Abstract
Conformational flexibility of proteins has been linked to their designated functions. Slow conformational fluctuations occurring at microsecond-millisecond time-scale, in particular, have recently attracted considerable interest in connection to the mechanism of enzyme catalysis. Computational methods are providing valuable insights into the connection between protein structure, flexibility and function. In this report, we present studies on identification and characterization of microsecond flexibility of ubiquitin, based on quasi-harmonic analysis (QHA) and normal mode analysis (NMA). The results indicate that the slowest 10 QHA modes, computed from 0.5 μs molecular dynamics ensemble, contribute over 78% of all motions. The identified slow movements show over 75% similarity with the conformational fluctuations observed in nuclear magnetic resonance ensemble and also agree with displacements in the set of X-ray structures. The slowest modes show high flexibility in β1-β2, α1-β3, and β3-β4 loop regions, with functional implications in mechanism of binding other proteins. NMA of ubiquitin structures was not able to reproduce the long time scale fluctuations as they were found to strongly depend on the reference structures. Further, conformational fluctuations coupled to the cis/trans isomerization reaction catalyzed by the enzyme cyclophilin A (CypA), occurring at the microsecond-millisecond time-scale, have also been identified and characterized based on QHA of conformations sampled along the reaction pathway. The results indicate that QHA covers the same conformational landscape as the experimentally observed CypA flexibility. Overall, the identified slow conformational fluctuations in ubiquitin and CypA indicate that the intrinsic flexibility of these proteins is closely linked to their designated functions.
Keywords: Conformational fluctuations, protein dynamics, enzyme catalysis, quasi-harmonic analysis, reaction coupled dynamics
I. INTRODUCTION
Proteins exhibit a wide range of conformational fluctuations, from subtle rearrangements of a few atoms to large-scale movements involving the entire protein. The time-scales for these internal protein motions range from femtoseconds (10−15 s) to microseconds and longer (>10−6 s).1,2 On one side of this range are repeated motions localized in small regions of the protein and involving a small number of atoms. These types of fast motions commonly known as vibrations (examples include bond stretching and angle bending) occur on femtosecond to picosecond time-scales. Motions such as rotations of side-chains and movement of flexible loops occur over longer time-scales, often spanning from a few nanoseconds to the sub-microsecond time-scale. On the other side of this range are the large conformational changes, involving multiple secondary structure elements or even spanning the entire protein, occurring on microsecond (or longer) time-scale. These slow, coherent, collective, and repeated conformational changes, also referred to as protein breathing motions, are different from the random conformational fluctuations.2 Recently, conformational motions in proteins and in particular the slow fluctuations, have attracted considerable interest due to their possible interconnection in enabling a protein to perform its designated function such as enzyme catalysis.1–12
Experimental techniques continue to provide information about protein motions at a variety of time-scales. Neutron scattering has been used to monitor fast thermal motions (picosecond to 100 picoseconds)13–15 nuclear magnetic resonance (NMR) experiments have provided information in the intermediate range (nanoseconds and longer);3,16,17 spin-echo neutron scattering on microsecond-millisecond range;18 hydrogen-deuterium exchange has been used to measure slower conformational changes occurring in proteins (milliseconds);19 and crystallography20 has revealed conformational changes between different states during reaction. Moreover, recent Mössbauer effect and neutron scattering experiments have indicated that the bulk solvent and hydration-shell fluctuations control protein motions and function.21–23 These experimental investigations of protein motions have faced an inherent problem; as mentioned above the internal motions of proteins occur at a wide range of time scales but the range of observed motions strongly depend on the relatively narrow time scale resolution window of the experimental method used.
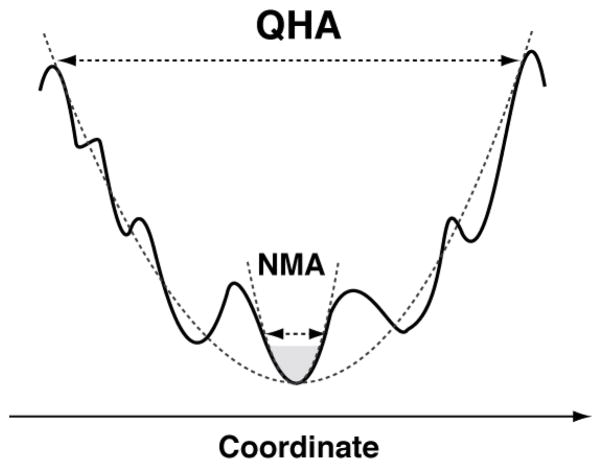
Theoretical and computational methods continue to provide new insights into protein motions, as well as the possible role of these motions in protein function such as enzyme catalysis.1,6,24 Particularly, normal mode analysis (NMA)25–28 and its various extensions29,30 have provided insights into the conformational fluctuations associated with individual protein structures, as well as flexibility intrinsically associated with the overall shape of proteins and its linkage to protein function. In NMA, a harmonic approximation of the potential energy landscape for a protein conformation in a local energy minimum (or its vicinity) is computed by diagonalization of the Hessian matrix.31 Even though NMA provides information about fast and slow protein motions, these are representative of conformational fluctuations observed in the vicinity of the reference structure (see Figure 1).32 Moreover, it is widely discussed that the slow conformational changes in the proteins show a large degree of anharmonicity.33–36 The harmonic approximation limits the use of NMA to small amplitude motions in the potential energy surface associated with a local minimum37 (corresponding to the gray region in Figure 1). Therefore, NMA is not well suited to study conformational changes associated with biochemical processes such as enzyme catalysis, which covers distant areas of the conformational energy landscape. Methods such as time-averaged normal coordinate analysis (TANCA)38 have been developed to overcome some of the inherent limitations of NMA. TANCA provides more reproducible modes by diagonalizing the time-averaged Hessian matrix. The fast motions that differ considerably between the reference structures are removed in TANCA. However, TANCA is computationally very expensive, as it requires calculation of the Hessian for each structure of molecular dynamics (MD) trajectory.
Figure 1. Conformational fluctuations identified by QHA and NMA are based on alternate ways to approximate potential energy surface.
NMA identifies motions in the vicinity of energy minimum, while QHA can identify motions that span distant areas of the energy surface.
Quasi-harmonic analysis (QHA)39 and related principal component analysis techniques,40,41 based on the eigenvalue decomposition of an ensemble of protein conformations, have provided a useful method for identifying motions particularly at long time-scales (see Figure 1). QHA captures the large-scale conformational fluctuations within a collection of protein conformations by diagonalizing the mass-weighted covariance matrix known as the atomic fluctuation matrix (Fαβ). For a system with N atoms, Fαβ is a 3N × 3N symmetric matrix, defined as shown below:
| (Eq. 1) |
where α and β represent the 3N degrees of freedom in Cartesian space; m is the mass of the atom and the quantity within 〈 〉 denotes an average over the ensemble of structures in MD simulation. The inverse square root of the eigenvalues determined by diagonalizing Fαβ represent the frequencies associated with protein eigenmodes. The eigenvectors represent the displacement vectors of the individual atoms. The lowest frequencies correspond to large-scale cooperative motions in the protein; the higher frequencies represent localized motions. For a system with N atoms there are 3N-6 internal modes; however, due to computational reasons typically only a limited number of slow modes (lowest frequencies) are computed. Note, QHA allows identification of protein motions at a variety of time-scales as the atomic fluctuation matrix can be computed from protein conformational sampled during a single MD simulation (short time-scale),42 a collection of MD trajectories (collectively representing long time-scale),43 or a set of conformations obtained using various sampling techniques (which could represent protein present in different stages during its functional cycle).
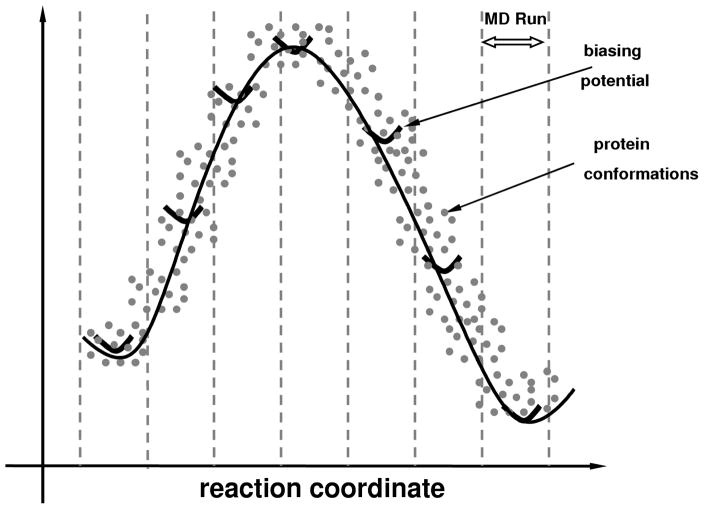
A significant advantage of QHA over other methods is that it allows identification of slow conformational fluctuations that span over distant areas of the protein energy landscape such as the reaction pathway during enzyme catalysis.5,43,44 MD simulations in combination with techniques such as umbrella sampling45 can be used to model the free energy landscape associated with enzyme reaction pathway as a function of a reaction coordinate. As depicted in Figure 2, QHA of the entire set of conformations sampled along the various sections of the reaction coordinates allows identification of conformational fluctuations at the reaction time-scale. Overcoming the limitations of NMA based approaches, QHA provides a methodology to explore protein motions that are associated with longer time-scales.
Figure 2. Methodology for identification of slow conformational fluctuations associated with an enzyme reaction.
