cell-to-cell variability could arise from the randomness intrinsic in the biochemical processes underlying gene expression. Alternatively, it could result from a pre-existing heterogeneity in the population, for example with regard to the expression level of critical TFs. IL-4 producers and non-producers are very similar with respect to (1) their levels of GATA-3 (Ouyang et al, 2000 and Figure 1C), c-Maf as well as nuclear NFAT upon stimulation (Guo et al, 2004), (2) the DNA accessibility at the il4 promoter and several regulatory sites (Guo et al, 2004), and (3) the DNA methylation state of the promoter (Tykocinski et al, 2005). A correlation between the probability of IL-4 expression and DNA demethylation in the first exon and intron of the il4 gene has been observed, but the gene can remain unexpressed despite being demethylated in this region and at the promoter (Tykocinski et al, 2005). Moreover, we have found that the ability to express IL-4 is not an imprinted trait in the Th2 cells used in this study, which agrees with earlier results (Hu-Li et al, 2001; Guo et al, 2004) and argues against differential DNA demethylation as the main cause for heterogeneity in IL-4 expression. Thus, the high variability observed for IL-4 expression seems to arise primarily from the stochasticity intrinsic in gene induction.
The molecular mechanisms responsible for expression variability are still unknown for most genes. Several studies that have focused on this question have suggested models of gene regulation with a similar structure to the one we use here, with the three main steps: gene activation, transcription, and translation (Raser and O'Shea, 2004; Golding et al, 2005; Bar-Even et al, 2006; Raj et al, 2006). As the rates of these processes vary significantly in different systems, it was found that each step of the model can be an important source of variability. In prokaryotes, it has been suggested that variability is mainly due to the low mRNA copy number, whose variations are then amplified by translation into proteins (Thattai and van Oudenaarden, 2001; Kaern et al, 2005). For IL-4, we exclude this possibility because of the high IL-4 mRNA levels that we have measured (∼100-fold more mRNA molecules of IL-4 than of the house-keeping gene HPRT).
In eukaryotes, the regulation of chromatin structure adds a complex layer of control that determines whether the general transcription machinery can access the promoter of a gene. Spontaneous transitions between an active and an inactive gene state can significantly increase the expression variability (Raser and O'Shea, 2004; Bar-Even et al, 2006), and give rise to transcriptional bursts that have also been observed experimentally (Chubb et al, 2006; Raj et al, 2006). In principle, the bimodal expression pattern of IL-4 could arise from such transitions, as long as mRNA and protein are sufficiently short lived to be completely removed from the cell between successive bursts (Pirone and Elston, 2004). In this scenario, only a fraction of cells would express the protein at a given point in time although each cell, at some point, would produce IL-4. Our data on IL-4 argue against this scenario because only a fraction of cells contains IL-4 protein even when secretion is blocked for an extended period of time (cf. Figure 1A).
By contrast, we describe and characterize a process that may be viewed as an extreme case of transcriptional bursting, where at most one prolonged transcription episode can occur per allele during a given stimulation. Such a burst occurs because of the superposition of two regulatory steps: probabilistic chromatin opening on the time scale of hours to days (kG, dG) and the transient availability of active TFs in the range of a few hours (tA). A bimodal expression pattern is observed because in the short-time window presented by antigen-dependent signaling, only a fraction of cells can switch to the active gene state (open chromatin) that allows IL-4 production. Therefore, the transient nature of the response, preventing the system from reaching the steady state, is central to understanding the origin of the expression variability.
Interestingly, the kinetic model of gene regulation we propose provides a unifying framework for the binary (digital) and graded (analog) modes of gene regulation. Although it has been suggested that genes may be controlled either in a binary or a graded manner (Biggar and Crabtree, 2001), our results show that these are not mutually exclusive regulatory patterns. Rather, the regulation of chromatin opening controls the fraction of expressing cells (binary mode), and the regulation of transcription initiation affects the expression level (graded mode). As a consequence, a TF that acts on both levels will give rise to mixed binary–graded responses, as shown here for NFAT1.
