Abstract
Archaea, which represent a large fraction of the phylogenetic diversity of organisms, are prokaryotes with eukaryote-like basal transcriptional machinery. This organization makes the study of their DNA-binding transcription factors (TFs) and their transcriptional regulatory networks particularly interesting. In addition, there are limited experimental data regarding their TFs. In this work, 3,918 TFs were identified and exhaustively analyzed in 52 archaeal genomes. TFs represented less than 5% of the gene products in all the studied species comparable with the number of TFs identified in parasites or intracellular pathogenic bacteria, suggesting a deficit in this class of proteins. A total of 75 families were identified, of which HTH_3, AsnC, TrmB, and ArsR families were universally and abundantly identified in all the archaeal genomes. We found that archaeal TFs are significantly small compared with other protein-coding genes in archaea as well as bacterial TFs, suggesting that a large fraction of these small-sized TFs could supply the probable deficit of TFs in archaea, by possibly forming different combinations of monomers similar to that observed in eukaryotic transcriptional machinery. Our results show that although the DNA-binding domains of archaeal TFs are similar to bacteria, there is an underrepresentation of ligand-binding domains in smaller TFs, which suggests that protein–protein interactions may act as mediators of regulatory feedback, indicating a chimera of bacterial and eukaryotic TFs’ functionality. The analysis presented here contributes to the understanding of the details of transcriptional apparatus in archaea and provides a framework for the analysis of regulatory networks in these organisms.
Keywords: transcription factors, protein families, archaeal genomes, evolution, gene regulation
Introduction
Regulation of gene expression at the transcriptional level is a ubiquitous and fine-tuned process observed in all cellular organisms. The ability to respond and adapt to environmental changes is defined by the cell’s repertoire of DNA-binding transcription factors (TFs) through interactions between the TFs and the cis-regulatory regions of their target genes in the form of a transcriptional regulatory network (Babu et al. 2004; Janga and Collado-Vides 2007). These TFs bind to the promoter regions of specific genes to, either positively or negatively, regulate expression. Due to the crucial role of TFs in coordinating the gene expression kinetics of a genome, they have been studied in many aspects, including mutational analysis, sequence comparisons, and elucidation of numerous 3D structures.
The identification of the TF repertoire in a genome sequence is a prerequisite to understanding the regulation of gene expression and, on a global scale, for the elucidation of regulatory networks. In this context, the organisms with the best studied transcriptional regulatory networks, where TFs have been identified, are the eukaryote Saccharomyces cerevisiae (Lee et al. 2002; Janga et al. 2008) and the bacteria Escherichia coli K12 (Babu and Teichmann 2003; Gama-Castro et al. 2008), Bacillus subtilis (Moreno-Campuzano et al. 2006; Sierro et al. 2008), and more recently Corynebacterium glutamicum (Brune et al. 2005; Brinkrolf et al. 2006). However, relatively, little is known about TFs and the transcriptional regulatory networks controlled by them in archaeal genomes, despite the fact that they represent a large fraction of the phylogenetic diversity of organisms. Furthermore, archaea are well suited as model organisms for eukaryotes because of the similarities they share in their information transfer machinery, due to a common ancestor, as proposed by the symbiotic theory (Martin and Muller 1998; Moreira and Lopez-Garcia 1998; Lopez-Garcia 1999; Martin et al. 2001; Esser and Martin 2007).
Archaea constitute one of the three cellular domains in the universal tree of life (Woese 1998) composed of organisms highly diverse in morphology, physiology, and natural habitats (Chaban et al. 2006; Clementino et al. 2007; Nam et al. 2008; Auguet et al. 2009). Organisms included in this cellular domain possess basal transcription machinery resembling that of eukaryotes. For instance, archaea include a TATA box promoter sequence, a TATA box–binding protein (TBP), a homologue of the transcription factor TFIIB (TFB), and a RNA polymerase (RNAp) containing between 8 and 13 subunits (Goede et al. 2006) (see supplementary fig. S1, Supplementary Material online). In contrast, archaeal messenger RNAs (mRNAs) are structurally similar to bacterial mRNAs, and, most importantly, the majority of identified TFs in archaeal organisms are homologous to bacterial activators and repressors (Kyrpides and Woese 1998; Bell 2005). Indeed, very few eukaryotic-like TFs were found to occur in archaea (Kruger et al. 1998). These observations raise different basic questions with regard to the mechanisms of transcriptional regulation and the manner by which bacterial-like TFs may interact or interfere with the components of the eukaryotic-like basal transcriptional machinery within an archaeal cell. It is for this reason that archaeal DNA-binding TFs represent an important class of proteins to explain the molecular mechanisms that underlie transcription regulation. Even though the ever-growing number of archaeal genome sequences reveals an increasing list of potential regulators (Coulson et al. 2007; Wu et al. 2008), archaeal transcriptional regulation is still poorly documented, and the most detailed and advanced studies have been performed with only a dozen TFs, mainly from the AsnC family (formerly feast/famine protein family) (see supplementary table S1, Supplementary Material online) (Napoli et al. 1999; Leonard et al. 2001; Bell 2005). Initial sequence analysis–based attempts using family-specific models from E. coli TFs resulted in a low proportion of bacterial-like TFs in archaea (Perez-Rueda et al. 2004; Coulson et al. 2007). One probable cause for this discrepancy could be that archaeal TF zregulatory repertoire includes additional classes of DNA-binding motifs not observed in E. coli, suggesting that our current knowledge on the repertoire of TFs in archaeal genomes is far from being complete. Importantly, comparative genomic analysis of archaea represents an opportunity to fill in this gap and is an indispensable step toward our understanding of gene regulation networks in prokaryotes and eukaryotes.
