Abstract
DNA hypomethylation was the initial epigenetic abnormality recognized in human tumors. However, for several decades after its independent discovery by two laboratories in 1983, it was often ignored as an unwelcome complication, with almost all of the attention on the hypermethylation of promoters of genes that are silenced in cancers (e.g., tumor-suppressor genes). Because it was subsequently shown that global hypomethylation of DNA in cancer was most closely associated with repeated DNA elements, cancer linked-DNA hypomethylation continued to receive rather little attention. DNA hypomethylation in cancer can no longer be considered an oddity, because recent high-resolution genome-wide studies confirm that DNA hypomethylation is the almost constant companion to hypermethylation of the genome in cancer, just usually (but not always) in different sequences. Methylation changes at individual CpG dyads in cancer can have a high degree of dependence not only on the regional context, but also on neighboring sites. DNA demethylation during carcinogenesis may involve hemimethylated dyads as intermediates, followed by spreading of the loss of methylation on both strands. In this review, active demethylation of DNA and the relationship of cancer-associated DNA hypomethylation to cancer stem cells are discussed. Evidence is accumulating for the biological significance and clinical relevance of DNA hypomethylation in cancer, and for cancer-linked demethylation and de novo methylation being highly dynamic processes.
Keywords: cancer, DNA methylation, DNA repeats, genomic sequencing, hypermethylation, hypomethylation
DNA hypomethylation: a ubiquitous feature of carcinogenesis
The first-described epigenetic changes in human cancer were losses of DNA methylation (m5C residues replaced by unmethylated C residues) reported in 1983. We observed this alteration of DNA methylation throughout the genome in various cancers versus a wide variety of normal tissues [1], and Feinberg and Vogelstein described hypomethylation of a few cancer-irrelevant gene regions in colon adenocarcinomas versus normal colonic epithelium [2,3]. It was apparent that metastases were even more susceptible to cancer-linked DNA hypomethylation than primary tumors. Many subsequent reports have confirmed the frequent overall genomic hypomethylation in cancers relative to control tissues (e.g., [4–9]).
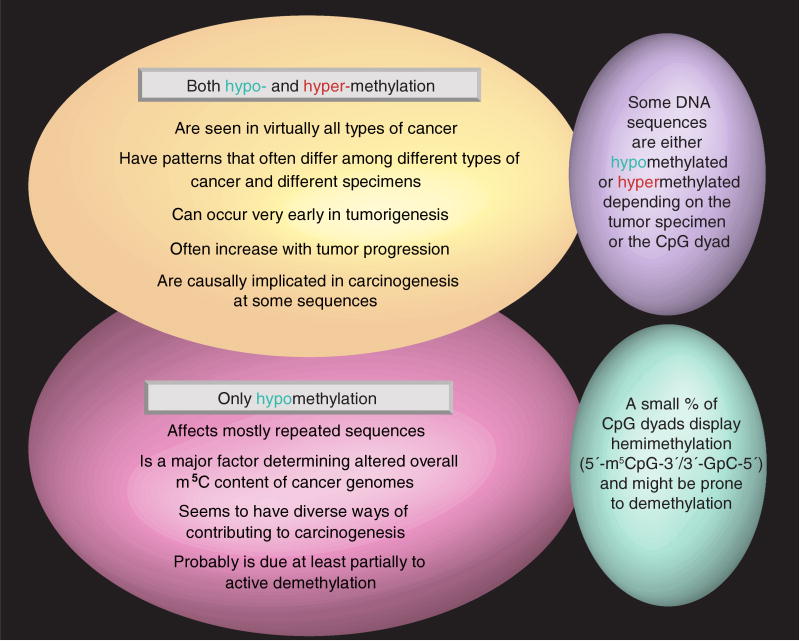
The opposite methylation change, hypermethylation of promoters of tumor suppressor genes, homeobox genes and other sequences, is also one of the most constant features of the cancer genome [10–15]. In spite of the prevalent increases in DNA methylation, overall deficiencies in the m5C content of DNA are found in almost every type of cancer, but not in all specimens (Figure 1). The following list contains just some of the types of tumors that have been shown to display overall losses in genomic m5C:
Figure 1. Similarilities and differences in cancer-associated hypo- and hyper-methylation of DNA.
Ovarian epithelial carcinomas versus the corresponding cystadenomas or various normal postnatal somatic tissues [16,17];
Prostate metastatic tumors versus normal prostate [4];
Leukocytes from B-cell chronic lymphocytic leukemia versus normal leukocytes [18];
Hepatocellular carcinomas versus matched nonhepatoma liver tissue [19];
Cervical cancer and high-grade dysplastic cervical lesions versus normal cervical tissue or low-grade dysplasia of the cervix [20];
Colon adenocarcinomas versus adjacent normal mucosa [21];
Wilms' tumors versus various normal postnatal somatic tissues [22].
Even a murine model of prostate cancer based upon early and late T-antigen transgenes expressed in a prostate-specific manner displays both satellite DNA hypomethylation and gene locus-specific hypermethylation in the tumors (∼2% of ∼1200 analyzed loci) [23].
Just as there are cancer-type specific differences in DNA hypermethylation patterns [12], so some DNA sequences are more or less susceptible to DNA hypomethylation depending on the kind of cancer [24]. Some types of cancer, for example, testicular germ cell seminomas, display an especially large amount of genomic hypomethylation. However, in the case of testicular germ cell seminomas, it could be the result of the cell of origin of the tumor having unusually hypomethylated DNA [25,26]. Moreover, for some DNA sequences, cancer-linked DNA hypermethylation can be seen in some specimens and hypomethylation seen in others, as discussed below.
Contribution of hypomethylation of DNA repeats to overall genomic hypomethylation
Hypomethylation of highly repeated DNA sequences [5,17,27,28], which comprise approximately half of the genome, is largely responsible for the global DNA hypomethylation that is observed so frequently in cancers (Figure 1), as we first demonstrated for satellite DNAs [29–31]. Tandem centromeric satellite α, juxtacentromeric (centromere-adjacent) satellite 2, the interspersed Alu and long interspersed elements (LINE)-1 repeats are the most frequently studied DNA cancer-hypomethylated repeats [17,27–34]. Although hypomethylation of centromeric and pericentromeric tandem repeats in cancers can be significantly associated with each other, sometimes one of these types of repeats is hypomethylated and the other is not [17,35,36].
Cancer-linked hypomethylation of gene regions
Hypomethylation of transcription regulatory regions in cancer seems to be much less frequent than hypermethylation of CpG islands overlapping promoters [5,14,23,27,37]. Nonetheless, some of the cancer-associated loss of DNA methylation encompasses gene regions, including transcription control sequences [38–45]. For example, the gene encoding the protease urokinase (PLAU/uPA) is overexpressed and hypomethylated in conjunction with tumor progression in breast cancers (grades 1, 2 and 3) and prostate cancers (benign prostate hyperplasia vs cancer) [46,47]. Decreased methylation of this gene that is implicated in breast cancer invasion was induced by incubation of a breast cancer cell line with 5-azadeoxycytidine and was associated with increased expression of this gene [48]. Among the genes displaying cancer-linked hypomethylation and transcriptional activation are cancer-testis genes, S100A4, mesothelin, claudin4, trefoil factor 2, maspin, PGP9.5, POMC and the heparinase gene [49–53]. Cancer-linked hypomethylation of gene promoters is often associated with decreases in overall genomic methylation [9,54] or satellite DNA hypomethylation [55].
DNA hypomethylation can be found early in carcinogenesis, but can also be associated with tumor progression
Regional hypermethylation can occur very early in tumorigenesis, but it can also be so strongly associated with tumor progression that it serves as a significant indicator of survival [56–58]. Similarly, hypomethylation of DNA sequences is often observed during the early stages of tumorigenesis or in abnormal non-neoplastic tissue, such as hyperplasia [21,29,38,59] (Figure 1). For example, age-related hypomethylation of certain DNA sequences appears to precede aneuploidy in a subset of gastrointestinal cancers [60].
Hypomethylation of DNA is generally more pronounced with tumor progression or the degree of malignancy [17,30,61,62], although this depends on the type of tumor and the individual specimens [35,59,63] (Figure 1). Among the examples of tumor progression-linked DNA hypomethylation is the transition from the chronic phase of chronic myeloid leukemia to blast crisis; this transition is significantly associated with hypomethylation of a CpG island in the LINE-1 repeat promoter, as determined by a quantitative methylation-specific PCR assay [64]. Overall, DNA hypomethylation is associated with the grade of cervical neoplasia [20], with multifocal versus unifocal hepatocellular carcinomas [65], and the disease stage, tumor size and histological grade for breast tumors [66]. For prostate adenocarcinomas, LINE-1 hypomethylation has a highly significant relationship with lymph node involvement [67]. For hepatocellular carcinomas, the degree of hypomethylation of either of two repetitive sequences was significantly correlated with postoperative recurrence of disease and was a better predictor than conventional factors [68]. Hypomethylation of both centromeric and juxtacentromeric satellite DNAs was significantly associated with tumor grade in ovarian carcinomas and was a highly significant marker of the risk for relapse and overall survival time [35]. Although more studies have revealed tumor progression-linked hypomethylation of DNA repeats, gene hypomethylation increasing with tumor progression has also been described [46,47].