We provide evidence that the probability of the il4 gene to transit from the inactive to the active state in a given stimulation increases progressively with the differentiation state of the cell. The data presented here suggest that this epigenetic gene transition requires the action of NFAT1, a TF induced by antigenic stimulation that is required for IL-4 transcription. NFAT1 enhances DNA accessibility of two regulatory sites in the il4 locus (Va and RHS7) (Avni et al, 2002; Lee and Rao, 2004), which come into close proximity with the il4 promoter in T cells (Spilianakis and Flavell, 2004). At least one of these regions (Va) contains an enhancer for IL-4 transcription and is more accessible in IL-4 producers than in non-producers, so that the opening of the Va region could be rate limiting for the initiation of transcription (Guo et al, 2004). The observation that the short-term memory described here has a similar half-life (∼3 days), as the accessible state of the il4 enhancer (between 1 day and 10 days (Guo et al, 2004), further supports this hypothesis. Interestingly, the il4 enhancer becomes increasingly accessible also in the resting state in the course of Th2 differentiation (Guo et al, 2004; Ansel et al, 2006). This might facilitate the formation of a multiprotein complex that mediates chromatin opening.
The transient memory for IL-4 expression of several days described here occurs on an intermediate time scale compared with, on the one hand, long-term epigenetic memory persisting for up to 106 cell generations (Rando and Verstrepen, 2007) and, on the other hand, the time scale of IL-4 mRNA turnover and protein secretion (∼1–2 h). The specific short-term memory for inducible IL-4 expression may serve a regulatory purpose by reinforcing an acute immune responses against a pathogen. At the same time, the limited duration of memory may safeguard against the harmful effect of unchecked cytokine production that can result in immunopathology (Corry and Kheradmand, 2009). Sigal et al (2006) have found ‘mixing times’ for protein expression of 20 h up to a few days in a human cancer cell line, with strong correlation between the two alleles of a gene. Thus, this phenomenon seems to be distinct from the memory for expression of an inducible gene described here.
The probability for the gene transition that we have estimated from experimental data is relatively low (kG∼0.05–0.5/h). It is unclear which molecular event could proceed with such low probability, as binding of chromatin remodelers to the DNA or the remodeling step itself happen on much faster time scales. The molecular mechanism of probabilistic regulation has been studied in detail for another cytokine, IFN-β, whose transcription is controlled by a rate-limiting step of chromatin rearrangement (Lomvardas and Thanos, 2002). This step was found to be controlled by a multiprotein complex assembling on the IFN-β promoter. As the factors that build up this complex are present at limiting concentrations, complex formation could well proceed with low probability. Similarly, such a complex might need to be formed on the il4 enhancer, where multiple factors bind (NFAT1, GATA-3, Brg1) and are required for opening of the chromatin. Complex formation might be facilitated through enhanced accessibility of the chromatin, resulting in an increased transition probability in further differentiated cells.
Another regulatory event that could proceed with low probability is the formation of long-range chromatin interactions. Such interactions were shown to be critical for the probabilistic regulation of IFN-β (Apostolou and Thanos, 2008), and they also occur on the il4 locus. Upon antigenic stimulation, the il4 promoter and enhancer are moved into close proximity to a genomic region ∼70 kb downstream of the il4 gene (SBS-C9) (Cai et al, 2006). Although functional implications of this observation have not yet been analyzed further, similar looping events have been found in transcriptional regulation of the well-studied β-globin gene (Palstra et al, 2003). Assuming that the formation of such a stable loop is initiated by the random interaction of two DNA elements, looping could proceed with rather low probability (Soutoglou and Misteli, 2007). Thus, multi-protein complex formation or long-range chromatin interaction could limit the il4 gene transition at the molecular level.