In the present study, an exhaustive analysis of gene sequences from 52 completely sequenced archaeal genomes to identify potential DNA-binding TFs was performed. In addition, a comparative analysis was carried out to deduce the distribution of TFs and their evolutionary families among the archaeal genome sequences. Using this repertoire of TFs, we show that 1) there is an underrepresentation of the number of TFs in these organisms compared with bacterial genomes, 2) a considerable number of TFs encode for short polypeptides with a significant fraction encoding for single-domain proteins, and 3) a high proportion of TFs are homologous between archaea and bacteria, mainly from the class clostridia of firmicutes.
Materials and Methods
List of Archaeal Genomes Analyzed in This Study
The archaeal genomes analyzed in this work are as follows (see supplementary table S2, Supplementary Material online, for a more detailed annotation of the genomes): Crenarchaea (C): Aeropyrum pernix K1, Caldivirga maquilingensis IC-167, Hyperthermus butylicus DSM 5456, Ignicoccus hospitalis KIN4/I, Metallosphaera sedula DSM 5348, Nitrosopumilus maritimus SCM1, Pyrobaculum aerophilum str. IM2, Pyrobaculum arsenaticum DSM 13514, Pyrobaculum calidifontis JCM 11548, Pyrobaculum islandicum DSM 4184, Staphylothermus marinus F1, Sulfolobus acidocaldarius DSM 639, Sulfolobus solfataricus P2, Sulfolobus tokodaii str. 7, Thermofilum pendens Hrk 5, Thermoproteus neutrophilus V24Sta; Euryarchaea (E): Methanocorpusculum labreanum Z, Methanoculleus marisnigri JR1, Methanopyrus kandleri AV19, Methanosaeta thermophila PT, Methanosarcina acetivorans C2A, Methanosarcina barkeri str. Fusaro, Methanosarcina mazei Go1, Methanosphaera stadtmanae DSM 3091, Methanospirillum hungatei JF-1, Methanothermobacter thermautotrophicus str. Delta H, Natronomonas pharaonis DSM 2160, Picrophilus torridus DSM 9790, Pyrococcus abyssi GE5, Pyrococcus furiosus DSM 3638, Pyrococcus horikoshii OT3, Thermococcus kodakarensis KOD1, Thermoplasma acidophilum DSM 1728, Thermoplasma volcanium GSS1, uncultured methanogenic archaeon RC-I, Methanocaldococcus jannaschii DSM 2661, Methanococcoides burtonii DSM 6242, Methanococcus aeolicus Nankai-3, Methanococcus maripaludis C5, Methanococcus maripaludis C6, Methanococcus maripaludis C7, Methanococcus maripaludis S2, Methanococcus vannielii SB, Archaeoglobus fulgidus DSM 4304, Candidatus Methanoregula boonei 6A8, Haloarcula marismortui ATCC 43049, Halobacterium salinarum R1, Halobacterium sp. NRC-1, Haloquadratum walsbyi DSM 16790, Methanobrevibacter smithii ATCC 35061; Korarchaeota (K): Candidatus Korarchaeum cryptofilum OPF8; Nanoarchaeum (N): Nanoarchaeum equitans Kin4-M.
Identification of DNA-Binding TFs
To identify and analyze the repertoire of TFs in 52 archaeal genome sequences, we used a combination of information sources and bioinformatics tools. First, 1,820 putative TFs were collected from Transcription Factor DB (Kummerfeld and Teichmann 2006), a database comprising computationally derived predictions of DNA-binding TFs using the SUPERFAMILY library and Pfam hidden Markov models (HMMs). From this data set, 223 proteins, annotated as transposases, invertases, and integrases, were manually excluded. In brief, this exclusion was based on sequence comparisons against the National Center for Biotechnology Information’s nonredundant (NR) protein database (E value = 10−3) by using Blast search followed by the identification of protein domains with CD-search (E value = 10−3) (Marchler-Bauer et al. 2007).
In the second phase, 90 family-specific HMMs previously reported for E. coli K12 (Perez-Rueda et al. 2004) and 57 family-specific HMMs for B. subtilis (Moreno-Campuzano et al. 2006) were used to scan the whole 52 archaeal genome sequences (E value threshold = 10−3), with the hmmsearch module from HMMer suite of programs (http://HMMER.wustl.edu). Briefly, these HMMs were constructed by using the previously identified TF families in E. coli K12 and B. subtilis as seeds, considering every protein family’s DNA-binding domain (DBD) sequences (around 60 amino acids). Proteins with less than 50% similarity in the DNA-binding region against their corresponding HMM were excluded. At this stage, 424 proteins were identified as potential TFs. This was an important step to explore potential TFs not identified in the first step and vice versa, that is, the coverage of superfamily and Pfam assignations correspond to approximately 70% of the universe of TFs, whereas the rest were complemented with these family-specific HMMs.
In the third phase, 70 new TFs were identified with HMMs constructed from 17 proteins annotated as TFs and not identified in previous searches. This step essentially involved retrieving these 17 TFs from Haloweb server (http://halo4.umbi.umd.edu/cgi-bin/haloweb/nrc1.pl?operation=nrc1), and using them as sequence seeds in Blast searches to retrieve homologous sequences from the NR database with an E value = 10−3. Redundancy was removed using CD-hit (Li and Godzik 2006) at 90%, and the potential DBD was identified with CD-search (Marchler-Bauer et al. 2007) (varying the E value from 10−3 to 10−1) in the remaining proteins. This region was then aligned using ClustalW, with parameters set to default and manually editing output. Finally, 14 HMMs were constructed with the HMMer suite of programs corresponding to the 17 proteins clustered by sequence similarity into 14 different groups. For two proteins, there was not enough information to construct a HMM as they appeared to be lineage specific and no homologues were identified.
In addition, a HMM corresponding to the helix-turn-helix (HTH) DNA-binding motif kindly provided by Yan (2006) was used to identify 686 HTH proteins in the archaeal genomes. This data set was also filtered to exclude those proteins described as transposases, ligases, synthases, synthetases, TFIIB, and TFIIE and those proteins identified in the previous phases, resulting in a total of 95 new probable TFs. Finally, Clusters of orthologous genes (COG) assignations associated to TFs in archaea were also used to retrieve new potential archaeal TFs. This resulted in 491 proteins, which were filtered and compared against the whole data set of predictions, but only 2 of them were found to be novel predictions.