Overview of relevance to cancer biology
Recently, there has been a surge in reports about cancer-associated DNA hypomethylation. These include genome-wide mapping of hypomethylation, investigating the roles of this hypomethylation in carcinogenesis, identifying and testing cancer markers, and designing and monitoring anticancer therapies. However, for a while, the importance of decreases in DNA methylation associated with carcinogenesis was overshadowed by the emphasis on cancer-linked hypermethylation of tumor-suppressor genes [69]. The biological significance of both decreases and increases in DNA methylation to cancer is underscored by their contribution to other human diseases, normal vertebrate development, bioengineering and by their susceptibility to environmental influences [33,70–80].
Some of the observed cancer-associated hypomethylation in gene regions contributes to the disease [5,53,81–83]. As described below, studies of cancer-linked DNA hypomethylation indicate that it is not simply a consequence of cancer-associated hypermethylation. Moreover, DNA methylation changes in cancers are much more dynamic than previously envisioned.
Some of the confounding factors in assessing cancer-associated changes in DNA methylation
Normal cell-type specificity, individual or age-related variations in DNA methylation & tumor purity
Cancer-linked DNA hypomethylation denotes less methylation relative to some control tissue(s), which should be specified. However, as we had demonstrated in collaboration with Charles Gehrke (formally Distinguished Emeritus Professor of Biochemistry, University of Missouri, Columbia, MO, USA, and recently deceased), there are tissue-specific differences, as well as species-specific differences, in the amounts and distribution of DNA methylation [84,85]. For example, normal human cerebellum DNA has 17% more m5C than heart DNA. In addition, tissues are generally composed of a heterogeneous mixture of cell types, which can differ in their genomic methylation. At the gene level, normal tissue-specific changes in promoter methylation could obscure oncogenesis-mediated changes if the appropriate control tissue is not used, for example, with the tumor-suppressor gene, SERPINB5 [86].
Even similar cell types may differ epigenetically because of derivation from different stem cell niches [87]. Moreover, a cancer may arise from a minor stem cell component or from independently transformed progenitor cells as part of a neoplastic field effect [88–90], and these might have an atypical methylation pattern (see below). Another factor confounding analysis of cancer-linked changes in DNA methylation is heterogeneity at the allelic level due to imprinting [91], or allelic differences that are independent of parent of origin [92].
Moreover, in quantitative studies of cancer-linked changes in DNA methylation, the percentage of cells from the tumor sample that are non-neoplastic may vary and affect the results if the tumor sample has not been microdissected. For some kinds of tumors, like Wilms' tumors and ovarian epithelial carcinomas, which consist mostly of neoplastic cells, this is not an important factor, but it is important for others, like breast adenocarcinomas.
In addition, there can be epigenetic variability among individuals. Increases or decreases in DNA methylation can occur with aging, and individual-specific differences in DNA methylation patterns have been found [93,94]. However, these are usually localized and subtle [85,95–97]. In some cancer studies, the variability of DNA methylation in non-neoplastic tissues is not adequately addressed by the proper choice of control tissue or by the use of a diverse set of control tissues. Nonetheless, numerous articles on cancer-linked changes in DNA methylation indicate that differences in DNA methylation in malignancies versus control tissues vastly exceed individual-specific differences in a given normal postnatal tissue [7-9,16-22].
Methods of analysis
There are many new methods for analyzing DNA methylation [28,37,98-101]. The types of alterations in DNA methylation that can be observed are strongly affected by the method of DNA methylation analysis and the choice of sequences analyzed; for example, normally unmethylated or partially methylated CpG islands [102] (Box 1). For example, in order to visualize both increases and decreases in methylation of the same DNA subregion, it is optimal for the DNA sequence in the control tissue to have an intermediate extent of methylation or some CpGs with highly conserved methylation and others with conservation of the unmethylated status [24].
Box 1. A summary of some of the salient spatial–temporal features of cancer DNA epigenetics.
At the level of several hundred base pairs: a high degree of site-to-site dependence
A neighboring-sites model rather that an independent-sites or context-dependent model best describes methylation changes in regions that are not under strong cancer-selection pressure.
Sometimes hypomethylated and hypermethylated CpG dyads are neighbors.
Some CpG dyads persist as hemimethylated sites.
At the level of genes, gene clusters or other large regions: long-range coupling
Selection pressures during tumorigenesis help shape DNA methylation patterns, for example, homogeneous hypermethylation of promoters of genes whose silencing facilitates cancer formation.
-
There are regions of long-range epigenetic changes containing either:
–Clusters of hypermethylated CpG islands;
–Pockets of hypomethylation; or
–Long tandem repeats with overall hyper- or hypomethylation.
Snapshots of a specific time & place
The detection of cancer-linked DNA methylation changes depends on: the method of analysis and choice of DNA sequences to be analyzed, the tumor specimen, tumor subregion, amount of ‘contaminating’ normal cells, the stage of carcinogenesis and the use of an appropriate normal tissue for comparison.
Usually only DNA sequences with an intermediate level of methylation in normal tissues will reveal both hypo- and hyper-methylation.
Another confounding factor in DNA epigenetics is the normal heterogeneity of DNA methylation patterns from molecule to molecule owing to the lack of precise preservation of methylation patterns in vivo. It is this heterogeneity that helps make epigenetics a major contributing factor to tumorigenesis. Some of the heterogeneity in the patterning of methylation changes of individual sequences, including laterally along the DNA and across the two strands of a given CpG dyad, evades detection in studies where the average methylation content at each examined CpG is determined. Such heterogeneous changes can be observed when individual DNA molecules are analyzed by molecular cloning [103-106].
Interplay between cancer-linked hypermethylation & hypomethylation
Cancer-linked hypo- & hypermethylation are generally independent, although most studied cancers display both changes in their genome
In a comprehensive study of satellite DNA hypomethylation, global DNA hypomethylation and hypermethylation of 55 gene loci in three types of ovarian epithelial tumors of different malignant potential and a variety of normal somatic tissues [17], we found that DNA hypomethylation and hypermethylation usually co-exist in the same tumor but in different sequences. Moreover, progressive changes in both promoter hypermethylation and hypomethylation of DNA repeats or global DNA hypomethylation can be seen in comparisons of premalignant lesions and certain types of cancer, cultured cell or animal models of tumor progression [62,107,108]. Even for tandemly repeated DNA sequences, the same cancer specimen can display hypomethylation of one type of repeat (satellite DNA) and hypermethylation of another (NBL2 and D4Z4 in subtelomeric regions or in the short arms of the acrocentric chromosomes) [24,109,110].
It had been proposed that there is cross-talk between demethylation and de novo methylation pathways during tumorigenesis, making one dependent on the other. For example, DNA demethylation might be used as a type of epigenetic repair in normal postnatal cells to attempt to compensate for physiologically inappropriate methylation of CpG islands overlapping promoters of tumor suppressor genes or polycomb-complex target genes. As for conventional DNA repair pathways, this epigenetic repair pathway might be only partially effective and might demethylate many more sequences than just the incorrectly methylated CpG islands. Alternatively, DNA hypomethylation might occur early in oncogenesis and be followed by hypermethylation [111] as a kind of increased, compensatory de novo methylation consequent to the genomic hypomethylation. This might be analogous to an Arabopsis antisense DNA methyltransferase (DNMT) gene, which caused gene hypermethylation despite an overall decrease in genomic cytosine methylation [112].
However, we demonstrated in the above-mentioned study of ovarian carcinomas [17] that DNA hypomethylation could not just be described as a consequence of DNA hypermethylation or vice versa. The degree of gene hypermethylation was associated with the degree of malignancy of ovarian epithelial tumors (benign, low-malignant potential or carcinomas). However, this association was independent of the association of satellite or global DNA hypomethylation with the degree of malignancy [17]. These results were consistent with previous less extensive analyses of other types of tumors [22,67] and with more recent studies [113]. In addition, in our analysis of ovarian tumors, no DNA hypomethylation variables were significantly associated with DNMT RNA levels after adjustment for multiple comparisons [17].
Nonetheless, both hyper- and hypo-methylation of DNA are linked to carcinogenesis and have some inter-relationship other than one being dependent on each other. They may have some step(s) in common in the poorly understood pathways that lead to these divergent epigenetic changes [105]. Indeed, cancer is generally characterized by the instability of the epigenome at the levels of DNA (methylation of cytosine) and chromatin (protein associations, modifications and chromatin conformation). Moreover, different types of cancer-linked epigenetic abnormalities can interact in various ways [90,114,115]. In addition, epigenetic abnormalities in cancer are occasionally associated with genetic changes, for example, methylation of the promoter of O6-methylguanine DNA methyltransferase and increased G–A mutations in glioblastomas [116].