A number of MD runs are used to sample the conformations along reaction pathway by using a suitable description of the reaction coordinate and umbrella sampling method. The biasing potentials or umbrella potentials (marked dark black curves) allow sampling of the higher energy regions. The set of conformations (each conformation is indicated by a gray dot) sampled in all MD simulations is used for generation of the free energy profile (black curve) as well as the construction of the covariance matrix for QHA.
Reaction coupled slow conformational fluctuations in a number of enzyme systems including cyclophilin A (CypA) has been successfully identified using the above approach.43,44 In CypA, identification of 3 reaction coupled slow conformational modes that are closely linked to enzyme mechanism has lead to the discovery of a network of coupled vibrations associated with the catalytic mechanism. Detailed characterization of conformational fluctuations in the cis/trans isomerization catalyzed by CypA showed the existence of a network of coupled motions within the protein affecting the catalytic step. These motions (termed as reaction coupled motions) extended all the way from the flexible surface loop regions to the active site of the protein, showing correlated motions even among residues that are located >15 Å away. More recently, we have also discovered that these conformational fluctuations identified with the use of QHA are preserved across enzymes folds catalyzing different types of chemical reactions: cis/trans isomerization by CypA; hydride transfer by dihydrofolate reductase (DHFR) and ribonuclease A (RNaseA). In these enzyme systems, the networks are formed by a series of hydrogen bonds and interactions are conserved across multiple species and show coupling to the enzyme mechanism at long time-scales. For CypA and DHFR, the presence of these networks has been independently verified by other computational studies and NMR investigations.12,46,47 Furthermore, QHA has also allowed identification and characterization of reaction-coupled flexibility in diverse members of an enzyme super-family that catalyze the common sub-step of hydride transfer.
QHA is emerging as an important method for identification of slow conformational fluctuations within proteins. However, the degree of certainty with which the motions identified by QHA at long time-scales (microsecond and longer) can reproduce the motions observed through experimental methods such as NMR/X-ray crystallography48,49 remains an important concern. The amount of conformational sampling that is required to observe the slow motions corresponding to experimentally available data is also of concern.50–52 Further, QHA does not provide any information about the time-scales of motions – only the relative direction and the magnitude of motions are available from the analysis. The eigenvalues provide relative information about the frequencies associated with the eigenmodes; however, in a number of cases the exact time-scale of the computed motions cannot be assessed definitively. For example, if the motions are computed by combining conformations sampled by several MD simulations based on different starting structures, information about the time-scale of the identified motions is missing.53,54
In this report, we present a systematic characterization of slow protein conformational fluctuations identified using QHA and NMA. Further, a methodology for identifying conformational fluctuations associated with an enzyme reaction is also discussed. Microsecond motions in ubiquitin, a small globular protein, are identified, characterized and compared to experimentally available microsecond ensemble of ubiquitin structures. The selection of ubiquitin is based on the availability of a number of conformations through NMR studies as well a large number of X-ray crystal structures. Recent studies by Lange and coworkers have provided insights into the structural heterogeneity of ubiquitin, characterized by NMR at the μs time-scale.49 Our results indicate that the computationally obtained slow motions can reliably reproduce the conformational fluctuations observed from experimental techniques. The use of multiple MD simulations based on different crystal structures accounting for the structural diversity in the protein allows the range of slow motions to be explored quickly. Further, the regions identified to have significant conformational flexibility identified by QHA correspond to the regions that show structural deviations in the different structures from X-ray as well as NMR. The utility of QHA is further analyzed by studying slow conformational changes associated with a reactive process such as the cis/trans isomerization reaction catalyzed by enzyme CypA. Detailed comparison shows that the identified motions are in agreement with the information available from experimental techniques (NMR). Detailed characterization of these computationally identified conformational fluctuations provides important insights in the interconnection between the protein structure, flexibility and function.
II. METHODS
A. Ubiquitin
Ubiquitin is found in all eukaryotes, known to have an important role in labeling proteins for degradation.55 Its structure is evolutionarily conserved, consisting of five β-strands arranged as an anti-parallel sheet interspersed with two α-helices located close to the N- and C-termini of the protein.56–58 Ubiquitin is known to bind diverse targets and therefore its flexibility associated with binding other proteins is of interest. Human ubiquitin (76 residues in a single chain) was selected for computationally identifying and characterizing the μs conformational flexibility due to the structural diversity available from different experimental techniques as well as μs flexibility as recently observed with NMR.49 The set of conformations in this NMR study included all of the structural heterogeneity observed from 46 X-ray crystallographic structures in which ubiquitin was complexed with diverse proteins. Moreover, it was observed that a linear combination of a small set of principal components was able to explain the pincer-like motion of residues involved in forming the binding interactions; and conformational selection, rather than induced-fit mechanism, was sufficient to explain all of the structural heterogeneity in ubiquitin binding dynamics.
The availability of a number of X-ray structure/NMR ensembles and structural deviations within these structures provides information on the conformational fluctuations when the protein samples the kinetically accessible parts of the energy landscape. The recently reported μs NMR refinement with orientational restraints (EROS) with 116 structures (PDB code: 2K39)49 for comparisons with the computational results. The 46 X-ray crystal structures of ubiquitin available in the PDB database59 were also used. These structures were aligned to the reference structure 1UBQ57 before simulation and analysis. The N-terminal residue 1 and the C-terminal tail consisting of residues 71–76 were excluded from our analysis due to the large displacements in the free ends, and for X-ray ensemble analysis only the heavy atoms were used.
System preparation and conformational sampling
8 different starting X-ray crystal structures, which covered the structural diversity of ubiquitin, were used as starting points for MD simulations (see supporting information for details). These starting structures were obtained from Protein Data Bank with the following access codes: 1UBQ; 1P3Q (chain U);57 1S1Q (chain B);60 1TBE (chain B);61 1YIW (chain A);62 2D3G (chain B);63 2FCQ (chain B);64 and 2G45 (chain B).65 Each of the 8 crystal structures was processed using the AMBER 8.0 simulation suite and the parm98 force-field.66,67 Note, we have previously verified the ability of parm98 force-field for its ability to reliably reproduce conformational fluctuations in proteins.44 Standard amino-acid residues were used to build the protein structures. After determining the protonation state for each amino-acid residue at pH 7.0, missing hydrogen atoms were added to the protein. The structures were placed in a rectangular box of SPC/E water,68 such that the distance between the protein and the side of water box was 10 Å.
The prepared systems were equilibrated using the following protocol. First, the water molecules were minimized using the steepest descent method for 500 steps, followed by conjugate gradients minimization until the root mean square (rms) of the gradients was less than 0.25 kcal/mol•Å. In the next step, the protein atoms were minimized using a similar procedure to release close contacts in crystal structures. A small MD simulation of 25.0 picoseconds (ps) with a gradual increase of the temperature in the system to about 300 K, followed by a 25.0 ps constant pressure simulation in which the water molecules were unrestrained to allow occupation of vacuous regions. Five additional steps of equilibration at constant volume were performed with each step consisting of an energy minimization (threshold 0.001 kcal/mol. Å) followed by a 5.0 ps MD run. In these steps, positional restraints were applied to solute atoms. For the first of the five step, the force constant was 100 kcal/mol. Å2, followed by a gradual scaling of 0.5 for subsequent steps. The final equilibration was performed without any positional restraints. Another MD simulation with a temperature ramp over 25.0 ps to readjust the temperature to 300 K followed by a 25.0 ps constant pressure MD step to fill any remaining voids was performed to reach the equilibrated structure.
All production runs were performed using the NVE ensemble, with periodic boundary conditions. The particle-mesh Ewald (PME) method was used for electrostatic interactions; a 10 Å cut-off for Lennard-Jones interactions and SHAKE was used for restricting motions of all covalent bonds with hydrogen atoms. All simulations were performed at 300 K and 1 atm pressure. The data from simulations were stored every 1 ps. A total of 62,500 snapshots were collected totaling 62.5 ns of time for each production runs. Collectively for the 8 systems, the total conformations sampled are referred to as the 0.5 μs MD ensemble.
Identification of protein motions
QHA was performed on the conformations sampled in the MD simulations; and a corresponding principal component analysis was performed with the 116 structures in EROS ensemble and the 46 X-ray crystal structures. For all the 8 MD simulations, the structures were fit to the reference structure of 1UBQ to remove rotational/translational movements. QHA modes were computed for 0.1 μs, 0.2 μs, 0.3 μs, 0.4 μs and 0.5 μs MD ensembles; for each analysis 1,250 structures (stored at regular intervals) from each of the 8 MD simulations, therefore a total of 10,000 structure, were used for each ensemble. Note that 0.1 μs, 0.2 μs, 0.3 μs, 0.4 μs and 0.5 μs MD ensembles are based on 12.5 ns, 25.0 ns, 37.5 ns, 50 ns and 62.5 ns individual MD trajectories respectively from each of the 8 MD simulations.
Normal Mode Analysis
Twelve structures from each simulation (at 5 ns, 10 ns, 15 ns, ..., 55 ns, 60 ns), therefore, a total of 96 structures from the eight simulations were analyzed with NMA. Prior to NMA, the starting structure was minimized using both Newton-Raphson and conjugate gradients minimization method until an RMS tolerance of 10−4 kcal/mol . Å−1 was reached. Solvent was excluded from the calculations and the nmode program.69 from the AMBER suite of programs was used to compute the normal modes for ubiquitin.