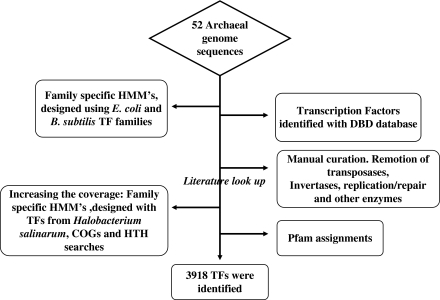
All data sets were finally compared and a total set of 3,918 proteins were compiled and used in this study as the final collection of TFs (see fig. 1 for a summary of the steps). This collection of proteins was classified into 75 families by using HMMs deposited in the Pfam DB (Finn et al. 2006) and searches with CD-search server (E value = 10−1) and aligned against their corresponding models by using the program hmmalign from HMMer.
FIG. 1.
Flowchart showing the different steps involved in the identification of high confidence set of archaeal TFs. Branch points on the vertical line from top to bottom correspond to the stage at which a particular step was taken in the process of obtaining a cleaner dataset.
Identification of Homologous DNA-Binding TFs in Bacteria and Eukarya
In order to identify TFs, which are homologous to the archaeal set, we compared the whole repertoire against 291 NR genome sequences (Moreno-Hagelsieb and Janga 2008), which included bacterial, archaeal, and eukaryotic sequences. A protein was considered as a homologue of a TF in a given genome if the alignment covered at least ≥60% of the query sequence with an E value ≤10−6.
Results and Discussion
Identification of DNA-Binding TFs in Archaea
To understand the distribution of TFs in 52 archaeal genomes (34 Euryarchaea, 16 Crenarchaeota, 1 Korarchaeota, and Nanoarchaeota each), we used a HMM-based strategy in two steps. In the first step, we used a battery of family-specific HMMs (see Materials and Methods for details) and DBD assignments characteristic of TFs to scan the archaeal genomes (see fig. 1 for a complete outline). These steps allowed the detection of 3,751 TFs in 52 genomes (see Materials and Methods for a complete list of genomes analyzed), including 53 of the 72 TFs (75%) from Halobacterium sp. NRC-1 described so far in the Haloweb server. Halobacterium sp. NRC-1 is one of the few archaea whose TF repertoire has been extensively analyzed, and thus, we used its TFs repertoire as a benchmark. In the second step, in order to increase the sensitivity, the 19 Halobacterium sp. NRC-1 TFs not identified in the first step were used as seeds for Blast searches against the NR database (E value cutoff = 10−3), and the matched proteins were used to build new HMMs for a second round of searches, identifying 70 new TFs. Additionally, archaeal genomes were scanned to look for HTH and COG annotations to identify new potential TFs not identified previously. Because it is known that HTH is one of the most prominent structure associated with TFs in prokaryotes (Perez-Rueda and Collado-Vides 2000, 2001), with at least 80% of the TFs containing this DNA-binding structure, we employed a specific HMM, which considers amino acid residue identity and solvent accessibility, constructed from a set of heterogeneous DNA-binding proteins with standard HTH motifs (Yan 2006). After manually excluding proteins that, although can bind to DNA, are unlikely to be TFs, 97 potential TFs that escaped our HMM-based searches were identified. This composite strategy allowed the detection of additional 167 potential archaeal TFs not identified previously and included all the 72 TFs described in Halobacterium sp. NRC-1. In total, a set of 3,918 potential TFs in 52 archaeal genomes were finally identified.
Although extensive survey performed in this work identified a large set of TFs widely distributed in archaea, it is still possible that some potential novel TFs escaped the search criteria or are missing because of their linage-specific nature, presumably due to de novo invention of TFs whose DNA-binding models are not included in our seed dataset.
Dissecting the Repertoire of TFs
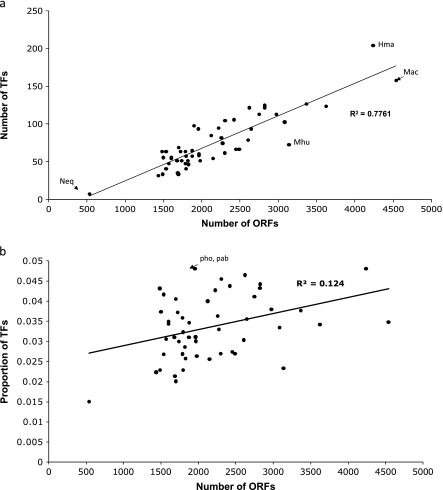
Comprehensive identification and characterization of the repertoire of TFs across archaeal genomes are the first step toward expanding the possibilities for exploration of their regulatory networks. Based on our predictions, we found that smaller archaeal genomes contain fewer TFs than larger ones, following a linear correlation (r2 = 0.82), as has been previously reported for bacteria (Perez-Rueda et al. 2004; fig. 2a). This finding might represent either an expansion or a contraction of the repertoire of TFs in archaea, as a consequence of adaptation to particular habitats or lifestyles. Although larger genomes might be harboring ampler repertoire of TFs to exploit diverse or more complex habitats, smaller genomes containing fewer regulators might be associated with specific niches. For instance, E. coli, which thrives on a large number of sugars, was found to harbor a higher number of TFs compared with B. subtilis, which is similar in genome size (Janga and Perez-Rueda 2009). Likewise, we found that the symbiotic hyperthermophile, N. equitans, has both a reduced genome and a lower proportion of TFs than other archaea, whereas Haloarcula marismortui, a chemoheterotrophic halophilic archaea, was found to have the highest proportion of TFs and Methanosarcina acetivorans (an aerobic chemolitho(aceto)autotrophic methanogen, nitrogen fixing) with one of the largest genomes contained the highest the number of TFs among archaeal genomes sequenced so far. An interesting case is that of Methanospirillum hungatei, a methanogenic archaea reported to have an unusual filamentous structure, which was found to have the lowest proportion of TFs after N. equitans among the archeael genomes studied. Complex lifestyles might require a higher proportion of genes and TFs to better orchestrate responses to changing environments, as is the case of Methanosarcina acetivorans that can form aggregate multicellular structures when passing from anaerobiosis to aerobiosis (Oelgeschlager and Rother 2008) or the case of Haloarcula marismortui, a halophilic archaea, which are generally described to be surprisingly different in its nutritional demands and metabolic pathways (Falb et al. 2008). In fact, the proportion of TFs in larger genomes is consistent with the hypothesis that an increase of genome complexity and physiological functionality is generally associated with a more complex regulation of gene expression (Woese 1998).