Tumor specificity of DNA hypomethylation
There is tumor-type specificity in cancer-associated DNA hypomethylation of a given sequence, just as there is for a gene region hypermethylation in cancer [24,117,118] (Figure 1). For example, we found that the extent of hypomethylation of juxtacentromeric satellite 2 DNA from chromosome 1 was often greater in ovarian carcinomas than in Wilms' tumors. Nonetheless, at least moderate levels of hypomethylation were frequent in both types of cancer [30,31,35,36,119].
Surprisingly, DNA sequences that display an intermediate level of methylation in normal tissues may exhibit opposite cancer-associated alterations depending on the tumor specimen or the tumor type [105]. For example, some ovarian epithelial carcinoma specimens exhibited strong overall hypermethylation of the NBL2 (a repeat found mostly on acrocentric chromosomes) and others had strong hypermethylation of this tandem repeat [24]. Such opposing changes in DNA methylation could be observed because the intermediate level of methylation of NBL2 in all control somatic tissues [24] allowed the determination of a decrease in methylation in some cancers and an increase in others. We also found opposite cancer-linked changes in methylation at a subtelomeric tandem repeat, D4Z4 [109].
Importantly, among 17 ovarian carcinomas and 44 examined Wilms' tumors, there was a significant correlation (p < 0.001) between the direction and degree of methylation change at D4Z4 and the dissimilar NBL2 [109]. This suggests that diverse sequences on different chromosomes can be recognized by the same demethylation or de novo methylation mechanisms during carcinogenesis. However, many of these cancers displaying hypermethylation of D4Z4 and NBL2 repeats had hypomethylation of satellite DNA, a very different type of tandem repeat [24]. This is similar to the findings of Katargin et al. on D4Z4 and satellite 2 methylation in cervical cancer [118]. We recently demonstrated that even satellite 2 DNA, which is highly susceptible to decreases in methylation in cancer, can exhibit focal hypermethylation in some cancers [106].
DNA methylation changes probably overshoot their biological targets in cancer & often spread over large genomic regions
A high-resolution tiling-array analysis of two entire chromosomes demonstrated that cancer-linked hypermethylation favors CpG islands and hypomethylation favors repeated sequences, including tandem repeats, interspersed repeats and segmental duplications [14], as we had inferred previously [5]. Most increases and decreases in DNA methylation are not strictly targeted to specific loci. Rather, they are found in many regions of the genome that are probably irrelevant to cancer [2,24,28]. This overshooting of cancer-linked DNA methylation changes to many more sites than are biologically relevant may parallel overshooting of tissue-specific alterations in DNA methylation during development [73]. For example, although aberrant promoter methylation is mostly associated with effects on transcription in cancer, sequences in the body of genes can also become hypermethylated without noticeable effects on transcription [15,108].
Recently, it has been observed that large regions of chromosomes may be coordinately methylated or demethylated in cancer [37,45,120] (Box 1). These apparently coordinated methylation changes might be related to the type, frequency, spacing of DNA repeats or to the presence of clusters of interrelated gene. However, cancer-linked DNA hypermethylation does not necessarily spread from repeats to neighboring sequences. Within a subtelomeric segmental duplication in several cancers, we have observed hypermethylation of the bulk of a tandem repeat array but hypomethylation of border sequences at the beginning of the repeat array and nonrepeated sequences proximal to the array [109].
Dynamic nature of cancer-linked DNA methylation changes
That a given DNA sequence can be susceptible to both increases and decreases in methylation during carcinogenesis is relevant to the one statistically significant association that we found between DNA hypomethylation and gene hypermethylation. This was an inverse relationship between hypomethylation of the juxtacentromeric satellite 2 DNA and hypermethylation of CDH13 in ovarian carcinomas and breast cancers [17,35]. This association might reflect waves of DNA demethylation during carcinogenesis that could affect some CpG-rich regions and reverse their cancer-linked de novo methylation [121,122].
The fluidity of epigenetic changes in cancer is also indicated by our above-mentioned findings that NBL2 and D4Z4 repeats could be extensively hypomethylated in some cancer specimens and hypermethylated in others [24,109]. Moreover, for NBL2 repeats, we showed that hypomethylated CpG dyads could be next to hypermethylated dyads [105]. These results and the low, but appreciable, frequency of stable hemimethylated CpG dyads in normal tissues and cancers [104–106,123] suggest that DNA methylation after differentiation is maintained by more dynamic mechanisms than generally assumed, rather than just by maintenance methylation. Similarly, Pfeifer et al. concluded that normal human cells have a dynamic methylation system from a study of bisulfite-mediated genomic sequencing of cloned DNA molecules from an X-linked gene in cultured cells with or without treatment with a DNA inhibitor [124].
The idea that the control of DNA methylation even in normal postnatal tissues involves de novo methylation and demethylation is gaining support [40,125–129]. In cancer cells, methylation and demethylation mechanisms appear to be much less regulated, which leads to DNA methylation instability in the directions of both gains and losses of methylation. This nonuniform loss of control of DNA methylation is seen in the metastatic tumor heterogeneity of promoter hypomethylation (LINE-1 interspersed repeat promoter) in prostate cancers [130]. Moreover, bidirectional changes in DNA methylation imply that DNA hypermethylation might spontaneously reverse during the course of carcinogenesis or be reversed in response to treatment.
Usually, studies of abnormal DNA methylation in cancers are snapshots of a specific time and place (Box 1). These analyses only reveal the epigenetic status of the analyzed portions of the genome in the tumor when and where it was sampled, and do not detect fluxes in DNA methylation changes during tumor development or progression. Selective advantages to certain alterations in DNA methylation during carcinogenesis, such as repressive methylation of promoters of tumor suppressor genes, probably help shape more uniform methylation patterns in cancer. Non-gene DNA repeats, like the tandem repeats mentioned above, may give insights into the nature of cancer-linked DNA methylation changes better than gene regions, because they are probably not subject to strong selection pressures for transcriptional silencing or activation during carcinogenesis, as are tumor-suppressor genes, polycomb-complex target genes and oncogenes. Moreover, their intermediate stages of methylation changes may be more stable because they reside in condensed regions of the genome.
Evidence for cancer-linked hypomethylation being causally involved in carcinogenesis
Nutrition & DNA methylation
Experiments involving DNA methylation inhibitors in vivo and in vitro [131–133] or analyzing DNMT-deficient mice [134,135] indicate that the loss of DNA methylation is important for oncogenesis. Studies of tumorigenesis in rodents fed methyl-deficient diets [70,136,137] are in accordance with a causative role for DNA hypomethylation in cancer. Global DNA hypomethylation could be induced in rats by dietary methyl deficiency and, if prolonged, was irreversible upon resumption of a normal diet [138]. This DNA hypomethylation was associated with formation of initiated hepatic foci. Evidence has been presented that diet can affect DNA methylation and thereby expression of some genes [139,140]. Moreover, for certain human cancers, inadequate folate intake is implicated as a risk factor, especially in combination with the C667T polymorphism in MTHFR [141]. However, a recent study showed that dietary folate deficiency in Ung-/- mice caused a significant decrease in DNA methylation in epithelial cells without inducing tumor formation [142]. Nonetheless, most of these nutritional studies argue for causal roles of DNA hypomethylation in carcinogenesis.
Mouse models with Dnmt deficiency or other genetic sources of loss of DNA methylation
Many animal models have been used for studying the effects of changes in DNA methylation on cancer formation but, as indicated above, the type of model greatly influences the results. DNA hypomethylation in ApcMin/+ mice induced by mutation (heterozygosity for a Dnmt1 mutation) and/or 5-azadeoxycytidine treatment (drug-induced demethylation) decreases, rather than increases, the number of intestinal polyps [143]. Experimentally induced DNA hypomethylation was associated with decreased progression of microadenomas to macroscopic tumor growths [144]. However, transgenic mice containing a hypomorphic mutant Dnmt1 allele and consequent DNA hypomethylation had increased formation of hepatocellular adenomas and carcinomas despite the decreased formation of macroscopic intestinal adenomas [135]. Transient decreases in Dnmt1 activity during murine development led to multiple tumors in association with the loss of imprinting [145]. A hypomorphic allele of Dnmt1 in mice resulted in thymic lymphomas that displayed frequent insertion of a transposable element into the Notch oncogene [146]. Genetic knockout of the chromatin remodeling protein Lsh in mouse transplantation experiments led to upregulation of retroviral elements and oncogenes, hypomethylation of DNA (especially of repeated sequences), impairment of normal erythropoeisis, and erythroleukemia [83]. The variety of tumor outcomes from animals in which DNA methylation levels are experimentally altered is likely to be dependent on the type of tumor that is studied, the genetic background of the animal, chromosome-epigenetic variations and the specific DNA sequences affected by the methylation changes [12,24,135].