Most studies on gene expression variability have investigated unicellular organisms, where fluctuations in protein abundance have been viewed as an unavoidable disturbance or suggested to offer a selective advantage in changing environments. In a multicellular organism, the highly stochastic regulation of a gene such as il4 could simply be a byproduct of tight regulation, allowing gene activation only with certain probability. However, there may be a distinct functional role of the stochastic mode of regulation uncovered here, as the bimodality in expression levels renders the response at the single-cell level unambiguous: the il4 gene is either ‘on’ or ‘off.’ IL-4 serves as an extracellular signal to induce antibody production and class switching in B cells (Stavnezer et al, 2008). Similarly to other cytokines that act as local signals (Busse et al, 2010), the IL-4-mediated interaction seems to take place between proximal pairs of Th and B cells (Kupfer et al, 1991). Binary IL-4 expression can ensure that each activated B cell receives a sufficient quantum of IL-4 to induce class-switch recombination (again an all-or-none process), where short-term memory can reinforce this acute response. Regulation of the il4 gene should then act at the population level by determining the number of IL-4 producers and hence activated B cells. Indeed, repeated antigen challenge triggers a more efficient response by increasing the fraction of IL-4-expressing Th2 cells (as seen in the comparison of 3-week versus 1-week cultured Th2 cells) while the level of expression per cell is changed very little. Thus, il4 gene regulation shows how molecular stochasticity giving rise to an on–off switch can serve a regulatory purpose.
Materials and methods
Mice, antibodies, chemical reagents
Balb/c, DO11.10 TCR-transgenic (wild-type) mice, and heterozygous il4wt/il4gfp mice (Hu-Li et al, 2001) were bred under specific pathogen-free conditions. Wild-type mice were used to generate Th2 cells in experiments shown in Figures 1, 3, 5, and 6; Supplementary Figures S1–S4, S13, and S15. Heterozygous il4wt/il4gfp mice were used to generate Th2 cells in experiments shown in Figures 1C, 2, and 4; Supplementary Figures S5–S13. Antibodies to IFN-γ (AN18.17.24), IL-12 (C17.8), IL-4 (11B11), CD3 (145-2C11), CD28 (37.51), CD4 (GK1.5), and CD62L (MEL14) were prepared from hybridoma supernatants. CsA was obtained from AWD, phorbol mysterate acetate (PMA), ionomycin, and BfA were supplied by Sigma-Aldrich.
Th2 cell culture
Naive CD4+ CD62L+ T cells were purified by magnetic sorting as described earlier (Assenmacher et al, 2002). Fresh splenocytes from 6- to 10-week-old mice were stained with a fluorescein-isothiocyanate-conjugated CD4-specific antibody and CD4-expressing cells were purified using the FITC-Multisort-Kit (Miltenyi Biotec) according to the manufacturer's instructions. Subsequently, naive T cells that express high levels of CD62L were purified using CD62L-Microbeads (Miltenyi Biotec). A purity of 97–99% was reached. For Th2 cell priming and differentiation, irradiated (30 Gy) splenocytes used as APC were cultured with the naive T cell in a ratio of 1:5 and a total cell density of 2 × 106 cells/ml. Cognate peptide OVA323−339 (0.5 μM), blocking antibodies to IFN-γ (10 μg/ml), and IL-12 (10 μg/ml) and recombinant IL-4 (30 ng/ml, Sigma-Aldrich) were added to the culture. For further differentiation, the cells were stimulated with the same protocol at Day 6 and eventually at Day 12 with addition of IL-2 (10 ng/ml, Sigma-Aldrich). For Th1 cell priming and differentiation, cognate peptide OVA323−339 (0.5 μM), blocking antibodies to IL-4 (10 μg/ml), and recombinant IL-12 (30 μg/ml) were added to the culture.