B. Cyclophilin A
Substrate free enzyme
The motions in the free enzyme were characterized based on 5 MD trajectories. The starting points for 4 of these trajectories were selected form the NMR ensemble (PDB code 1OCA)70 and an X-ray crystal structure (PDB code 1AWQ)71 with the substrate excluded. AMBER’s parm98 force-field was used for simulations. Each of these structures was simulated in explicit solvent and equilibrated following the procedure already described for ubiquitin simulations. A total of 10 000 conformations corresponding to the total 10 ns sampling (stored after every 1 ps over 2.0 ns for each MD trajectory) were used for the QHA analysis.
Modeling the enzyme reaction
The motions within cyclophilin A associated with the enzyme reaction are known to occur on the μs-ms time-scale. A combination of 37 simultaneous MD simulations was used to characterize the conformational fluctuations associated with the cis/trans isomerization reaction catalyzed by the enzyme. As illustrated in Figure 2, the methodology samples the reaction pathway with the use of reaction coordinate (amide bond dihedral angle) into discrete intervals. Individual MD runs are then used to sample the potential of mean force (PMF) for the free energy profile (FEP) as a function of the chosen reaction coordinate. In the case of CypA, the counter clock-wise rotation along the peptide bond of the substrate was chosen as the reaction coordinate. A discrete interval of 5° rotations for the peptide bond was chosen to sample the reaction pathway. The details of the simulations set-up and production runs have been published previously.43,44
CypA end-states only
QHA modes were also computed for an ensemble of enzyme complex conformation with substrate in reactant and product only (end-states). Extended MD trajectories of 5ns each for the reactant and product states were generated with a total of 8,000 conformations used for the QHA modes computation.
III. RESULTS
Identification of slow conformational flexibility of ubiquitin
Slow conformational fluctuations in ubiquitin were identified with QHA of the conformations that sampled the potential energy surface during the MD trajectories. In the most straight forward way, these fluctuations could be identified by generating a microsecond trajectory followed by the QHA of the system snapshots collected during the simulation. However, currently computing microsecond trajectories of even small proteins is computationally expensive and time-prohibitive as it would require several months of simulation run-time even with best computer hardware. As an alternate approach, for this study 8 independent MD simulations each based on a different starting X-ray crystal structure and 62.5 ns in duration were generated. The total set of conformations used for QHA corresponds to 0.5 μs sampling of the potential energy surface (obtained by combining all the 8 MD simulations). This approach leads to an obvious question: does the analysis of this total ensemble provide information about ubiquitin flexibility at microsecond or the nanosecond time-scale? We provide an answer to this important question below after the detailed characterization of the QHA modes and comparison with the experimental information.
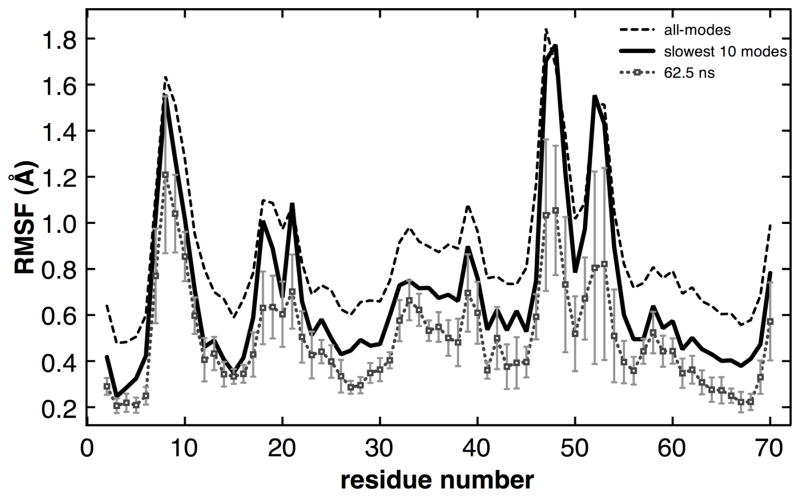
Figure 3 depicts the ubiquitin backbone flexibility as identified based on the individual MD trajectories as well as the 0.5 μs MD ensemble. An indication of the backbone flexibility of ubiquitin is provided by the root mean square fluctuations (RMSF) as computed from the 0.5 μs ensemble of MD structure. RMSF includes the complete set of motions; however, the motivation of this study is to characterize the slow conformational flexibility. Therefore, slow conformational fluctuations were identified by computing QHA modes from a set of 10,000 conformations in the individual 62.5ns MD trajectories as well as the entire 0.5 μs MD ensemble. The modes computed from the 0.5 μs MD ensemble are referred to as QHA0.5μs in the remaining text of this report. As depicted in Figure 3, QHA analysis of individual 62.5 ns MD trajectories can identify the mobile regions qualitatively; however, are unable to correctly characterize the range of motions (displacements) due to limited coverage of the energy landscape. An aggregation of the slowest 10 QHA0.5μs modes (obtained by summing up the atomic displacements in the modes) indicates that they are sufficient to capture the majority of fluctuations in the most mobile region of ubiquitin when compared to all motions; in particular, these slowest modes show similar displacements in the most mobile regions of ubiquitin backbone. Note the slowest modes are defined by the low frequencies (eigenvalues) corresponding to the modes. Quantitative estimates indicate that slow 10 modes represent 78.4 % of the flexibility reflected in the 0.5 μs ensemble (slowest 20 modes capture 87% and slowest 50 modes capture 94% of flexibility). Therefore, the slowest 10 modes were selected for detailed analysis and characterization. The advantage of using the slowest 10 modes instead of the total RMSF (root mean square fluctuations) is that it allows characterization of the slow conformational fluctuations by removing the fast motions.
Figure 3. Ubiquitin backbone flexibility as characterized by 0.5 μs and 62.5 ns conformation sampling.
The gray curve corresponds to an average of 3 individual MD trajectories (based on the 1P3Q, 1S1Q and 1UBQ crystal structures) each 62.5 ns in duration, while the solid black curve corresponds to a total of 0.5 μs sampling from 8 separate MD simulations. Flexibility as quantified by the slowest 10 QHA modes (solid black curve) is compared to total RMSF of 0.5 μs MD ensemble (dashed black curve), which corresponds to all modes.
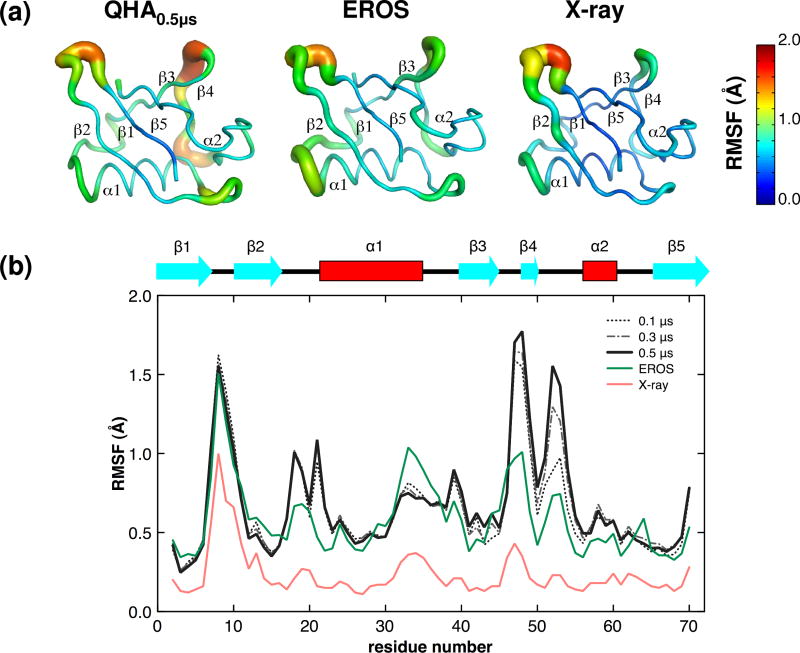
Ubiquitin shows considerable flexibility at the microsecond time-scale (see Figure 4). In addition to the QHA0.5μs, ubiquitin flexibility was also characterized based on the structures from the X-ray and the NMR ensembles. The modes based on 46 X-ray structures and 116 structures in the NMR ensemble (EROS) represent the principal component modes computed in a similar way to the QHA. Note, that the EROS ensemble corresponds to the microsecond time-scale as defined by the resolution of the NMR experiments.49 Figure 4 (a) depicts slow conformational fluctuations of ubiquitin as an aggregate of the top 10 slow modes from the QHA0.5μs, EROS and X-ray. The microsecond scale flexibility is observed in three major regions: β1-β2 hairpin; α1-β3 loop and β3-α2 loop. The computational results agree with the EROS results, except for the β3-α2 loop, where the simulations indicate higher flexibility; also computational simulations show additional flexibility in the β2-α1 region (see further discussion below). Figure 4 (b) depicts that computationally even at 0.1 μs there is qualitative agreement within the experimentally observed flexibility as the regions of high flexibility are similar. However, with additional sampling of the potential energy surface (0.2 μs, 0.3 μs, 0.4μs and 0.5 μs), the amplitude of the motions in the flexible regions of the protein becomes enhanced and is able to qualitatively reproduce the experimentally observed backbone flexibility in the microsecond NMR ensemble. While NMR ensemble agrees with computationally characterized flexibility, the X-ray ensemble shows subdued motions. This is potentially an artifact as the amplitude of fluctuations observed are based on simulations of unbound ubiquitin in solution, compared to the ubiquitin from X-ray ensembles where it is present in complex with other proteins.72 This has also been reported previously in the comparison of X-ray structure with the NMR ensemble.49 Particularly, the residues Ile44 and His68 show close contacts with binding partners in the X-ray structures; in simulations where the binding partners are absent, these regions show increased flexibility.