Gene hypomethylation contributing to tumor formation & tumor progression
Hypomethylation of gene regions, which has been implicated in various diseases [147], may foster the formation and progression of tumors in diverse ways. Some of the gene-regulatory regions that display cancer-linked hypomethylation have correlated increases in transcription [5,82]. Hypomethylation and overexpression of some imprinted genes, including the IGF-II and H19 genes, are implicated in carcinogenesis [145,147,148]. Gene region hypomethylation could contribute to oncogenesis by affecting classical oncogenes, but evidence suggests a greater involvement in the activation of genes associated with tumor invasion or metastasis [48].
The synuclein-γ gene is among the cancer invasion and metastasis genes that seems to be a target of the oncogenic effects of DNA demethylation in a wide variety of cancers [81,149–151]. It is a marker of metastasis and can be mechanistically linked to tumor progression. Among the pathways by which its tumor progression- and demethylation-linked overexpression may influence tumor progression are its roles in neurofilament integrity, signaling pathways [301] and proliferation [81]. Another gene target of tumor progression-linked DNA methylation changes is drug-resistance genes. The increasing destabilization of DNA methylation patterns that characterizes much tumor progression may interfere with cancer treatment by increasing the drug resistance of cancer cells through promoter region hypomethylation for some genes and hypermethylation for others [152].
Important insight into DNA methylation changes and oncogenesis came from a study of global DNA hypomethylation and estrogen receptor gene hypermethylation observed in colorectal adenomatous polyps and adenocarcinomas, but not at the earlier hyperplastic polyp stage [153]. These methylation abnormalities were more reversible by treating patients with a nonsteroidal anti-inflammatory drug (celecoxib, Pfizer, Inc., NY, USA) at the adenomatous polyp stage than at the adenocarcinoma stage. This suggests an epigenetic progression that might involve chromatin changes subsequent to DNA methylation changes.
Hypomethylation of DNA repeats contributing to carcinogenesis & chromosomal rearrangements
The hypomethylation of interspersed repeats and tandem repeats might promote tumor formation or progression by fostering DNA rearrangements [31,134,154–158]. For example, there is an association between hypomethylation of satellite DNA at the juxtacentromeric heterochromatin of chromosomes 1 and 16 and chromatin decondensation and rearrangements in these regions in lymphoid cells. This is seen in cells from patients with the immunodeficiency, centromeric region instability, facial anomalies (ICF) syndrome [72,159,160], and in certain human cell lines [161,162]. In cancer, pericentromeric rearrangements involving these regions are over-represented and often manifest as unbalanced translocations that can promote tumor formation by affecting the copy number of genes relevant to cancer [163,164]. Murine embryonic stem cells show an increase in chromosomal translocations upon knockout of Dnmt1 [156]. Associations between DNA hypomethylation and chromosomal rearrangements in human cancer have been observed [157,165]. There is also cause-and-effect evidence for DNA hypomethylation predisposing to chromosomal rearrangements [154,155,158]. The carcinogenesis-associated role of DNA hypomethylation in activation of transposable elements or endogenous viruses [146] is another example of its role in promoting cancer-predisposing or tumor progression-linked genomic rearrangements. However, the amount of cancer-linked DNA hypomethylation in individual cancer cells versus the frequency of chromosomal rearrangements in those cells suggest other pathways for DNA hypomethylation favoring tumorigenesis and tumor progression, in addition to the enhancement of rearrangements [36].
Indirect effects of hypomethylation of DNA repeats on gene expression
We have proposed another pathway for hypomethylation of DNA repeats contributing to carcinogenesis that involves regulating, in trans, the expression of genes by altering the sequestration of transcription control proteins at DNA repeats. Given the highly reiterated nature of satellite DNA (Alu repeats and LINE-1 elements), these sequences have high potential to act as holding or delivery docks for transcription control proteins. These four categories of DNA repeats are very frequently subject to cancer-linked DNA hypomethylation [5,27,28]. Because DNA methylation affects the DNA binding of many proteins [166–169], transcription control proteins could be sequestered by DNA repeats in a methylation-responsive fashion. This would provide a novel, possibly tissue-specific, pathway by which hypomethylation of DNA repeats could impact gene expression and carcinogenesis.
Examples of mammalian transcription regulators that bind selectively to centromeric or juxtacentromeric heterochromatin by DNA–protein and/or protein–protein interactions are accumulating: Ikaros; ATRX; heat shock factor 1; C/EBPα, C/EBPβ and C/EBPδ; DNMT1, DNMT3A and DNMT3B (which can repress transcription independently of their methylating activities); the MeCP proteins MBD1, MBD2 and MeCP2; HDAC1 and 2; and HP1-associated proteins like TIF1 β [170–178]. Heat shock factor 1 localizes to 9qh and the centromeric regions of chromosomes 12 and 15 upon heat shock [172], with induction of transcription of satellite 3 at chromosome 9 [179]. The transcription is associated with the co-localization of a transcription factor to this heterochromatic region. Additional pathways for transcription-regulatory changes in methylation of nongene DNA repeats may be through alterations in subnuclear compartmentalization of chromatin [27,180–183] or altering transcription of noncoding RNAs [184–187].
Cancer stem cell hypothesis is compatible with demethylation during carcinogenesis
Can the term ‘demethylation’ be applied to the origin of the pervasive hypomethylation of DNA in cancer? In the context of DNA epigenetics, demethylation means the exchange of C residues for m5C residues without implying the mechanism. Instead of DNA demethylation during carcinogenesis, cancer-associated hypomethylation might have been due to selection for a pre-existing hypomethylated state of many DNA sequences in cancer stem cells that give rise to tumors. Studies of stem cells and early embryonic cells clearly indicate that most cancer-linked hypomethylation of DNA repeats, the predominant sequence subject to this hypomethylation, can be described as carcinogenesis-associated demethylation (see below). Moreover, as mentioned above, the frequent finding that increased tumor progression is associated with increased DNA hypomethylation indicates that demethylation must be occurring during tumor progression. Some of the other arguments for DNA demethylation during carcinogenesis follow.
While there is renewed interest in the concept of cancer stem cells, including epigenetic evidence for such cells [188,189], the identification of cancer-initiating cells as normal stem cells is still controversial [190]. Even if normal stem cells are the source of many cancers, it is clear that epigenetic changes in DNA and chromatin are part of the early stages of carcinogenesis [190]. Although DNA methylation changes have been observed in a recent genome-wide comparison of DNA methylation in mouse embryonal stem (ES) cells and ES-derived neural precursor cells, there was only approximately 2% demethylation (which is much less than the levels of global demethylation of the genome routinely observed in cancer [5]) and de novo methylation of CpGs [191].
Importantly, studies of satellite DNA methylation in normal differentiation precursor cells do not support the hypothesis that satellite DNA hypomethylation in cancer is a pre-existing condition in normal stem cells giving rise to tumors. Wild-type mouse embryonic stem cells have normal high levels of satellite DNA methylation [192]. Furthermore, by the early preimplantation stage, murine embryoblasts have remethylated satellite DNA sequences following extensive demethylation in the earliest stages of embryogenesis [193,194]. In addition, our comparison of satellite DNA methylation in Wilms' tumors to that in nephrogenic rests, embryonic kidney precursors of Wilms' tumors, indicated neither satellite DNA hypomethylation nor the promoter hypermethylation in the nephrogenic rests [22]. Similarly, Fanelli et al. found that satellite 2 DNA from glioblastoma-derived brain tumor stem cells was hypomethylated to a similar extent when compared with either normal human astrocytes or normal neural progenitor cells [89]. These findings are consistent with cancer stem cell populations from tumors having undergone DNA demethylation during their generation. Therefore, altered DNA methylation in cancer does not simply reflect the methylation status of the original cancer progenitor cell(s).
What is known & not known about the mechanism of DNA demethylation
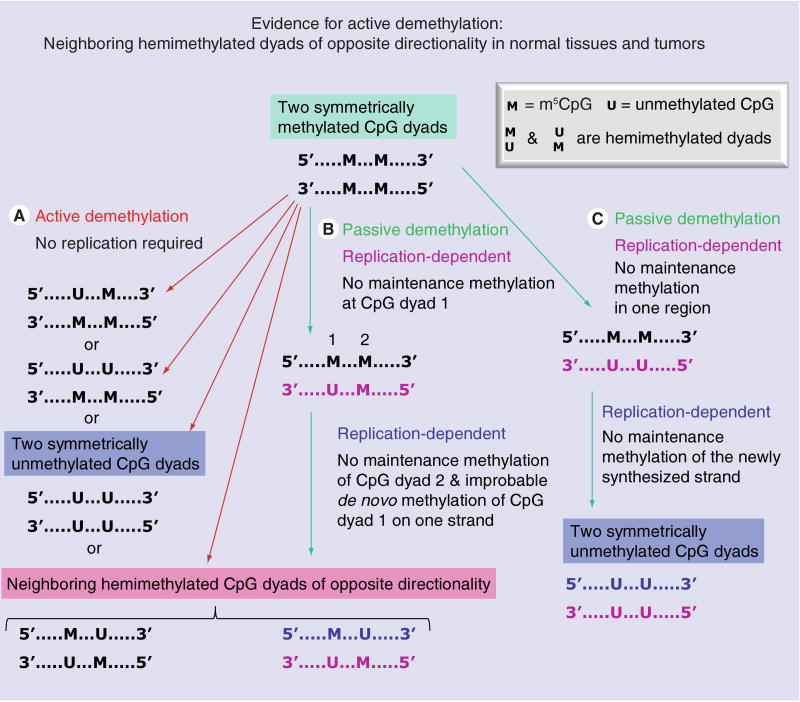
Many studies suggest that there is active DNA demethylation in mammalian cells [40,125,195] rather than just passive demethylation owing to a failure of maintenance methylation (Figure 2). Active demethylation probably first occurs on one strand at a given dyad [40,123]. In murine and bovine zygotes even before DNA replication, there is active demethylation of paternally inherited DNA, which includes demethylation of DNA repeats [196–199]. However, the mechanism is still not clear.