FIG. 2.
a) Distribution of TFs identified in 52 archaeal genomes. Nanoarchaeum equitans (Neq), Haloarcula marismortui (Hma), Methanospirillum hungatei (Mhu), and Methanosarcina acetivorans C2A (Mac) are indicated as a reference. On x axis, genomes are sorted from smallest to largest size and on y axis the number of TFs is plotted. A linear regression was calculated using the Pearson correlation (r2) between the number of genes and the total number of TFs. b) Proportion of TFs in all the archaeal genomes. Proportion of TFs was calculated as the fraction of ORFs encoding for TFs and plotted against the total number of ORFs for each genome. Pyrococcus horikoshii (pho) and Pyrococcus abyssi (pab) are indicated as a reference. On x axis, genomes are sorted from smallest to largest size and on y axis, the fraction of TFs is plotted.
In this context, the number of predicted TFs in archaea is variable (see supplementary table S2, Supplementary Material online), ranging from 8 in the archaeon with the smallest sequenced genome (N. equitans) to up to 158 TFs in the largest genome, Methanosarcina acetivorans C2A. A closer look into the normalized distribution of TFs calculated as the proportion of the genes coding for TFs gave further insights into the evolution of TFs in the context of their genome size and lifestyles. For instance, as shown in figure 2b, less than 5% of the open reading frames (ORFs) in most archaeal genomes are devoted to gene regulation in contrast to about 8–10% observed in bacterial genomes with similar number of ORFs (Perez-Rueda and Collado-Vides 2000, 2001). Indeed, larger archaeal genomes, such as Methanosarcina acetivorans and Haloarcula marismortui, with similar number of ORFs to E. coli K12, encode a lesser proportion of TFs (4.8%, 3.5%, and 8%, respectively). Thus, the TF repertoire observed in archaea is much more similar to bacteria associated with gene loss events, such as intracellular pathogens and endosymbionts (3.9% in average). Notable exceptions are Pyrococcus horikoshi and Pyrococcus abyssi, two small genomes containing 4.8% and 5.1% of TFs, respectively, comparable with the proportion of TFs in larger archaeal genomes. In contrast, N. equitans, which was found to follow the trend in figure 2a, exhibited a clear deviation when proportion of genes coding for TFs was compared against genome size.
Although this intriguingly low proportion of TFs in archaea compared with bacteria could be partially explained due to our inability to identify those lineage or organism-specific TFs, it is also possible to suggest that other regulatory strategies in this cellular domain might be compensating for this underrepresentation. These could involve, for example, formation of alternative TBP–TFB–RNAp complexes, with the possibility of interactions with different accessory factors (Baliga et al. 2000; Facciotti et al. 2007). However, the existence of new classes of TFs not explored here or archaeal-specific regulatory mechanisms cannot be excluded to be responsible for this trend. For instance, it has been shown recently from a global analysis of translationally regulated genes in Halobacterium salinarum and Halobacterium volcanii that 20% and 12% of all genes in these genomes show growth phase–dependent differential translational regulation (Lange et al. 2007). However, the overlap between the two sets was found to be negligible, indicating that archaeal organisms may use differential translational control for regulation of gene expression, adding a layer of regulatory complexity at posttranscriptional level (Mittal et al. 2009). Therefore, regulatory strategies that are found exclusively in archaea or those that are exploited to a greater extent in archaea compared to bacteria might be responsible for these differences.
Archaeal Genomes Encode a Large Proportion of Small TFs
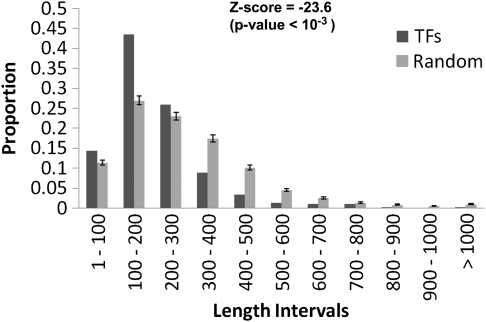
Transcription regulation in archaea appears to be a chimera, with general TFs being clearly eukaryote-like and candidates for regulating specific responses being bacterial like (Aravind and Koonin 1999). We found that a large proportion (43.5%) of TFs in the archaeal genomes were small in size (100–200 amino acids). In contrast, 42% of the bacterial TFs have between 200 and 300 amino acids (vs. 26.5% of the archaeal TFs with this length). Nonetheless, 287 large TFs with amino acid length greater than 400, corresponding to about 2.3%, were identified in the archaeal repertoire (fig. 3). To determine the significance of these findings, we randomly sampled 1,000 collections of 3,918 proteins from the archaeal genome sequences and compared their lengths with those observed in TFs. As the distribution of average length of proteins in the random samples followed a normal distribution, a Z score was used as a test statistic. Z score was calculated as the number of standard deviations the observed value (average length of an archaeal TF) is away from the mean of the 1,000 random collections. This is obtained as the ratio of the difference between the observed, x, and the random expected, μ, values to the standard deviation, σ, that is, Z = (x − μ)/σ. P value was defined as the fraction of the 1,000 random collections that showed an average length greater than or equal to what was observed in the archaeal TF collection. Using this approach for the TF population, a Z score of −23.6 (corresponding to a P value <10−3) was found, indicating that TFs in archaea tend to be significantly smaller than the overall proteome. In contrast, the repertoire of TFs in E. coli K12 does not exhibit such a tendency compared with the rest of the proteome (see supplementary fig. S2, Supplementary Material online). In fact, a higher proportion of TFs in E. coli are generally longer compared with other proteins, indicating that archaeal TFs are indeed encoded as small genes. To test whether this observation is more general, we compared the lengths of archaeal TFs against a complete set of bacterial TFs available from the DBD database (Kummerfeld and Teichmann 2006). We found that archaeal TFs showed significantly lower lengths compared with bacterial ones (median size of 179 vs. 236 amino acids, P < 2.2 × 10−16, Wilcoxon