Figure 4. Microsecond conformational fluctuations in ubiquitin as identified by QHA0.5μs, NMR (EROS) and X-ray ensembles.
(a) Normalized inverse frequency weighted positional fluctuations for Cα atoms are shown in a tube-like representation; thicker tubes represent large-scale fluctuations and thinner tubes represent lower fluctuations. The actual magnitude of the fluctuation is also color-coded, red indicating regions with highest conformational flexibility and blue representing regions within the protein that are relatively less flexible. (b) Comparison of the ubiquitin backbone fluctuation at the microsecond time-scale. Inverse frequency weighted positional fluctuations in ubiquitin. The positional fluctuations from MD ensembles are shown compared with the microsecond scale NMR and X-ray ensemble. The secondary structure of ubiquitin is overlaid on top of the plot for ease of visualization and identifying regions that show high flexibility.
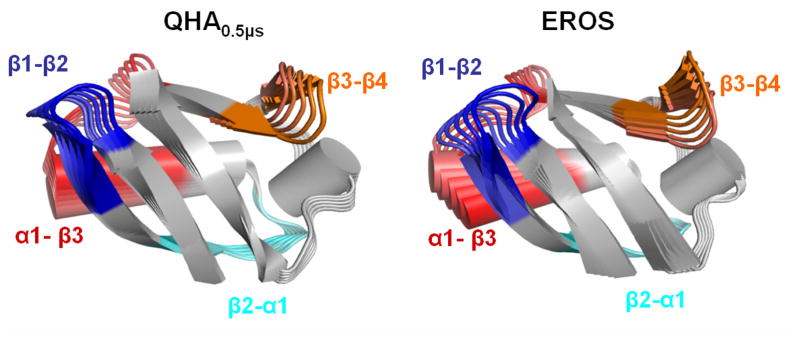
Individual slow modes from the QHA0.5μs and EROS ensembles provide interesting insights into intrinsic large scale motions of ubiquitin with a potentially functional role. Figure 5 shows a movie like representation of the slowest mode from the two ensembles; this mode shows an opening/closing motion associated with the binding site of ubiquitin.73 As also previously reported, this mode involves the “pincer-like” motion of α1-β3, β1-β2 and β3-β4 loop regions. QHA0.5μs also reveals large motions in the β2-α1 regions and to smaller extent in the β3-α2 loop region (see supporting information for animated movies of the modes). These two loops show highly correlated movements with to the opening/closing motions. The motion of residues Ile44, Lys63 and His68 that make close contacts with the binding proteins are observed to be restricted in this mode, which also agrees with the findings from the study of the EROS ensemble. A comparison of other slow modes indicates that the magnitude and the directions of motions in modes 2-10 from QHA0.5μs and EROS ensembles were also observed to be similar. As mentioned above, it was found that the slowest 10 modes contribute >78% of the ubiquitin flexibility, therefore, similarity in the slowest 10 modes between the computational and experimentally calculated modes indicates overall similar microsecond flexibility of ubiquitin. Overall, similarity in flexible regions and the displacement amplitudes within modes from the MD simulations and experimental ensembles (EROS) indicates that QHA is able to reproduce the conformational fluctuations of ubiquitin.
Figure 5. Comparison of the slowest mode of ubiquitin at microsecond time-scale based on QHA0.5μs (computational) and EROS ensemble (experimental).
The modes are depicted in a movie like fashion, with subsequent conformations shown in lighter colors. Four regions of the protein are highlighted with different colors and labeled; these regions indicate large displacements. These motions involve a pincer-like motion involving β1-β2, C-terminal part of α1 helix and α1-β3 loops. The amplitudes of the motions are arbitrarily scaled for visualization, however, see text for comparison of the coverage of conformational landscape.
The ability of QHA0.5μs to reproduce the experimentally observed ubiquitin flexibility at microsecond time-scale was characterized in further detail. In order to perform a quantitative comparison, the large-scale fluctuations computed using QHA on the total 0.5 μs MD simulations and EROS ensemble were compared by calculation of the overlap for the slowest modes. The overlap between two sub-spaces spanned by the eigenvectors is defined by Hess’s metric74 as follows:
| (Eq. 2) |
where D1 and D2 represents the number of eigenvectors considered from each of the ensembles; represents the ith eigenvector from each of the ensembles. γ indicates the degree of similarity of motions between the two ensembles, with a value of 1 indicating identical motions. Based on the observation that slowest 10 modes are able to qualitatively reproduce the flexibility in most mobile region of the proteins, we computed the overlap for slowest 10 modes. As indicated in Table 1, the QHA modes computed based on 0.1 μs, 0.2 μs, 0.3 μs and 0.4 μs MD sampling show close to 80% overlap, indicating that the large scale conformation fluctuations are reproducible across various sections of the MD sampling. This also provides an indication of the robustness of QHA methodology in identifying slow conformational fluctuations.
Table 1. Similarity of motions spanned by QHA, EROS and X-ray ensembles for ubiquitin.
Each entry in the table is computed via Equation 2, for the slowest 10 modes determined from each of the ensembles from the MD simulation.
| 0.1 μs | 0.2 μs | 0.3 μs | 0.4 μs | 0.5 μs | |
|---|---|---|---|---|---|
| 0.1 μs | 0.872 | 0.795 | 0.774 | 0.755 | |
| 0.2 μs | 0.880 | 0.855 | 0.783 | ||
| 0.3 μs | 0.980 | 0.880 | |||
| 0.4 μs | 0.895 | ||||
| EROS | 0.748 | 0.747 | 0.749 | 0.751 | 0.747 |
| XRAY | 0.405 | 0.396 | 0.362 | 0.364 | 0.349 |
A corresponding calculation of overlaps of the modes from MD simulation with that of EROS also indicates about 75% agreement between the two ensembles (see Table 1). Note that the window of previous NMR investigations has been very broadly defined on the microsecond time-scale. X-ray ensemble shows lower degree of similarity; as mentioned before this is possibly due to the comparison of bound ubiquitin in X-ray complexes with apo-ubiquitin. The largest differences in flexibility between the computational and EROS ensembles are observed in the β2-α1 and β3-α2 loops. It is possible that the 0.5 μs scale simulations have allowed β2-α1 and β3-α2 loop regions to explore areas within the potential energy surface that were not accessible during the window of the EROS or the X-ray ensembles, particularly the higher energy regions of the conformational energy landscape that were not accessible to EROS/X-ray ensemble structures (see further discussion below on conformational sampling of various regions of the conformational energy landscape). Additionally, it is also possible that the force-field overestimates the flexibility of these two regions in the MD simulations, or that the NMR investigations have underestimated the flexibility of the protein. Table 1 and the information in Figure 4(b) also indicate that the extent of conformational fluctuations improves with additional sampling between the 0.1–0.5 μs ensembles. The comparison of the overlap between these ensembles (see last column Table 1) indicates convergence toward the conformational flexibility at the microsecond time-scale. Additional sampling of the conformational landscape may lead to changes in the overlaps; however, as discussed below these changes are not expected to be qualitatively much different.
