Abstract
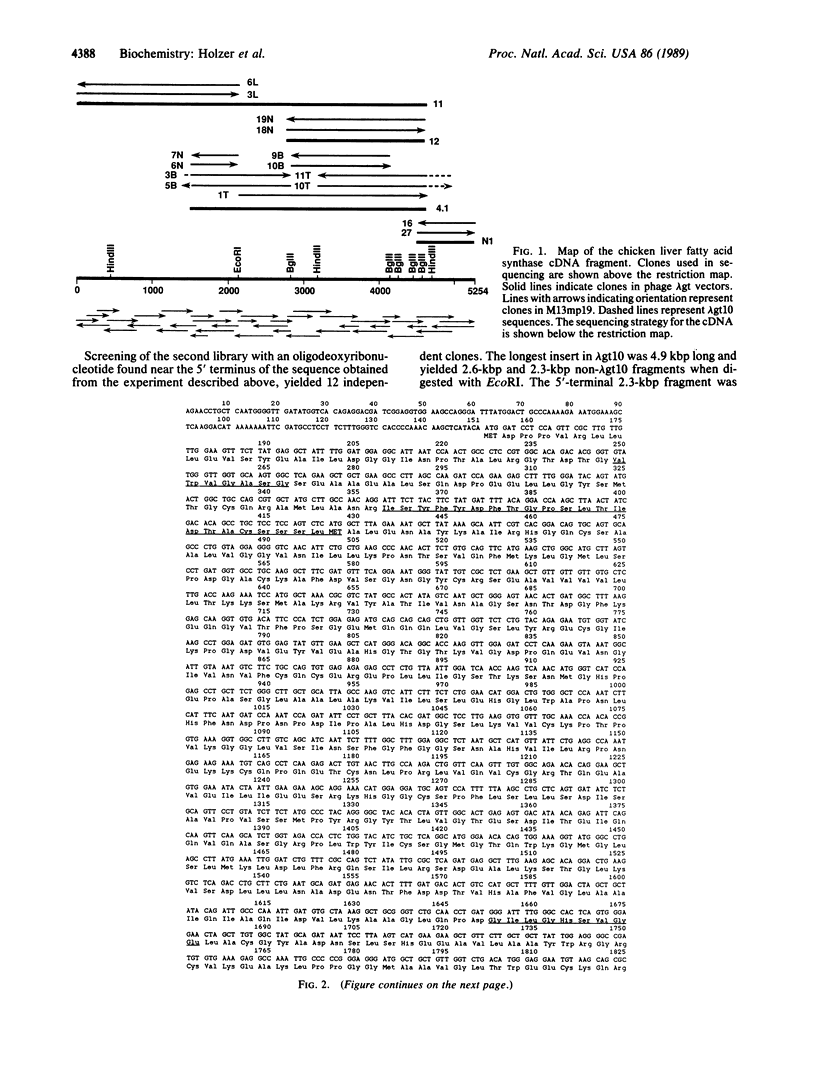
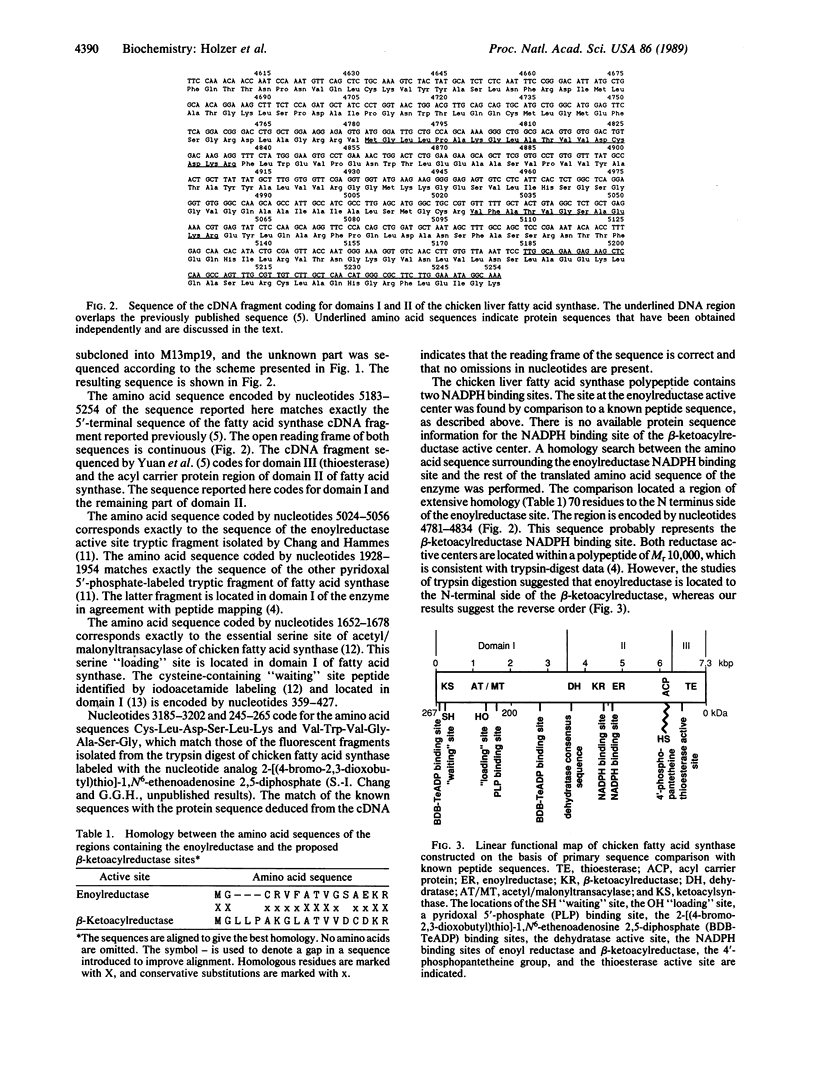
The complete amino acid sequence of chicken liver fatty acid synthase [acyl-CoA:malonyl-CoA C-acyltransferase (decarboxylating, oxoacyl- and enoyl-reducing, and thioester-hydrolyzing), EC 2.3.1.85] has been determined from the corresponding cDNA sequence. A 5.3-kilobase-pair (kbp) region of cDNA coding for chicken fatty acid synthase has been cloned and sequenced that is contiguous to the 2.3-kbp region previously sequenced [Yuan, Z., Liu, W. & Hammes, G.G. (1988) Proc. Natl. Acad. Sci. USA 85, 6328-6331]. The cDNA codes for the remaining 1677 amino acids of the previously unsequenced region of the protein. The amino acid sequence contains peptides known to be associated with the NADPH binding site of the enoylreductase active center, the acetyl/malonyltransacylase active site, the "waiting" site containing cysteine, and a pyridoxal 5'-phosphate binding site. Locations of the NADPH binding site of the beta-ketoacylreductase active site and of the dehydratase active site are proposed on the basis of protein sequence homologies to catalytic sites in other enzymes. The molecular weight of the complete polypeptide chain is 267,288. A linear functional map of the chicken fatty acid synthase derived from its primary sequence is presented.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang S. I., Hammes G. G. Amino acid sequences of substrate-binding sites in chicken liver fatty acid synthase. Biochemistry. 1988 Jun 28;27(13):4753–4760. doi: 10.1021/bi00413a026. [DOI] [PubMed] [Google Scholar]
- Chirala S. S., Kuziora M. A., Spector D. M., Wakil S. J. Complementation of mutations and nucleotide sequence of FAS1 gene encoding beta subunit of yeast fatty acid synthase. J Biol Chem. 1987 Mar 25;262(9):4231–4240. [PubMed] [Google Scholar]
- Datta P., Goss T. J., Omnaas J. R., Patil R. V. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci U S A. 1987 Jan;84(2):393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristensky B., Lis J., Wu R. Portable microcomputer software for nucleotide sequence analysis. Nucleic Acids Res. 1982 Oct 25;10(20):6451–6463. doi: 10.1093/nar/10.20.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes G. G. Fatty acid synthase: elementary steps in catalysis and regulation. Curr Top Cell Regul. 1985;26:311–324. doi: 10.1016/b978-0-12-152826-3.50030-9. [DOI] [PubMed] [Google Scholar]