For Th2 cell culture used in short-term restimulations, after 6 days of Th2 priming conditions (Day 0 in the text), the populations were cultured in the presence of neutral costimulatory signals (IL-2 (10 ng/ml), anti-CD3/28 antibodies and blocking antibodies to IL-4 (10 μg/ml), IFN-γ (10 μg/ml), and IL-12 (10 μg/ml)) for 24 h. To return to the resting state, the cells were transferred onto plates without anti-CD3 and rested overnight. After 1, 2, 3, 4, or 5 days, a fraction of the cells were restimulated with PMA/ionomycin/BfA.
T-cell stimulation, intracellular cytokine staining, cell labeling, and ELISA
Before stimulation, dead cells were removed by gradient centrifugation using Histopaque-1083 (Sigma-Aldrich). Cells were stimulated with 10 ng/ml PMA and 1 μg/ml ionomycin in a concentration of 2 × 106 cells/ml. Where indicated, BfA (5 μg/ml) was added to block secretion after 1 h of stimulation. For IL-4 staining, cells were fixed for 15 min in 2% formaldehyde. In the presence of 0.5% saponin, the cells were stained intracellularly with antibodies to CD4 and IL-4 (BD). In parallel, an aliquot of cells was stained with isotype control antibodies. For Gata-3 staining, Gata-3 antibody and Foxp3 fixation and permeabilization buffer (Ebioscience) were used according to the manufacturer's instructions. For DDAO-SE labeling, cells were washed in PBS, resuspended in 2.5 μM DDAO-SE (Invitrogen) at concentration of 107 cells/ml, and incubated for 5 min at ice. Data were measured by flow cytometry using a FACS Calibur (BD) and analyzed using FlowJo software (Tree Star). To quantify IL-4 in the supernatant, IL-4 duo set (R&D) was used according to the manufacturer's instructions.
IL-4 secretion assay
One-week-differentiated Th2 cells were stimulated with antibodies specific for CD3 (plate-bound 1 μg/ml), CD28 (1 μg/ml), and in the presence of blocking antibodies to IL-4 (10 μg/ml) for 3.5 h. Cells were harvested and washed. IL-4 bound to IL-4 receptors were blocked with purified anti-IL-4 antibody (BVD6-24G2; 400 μg/ml) for 5 min on ice in 0.5% BSA in PBS. Then, the IL-4 secretion assay (Miltenyi Biotec) (Ouyang et al, 2000) was performed according to the manufacturer's instructions. Briefly, labeling of the cell surface with an IL-4 capture matrix was followed by a 30-min secretion phase. Then, IL-4 bound to the cell surface was stained with a PE-coupled antibody, and the positive and negative cells were purified by FACS sorting. Control staining performed to ensure the specificity of the assay can be found in Supplementary Figure S6.
Positive and negative populations were stimulated for additional 20 h with anti-CD3/28 antibodies in the presence of neutral costimulatory signals (IL-2 (10 ng/ml), anti-CD3/28 antibodies and blocking antibodies to IL-4 (10 μg/ml), IFN-γ (10 μg/ml), and IL-12 (10 μg/ml)). To return to the resting state, the cells were transferred to a plate without anti-CD3 and rested overnight. After 1, 2, 3, 4, or 5 days, a fraction of the cells were restimulated with PMA/ionomycin/BfA in the presence of the IL-4-specific antibody (24G2, 100 μg/ml) that impairs the binding of IL-4 to the capture matrix remaining on the cell surface. After stimulation, the cells were fixed for intracellular staining and incubated with unlabeled anti-IL-4-specific antibody (11B11) to saturate surface-bound IL-4. Then, IL-4 was stained intracellularly with an APC-labeled IL-4 antibody (11B11, BD).