The ability of the slow modes to cover the conformational landscape was also characterized, based on the calculation of projections along the MD trajectories and NMR ensemble. The projections from each of the ensembles were calculated using:
| (Eq. 3) |
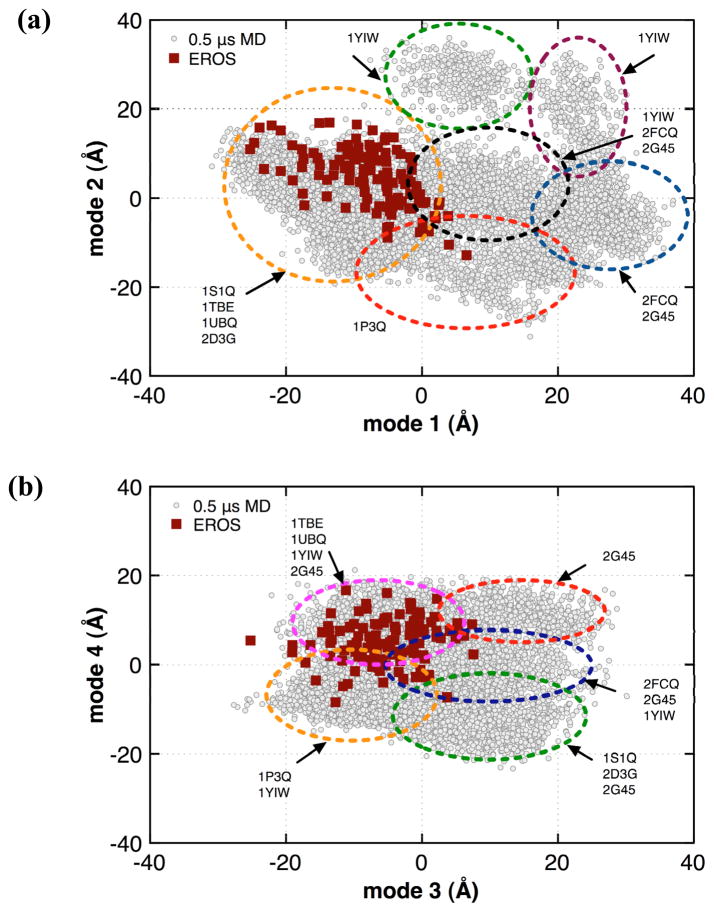
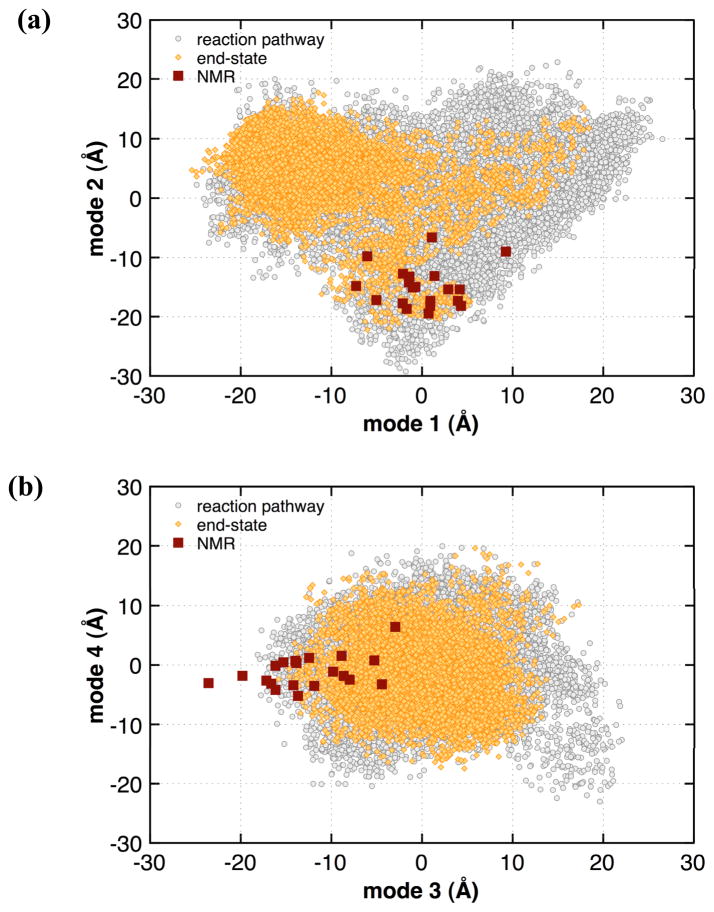
where qi (t) is the projection of the tth snapshot in the trajectory; x(t) is the corresponding positional vector, 〈 x 〉 the mean positional vector and vi represents the ith eigenvector. The extent of overlap between the projections from the slow modes has been previously used in literature to characterize the sampling of the conformational energy landscape in computational methods as well as the structural deviations observed in experimental techniques (including NMR and X-ray ensembles).42,49,54 Within the projections of the slowest two modes (see Figure 6), it was observed that the EROS ensemble is entirely covered by the combined MD simulation. This indicates that 0.5 μs aggregate MD simulations were sufficient to cover all the states visited by EROS ensembles. In addition, MD simulations are also able to cover certain areas of the potential energy landscape not observed from the experimental ensembles (explaining the larger motions sampled by MD). This is especially evident at the right side of Figure 6 (a), which is markedly devoid of any EROS related structures. Similarly, other slower modes also indicate that the MD simulation covers all of the conformations from the EROS ensemble [Figure 6 (b)], indicating a widespread agreement between the subspaces spanned by MD and EROS ensembles.
Figure 6. Projections of the slowest modes from μs MD ensemble and experimental EROS for (a) mode1 vs. mode2 and (b) mode3 v. mode4.
Gray circles represent the ensemble of structures from MD simulations with red squares represent EROS structures. The slowest 4 modes computed with QHA0.5μs were used calculating the projections for structures from all 8 MD simulations and the NMR ensemble. As marked with ellipses, the projections can be separated into 6 and 5 clusters; one or more of which are sampled by individual trajectories. Note the computed projections represent summation over all atoms in the proteins.
The use of 8 separate MD simulations based on different starting structures allows characterization of μs flexibility with relatively short trajectories (62.5 ns each). As is depicted in Figure 6, the slowest QHA0.5μs modes cover the part of the landscape that is also explored by the EROS ensemble. The projection of the slowest QHA0.5μs modes on the MD and NMR structures indicate that starting MD trajectories from different structures allows quick sampling of the neighboring area of the landscape, while it could be envisioned that the longer trajectories would allow sampling of other distant areas of the landscape. From these projections, up to 6 areas or clusters are identified and marked by the ellipses in Figure 6. These clusters correspond to areas of the landscape visited by MD simulations. Note that some individual trajectories visited more than one cluster (for example 1YIW and 2G45) with the intersection point or overlap between the ellipses corresponding to higher energy states or transition points. Any of the 8 ubiquitin MD simulations less than 50 ns did not show the presence of these conformational states or transition points. The use of multiple MD simulations allows quick sampling of the different clusters, which would otherwise take much longer simulations time. An interesting observation from these plots, indicate that the projections from NMR ensemble are located in one or more clusters; which are also most visited area by the various MD trajectories. This common area of the plot is the most visited area by multiple MD simulations. This could possibly indicate the presence of a more populated conformational sub-state of ubiquitin that is visited both during the computational (MD) and experimental (NMR) methods. These plots also provide an indication that the 0.5 μs MD ensemble has covered the extent of conformational landscape accessible by ubiquitin at the microsecond time-scale. Additionally, the coverage of larger portion of the landscape (beyond what is explored by NMR) provides an explanation of higher flexibility indicated by the MD simulations. These observations are consistent with the overlaps listed in Table 1. Further MD sampling would lead to filling out the gaps in these plots (sampling of the higher energy transition points); therefore, qualitatively any significant differences in the slow conformational fluctuations are not expected.
Comparison of QHA0.5μs with NMA
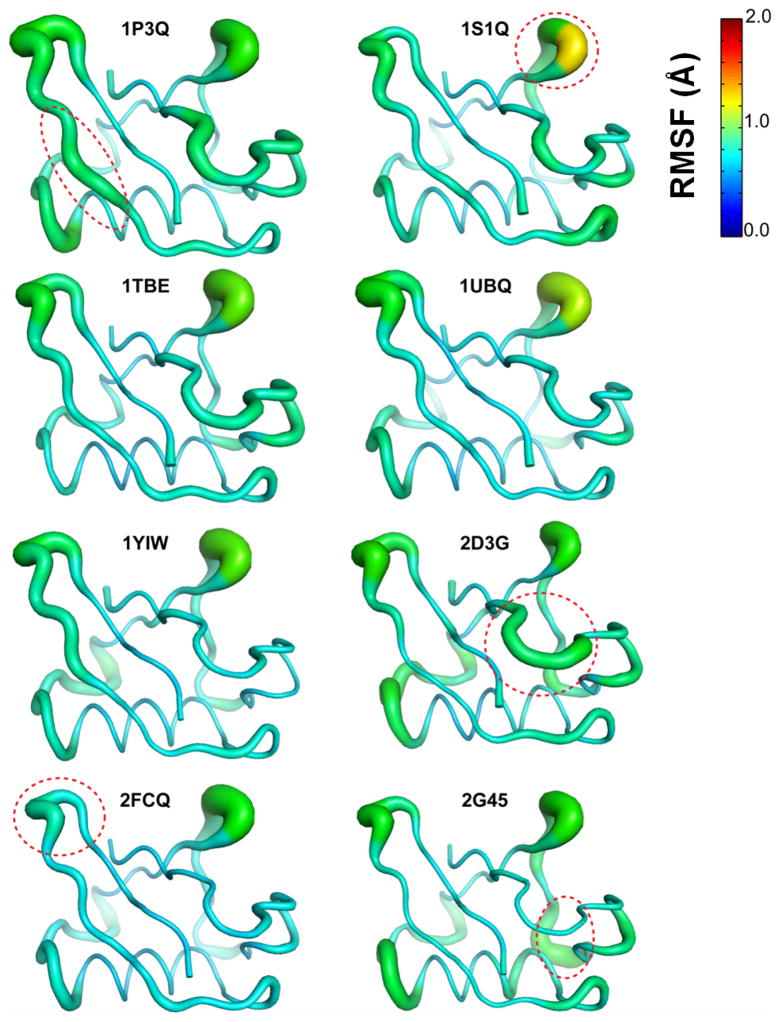
NMA provides a quadratic approximation of the potential energy surface based on a reference structure; and is well suited to study motions close to a local energy minimum (see Figure 1). NMA modes were computed for 12 structures each separated by 5 ns for each of 8 MD simulations, therefore, a total of 96 set of NMA modes were obtained (see Methods section for more details). The results are summarized in Figure 7 and Figure 8. As depicted in Figure 7, various ubiquitin regions show high flexibility; however; there are considerable differences in the regions of larger flexibility between different structures. The high-lighted regions particularly show flexibility that is not reproduced in other structures. Even though the regions of flexibility are similar to the QHA0.5μs; the magnitude of displacement is much smaller for NMA modes when compared to the QHA modes, as is visible by comparison with colored regions in Figure 4 (note that the color-bars are on same scale for the two figures). This difference possibly exists as NMA characterizes motions that explore the immediate vicinity of the local minimum of the reference structure (gray area in Figure 1) whereas QHA of 0.5 μs ensemble indicates sampling of a larger portion of the conformation energy landscape.26 The conformational flexibility identified by NMA shows considerably lower agreement with the EROS ensemble as well (see Table 2); therefore, is much less able to reproduce the experimentally observed ubiquitin flexibility.
Figure 7. Average RMS fluctuations determined based on NMA of ubiquitin, based on 8 structures used in this study.
The displacement vectors for the slowest 10 modes were aggregated (by summing all atomic displacements in the modes) and averaged for 12 conformations. Note, that the fluctuations are colored on the same scale as results of QHA analysis (Figure 4). The circled areas indicate regions with flexibility which is not reproduced in other structures. These results show lower amplitudes and less reliable modes than QHA.