- Mattick J. S., Tsukamoto Y., Nickless J., Wakil S. J. The architecture of the animal fatty acid synthetase. I. Proteolytic dissection and peptide mapping. J Biol Chem. 1983 Dec 25;258(24):15291–15299. [PubMed] [Google Scholar]
- Noda C., Ito K., Nakamura T., Ichihara A. Primary structure of rat liver serine dehydratase deduced from the cDNA sequence. FEBS Lett. 1988 Jul 18;234(2):331–335. doi: 10.1016/0014-5793(88)80110-8. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Miller D. A., Dunn T., Su Y., Burcham J. M., Peraino C., Fujioka M., Babcock K., Pitot H. C. Isolation and nucleotide sequence of the cDNA for rat liver serine dehydratase mRNA and structures of the 5' and 3' flanking regions of the serine dehydratase gene. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5809–5813. doi: 10.1073/pnas.85.16.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz E., Schmitt W. Sequence of Escherichia coli D-serine dehydratase. Location of the pyridoxal-phosphate binding site. FEBS Lett. 1981 Nov 2;134(1):57–62. doi: 10.1016/0014-5793(81)80550-9. [DOI] [PubMed] [Google Scholar]
- Schweizer M., Roberts L. M., Höltke H. J., Takabayashi K., Höllerer E., Hoffmann B., Müller G., Köttig H., Schweizer E. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol Gen Genet. 1986 Jun;203(3):479–486. doi: 10.1007/BF00422073. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Wakil S. J. Isolation and mapping of the beta-hydroxyacyl dehydratase activity of chicken liver fatty acid synthase. J Biol Chem. 1988 Nov 5;263(31):16225–16229. [PubMed] [Google Scholar]
- Tsukamoto Y., Wong H., Mattick J. S., Wakil S. J. The architecture of the animal fatty acid synthetase complex. IV. Mapping of active centers and model for the mechanism of action. J Biol Chem. 1983 Dec 25;258(24):15312–15322. [PubMed] [Google Scholar]
- Wakil S. J., Stoops J. K., Joshi V. C. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yuan Z. Y., Hammes G. G. Elementary steps in the reaction mechanism of chicken liver fatty acid synthase. Acylation of specific binding sites. J Biol Chem. 1985 Nov 5;260(25):13532–13538. [PubMed] [Google Scholar]
- Yuan Z. Y., Liu W., Hammes G. G. Molecular cloning and sequencing of DNA complementary to chicken liver fatty acid synthase mRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6328–6331. doi: 10.1073/pnas.85.17.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]