mRNA extraction and quantification by real-time PCR
Total RNA was isolated from 0.5 to 1 × 106 T cells using Nucleospin RNA II (Macherey-Nagel) according to the manufacturer's instructions. With TaqMan Reverse Transcription Reagents (Applied Biosystem) 300–400 ng of total RNA was transcribed to cDNA in a total volume of 20 μl. Real-time PCR was performed using a LightCycler 2.0 Instrument (Roche), in a total volume of 5 μl with LightCycler FastStart DNA Master SYBR Green I (Roche). The reactions were performed in the presence of 3 mM (HPRT) or 5 mM (IL-4) MgCl2. DNA was denatured for 10 min at 96°C, followed by 42 cycles of melting (15 s, 96°C), primer annealing (12 s, 65°C) and elongation (12 s, 72°C). The specificity of each reaction was controlled by a melting curve measured through a ramp from 60 to 96°C with 0.1 C°/s. The IL-4 primer sequences used were 5′-ACGGAGATGGATGTGCCAAACGTC-3′ and, 5′-CGAGTAATCCATTTGCATGATGC-3′, sequences for HPRT have been described earlier (Niesner et al, 2008).
Estimation of mRNA and protein kinetics
Average lifetimes of mRNA and protein were inferred from the decay rate of the mRNA levels and from the delay between mRNA and protein signals (for further details refer to Supplementary Figure S3). The degradation rates of mRNA and protein are the inverse of the respective lifetimes. To estimate the IL-4 protein production in absolute numbers, we used the IL-4 protein accumulation values in pg/ml, from ELISA at hours 2, 3, and 4 (peak of IL-4 protein production) after restimulation from 2-week-differentiated Th2 cells (two independent experiments). We calculated the amount of IL-4 protein produced per cell [pg/cell], by rescaling the values to the cell concentration used in the stimulation (2 × 106 cells/ml) and to the fraction of IL-4 positive cells (refer to Supplementary Figure S1) at the indicated time. To convert the weight to the absolute number of proteins, we used the weight of the IL-4 protein, ∼14 kDa (kDa=1.6 × 10−24 g). To obatin the number of IL-4 protein produced per activated cell in 1 h, we calculated the increment during hours 3 and 4 (mean and s.d. are presented in Table I). To estimate the mean mRNA (relative to HPRT), we used the data from RT–PCR at hour 2, 2.5, and 3 (peak of IL-4 mRNA production) after restimulation (three independent experiments). The relative level per producing cell was found by rescaling to the fraction of IL-4 positive cells at the indicated time (mean and s.d. are presented in Table I). To estimate the absolute transcription rate in a producing cell (no/min), the mean mRNA level was divided by the lifetime of mRNA and multiplied by the mean number of HPRT molecules per cell, assumed to be equal to 20. To estimate the absolute translation rate in a producing cell (no/mRNA/min), we divided the estimated mean protein production per hour by the estimated mean number of mRNA molecules.
Table 1. Estimated rate constants used in the model of IL-4 expression.
| Experimental quantification | Parameter estimates (mean) | Parameter estimates (error) |
|---|---|---|
| mRNA level (rel. to HPRT) | 160-fold/cell | ±60-fold/cell (s.d.) |
| mRNA average lifetime, τR | 90 min | <120 min |
| Protein production rate | 0.75 × 106 protein/h/cell | ±0.17 × 106 protein/h/cell (s.d.) |
| Protein average lifetime, τP | 45 min | [35, 60] min (95% CI) |
| Experimentally estimated rate constants | ||
| Transcription rate, kR | 40 mRNAs/mina | ±15 mRNAs/mina (s.d.) |
| Translation rate, kP | 4 proteins/mRNA/mina | ±0.9 proteins/mRNA/mina (s.d.) |
| mRNA degradation rate, dR | 0.7/h | >0.5/h |
| Protein secretion rate, dP | 1.4/h | [1, 1.7]/h (95% CI) |
| Stochastic parameters derived from model fitting | ||
| Chromatin opening rate, kG | ||
| 1-week-differentiated cells | 0.042/h | [0.030, 0.054]/h (95% CI) |
| 2-week-differentiated cells | 0.23/h | [0.20, 0.27]/h (95% CI) |
| 3-week-differentiated cells | 0.31/h | [0.24, 0.4]/h (95% CI) |
| Chromatin closing rate, dG | 0.015/h | <0.06/h (95% CI) |
| Mean TF occupancy, <tA> | 1.3 h | [1.2, 1.4] h (95% CI) |
| aAssuming 20 HPRT mRNAs per cell. | ||
| CI, confidence interval (estimated through profile-likelihood method). | ||
Stochastic model of IL-4 expression upon stimulation
This model considers a population of cells, where each cell carries two copies of a considered gene. At time t=0, each cell has: protein and mRNA values equal to 0 and both the copies of the gene in the closed state. The TF occupancy at the binding site is approximated in rapid equilibrium and unsaturated, so that OTF(t)∝TF(t) and can be rescaled to an initial value OTF(0)=1 (see ‘Mathematical appendix 2’ in Supplementary information).