Figure 2. A comparison of active and passive pathways for DNA demethylation and, in particular, for generating neighboring hemimethylated CpG dyads with opposite polarity.
This figure demonstrates that it is unlikely that inhibition of passive demethylation is responsible for producing such DNA methylation patterns, which were observed in both cancers and normal tissues. Although neighboring, oppositely-oriented hemimethylated dyads (pink highlighting) were seen only infrequently, they were found in three different hairpin sequencing studies of DNA methylation and in normal tissues as well as cancer samples [104–106]. We conclude that active demethylation is responsible for at least some of the demethylation in vivo.
The best evidence for active demethylation comes from studies of Arabidopsis, which demonstrate that a DNA glycosylase hydrolyzes the m5C-deoxyribose bond and then the enzyme's lyase activity nicks the backbone at the resulting apyrimidinic site. This is followed by completion of base excision repair with incorporation at the formerly methylated C site of an unmethylated C residue [200]. There has been evidence for various pathways for active demethylation in vertebrates as follows:
An m5C DNA glycosylase [195] for m5C removal as the free base by base-excision repair [195];
A demethylase that catalyzes the release of the 5-methyl group from m5C [201];
A growth arrest protein, GADD45A [125], that may function in nucleotide excision repair to release m5C as a nucleotide [125,126];
DNMT3A or DNMT3B for catalysis of the oxidative deamination of the C4 position of m5C to give a T residue replaced with an unmethylated C residue by mismatch repair [126,128].
There is controversy about each of these pathways [126,202]. Moreover, the existence of active demethylation in mammalian cells has recently been questioned [203].
Our analyses of symmetrical and asymmetrical methylation tandem DNA repeats in normal tissues and cancers give evidence in support of active demethylation in vivo in normal cells, where it may counteract aberrant de novo methylation, and in tumor cells, where it would create at least some of the cancer-linked hypomethylation. We used bisulfite-based hairpin genomic sequencing, which distinguishes CpG dyads that are symmetrically methylated, symmetrically unmethylated or hemimethylated (Figure 2) on individual DNA fragments whose strands are ligated together during the analysis [104]. In diverse normal somatic tissues, we found most CpG dyads highly methylated and a few mostly unmethylated in a 0.2-kb subregion of the 1.4-kb NBL2 repeat unit [105]. Compared with these somatic tissues, ovarian carcinomas and Wilms' tumors displayed hypomethylated CpG dyads next to hypermethylated CpG dyads in this subregion, suggestive of dynamic changes in DNA methylation. Approximately 2–6% of the CpG dyads were hemimethylated (Figures 1 & 2) in control tissues and cancers. These percentages are high enough to suggest that they are fairly stable structures. The dense packing of heterochromatic tandem repeats may prolong the lifetime of hemimethylated intermediates in cancer-linked DNA demethylation, thereby providing longer-lived intermediates in demethylation.
Much of the cancer-associated hypomethylation seen in NBL2 (1.4-kb repeat unit) and in satellite 2 DNA (1.3-kb higher-order repeat unit) was highly focal [105,106]. This was especially apparent in the runs of hemimethylated dyads overrepresented in the ovarian carcinomas and Wilms' tumors. Maintenance methylation is quite processive [204–206]. Therefore, if cancer-linked hypomethylation were owing to the inhibition of maintenance methylation, almost all of the hypomethylation in the examined 0.2-kb subregions would have been expected to occur in long runs, contrary to our findings [105,106].
Most importantly, some of the hemimethylated dyads in normal tissues and tumors in NBL2 and satellite 2 [105,106] were present in runs of the opposite orientation (Figure 2). While a local failure of maintenance methylation on the nascent strand could generate a hemimethylated dyad from a symmetrically methylated dyad, a simple failure of maintenance methylation cannot explain two nearby hemimethylated dyads with their m5C residues on opposite strands (Figure 2). We conclude that there was at least some active demethylation in the normal tissues and in the cancers at these repeats.
In addition, some of the hairpin sequencing DNA clones from the tandem repeats gave evidence for nonrandom clustering of identically unmethylated or hemimethylated sites [105,106]. This suggests localized spreading of DNA demethylation. DNA hypermethylation appears to be able to spread in cis [120,207]. There has been less analysis of the possible spreading of demethylation, which might involve demethylation on one strand at a single CpG dyad [40], followed by multiple, symmetrical demethylation events. The hypothesized spreading of active demethylation might occur by a hopping or sliding mechanism over small distances along a DNA molecule in analogy to how uracil DNA glycosylase (UNG) appears to catalyze the formation of abasic sites over regions less than 100 bp [208]. The spreading of UNG-mediated removal of uracil from DNA can even involve bases on opposite strands [208], as might be occurring during cancer-linked demethylation (Figure 2). Consistent with the UNG study and with evidence for involvement of base-excision repair in DNA demethylation [128], the longest runs of consecutive hemimethylated dyads that we have observed were five CpG dyads extending over about 60 bp in the cancers. The initial cancer-associated demethylation of clustered CpGs might subsequently seed more extensive spreading of demethylation to give rise to the fraction of cancer DNA clones that were derived from completely demethylated DNA sequences.
In a statistical modeling study based upon our hairpin sequencing data from NBL2 and satellite 2 in ovarian carcinomas, we considered three stochastic models for the spreading of methylation patterns [209]. The CpG dyad sites might independently assume their methylation status (independent sites model; Box 1). Alternatively, they might be subject to a ‘snowball effect’ of initial de novo or demethylation leading to fairly homogeneous methylation changes with the positions of such changes being random within the region (context-dependent model). The third model (neighboring sites model) depends upon stochastic growth or shrinkage of regions of epigenetic change, with these changes highly dependent on neighboring sites on both sides of an individual dyad. Our results on these tandem repeats do not fit the first model and best fit the third one. Our data supporting the neighboring sites model is consistent with demonstrated preferences of mammalian DNMTs for specific sequences [210].
Niehrs recently summarized many other studies that offer evidence for active demethylation in animal and plant cells and persuasively argues for maintenance of DNA methylation patterns involving a dynamic equilibrium of active demethylation and de novo methylation in addition to maintenance methylation [126]. He proposes a two-step model for active, strand-specific demethylation of DNA that would involve two different repair pathways in each step to convert m5C residues to T residues at an m5CpG dyad and product inhibition after the first step. This would allow repair of the first strand before repair begins on the second strand, and thereby avoid the introduction of deleterious double-strand breaks. The model fits well with our data on strand asymmetry of hemimethylated sites at the NBL2 repeat in cancers [105]. I further propose that the apparently persistent hemimethylation that we observed at some CpG sites could be due to delays in completing the first rate-limiting repair-demethylation step, for example, due to inappropriate dephosphorylation of a 5′-phosphate end at the site of repair.
Whatever the exact mechanism by which DNA demethylation occurs, it is likely to have diverse proximate causes. There could be decreases in DNMT isoforms, their subcellular distribution or their recruitment to DNA, although the often simultaneous development of cancer-linked demethylation and de novo methylation has to be explained. Among the newly discovered triggers for changes in DNMT3A and DNMT3B levels is microRNA-29b [211]. With respect to modulating chromatin interactions of Dnmts/DNMTs, it has been proposed that the SNP2 chromatin remodeling factor Lsh/LSH interacts with Dnmt3b/DNMT3B to recruit it to certain DNA sequences [83]. Deletion of Lsh causes widespread genomic demethylation, especially in DNA repeats [212] and erythroleukemia [83], as mentioned above. DNMT3B, which has many isoforms [213–215], is especially associated with the methylation of various tandem DNA repeats, including NBL2, satellite 2 and D4Z4 [71,216]. Certain isoforms of DNMT3B might be altered during carcinogenesis such that hypomethylation of a subset of DNA sequences results. In addition, DNMTs have many specific protein-binding partners, which could influence their activity during carcinogenesis. For example, there is evidence that DNMT1 binds to PARP-1 after PARP-1 automodifies itself by poly(ADP-ribosyl)ation, and that this results in inhibition of DNA methylation [217]. In addition, viral and bacterial infections can alter DNA epigenetics, including inducing global DNA hypomethylation by various pathways [218].