test; see supplementary fig. S3, Supplementary Material online). Because three of the abundant families, ArsR, AsnC, and HTH_3, were found to be composed of small proteins contributing to about 40% of the total TF repertoire (see below), to exclude the possibility that these large families are indeed responsible for this tendency, we excluded this set of TFs from the complete collection and compared their length distribution with bacterial TFs. This comparison clearly revealed that independent of these large families archaeal TFs show smaller lengths compared with bacterial ones (median size of 190 vs. 236 amino acids, P < 2.2 × 10−16, Wilcoxon test; see supplementary fig. S3, Supplementary Material online). These observations raise the question, if archaeal TFs are shorter than bacterial TFs, do they also encode for smaller number of domains? To address this, we compared the number of domains archaeal TFs possess in comparison with those seen for bacterial ones by obtaining all those TFs for which superfamily domain assignments were available (Madera et al. 2004). Of the 2,621 archaeal TFs for which domain assignments were available, we found that 1,963 comprised single-domain proteins (∼75%), whereas single domain containing TFs in bacteria comprised 50% of the total data set analyzed. Further analysis of the distributions of the number of domains in TFs of both the major kingdoms of life unambiguously revealed that archaeal TFs encode for lesser number of domains independent of the exclusion of the large archaeal families (P < 2.2 × 10−16, Wilcoxon test). These results clearly unveil that archaeal TFs comprise a significant proportion of single-domain proteins. One possibility is that most of these one-domain proteins encode for a DBD and might not contain a ligand-binding domain, suggesting that although archaeal TFs contain DBDs similar to bacteria, their mechanism of action might be similar to eukaryotic TFs. In light of these observations, it is possible to hypothesize that archaeal TFs although similar in sequence recognition domains with bacteria (discussed below) might be similar to eukaryotic TFs in mechanistic sense.
FIG. 3.
Distribution of amino acid sequence lengths for TFs. On x axis, the intervals of protein size are shown and on y axis, the normalized frequency of TFs per interval is shown. Thousand groups of 3,918 protein sequences were randomly retrieved from archaeal genome sequences to compare the length distribution of TFs against other protein-coding genes. In each length internal, bars marked as random represent the proportion of proteins in an interval ± their standard deviations from the average in the random samples.
The high proportion of small TFs in archaea together with the observation that most archaea have few TFs per genome also suggests a dense combinatorial interplay of TFs for mediating regulation. These data support various possible scenarios namely 1) regulation similar to bacteria, where homodimers can regulate gene expression; 2) formation of different oligomeric assemble forms affected by the interaction with metabolites associated to a particular metabolic state, that is, the formation of oligomers with different sizes, that is, dimers, tetramers, octamers, and so on, as has been observed for the members of the AsnC family (with an average length of around 160 amino acids), whose small TFs can form dimers, tetramers, or octamers with differing regulatory functions (Koike et al. 2004), such as FL11 of Pyrococcus sp., which can form a disc or a chromatin-like cylinder upon interaction of two peptides and TrmB of Pyrococcus furiosus, which is tetrameric at ambient temperature and octameric in the presence of its inducer (maltotriose or maltose) (Lee et al. 2005; Krug et al. 2006); 3) binding of the same protein to a broad spectrum of compounds or ligands, enhancing its activity under different metabolic states, such as TrmB that binds maltose, sucrose, maltotriose, and trehalose compounds in decreasing order of affinity (Koike et al. 2004; Lee et al. 2005); and 4) alternative physical interactions or cocomplex memberships with TBP–TFB–RNAp can also be modulating the structure of the regulatory network in archaea similar to eukarya. In this regard, Facciotti et al. found with protein coimmunoprecipitation, ChIP-Chip, global transcriptional factor (GTF) perturbation and knockout, and measurement of transcriptional changes that global transcriptional factors can associate to nearly half of all putative promoters and show evidence for at least 7 of the 42 possible functional GTF pairs (Baliga et al. 2000; Facciotti et al. 2007).
Phylogenetic Distribution of TFs in Archaea
It has been previously proposed that DNA-binding TFs can be grouped into families based on their amino acid sequence similarity (Perez-Rueda and Collado-Vides 2000). In order to determine the number of TF families associated with archaeal genomes, all the 3,918 DNA-binding TFs were grouped into 75 families according to the Pfam database (Finn et al. 2006). As elaborated below, we explored the familial abundance in the archaeal genomes and the relative contribution of each family to the proteome size and overall proportion of TFs. This analysis also enabled us to determine the families that are shared between archaea, bacteria, and eukarya and the main functions of these families.
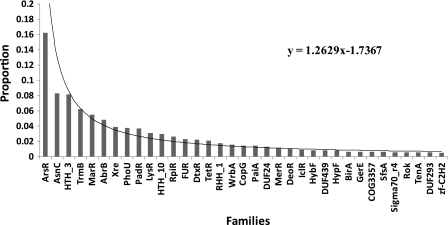
The population of TF families was found to follow a power-law distribution, with 13 families containing more than 100 members each, representing 71% of the whole TF repertoire (fig. 4). The top three most populated families are ArsR (721 TFs), the HTH_3 (361 TFs), and the AsnC (367 TFs), whereas other ten families contained between 101 and 276 TFs. About 49 families comprised less than 30 TFs each, representing in total ∼11% of the TF repertoire. Previous analysis (Moreno-Campuzano et al. 2006; Janga and Perez-Rueda 2009) suggests that global regulators (GRs) in bacteria usually belong to small families; however, in Archaea apparently, this is not the case, at least for the GRs identified so far. For instance, ArsR and TrmB were found to belong to two large families with 721 and 276 members, respectively.