For t>0: (1) OTF has an initial costant phase tA, whose duration is a random variable different for each cell and exponentially distributed 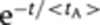 (see ‘Mathematical appendix 3’ in Supplementary information). After the initial constant phase, the following decay phase of OTF was assumed for simplicity equal to
(see ‘Mathematical appendix 3’ in Supplementary information). After the initial constant phase, the following decay phase of OTF was assumed for simplicity equal to 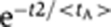 , so that the entire OTF kinetics depends on a single parameter <tA>, which has to be estimated by the fitting procedures. (2) Each allele switches stochastically from the closed to the open gene state with a probability rate kGOTF(t). The switch from the open to the closed state happens with a probability rate dG.
, so that the entire OTF kinetics depends on a single parameter <tA>, which has to be estimated by the fitting procedures. (2) Each allele switches stochastically from the closed to the open gene state with a probability rate kGOTF(t). The switch from the open to the closed state happens with a probability rate dG.
In the open state, the production/degradation of mRNA and the production/secretion of proteins of each allele are described by the following system of differential equations:
In the simulation, kR, kP, dR, and dP were set to the experimentally estimated values (Table I).
Using Matlab software (MathWorks, see online material), for each cell we extracted tA and the times of closed/open switching for each allele from the related exponential distributions. Then, we solved numerically the equations for mRNA and protein levels for both alleles, whose sum gives the values for each cell. To find the best-fitting parameters and the 95% confidence interval, we used a simulated annealing strategy and profile-likelihood method, respectively (Supplementary Figures S4 and S15).
Stochastic model of IL-4 induction upon second stimulation
This model considers a population of the cells carrying the gene in two possible states, open or closed, and two subsequent stimulations assessed at different time intervals. Before the first stimulation, all the cells have the gene in the closed state. At the end of the first stimulation (Day 0), the fraction of positive cells determines the gene-opening probability upon stimulation ρ+. After the first stimulation, the cells rest for a variable period t before the second stimulation. During this resting phase, the open genes can switch back to the closed state with a constant probability rate δG, and the fraction of open genes in the resting population decays as 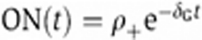 . Upon a second stimulation assessed after a resting phase t, the sum of the fraction of open genes from the first stimulation, ON(t), and the fraction of cells that switches from the closed to the open state during the second stimulation, ρ+ (1−ON (t)), gives the total fraction of positive cells upon second stimulation
. Upon a second stimulation assessed after a resting phase t, the sum of the fraction of open genes from the first stimulation, ON(t), and the fraction of cells that switches from the closed to the open state during the second stimulation, ρ+ (1−ON (t)), gives the total fraction of positive cells upon second stimulation 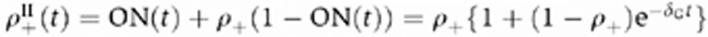 .
.
Supplementary Material
This archive contains the Matlab scripts for the simulations of the IL-4 stochastic model.