How can cancer-linked hypomethylation affect the detection & treatment of cancer?
Diagnosis & prognosis
The association of global or focal DNA hypomethylation with the early stages of carcinogenesis or with tumor progression [1,7,8,20,35,52,55,61,62,67,68,105,149,151,219–221] provides cancer markers that should be very useful in the clinic. In addition, knowledge of which metastasis genes are activated by hypomethylation will be necessary for development of personalized treatment programs that are molecularly based. Translation of research findings to the clinical setting will be facilitated by the recent developments in high-throughput and high-resolution methods for DNA methylation analysis [28,37,98].
Cancer demethylation treatment
DNA methylation inhibitors, including, 5-aza-2′-deoxycytidine (decitabine), 5-azacytidine, 5,6-dihydro-5-azacytidine, zebularine and RNA interference (RNAi) and antisense inhibitors of DNMT1 are being used as cancer chemotherapeutic agents or in clinical trials [222–224]. For solid tumors, usually little or no clinical efficacy, and often no disease stabilization, is seen, and many toxic effects are observed [225–227]. 5-azacytidine and decitabine have had the most success as cancer chemotherapeutic agents for patients with myelodysplastic syndromes (MDS), acute myeloid leukemia and chronic myelogenous leukemia [228–232]. Clinical efficacy for this type of therapy applied to MDS patients has been increased by using optimized treatment schedules that take into account the mechanism of action of demethylating agents [230]. Combination therapies involving DNA demethylating agents have been tried for various cancers and are being refined with differing success rates [231,233–235].
In addition, it has been proposed that the specificity of cancer-testis genes for some cancers could be the basis of a cancer vaccine [236,237]. Treatment of cancer cell lines with decitabine demonstrated that cancer-testis genes are especially sensitive to activation by this agent [238], which suggested that combination therapy with this drug followed by treatment with a vaccine for cancer-testis genes might be efficacious [239]. However, heterogeneous expression of the antigenic products of these genes by cancer cells in some tumors might limit the usefulness of this approach [240].
Mechanism of action of cancer demethylating chemotherapeutic agents
A total of five out of seven patients with MDS who were treated with decitabine displayed decreases of up to 70% in global methylation of bone marrow cells [241]. Major decreases in DNA methylation owing to therapy with these DNA demethylating drugs can be largely reversed after treatment [232,241,242]. Several studies have demonstrated that p15 promoter hypermethylation was reversed by decitabine alone or in combination with the histone deacetylase inhibitor sodium phenybutyrate [234,243,244]. However, the effectiveness of DNA methylation inhibitors as cancer chemotherapeutic agents is partly owing to cytotoxic effects unrelated to their DNA-demethylating activity [245–248]. Decitabine and 5-azacytidine are DNA replication inhibitors as well as inhibitors of DNMT activity upon incorporation into DNA [246,249].
Caution is warranted
There is insufficient long-term survival data on cancer patients treated with DNA methylation inhibitors to evaluate the possible long-term risks associated with these treatments. In considering modalities for therapy for various types of patients, the pro-oncogenic potential of DNA hypomethylation should not be overlooked [250]. This includes associations of DNA hypomethylation with tumor progression, demethylation of promoters of genes that may contribute toward oncogenesis and increased chromosomal rearrangements, as well as experiments implicating DNA hypomethylation as a causal agent in carcinogenesis or tumor progression. Some other cancer therapies also have proven oncogenic potential. Nonetheless, the close association of DNA hypomethylation with oncogenesis and the finding that DNA hypomethylation often becomes more pronounced upon tumor progression, should be factored into clinical decisions, especially if other effective therapies are available for the type of cancer in question.
Conclusion
There is a hierarchy of patterns of DNA epigenetic changes in cancer, starting from the local level of individual CpG dyads to subregions of several hundred base pairs, several thousand base pairs and large regions of up to 1 million base pairs. These changes are the result of aberrant de novo methylation and demethylation, which probably includes active demethylation through a hemimethylated intermediate. Both demethylation and de novo methylation occur early in tumorigenesis, but often increase with tumor progression. Given the evidence causally linking DNA hypomethylation to cancer and the near ubiquity of cancer-associated DNA hypomethylation, it is likely that this loss of methylation plays important roles in tumor formation and progression which underscore the need for caution in design of DNA demethylation strategies to counteract hypermethylation-dependent silencing of tumor suppressor and homeobox genes.
Future perspective
Over the next 5–10 years, there will probably be many clinical tests in use that are based upon DNA hypomethylation markers for cancer detection and prognosis. In addition, in the era of personalized cancer medicine that is soon to be commonplace, DNA hypomethylation markers and oncogene targets will be examined for their methylation status to characterize cancers and design the best treatment for a given tumor and individual. Eventually, treatments will be developed for reversing cancer-causing gene-promoter hypermethylation that will be targeted to specific gene regions. This will replace the current blunt-force approaches to genome-wide demethylation that may be causing tumor progression as a late side effect. Lastly, the mechanisms for cancer-associated de novo methylation and demethylation will be elucidated with convincing detail, and lead to new kinds of epigenetic-based medicine.
Executive summary.
Overview
DNA hypomethylation is a ubiquitous feature of carcinogenesis.
Most cancer-linked hypomethylation is in repeated DNA sequences, but gene regions show some too.
DNA hypomethylation can be found early in carcinogenesis, but also is often associated with tumor progression.
There are confounding factors to consider in assessing cancer-associated changes in DNA methylation.
DNA hypomethylation patterns can be tumor-specific just as DNA hypermethylation patterns are.
Interrelationships of cancer-linked hypo- & hyper-methylation
Cancer-linked hypo- and hyper-methylation are generally independent although most studied cancers display both changes in their genome.
Some DNA sequences display cancer-linked hyper- and hypo-methylation.
Cancer-linked DNA methylation changes can be dynamic.
DNA hypomethylation: mechanisms
The cancer stem cell hypothesis is compatible with demethylation during carcinogenesis.
Hypomethylation of DNA repeats can promote carcinogenesis by increasing DNA recombination and by direct and indirect effects on gene expression.
There is evidence for active demethylation in normal tissues and cancers, although the mechanism is still unclear.
The patterns of methylation in CpG dyads in repeated sequences favor a ‘neighboring sites’ model for DNA methylation changes.
Clinical applications of research on DNA hypomethylation in cancer
Diagnosis and prognosis: Repeated DNA sequences that are frequently hypomethylated in cancer can provide markers for cancer diagnosis and prognosis, and are even more sensitive markers than unique sequences that are subject to cancer-linked DNA hypermethylation.
Treatment: DNA demethylation therapy alone or in combination with other therapies is used for certain leukemias and myelodysplastic syndromes.
Cautions: Treatment that involves demethylating DNA may inadvertently foster metastasis; methods should be developed for targeting the demethylation to promoters of tumor suppressor genes.
Footnotes
In memoriam: This article is dedicated to the memory of Charles Gehrke, PhD and Professor of Biochemistry, University of Missouri, Columbia. His expertise in analytical biochemistry and gracious willingness to collaborate with me, starting in 1979, when I was very much his junior, were critical to launching my career in the then nonexistent field of cancer epigenetics [1,84].
Financial & competing interests disclosure: The author is supported in part by grant R01 NS048859 from the National Institutes of Health and a grant from the Louisiana Cancer Research Consortium. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 4.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 5.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 6.de Capoa A, Musolino A, Della Rosa S, et al. DNA demethylation is directly related to tumour progression: evidence in normal, pre-malignant and malignant cells from uterine cervix samples. Oncol Rep. 2003;10:545–549. [PubMed] [Google Scholar]
- 7.Brothman AR, Swanson G, Maxwell TM, et al. Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Cancer Genet Cytogenet. 2005;156:31–36. doi: 10.1016/j.cancergencyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Seifert HH, Schmiemann V, Mueller M, et al. In situ detection of global DNA hypomethylation in exfoliative urine cytology of patients with suspected bladder cancer. Exp Mol Pathol. 2007;82(3):292–297. doi: 10.1016/j.yexmp.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 10.Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 11.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 12.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumor-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 13.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 14.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen C, Liang G, Nguyen TT, et al. Susceptibility of nonpromoter CpG islands to de novo methylation in normal and neoplastic cells. J Natl Cancer Inst. 2001;93:1465–1472. doi: 10.1093/jnci/93.19.1465. [DOI] [PubMed] [Google Scholar]
- 16.Cheng P, Schmutte C, Cofer KF, Felix JC, Yu MC, Dubeau L. Alterations in DNA methylation are early, but not initial, events in ovarian tumorigenesis. Br J Cancer. 1997;75:396–402. doi: 10.1038/bjc.1997.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M, Woods C, Yu M, et al. Quantitative analysis of association between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlfors J, Hiltunen H, Heinonen K, Hamalainen E, Alhonen L, Janne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992;80:2074–2080. [PubMed] [Google Scholar]
- 19.Lin CH, Hsieh SY, Sheen IS, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61:4238–4243. [PubMed] [Google Scholar]
- 20.Kim YI, Giuliano A, Hatch KD, et al. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 22.Ehrlich M, Jiang G, Fiala ES, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]; • Embryonic kidney precursors (nephrogenic rests) of Wilms' tumors have neither satellite DNA hypomethylation nor the promoter hypermethylation seen in the Wilms' tumors.