FIG. 4.
Abundance of TF families in archaeal genomes. Proportion of TFs in each family was calculated as the fraction of total TFs identified that belonged to a particular family. The families are displayed from largest to smallest size. Families with less than 20 members were not displayed as they corresponded to less than 6% of the total dataset.
Figure 5 shows that four families are universally distributed across the four archaeal divisions (Crenarchaea, Euryarchaea, Nanoarchaea, and Korarchaea) namely: the HTH_3 (a family of putative activator proteins), AsnC (associated with global regulation of amino acid biosynthesis), TrmB (maltose-specific regulation), and ArsR (detoxification process). These families might belong to the ancestral core of TFs in archaea. A second group of families (PhoU and RpiR) was detected in all archaeal genomes, with the exception of the endosymbiont, N. equitans, and hence can also be considered as part of the archaeal TF core set. These families are mainly putative regulators of phosphate uptake (PhoU) and sugar metabolism (RpiR). Based on these findings, it is possible to suggest that archaea from new divisions might carry on TFs from these universal families, potentially regulating central metabolic processes, as might be the case with the last common ancestor of archaea. Some families such as TrpR were found exclusively in Metallosphaera sedula, and CopY was found in diverse Halobacterium strains suggesting that they might have been transferred laterally from bacteria to archaea.
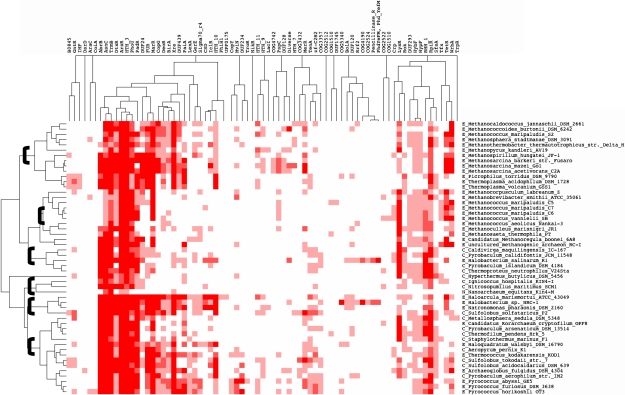
FIG. 5.
Clustering of TF families and archaeal genomes. A hierarchical centroid linkage-clustering algorithm was applied with uncentered correlation as the similarity measure and complete linkage (Eisen et al. 1998). Brackets indicate the clusters identified by using a correlation value ≥0.6. Nomenclature is as follows: Crenarchaea (C); Euryarchaea (E); Korarchaeota (K), and Nanoarchaeum (N).
It is possible to speculate from this data that abundant families like ArsR, AsnC, or HTH_3 might be a consequence of the lifestyles and a response to the deficit of TFs, that is, archaea might have expanded certain families associated with small sizes, to generate a plethora of combinatorial possibilities to regulate their gene expression. It is noteworthy to mention in this context that these three families contribute to around 40% of the total TFs with length between 100 and 200 amino acids.
In order to understand the similarity of TF repertoires per family among the archaeal genomes, a hierarchical centroid linkage-clustering algorithm (Eisen et al. 1998) was applied with uncentered correlation as the similarity measure. The clustering results were visualized using the treeview program (Saldanha 2004). From this clustering, six groups of archaea sharing a common set of TFs were identified (based on a node correlation value ≥0.6), whereas three organisms could not be included in any cluster and were hence considered as orphans (see fig. 5). It is evident from this analysis that these six clusters reflect the major taxonomic positions of the organisms analyzed, although some exceptions could be observed. The TF repertoire also reflects the main lifestyle of archaea, such as the first cluster that includes mainly methanogenic archaea (such as Methanocaldococcus jannaschii and Methanococcus maripaludis S2 among others). The intermixing of organisms in some clusters might be a consequence of lateral gene transfer events, as has been suggested for archaea included in the fourth cluster, that is, N. equitans (Nanoarchaeum) and I. hospitalis (Desulfurococcales) (Podar et al. 2008).
Comparison of the TF Repertories of Bacteria and Archaea
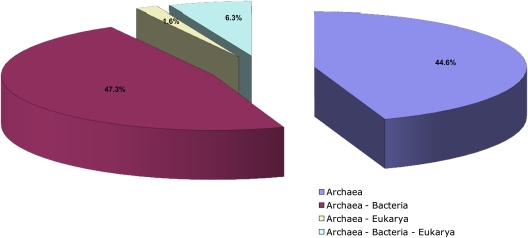
It has been proposed that bacteria and archaea share a great similarity at gene regulatory level (Aravind and Koonin 1999), with archaeal TFs clearly being bacterial like, whereas their basal transcriptional machinery clearly associated to eukarya. Thus, to understand the degree of conservation of TFs between archaea, bacteria, and eukarya, the probable homologues of the repertoire of transcriptional regulators were identified (see Materials and Methods). From this analysis, it was found that 53% of the 3,918 archaeal TFs exhibit at least one homologue in bacterial genomes (fig. 6). In particular, archaea and clostridia share TFs from the families HTH_3, Xre, and Rrf2, whereas TFs from the families DeoR, IclR, and cold shock are shared with several actinobacteria and some gammaproteobacteria. Another 45% of the 3,918 TFs were clearly identified as archaeal specific, whereas other 6% exhibited homology with bacterial and eukaryotic TFs and about 2% exhibited homology with only eukaryotes (mainly with Ascomycetes) possibly suggesting a lateral gene transfer. This reinforces the notion that TFs of bacteria and archaea share a common ancestry and highlight a close relationship between the TFs from archaea and firmicutes, pointing evidence to drive experiments that can confirm if they share a functional relatedness as well.