Table of Contents- Mathematical appendix 1: Post-transcriptional memory time- Mathematical appendix 2: Transcription factor activity and occupancy of the binding site- Mathematical appendix 3: Fraction and mean expression value of IL-4 positive cells- Supplementary figures legends- Supplementary information references- Figure S1- Figure S2- Figure S3- Figure S4- Figure S5- Figure S6- Figure S7- Figure S8- Figure S9- Figure S10- Figure S11- Figure S12- Figure S13- Figure S14- Figure S15
Acknowledgments
We thank Danilo Dongiovanni, Michael Floßdorf, Maciej Dobrzynski, and Nils Blüthgen for help with statistical methods; Inka Albrecht for useful discussions and technical support; Jenny Kirsch and Toralf Kaiser for help with the cell sorting facility at DRFZ; Riccardo Spina, for help with graphical design; Nils Blüthgen, Maciej Dobrzynski, Michael Floßdorf, Stefan Legewie, and Antonio Z Politi for critical reading of the paper. This work was supported by DFG-SFB 618, the BMBF FORSYS-Partner programme, the Helmholtz Alliance for Systems Biology/SBCancer (grants to TH) and the Volkswagen Foundation (Lichtenberg professorship to ML). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agarwal S, Avni O, Rao A (2000) Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12: 643–652 [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular Biology of the Cell, 4th edn, New York: Garland [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24: 607–656 [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D (2008) Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96 [DOI] [PubMed] [Google Scholar]
- Assenmacher M, Lohning M, Radbruch A (2002) Detection and isolation of cytokine secreting cells using the cytometric cytokine secretion assay. Curr Protoc Immunol Chapter 6: Unit 6 27 [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A (2002) T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3: 643–651 [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR (2001) Cell signaling can direct either binary or graded transcriptional responses. EMBO J 20: 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix M, Locksley RM (1998) Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science 281: 1352–1354 [DOI] [PubMed] [Google Scholar]
- Blake WJ, KAErn M, Cantor CR, Collins JJ (2003) Noise in eukaryotic gene expression. Nature 422: 633–637 [DOI] [PubMed] [Google Scholar]
- Busse D, De la Rosa M, Hobiger K, Thurley K, Flossdorf M, Scheffold A, Höfer T (2010) Competing feedback loops shape IL-2 signaling between helper and regulatory T cells in cellular microenvironments. Proc Natl Acad Sci USA 107: 3058–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Calado DP, Paixão T, Holmberg D, Haury M (2006) Stochastic monoallelic expression of IL-10 in T cells. J Immunol 177: 5358–5364 [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S (2008) Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453: 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry DB, Kheradmand F (2009) Toward a comprehensive understanding of allergic lung disease. Trans Am Clin Climatol Assoc 120: 33–48 [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G (2008) Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science 321: 1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB (2004) Noise minimization in eukaryotic gene expression. PLoS Biol 2: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Guo L, Hu-Li J, Paul WE (2004) Probabilistic regulation of IL-4 production in Th2 cells: accessibility at the Il4 locus. Immunity 20: 193–203 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC (2002) MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol 3: 30–40 [DOI] [PubMed] [Google Scholar]
- Hegazy A, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, Flatz L, Pinschewer D, Radbruch A, Löhning M (2010) Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 32: 116–128 [DOI] [PubMed] [Google Scholar]
- Hofer T, Nathansen H, Lohning M, Radbruch A, Heinrich R (2002) GATA-3 transcriptional imprinting in Th2 lymphocytes: a mathematical model. Proc Natl Acad Sci USA 99: 9364–9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17: 2205–2232 [DOI] [PubMed] [Google Scholar]
- Holländer GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, Terhorst C, Wishart W, Golan DE, Bhan AK, Burakoff SJ (1998) Monoallelic expression of the interleukin-2 locus. Science 279: 2118–2121 [DOI] [PubMed] [Google Scholar]
- Hu-Li J, Pannetier C, Guo L, Lohning M, Gu H, Watson C, Assenmacher M, Radbruch A, Paul WE (2001) Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity 14: 1–11 [DOI] [PubMed] [Google Scholar]