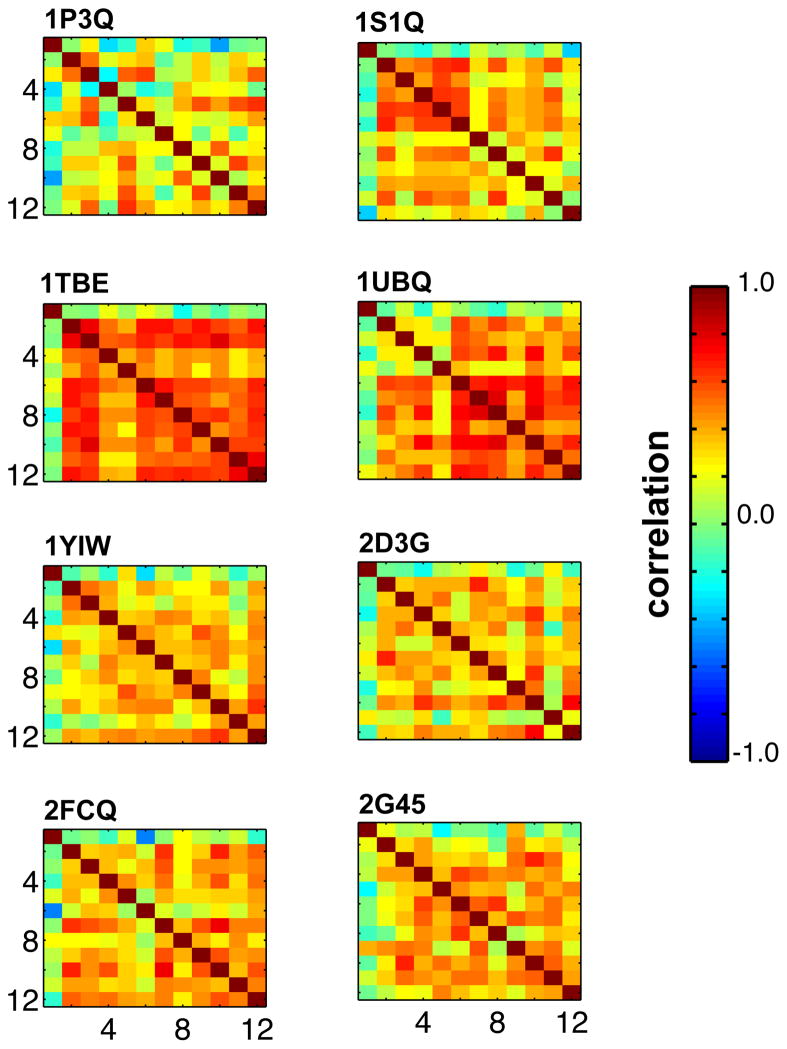
Figure 8. Correlation of positional fluctuations as computed by NMA for structures along the 8 MD trajectories.
The 8 plots indicate the 8 MD systems used in this study. NMA was performed on 12 structures in each trajectory separated by 5ns (5ns, 10ns, …, 60ns) and the degree of correlations of normal modes between the 12 structures are shown as a matrix. The degree of correlation was obtained as taking a dot product of all eigenmodes following by normalization. These results indicate change in NMA modes over the course of the MD trajectory, except for 1TBE and 1UBQ that seem to be sampling in the nearby areas.
Table 2. Correlation of positional fluctuations between NMA, QHA and EROS modes.
The comparison represents the correlation between inverse frequency weighted top 10 modes. NMA shows lower agreement with EROS as compared to QHA results.
| Normal Mode Analysis |
||||
|---|---|---|---|---|
| EROS | 1P3Q | 1TBE | 2G45 | |
| QHA0.5μs | 0.713 | 0.641 | 0.490 | 0.441 |
| EROS | 0.607 | 0.624 | 0.436 | |
Characterization of the correlation between the NMA modes indicates several interesting aspects of ubiquitin flexibility. As depicted in Figure 8, the NMA show considerable variation during the MD trajectory as indicated by considerably lower correlations with the other structures in the same MD simulation. For 1P3Q, 1YIW and 2G45 in particular as the trajectory evolved the lower correlation indicates changes in normal modes. This is also possibly an indication of the MD simulation sampling different areas of the energy landscape. However, in the case of 1TBE and 1UBQ the normal modes show a larger correlation, possibly indicate of MD simulation sampling the neighboring areas of the energy landscape. It is interesting to note that these observations are similar to the one mentioned above for QHA.
Cyclophilin A
Reaction coupled flexibility of CypA at millisecond time-scale
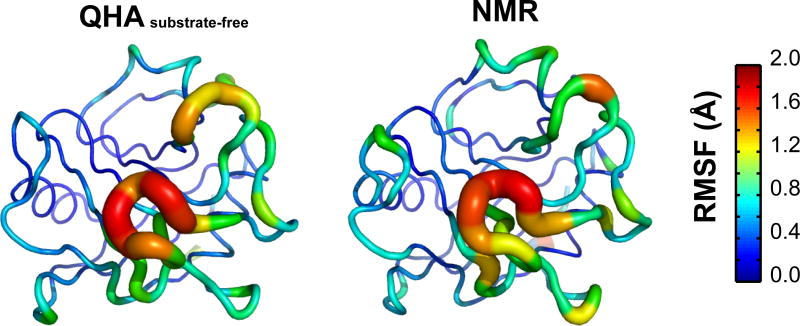
We have previously reported the reaction coupled flexibility in cyclophilin A.1,10,43,44 The slow modes were identified based on 18,500 enzyme-substrate conformations sampled along the reaction pathway. The amide bond dihedral angle was used as reaction coordinate and the reaction was slowly mapped from the reactant to the product state. QHA was used to compute the modes and the coupling to the reaction was defined by the amount of variation in the amide bond dihedral angle (reaction coordinate) in each mode. Figure 9 depicts the aggregated flexibility of CypA at microsecond-millisecond time-scale coupled to the reaction. The results show the presence of large flexibility both in regions close to the active-site (substrate is indicated by ball-and-stick substrate in the figure) as well as in the distal areas of CypA. In particular the regions 12–15, 68–76, 78–84, 101–107, 118–125 and 136–152 show large flexibility along the reaction pathway. Previously, we have reported the top 3 reaction coupled modes which were characterized in detail; and these modes were independently reproduced in different 4 X-ray structures of human CypA catalyzing entirely different substrates. Qualitatively the regions of flexibility agree with NMR spin relaxation studies as reported by Kern and coworkers. 12 An NMR ensemble for human CypA corresponding to the cis/trans isomerization reaction is not available to perform analysis similar to ubiquitin as reported above. However, an NMR ensemble for substrate free CypA is available with 20 structures (PDB code: 1OCA).70 Therefore, comparison of flexibility in substrate-free CypA was performed. The substrate free simulations of CypA and the NMR ensemble show similar conformational flexibility. As depicted in Figure 10, the regions showing large conformational fluctuations include residues 12–15, 43–51, 66–84, 101–106, 120–126 and 146–151. The QHA modes were computed with 5 MD simulations, each 2.0 ns in duration, therefore, the total conformational sampling only corresponds to 10.0 ns. However, even at the nanosecond time-scale (left) the results qualitatively show agreement with the NMR ensemble (right). Based on the insights from the ubiquitin results presented above, it can be envisioned that with more sampling there would be quantitative agreement in the observed flexibility as well.
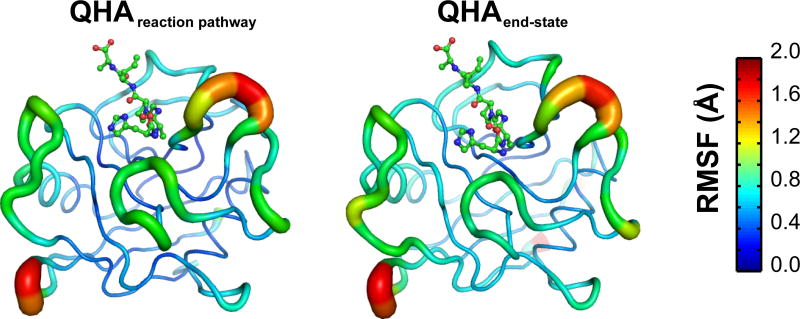
Figure 9. Reaction coupled flexibility in the enzyme CypA.
Inverse frequency weighted positional fluctuations for the top 10 reaction coupled modes based on QHA of conformations from (a) entire reaction pathway (b) end-states only (reactant + product). Substrate is shown in ball-and-stick representation, while the enzyme is shown as cartoon. Similar results indicate that the end-state results qualitatively agree with the full reaction pathway.
Figure 10. Conformational fluctuations of substrate-free cyclophilin A.
from MD simulation and NMR ensemble (1OCA). The inverse frequency weighted positional fluctuations for the slowest 10 modes for CypA are depicted.
The extent of conformation fluctuations observed within CypA during the course of reaction pathway was further characterized by computing the projections of the slowest modes. The projections from the slow modes of QHA from reaction path sampling and NMR ensembles indicate that the structural heterogeneity in the reaction path sampling as well as the substrate-free MD covered and even exceeded the NMR ensemble (see Figure 11). The presence of additional structures within the reaction path-sampling ensemble indicates that it covered a larger area within the potential energy landscape, which includes the NMR ensemble as a subset. Even though the NMR projections are localized to a section of the projection maps, it provides an indication that the reaction-coupled modes are sampled by the NMR ensemble. Note that the NMR ensemble does not have the substrate bound; however, as previously reported these reactions promoting motions are intrinsically associated with the CypA structure and mechanism.12,44 In our calculations, the overlap between the reaction path sampling and substrate-free MD was found to be 0.65.