FIG. 6.
Distribution of archaeal TFs shared by the three cellular domains, archaea, bacteria, and eukarya. Pie chart showing the distribution of archaeal TF homologues identified in different domains of life; Blast searches were performed between all TFs previously identified against total sequences of bacterial and eukaryotic genomes. A protein was considered as homologue if the alignment covered at least ≥60% of the query sequence, with an E value ≤10−6.
Archaeal TFs Are Predominantly Comprised Bacterial DBDs
An important aspect of TFs is their ability to organize into multidomain proteins and hence understanding them in a structural context can provide important clues about how they coordinate regulation. Therefore, the repertoire of archaeal TFs was analyzed using the library of HMMs deposited in superfamily database (Madera et al. 2004). From this analysis, we found that the most abundant DBD in these TFs is the winged helix DBD, detected in 45% of the total set. The second most abundant binding domain corresponds to the lambda repressor-like DBD (∼15%). This result is similar to that previously observed for the repertoire of bacterial TFs, reinforcing the notion of common ancestry in the transcriptional regulatory machinery of prokaryotes (Aravind and Koonin 1999; Aravind et al. 2005). Alternative DBDs, such as integration host factor-like DBD, PhoU-like domain, nucleic acid–binding domain associated to cold shock proteins or zinc-finger domains, were also identified, although in lower proportions (corresponding to around 12% of the total TFs). Several of these domains were also identified in bacterial TFs. Zinc fingers represent an intriguing result because this class of proteins has been found exclusively in eukaryotic transcriptional proteins.
Most TF families have been found to undergo lineage-specific duplications resulting in the accumulation of particular families in some microbial species, such as LysR family in E. coli (45 TFs; Janga and Perez-Rueda 2009) or ArsR in Methanosarcina acetivorans C2A (48 TFs). Indeed, this hypothesis is consistent with the more general notion that a genome evolves from a set of precursor genes to a mature size by gene duplications and increasing modifications (Yanai et al. 2000; Koonin et al. 2002). Therefore, the domain organization and more generally the properties of the TF repertoire described for archaeal genomes in this study open diverse questions like, if the evolution of regulatory networks in archaea is different to that observed in E. coli, B. subtilis, and/or other biological systems (Aravind and Koonin 1999; Koike et al. 2004; Lee et al. 2005; Lozada-Chavez et al. 2006; Janga et al. 2008, 2009; Perez and Groisman 2009).
Conclusions
In this study, 52 archaeal genome sequences representing a plethora of lifestyles were analyzed to identify the repertoire of proteins involved in controlling the gene expression. Given the fact that there is currently no archaeal genome, which is completely characterized at the level of transcriptional regulation, the repertoire of TFs and the conclusions presented here can be a good starting point in understanding transcriptional regulatory networks in archaeal genomes. In particular, because the archaeal genomes studied here are from different taxa, the results presented here should be valid with high confidence for a wide range of archaea.
Our analysis suggests that although there is a correlation between the number of TFs and genome size, there is also a deficit for TFs in all the archaeal genomes, indicating that this deficit in TFs, and hence, regulatory plasticity is possibly supplemented by their ability to form different assembly structures by small-sized TFs found to be enriched in archaea. We also note that there is an important fraction of transcriptional regulators common to archaea and bacteria. The distribution of TF families common to prokaryotes shows an ancient evolution of transcriptional machinery in bacteria and archaea. We found that the number of TF families is distributed almost homogeneously among all archaea, although there are a small proportion of them that are overrepresented in all archaea but not in bacteria. Further research is necessary to determine the physiological function of such species-specific or shared transcriptional regulators. Nevertheless, the analysis presented here will provide a basis for understanding the organization and evolution of regulatory networks in archaea.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
E.P.R. was financed by a grant (ASTF 224-2005) from EMBO and by a grant (IN-217508) from DGAPA-UNAM. E.P.R. thanks Lorenzo Segovia, Claudia Martinez-Anaya, and Javier Diaz-Mejia for their helpful comments in the preparation of the manuscript and Rosa Maria Gutierrez in the clustering analysis. S.C.J. acknowledges financial support from MRC Laboratory of Molecular Biology and Cambridge Commonwealth Trust. We would also like to thank Nitish Mittal and AJ Venkatakrishnan for critically reading the manuscript and providing helpful comments. TF predictions for all the archaeal genomes analyzed in this study can be obtained from the Web site http://www.mrc-lmb.cam.ac.uk/genomes/sarath/Archtfs/ or upon request from the authors.