- Hume DA (2000) Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood 96: 2323–2328 [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Kelly BL, Locksley RM (2000) Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol 165: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Kupfer A, Mosmann TR, Kupfer H (1991) Polarized expression of cytokines in cell conjugates of helper T cells and splenic B cells. Proc Natl Acad Sci USA 88: 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Rao A (2004) Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci USA 101: 16010–16015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh C, Carew JA, Kim J, Hogan PG, Rao A (1996) T-cell receptor stimulation elicits an early phase of activation and a later phase of deactivation of the transcription factor NFAT1. Mol Cell Biol 16: 3945–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D (2002) Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110: 261–271 [DOI] [PubMed] [Google Scholar]
- Losick R, Desplan C (2008) Stochasticity and cell fate. Science 320: 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesner U, Albrecht I, Janke M, Doebis C, Loddenkemper C, Lexberg MH, Eulenburg K, Kreher S, Koeck J, Baumgrass R, Bonhagen K, Kamradt T, Enghard P, Humrich JY, Rutz S, Schulze-Topphoff U, Aktas O, Bartfeld S, Radbruch H, Hegazy AN et al. (2008) Autoregulation of Th1-mediated inflammation by twist1. J Exp Med 205: 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Openshaw P, Murphy EE, Hosken NA, Maino V, Davis K, Murphy K, O'Garra A (1995) Heterogeneity of intracellular cytokine synthesis at the single-cell level in polarized T helper 1 and T helper 2 populations. J Exp Med 182: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM (2000) Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12: 27–37 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Pirone JR, Elston TC (2004) Fluctuations in transcription factor binding can explain the graded and binary responses observed in inducible gene expression. J Theor Biol 226: 111–121 [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ (2007) Timescales of genetic and epigenetic inheritance. Cell 128: 655–668 [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG (1997) Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 15: 707–747 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Lohning M, Radbruch A (1999) Instruction for cytokine expression in T helper lymphocytes in relation to proliferation and cell cycle progression. J Exp Med 190: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere I, Sunshine MJ, Littman DR (1998) Regulation of IL-4 expression by activation of individual alleles. Immunity 9: 217–228 [DOI] [PubMed] [Google Scholar]
- Schulz EG, Mariani L, Radbruch A, Hofer T (2009) Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity 30: 673–683 [DOI] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U (2006) Variability and memory of protein levels in human cells. Nature 444: 643–646 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T (2007) Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev 17: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE (2008) Mechanism and regulation of class switch recombination. Annu Rev Immunol 26: 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 98: 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, Pannetier C, Grutz G, Walter J, Paul WE, Radbruch A (2005) A critical control element for interleukin-4 memory expression in T helper lymphocytes. J Biol Chem 280: 28177–28185 [DOI] [PubMed] [Google Scholar]
- Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DI (1995) Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA 92: 7125–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW (2006) Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol 177: 4426–4435 [DOI] [PubMed] [Google Scholar]
- Zhu J, Paul WE (2008) CD4 T cells: fates, functions, and faults. Blood 112: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This archive contains the Matlab scripts for the simulations of the IL-4 stochastic model.
Table of Contents- Mathematical appendix 1: Post-transcriptional memory time- Mathematical appendix 2: Transcription factor activity and occupancy of the binding site- Mathematical appendix 3: Fraction and mean expression value of IL-4 positive cells- Supplementary figures legends- Supplementary information references- Figure S1- Figure S2- Figure S3- Figure S4- Figure S5- Figure S6- Figure S7- Figure S8- Figure S9- Figure S10- Figure S11- Figure S12- Figure S13- Figure S14- Figure S15