Figure 11. Projection of MD and NMR structures on the modes coupled to the reaction catalyzed by CypA.
(a) mode 1 vs mode 2 (b) mode 3 vs mode 4. The grey open circles correspond to the projections from the set of 18,500 conformations sampled along the reaction pathway; red squares correspond to the NMR structures. The yellow filled circles correspond to the end-states MD only. Note the computed projections represent summation over all atoms in the protein.
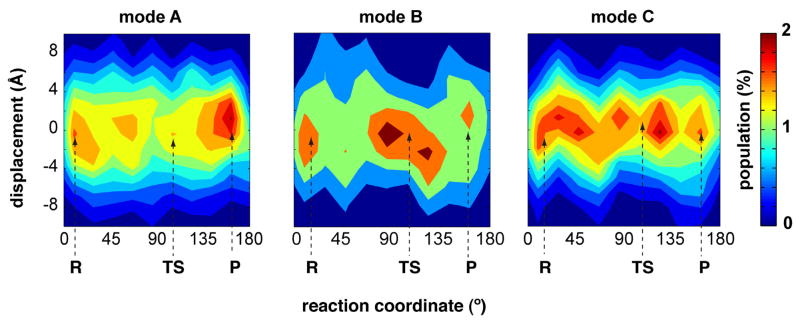
The time-scale of the identified reaction coupled conformational fluctuations in CypA has been suggested to the time-scale of reaction, which corresponds to the μs-ms time-scale. However, the total MD sampling performed for reaction pathway is only on the order of nanoseconds. The formulation of QHA loses the information about time (see Eq. 1), moreover the combination of different MD simulations along the reaction pathway adds uncertainty to the time-scale of the QHA modes. The eigenvalues associated with the modes obtained only provide relative information between the modes. Therefore, the top most reaction coupled modes were characterized for the behavior along the reaction profile as well as to obtain information the on the time-scale of the periodicity. Figure 12 depicts the projections of conformational snapshots on the top most reaction coupled modes. The results indicate a periodic behavior at the time-scale of the reaction (μs-ms). Therefore, the observed slow conformational fluctuations from the QHA of conformational snap-shots along the reaction pathway correspond to the μs-ms time-scale. Note that the significant similarity between the ubiquitin flexibility computed from QHA0.5μs and EROS ensemble indicate presence of μs conformational fluctuations in proteins.
Figure 12. Displacement (averaged projection) as a function of reaction coordinate for the three top most modes coupled to the reaction in enzyme CypA.
The occurrence of maxima within these plots provides indication of time scale of the conformational changes.
Comparing reaction path coupled and end-states flexibility
Identification of the reaction coupled flexibility is computationally expensive as it requires sampling the entire reaction pathway; moreover, this methodology can be considered limiting if suitable description of the reaction coordinate is not available. As an alternate methodology, we investigated the QHA modes obtained from the set of CypA-substrate conformations in the end states only (reactant and product states). Comparison of the aggregated flexibility based on the slowest 10 reaction coupled modes (see Figure 9) as well as the projections of the modes computed from the reaction pathway and the end states only (reactant + product) indicate that the results are qualitatively similar (see Figure 11). A detailed comparison of the individual motions from the two simulations as well as the overlap between the modes further indicated that the slowest reaction coupled modes from the reaction path sampling and end-state simulations were similar. These results indicate that the intrinsic flexibility of CypA does not change; however, sampling over the reaction pathway provides the complete extent of conformational fluctuations that are sampled by the enzyme.
It is interesting to note that the end-state MD showed two densely sampled regions within the landscape whereas the reaction path sampling was more uniform. As could be somewhat expected, the end-state structures sampled states did not entirely overlap with the reaction path sampling. Based on these observations, it appears that the information contained in the end-state simulations is only qualitatively representative of the conformational flexibility within the protein during the course of the reaction step. However this information is not sufficient to provide quantitative estimates of the actual motions (displacements) along the course of the reaction. Nonetheless, the end-state QHA modes provide a quick way for identification of the reaction coupled flexibility. It should be cautioned that if the end-states differ considerably, and there is structural rearrangement in the protein and the end states sample different parts of the phase space, then the qualitatively insight obtained may not be representative of the reaction pathway.
IV. CONCLUSIONS AND SUMMARY
Proteins exhibit motions at a wide rage of time-scales and an increasing body of evidence suggests the possible link between the slow conformational fluctuations and protein function such as enzyme catalysis. In this paper, we have used computational methods to identify and characterize slow conformational fluctuations in proteins. The flexibility of a soluble protein, ubiquitin, sampling the conformational energy landscape and the flexibility coupled to the peptidyl-prolyl cis/trans isomerization catalyzed by enzyme cyclophilin A have been identified. The characterization of these slow motions identified using computationally methods are in agreement with the flexibility associated with NMR ensembles and the structural deviations in a collection of X-ray structures.
The microsecond flexibility in ubiquitin was characterized using QHA of the conformations generated along 8 MD trajectories starting from 8 different crystal structures. Overall, the total ensemble corresponded to 0.5 μs sampling. In addition to significantly reducing the run time of the simulations by performing these 8 runs simultaneously, this approach enables the sampling of separate areas of the energy surface due to starting from differences in crystal structures. The characterization of the slow conformations indicated that the top 10 modes accounted for over 78% of flexibility observed at the microsecond time-scale. The slowest mode indicated “pincer-like” motion, as was previously identified using microsecond NMR. This motion has been implicated in ubiquitin binding to other proteins in solution. The identified motions were compared to the structural deviations observed in the collection of ubiquitin structures in complex with other proteins, as well as the recently characterized NMR ensemble. The degree of similarity between NMR ensemble flexibility and the QHA0.5μs modes was observed to be close to 0.75 (for the top 10 modes), indicating a significant agreement in the nature of the slow motions at microsecond time-scale. Further, the coverage of the conformational landscape by the computationally and experimentally identified flexibility was observed to be similar, even though individual trajectories sampled only 62.5 ns. The use of multiple trajectories allows for most of the conformational flexibility at the μs time-scale to be reproduced. Therefore, we propose that combination of multiple trajectories provides an efficient method to explore long scale conformational fluctuations. Other investigations have also reported similar observations.41,75
Normal modes, which represent a quadratic approximation of the energy landscape near a local energy minimum, indicated similar flexibility in ubiquitin for a collection of structures. However, the motions from NMA were only able to qualitatively reproduce the protein regions with large displacements. The magnitude and the direction of the motions differed considerably with the starting structures, and showed low overlap (less that 50%) with the NMR ensemble. Therefore, it appears that the motions corresponding to the μs time-scale are less likely to be fully characterized with NMA and related methods.
Microsecond-millisecond (μs-ms) flexibility coupled to the reaction catalyzed by enzyme CypA was identified by QHA of the enzyme-substrate conformations sampled along the reaction pathway. In combination of reaction pathway sampling such an umbrella sampling, QHA provides a unique method of identifying slow conformational flexibility coupled to the enzyme mechanism. Similar to the case of ubiquitin these modes also show agreement with the experimentally determined protein flexibility. Further, as we previously reported these modes play a promoting role in the cis/trans isomerization reaction by the CypA. For enzyme catalyzed reactions, the reaction coupled flexibility can be approximated by QHA of conformations obtained by a combination of the reactant and the product states only. This allows overcoming a limitation of this methodology, as it requires sampling along the reaction pathway. The slow conformation fluctuations based on the entire reaction pathway and the end-states only were qualitatively similar for the CypA catalyzed reaction. The coverage of conformational landscape was similar for the first 4 modes coupled to the reaction. From a mechanistic point of view, it could be rationalized that when the reactant and product states of the enzyme do not show large conformational changes involving rearrangements of secondary structures, sampling just the reactant and product states provided qualitative information about the large-scale motions involved in the catalytic sub-step.
Overall the characterization of μs-ms conformational fluctuations of ubiquitin and CypA indicates that flexibility is closely related to their designated function. Even though the role of protein flexibility in function is topic of an ongoing debate;76 similar to the observations reported here, increasing evidence from other groups also suggests a close link between protein conformational fluctuations and enzyme catalysis.77–79 Computational methods continue to provide valuable insights into protein motions. The results presented here indicate that QHA is a robust method for identifying the slow conformational fluctuations at long time-scales. Recent investigations from our group have indicated that the reaction promoting slow conformational fluctuations are conserved as a part of enzyme folds catalyzing different chemistries such as peptide bond isomerization, hydride transfer and single strand RNA hydrolysis.80 Further, we have also discovered that the reaction coupled flexibility in diverse members of an enzyme superfamily, catalyzing the common sub-step of hydride transfer, is entirely conserved.81 Overall these results indicate that enzyme structure encodes dynamics and together structure-dynamics encode function.
Supplementary Material
Acknowledgments
PKA acknowledges the financial support by ORNL’s LDRD fund and the National Institutes of Health (R21GM083946). AR’s acknowledges the Advanced Short-Term Research Opportunity (from ORISE) program. This research used resources of the National Center for Computational Sciences ORNL under the Director’s Discretionary allocation (project: BIP003), which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725.