References
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet JC, Barberan A, Casamayor EO. Global ecological patterns in uncultured archaea. ISME J. 2009;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Babu MM, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- Bell SD. Archaeal transcriptional regulation—variation on a bacterial theme? Trends Microbiol. 2005;13:262–265. doi: 10.1016/j.tim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Brinkrolf K, Brune I, Tauch A. Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Genet Mol Res. 2006;5:773–789. [PubMed] [Google Scholar]
- Brune I, Brinkrolf K, Kalinowski J, Puhler A, Tauch A. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics. 2005;6:86. doi: 10.1186/1471-2164-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Ng SY, Jarrell KF. Archaeal habitats—from the extreme to the ordinary. Can J Microbiol. 2006;52:73–116. doi: 10.1139/w05-147. [DOI] [PubMed] [Google Scholar]
- Clementino MM, Fernandes CC, Vieira RP, Cardoso AM, Polycarpo CR, Martins OB. Archaeal diversity in naturally occurring and impacted environments from a tropical region. J Appl Microbiol. 2007;103:141–151. doi: 10.1111/j.1365-2672.2006.03230.x. [DOI] [PubMed] [Google Scholar]
- Coulson RM, Touboul N, Ouzounis CA. Lineage-specific partitions in archaeal transcription. Archaea. 2007;2:117–125. doi: 10.1155/2006/629868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Martin W. Supertrees and symbiosis in eukaryote genome evolution. Trends Microbiol. 2007;15:435–437. doi: 10.1016/j.tim.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Facciotti MT, Reiss DJ, Pan M, et al. (11 co-authors) General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci U S A. 2007;104:4630–4635. doi: 10.1073/pnas.0611663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. Metabolism of halophilic archaea. Extremophiles. 2008;12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, et al. (13 co-authors) Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, et al. (19 co-authors) RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede B, Naji S, von Kampen O, Ilg K, Thomm M. Protein-protein interactions in the archaeal transcriptional machinery: binding studies of isolated RNA polymerase subunits and transcription factors. J Biol Chem. 2006;281:30581–30592. doi: 10.1074/jbc.M605209200. [DOI] [PubMed] [Google Scholar]
- Janga SC, Collado-Vides J. Structure and evolution of gene regulatory networks in microbial genomes. Res Microbiol. 2007;158:787–794. doi: 10.1016/j.resmic.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janga SC, Collado-Vides J, Babu MM. Transcriptional regulation constrains the organization of genes on eukaryotic chromosomes. Proc Natl Acad Sci U S A. 2008;105:15761–15766. doi: 10.1073/pnas.0806317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janga SC, Perez-Rueda E. Plasticity of transcriptional machinery in bacteria is increased by the repertoire of regulatory families. Comput Biol Chem. 2009;33:261–268. doi: 10.1016/j.compbiolchem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Janga SC, Salgado H, Martinez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009;37:3680–3688. doi: 10.1093/nar/gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H, Ishijima SA, Clowney L, Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc Natl Acad Sci U S A. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- Krug M, Lee SJ, Diederichs K, Boos W, Welte W. Crystal structure of the sugar binding domain of the archaeal transcriptional regulator TrmB. J Biol Chem. 2006;281:10976–10982. doi: 10.1074/jbc.M512809200. [DOI] [PubMed] [Google Scholar]
- Kruger K, Hermann T, Armbruster V, Pfeifer F. The transcriptional activator GvpE for the halobacterial gas vesicle genes resembles a basic region leucine-zipper regulatory protein. J Mol Biol. 1998;279:761–771. doi: 10.1006/jmbi.1998.1795. [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B alpha-beta-delta subunit families. Proc Natl Acad Sci U S A. 1998;95:3726–3730. doi: 10.1073/pnas.95.7.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Zaigler A, Hammelmann M, Twellmeyer J, Raddatz G, Schuster SC, Oesterhelt D, Soppa J. Genome-wide analysis of growth phase-dependent translational and transcriptional regulation in halophilic archaea. BMC Genomics. 2007;8:415. doi: 10.1186/1471-2164-8-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Moulakakis C, Koning SM, Hausner W, Thomm M, Boos W. TrmB, a sugar sensing regulator of ABC transporter genes in Pyrococcus furiosus exhibits dual promoter specificity and is controlled by different inducers. Mol Microbiol. 2005;57:1797–1807. doi: 10.1111/j.1365-2958.2005.04804.x. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, et al. (21 co-authors) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Leonard PM, Smits SH, Sedelnikova SE, Brinkman AB, de Vos WM, van der Oost J, Rice DW, Rafferty JB. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P. DNA supercoiling and temperature adaptation: a clue to early diversification of life? J Mol Evol. 1999;49:439–452. doi: 10.1007/pl00006567. [DOI] [PubMed] [Google Scholar]
- Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. (25 co-authors) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Mittal N, Roy N, Babu MM, Janga SC. Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc Natl Acad Sci U S A. 2009;106:20300–20305. doi: 10.1073/pnas.0906940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- Moreno-Campuzano S, Janga SC, Perez-Rueda E. Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes—a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Janga SC. Operons and the effect of genome redundancy in deciphering functional relationships using phylogenetic profiles. Proteins. 2008;70:344–352. doi: 10.1002/prot.21564. [DOI] [PubMed] [Google Scholar]
- Nam YD, Chang HW, Kim KH, Roh SW, Kim MS, Jung MJ, Lee SW, Kim JY, Yoon JH, Bae JW. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol. 2008;46:491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- Napoli A, van der Oost J, Sensen CW, Charlebois RL, Rossi M, Ciaramella M. An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus which binds to its own promoter. J Bacteriol. 1999;181:1474–1480. doi: 10.1128/jb.181.5.1474-1480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager E, Rother M. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol. 2008;190:257–269. doi: 10.1007/s00203-008-0382-6. [DOI] [PubMed] [Google Scholar]
- Perez JC, Groisman EA. Evolution of transcriptional regulatory circuits in bacteria. Cell. 2009;138:233–244. doi: 10.1016/j.cell.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rueda E, Collado-Vides J. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J Mol Evol. 2001;53:172–179. doi: 10.1007/s002390010207. [DOI] [PubMed] [Google Scholar]
- Perez-Rueda E, Collado-Vides J, Segovia L. Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput Biol Chem. 2004;28:341–350. doi: 10.1016/j.compbiolchem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Podar M, Anderson I, Makarova KS, et al. (27 co-authors) A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008;9:R158. doi: 10.1186/gb-2008-9-11-r158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. The universal ancestor. Proc Natl Acad Sci U S A. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang S, Bai J, Shi L, Li D, Xu Z, Niu Y, Lu J, Bao Q. ArchaeaTF: an integrated database of putative transcription factors in archaea. Genomics. 2008;91:102–107. doi: 10.1016/j.ygeno.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Yan C. Proceedings of IEEE International Conference on Granular Computing. IEEE Xplore: Los Alamitos (CA); 2006. A hidden Markov model approach to model protein sequence and structural information: identification of helix-turn-helix DNA-binding motif; pp. 385–388. [Google Scholar]
- Yanai I, Camacho CJ, DeLisi C. Predictions of gene family distributions in microbial genomes: evolution by gene duplication and modification. Phys Rev Lett. 2000;85:2641–2644. doi: 10.1103/PhysRevLett.85.2641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.