- 23.Morey SR, Smiraglia DJ, James SR, et al. DNA methylation pathway alterations in an autochthonous murine model of prostate cancer. Cancer Res. 2006;66:11659–11667. doi: 10.1158/0008-5472.CAN-06-1937. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama R, Qi L, Tsumagari K, et al. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol Ther. 2005;4:440–448. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- 25.Gama-Sosa MA, Wang RY, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smiraglia DJ, Rush LJ, Fruhwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83:296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan A, Ji W, Zhang XY, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Qu G, Dubeau L, Narayan A, Yu M, Ehrlich M. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mut Res. 1999;423:91–101. doi: 10.1016/s0027-5107(98)00229-2. [DOI] [PubMed] [Google Scholar]
- 31.Qu G, Grundy PE, Narayan A, Ehrlich M. Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genet Cytogenet. 1999;109:34–39. doi: 10.1016/s0165-4608(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 32.Florl AR, Steinhoff C, Muller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol. 2009;22:442–449. doi: 10.1038/modpathol.2008.203. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez J, Vives L, Jorda M, et al. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widschwendter M, Jiang G, Woods C, et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 36.Ehrlich M, Hopkins N, Jiang G, et al. Satellite hypomethylation in karyotyped Wilms tumors. Cancer Genet Cytogenet. 2003;141:97–105. doi: 10.1016/s0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- 37.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 39.Wasson GR, McGlynn AP, McNulty H, et al. Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr. 2006;136:2748–2753. doi: 10.1093/jn/136.11.2748. [DOI] [PubMed] [Google Scholar]
- 40.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Lindsey JC, Lusher ME, Anderton JA, Gilbertson RJ, Ellison DW, Clifford SC. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br J Cancer. 2007;97:267–274. doi: 10.1038/sj.bjc.6603852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grunau C, Brun ME, Rivals I, et al. BAGE hypomethylation, a new epigenetic biomarker for colon cancer detection. Cancer Epidemiol Biomarkers Prev. 2008;17:1374–1379. doi: 10.1158/1055-9965.EPI-07-2656. [DOI] [PubMed] [Google Scholar]
- 44.Ortmann CA, Eisele L, Nuckel H, et al. Aberrant hypomethylation of the cancer-testis antigen PRAME correlates with PRAME expression in acute myeloid leukemia. Ann Hematol. 2008;87:809–818. doi: 10.1007/s00277-008-0514-8. [DOI] [PubMed] [Google Scholar]
- 45.Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68:8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pakneshan P, Tetu B, Rabbani SA. Demethylation of urokinase promoter as a prognostic marker in patients with breast carcinoma. Clin Cancer Res. 2004;10:3035–3041. doi: 10.1158/1078-0432.ccr-03-0545. [DOI] [PubMed] [Google Scholar]
- 47.Pulukuri SM, Estes N, Patel J, Rao JS. Demethylation-linked activation of urokinase plasminogen activator is involved in progression of prostate cancer. Cancer Res. 2007;67:930–939. doi: 10.1158/0008-5472.CAN-06-2892. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Ateeq B, Unterberger A, Szyf M, Rabbani SA. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 2008;10:266–278. doi: 10.1593/neo.07947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosty C, Ueki T, Argani P, et al. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- 52.Lee YM, Lee JY, Kim MJ, et al. Hypomethylation of the protein gene product 9.5 promoter region in gallbladder cancer and its relationship with clinicopathological features. Cancer Sci. 2006;97:1205–1210. doi: 10.1111/j.1349-7006.2006.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye L, Li X, Kong X, et al. Hypomethylation in the promoter region of POMC gene correlates with ectopic overexpression in thymic carcinoids. J Endocrinol. 2005;185:337–343. doi: 10.1677/joe.1.05963. [DOI] [PubMed] [Google Scholar]
- 54.Kaneda A, Tsukamoto T, Takamura-Enya T, et al. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Sci. 2004;95:58–64. doi: 10.1111/j.1349-7006.2004.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunau C, Sanchez C, Ehrlich M, et al. Frequent DNA hypomethylation in the human juxtacentromeric BAGE loci in cancer. Genes Chrom Cancer. 2005;43:11–24. doi: 10.1002/gcc.20155. [DOI] [PubMed] [Google Scholar]
- 56.Salem C, Liang G, Tsai YC, et al. Progressive increases in de novo methylation of CpG islands in bladder cancer. Cancer Res. 2000;60:2473–2476. [PubMed] [Google Scholar]
- 57.Fruhwald MC, O'Dorisio MS, Dai Z, et al. Aberrant promoter methylation of previously unidentified target genes is a common abnormality in medulloblastomas – implications for tumor biology and potential clinical utility. Oncogene. 2001;20:5033–5042. doi: 10.1038/sj.onc.1204613. [DOI] [PubMed] [Google Scholar]
- 58.Brock MV, Gou M, Akiyama Y, et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res. 2003;9:2912–2919. [PubMed] [Google Scholar]
- 59.Jackson K, Yu M, Arakawa K, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K, Suzuki I, Leodolter A, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Repetitive DNA hypomethylation in the advanced phase of chronic myeloid leukemia. Leuk Res. 2008;32:487–490. doi: 10.1016/j.leukres.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 62.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009 doi: 10.1002/path.2596. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 63.Markl ID, Cheng J, Liang G, Shibata D, Laird PW, Jones PA. Global and gene-specific epigenetic patterns in human bladder cancer genomes are relatively stable in vivo and in vitro over time. Cancer Res. 2001;61:5875–5884. [PubMed] [Google Scholar]
- 64.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 65.Shen L, Fang J, Qiu D, et al. Correlation between DNA methylation and pathological changes in human hepatocellular carcinoma. Hepatogastroenterology. 1998;45:1753–1759. [PubMed] [Google Scholar]
- 66.Soares J, Pinto AE, Cunha CV, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- 67.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 68.Itano O, Ueda M, Kikuchi K, et al. Correlation of postoperative recurrence in hepatocellular carcinoma with demethylation of repetitive sequences. Oncogene. 2002;21:789–797. doi: 10.1038/sj.onc.1205124. [DOI] [PubMed] [Google Scholar]
- 69.Ehrlich M. The controversial denouement of vertebrate DNA methylation research. Biochemistry (Mosc) 2005;70:568–575. doi: 10.1007/s10541-005-0150-z. [DOI] [PubMed] [Google Scholar]
- 70.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132:S2401–S2405. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 71.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 72.Gisselsson D, Shao C, Tuck-Muller C, et al. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma. 2005;114:118–126. doi: 10.1007/s00412-005-0343-7. [DOI] [PubMed] [Google Scholar]
- 73.Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- 74.Kersh EN, Fitzpatrick DR, Murali-Krishna K, et al. Rapid demethylation of the IFN-γ gene occurs in memory but not naive CD8 T cells. J Immunol. 2006;176:4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 75.Chang HS, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147:5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- 76.Murayama A, Sakura K, Nakama M, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2006;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 78.Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 80.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein γ gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 82.Ogishima T, Shiina H, Breault JE, et al. Increased heparanase expression is caused by promoter hypomethylation and upregulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 83.Fan T, Schmidtmann A, Xi S, et al. DNA hypomethylation caused by Lsh deletion promotes erythroleukemia development. Epigenetics. 2008;3:134–142. doi: 10.4161/epi.3.3.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ehrlich M, Gama-Sosa M, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gama-Sosa MA, Midgett RM, Slagel VA, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740:212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- 86.Khalkhali-Ellis Z. Maspin: the new frontier. Clin Cancer Res. 2006;12:7279–7283. doi: 10.1158/1078-0432.CCR-06-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim JY, Siegmund KD, Tavare S, Shibata D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 2005;3:10. doi: 10.1186/1741-7015-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eads CA, Lord RV, Kurumboor SK, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 89.Fanelli M, Caprodossi S, Ricci-Vitiani L, et al. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene. 2008;27:358–365. doi: 10.1038/sj.onc.1210642. [DOI] [PubMed] [Google Scholar]
- 90.Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol. 2009;4:229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- 91.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 92.Yamada Y, Watanabe H, Miura F, et al. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–266. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 94.Flanagan JM, Popendikyte V, Pozdniakovaite N, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tra J, Kondo T, Lu Q, Kuick R, Hanash S, Richardson B. Infrequent occurrence of age-dependent changes in CpG island methylation as detected by restriction landmark genome scanning. Mech Ageing Dev. 2002;123:1487–1503. doi: 10.1016/s0047-6374(02)00080-5. [DOI] [PubMed] [Google Scholar]
- 96.Issa JP. Age-related epigenetic changes and the immune system. Clin Immunol. 2003;109:103–108. doi: 10.1016/s1521-6616(03)00203-1. [DOI] [PubMed] [Google Scholar]
- 97.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem. 2009;55:1471–1483. doi: 10.1373/clinchem.2008.121962. [DOI] [PubMed] [Google Scholar]
- 99.Sepulveda AR, Jones D, Ogino S, et al. CpG methylation analysis – current status of clinical assays and potential applications in molecular diagnostics: a report of the association for molecular pathology. J Mol Diagn. 2009;11:266–278. doi: 10.2353/jmoldx.2009.080125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu T, Burke B, Bunce K, Surti U, Allen Hogge W, Peters DG. A microarray-based approach for the identification of epigenetic biomarkers for the noninvasive diagnosis of fetal disease. Prenat Diagn. 2009;29(11):1020–1030. doi: 10.1002/pd.2335. [DOI] [PubMed] [Google Scholar]
- 101.Rodenhiser DI, Andrews J, Kennette W, et al. Epigenetic mapping and functional analysis in a breast cancer metastasis model using whole-genome promoter tiling microarrays. Breast Cancer Res. 2008;10:R62. doi: 10.1186/bcr2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azhikina T, Gainetdinov I, Skvortsova Y, Sverdlov E. Methylation-free site patterns along a 1-Mb locus on Chr19 in cancerous and normal cells are similar. A new fast approach for analyzing unmethylated CCGG sites distribution. Mol Genet Genomics. 2006;275:615–622. doi: 10.1007/s00438-006-0111-2. [DOI] [PubMed] [Google Scholar]
- 103.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laird CD, Pleasant ND, Clark AD, et al. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci USA. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nishiyama R, Qi L, Lacey M, Ehrlich M. Both hypomethylation and hypermethylation in a 0.2-kb region of a DNA repeat in cancer. Mol Cancer Res. 2005;3:617–626. doi: 10.1158/1541-7786.MCR-05-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]; • NBL2 tandem repeats are extensively hypomethylated in some cancer specimens and hypermethylated in others, and have hypermethylated CpG dyads near hypomethylated dyads.