Footnotes
Supporting information available: Summary of structural heterogeneity in the 8 X-ray crystal structures and animation movies of the ubiquitin slow conformational fluctuations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Agarwal PK. Microb Cell Fact. 2006;5:2. doi: 10.1186/1475-2859-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon WR, Benkovic SJ. J Biol Chem. 1998;273:26257. doi: 10.1074/jbc.273.41.26257. [DOI] [PubMed] [Google Scholar]
- 3.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. Nature. 2007;450:913. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 4.Hammes GG. Biochemistry. 2002;41:8221. doi: 10.1021/bi0260839. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal PK, Billeter SR, Rajagopalan PTR, Benkovic SJ, Hammes-Schiffer S. Proc Natl Acad Sci U S A. 2002;99:2794. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammes-Schiffer S. Biochemistry. 2002;41:13335. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- 7.Caratzoulas S, Mincer JS, Schwartz SD. J Am Chem Soc. 2002;124:3270. doi: 10.1021/ja017146y. [DOI] [PubMed] [Google Scholar]
- 8.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Science. 2002;295:1520. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 9.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal PK. J Am Chem Soc. 2005;127:15248. doi: 10.1021/ja055251s. [DOI] [PubMed] [Google Scholar]
- 11.Henzler-Wildman K, Wolf-Watz M, Thai V, Kern D. Protein Sci. 2004;13:104. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 12.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Nature. 2005;438:117. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 13.Bellissent-Funel MC, Daniel R, Durand D, Ferrand M, Finney JL, Pouget S, Reat V, Smith JC. J Am Chem Soc. 1998;120:7347. [Google Scholar]
- 14.Bennett B, Langan P, Coates L, Mustyakimov M, Schoenborn B, Howell EE, Dealwis C. Proc Natl Acad Sci U S A. 2006;103:18493. doi: 10.1073/pnas.0604977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaccai G. Science. 2000;288:1604. doi: 10.1126/science.288.5471.1604. [DOI] [PubMed] [Google Scholar]
- 16.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Proc Natl Acad Sci U S A. 2002;99:5247. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wand AJ. Nat Struct Biol. 2001;8:926. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 18.Bellissent-Funel MC, Zanotti JM, Chen SH. Faraday Discuss. 1996;103:281. [Google Scholar]
- 19.Zavodszky P, Kardos J, Svingor A, Petsko GA. Proc Natl Acad Sci U S A. 1998;95:7406. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller WT. Acta Crystallogr D. 2005;61:33. doi: 10.1107/S0907444904025855. [DOI] [PubMed] [Google Scholar]
- 21.Daniel RM, Dunn RV, Finney JL, Smith JC. Annu Rev Biophys Biomol Struct. 2003;32:69. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 22.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Proc Natl Acad Sci U S A. 2002;99:16047. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenimore PW, Frauenfelder H, McMahon BH, Young RD. Proc Natl Acad Sci U S A. 2004;101:14408. doi: 10.1073/pnas.0405573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. Science. 2004;303:186. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 25.Brooks B, Karplus M. Proc Natl Acad Sci U S A. 1983;80:6571. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Q, Bahar I. Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. Chapman and Hall/CRC; London: 2006. [Google Scholar]
- 27.Levitt M, Sander C, Stern PS. J Mol Biol. 1985;181:423. doi: 10.1016/0022-2836(85)90230-x. [DOI] [PubMed] [Google Scholar]
- 28.Van Wynsberghe AW, Cui Q. Structure. 2006;14:1647. doi: 10.1016/j.str.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Yang LW, Eyal E, Chennubhotla C, Jee J, Gronenborn AM, Bahar I. Structure. 2007;15:741. doi: 10.1016/j.str.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rader SD, Agard DA. Protein Sci. 1997;6:1375. doi: 10.1002/pro.5560060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyashita O, Onuchic JN, Wolynes PG. Proc Natl Acad Sci U S A. 2003;100:12570. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J. Structure. 2005;13:373. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Di Pace A, Cupane A, Leone M, Vitrano E, Cordone L. Biophys J. 1992;63:475. doi: 10.1016/S0006-3495(92)81606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel RM, Smith JC, Ferrand M, Hery S, Dunn R, Finney JL. Biophys J. 1998;75:2504. doi: 10.1016/S0006-3495(98)77694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel RM, Finney JL, Reat V, Dunn R, Ferrand M, Smith JC. Biophys J. 1999;77:2184. doi: 10.1016/S0006-3495(99)77058-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tournier AL, Smith JC. Phys Rev Lett. 2003;91:208106. doi: 10.1103/PhysRevLett.91.208106. [DOI] [PubMed] [Google Scholar]
- 37.Barton NP, Verma CS, Caves LSD. J Phys Chem B. 2002;106:11036. [Google Scholar]
- 38.Noid DW, Fukui K, Sumpter BG, Yang C, Tuzun RE. Chem Phys Lett. 2000;316:285. [Google Scholar]
- 39.Karplus M, Kushick JN. Macromolecules. 1981;14:325. [Google Scholar]
- 40.Kitao A, Go N. Curr Opin Struct Biol. 1999;9:164. doi: 10.1016/S0959-440X(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 41.Caves LSD, Evanseck JD, Karplus M. Protein Sci. 1998;7:649. doi: 10.1002/pro.5560070314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeffrey DE, Leo SDC, Martin K. Protein Sci. 1998;7:649. [Google Scholar]
- 43.Agarwal PK. Proteins. 2004;56:449. doi: 10.1002/prot.20135. [DOI] [PubMed] [Google Scholar]
- 44.Agarwal PK, Geist A, Gorin A. Biochemistry. 2004;43:10605. doi: 10.1021/bi0495228. [DOI] [PubMed] [Google Scholar]
- 45.Shankar K, John MR, Djamal B, Robert HS, Peter AK. J Comput Chem. 1992;13:1011. [Google Scholar]
- 46.Garcia-Viloca M, Truhlar DG, Gao JL. Biochemistry. 2003;42:13558. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Goodey NM, Benkovic SJ, Kohen A. Proc Natl Acad Sci U S A. 2006;103:15753. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GarcÌa AE. Phys Rev Lett. 1992;68:2696. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 49.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KFA, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Science. 2008;320:1471. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 50.Andrea A, Marc AC, Di Nola A. Proteins. 1999;36:419. [Google Scholar]
- 51.Daggett V. Curr Opin Struct Biol. 2000;10:160. doi: 10.1016/s0959-440x(00)00062-2. [DOI] [PubMed] [Google Scholar]
- 52.Clarage JB, Romo T, Andrews BK, Pettitt BM, Phillips GN. Proc Natl Acad Sci U S A. 1995;92:3288. doi: 10.1073/pnas.92.8.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balsera MA, Wriggers W, Oono Y, Schulten K. J Phys Chem. 1996;100:2567. [Google Scholar]
- 54.Lange OF, Grubmuller H. J Phys Chem B. 2006;110:22842. doi: 10.1021/jp062548j. [DOI] [PubMed] [Google Scholar]
- 55.Pickart CM. Annu Rev Biochem. 2001;70:503. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 56.Di Stefano DL, Wand AJ. Biochemistry. 1987;26:7272. doi: 10.1021/bi00397a012. [DOI] [PubMed] [Google Scholar]
- 57.Vijaykumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 58.Weber PL, Brown SC, Mueller L. Biochemistry. 1987;26:7282. doi: 10.1021/bi00397a013. [DOI] [PubMed] [Google Scholar]
- 59.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Mol Cell. 2004;13:783. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 61.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. J Mol Biol. 1994;236:601. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 62.Duhee B, George IM, Valentina T, Anthony AK, Stephen BK. Angew Chem Int Edit. 2005;44:3852. [Google Scholar]
- 63.Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Nat Struct Mol Biol. 2006;13:272. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 64.Bang D, Gribenko AV, Tereshko V, Kossiakoff AA, Kent SB, Makhatadze GI. Nat Chem Biol. 2006;2:139. doi: 10.1038/nchembio766. [DOI] [PubMed] [Google Scholar]
- 65.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. Cell. 2006;124:1197. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 66.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Comp Phys Comm. 1995;91:1. [Google Scholar]
- 67.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Single UC, Weiner PK, Kollman P. AMBER. Vol. 7. University of California; San Francisco: 2002. [Google Scholar]
- 68.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327. [Google Scholar]
- 69.Nguyen DT, Case DA. J Phys Chem. 1985;89:4020. [Google Scholar]
- 70.Ottiger M, Zerbe O, Guntert P, Wuthrich K. J Mol Biol. 1997;272:64. doi: 10.1006/jmbi.1997.1220. [DOI] [PubMed] [Google Scholar]
- 71.Vajdos FE, Yoo SH, Houseweart M, Sundquist WI, Hill CP. Protein Sci. 1997;6:2297. doi: 10.1002/pro.5560061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Best RB, Lindorff-Larsen K, DePristo MA, Vendruscolo M. Proc Natl Sci Acad USA. 2006;103:10901. doi: 10.1073/pnas.0511156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kitahara R, Yokoyama S, Akasaka K. J Mol Biol. 2005;347:277. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 74.Hess B. Physical Review E. 2000;62:8438. doi: 10.1103/physreve.62.8438. [DOI] [PubMed] [Google Scholar]
- 75.Cheng XL, Ivanov I, Wang HL, Sine SM, McCammon JA. Biophys J. 2007;93:2622. doi: 10.1529/biophysj.107.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olsson MHM, Parson WW, Warshel A. Chem Rev. 2006;106:1737. doi: 10.1021/cr040427e. [DOI] [PubMed] [Google Scholar]
- 77.Boehr DD, Dyson HJ, Wright PE. Chem Rev. 2006;106:3055. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 78.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Biochemistry. 2008;47:3317. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 79.Klinman JP. Chem Phys Lett. 2009;471:179. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramanathan A, Agarwal PK. 2009 Submitted. [Google Scholar]
- 81.Ramanathan A, Agarwal PK. 2009 Submitted. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.