- 106.Shao C, Lacey M, Dubeau L, Ehrlich M. Hemimethylation footprints of DNA demethylation in cancer. Epigenetics. 2009;4:165–175. doi: 10.4161/epi.4.3.8277. [DOI] [PubMed] [Google Scholar]; • A satellite DNA sequence can have focal hypermethylation as well as overall hypomethylation in cancers and nonrandom runs of hemimethylated dyads.
- 107.Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009;69:5251–5258. doi: 10.1158/0008-5472.CAN-08-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res. 2008;6:1365–1374. doi: 10.1158/1541-7786.MCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsumagari K, Qi L, Jackson K, et al. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36:2196–2207. doi: 10.1093/nar/gkn055. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Sequences exhibiting cancer-linked hypermethylation can be adjacent to sequences displaying cancer-linked hypomethylation.
- 110.Choi SH, Worswick S, Byun HM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125:723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pogribny IP, Miller BJ, James SJ. Alterations in hepatic p53 gene methylation patterns during tumor progression with folate/methyl deficiency in the rat. Cancer Lett. 1997;115:31–38. doi: 10.1016/s0304-3835(97)04708-3. [DOI] [PubMed] [Google Scholar]
- 112.Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 113.Lee HS, Kim BH, Cho NY, et al. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15:812–820. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- 114.Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4–20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- 115.Deng G, Nguyen A, Tanaka H, et al. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. Int J Cancer. 2006;118:2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- 116.Nagarajan RP, Costello JF. Molecular epigenetics and genetics in neuro-oncology. Neurotherapeutics. 2009;6:436–446. doi: 10.1016/j.nurt.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Katargin AN, Pavlova LS, Kisseljov FL, Kisseljova NP. Hypermethylation of genomic 3.3-kb repeats is frequent event in HPV-positive cervical cancer. BMC Med Genomics. 2009;2:30. doi: 10.1186/1755-8794-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 120.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16(Spec 1):R88–95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- 121.Cheng CW, Wu PE, Yu JC, et al. Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene. 2001;20:3814–3823. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 122.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 123.Saluz HP, Jiricny J, Jost JP. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci USA. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pfeifer GP, Steigerwald SD, Hansen RS, Gartler SM, Riggs AD. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Evidence for a dynamic methylation system of a study of cloned DNA molecules from an X-linked gene in cultured cells with or without treatment with a DNA inhibitor.
- 125.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 126.Niehrs C. Active demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]; •• This excellent review of the past literature carefully weighs the evidence for active demethylation in verterbrates and provides a cogent model.
- 127.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]; • DNMT3A or DNMT3B catalyzing the oxidative deamination of the C4 position of m5C to give a T residue that is excised by mismatch repair and replaced with an unmethylated deoxycytidine monophosphate residue.
- 129.Riclet R, Chendeb M, Vonesch JL, et al. Disruption of the interaction between transcriptional intermediary factor 1{β} and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol Biol Cell. 2009;20:296–305. doi: 10.1091/mbc.E08-05-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yegnasubramanian S, Haffner MC, Zhang Y, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carr BI, Reilly G, Smith SS, Winberg C, Riggs A. The tumorigenicity of 5-azacytidine in the male Fisher rat. Carcinogenesis. 1984;5:1583–1590. doi: 10.1093/carcin/5.12.1583. [DOI] [PubMed] [Google Scholar]
- 132.Thomas GA, Williams ED. Production of thyroid tumours in mice by demethylating agents. Carcinogenesis. 1992;13:1039–1042. doi: 10.1093/carcin/13.6.1039. [DOI] [PubMed] [Google Scholar]
- 133.Denda A, Rao PM, Rajalakshmi S, Sarma DS. 5-azacytidine potentiates initiation induced by carcinogens in rat liver. Carcinogenesis. 1985;6:145–146. doi: 10.1093/carcin/6.1.145. [DOI] [PubMed] [Google Scholar]
- 134.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 135.Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Transgenic mice containing a hypomorphic mutant Dnmt1 allele linked to DNA hypomethylation had increased formation of hepatocellular adenomas plus carcinomas despite the decreased formation of macroscopic intestinal adenomas.
- 136.Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26:185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 137.Pogribny IP, Basnakian AG, Miller BJ, Lopatina NG, Poirier LA, James SJ. Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res. 1995;55:1894–1901. [PubMed] [Google Scholar]
- 138.Pogribny IP, Ross SA, Wise C, et al. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 139.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:S2393–S2400. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 140.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Toffoli G, De Mattia E. Pharmacogenetic relevance of MTHFR polymorphisms. Pharmacogenomics. 2008;9:1195–1206. doi: 10.2217/14622416.9.9.1195. [DOI] [PubMed] [Google Scholar]
- 142.Linhart HG, Troen A, Bell GW, et al. Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology. 2009;136:227–235. doi: 10.1053/j.gastro.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Laird PW, Jackson-Grusby L, Fazeli A, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 144.Lin H, Yamada Y, Nguyen S, et al. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, 3rd, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 146.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]; • A hypomorphic allele of Dnmt1 in mice resulted in thymic lymphomas that displayed frequent insertion of a transposable element into the Notch oncogene.
- 147.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–2261. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tycko B. Genomic imprinting and human neoplasia. In: Ehrlich M, editor. DNA and Alterations in Cancer: Genetic and Epigenetic Alterations. Eaton Publishing; MA, USA: 2000. pp. 333–349. [Google Scholar]
- 149.Czekierdowski A, Czekierdowska S, Wielgos M, Smolen A, Kaminski P, Kotarski J. The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-γ (SNCG) in ovarian cancer. Neuro Endocrinol Lett. 2006;27:381–386. [PubMed] [Google Scholar]
- 150.Zhao W, Liu H, Liu W, et al. Abnormal activation of the synuclein-γ gene in hepatocellular carcinomas by epigenetic alteration. Int J Oncol. 2006;28:1081–1088. [PubMed] [Google Scholar]
- 151.Liu H, Liu W, Wu Y, et al. Loss of epigenetic control of synuclein-γ gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65:7635–7643. doi: 10.1158/0008-5472.CAN-05-1089. [DOI] [PubMed] [Google Scholar]
- 152.Chekhun VF, Lukyanova NY, Kovalchuk O, Tryndyak VP, Pogribny IP. Epigenetic profiling of multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals novel hyper- and hypomethylated targets. Mol Cancer Ther. 2007;6:1089–1098. doi: 10.1158/1535-7163.MCT-06-0663. [DOI] [PubMed] [Google Scholar]
- 153.Shen R, Tao L, Xu Y, Chang S, Van Brocklyn J, Gao JX. Reversibility of aberrant global DNA and estrogen receptor-α gene methylation distinguishes colorectal precancer from cancer. Int J Clin Exp Pathol. 2009;2:21–33. [PMC free article] [PubMed] [Google Scholar]; • Global DNA hypomethylation and estrogen receptor-α gene hypermethylation seen in colorectal tumors were more reversible by treating patients with a nonsteroidal anti-inflammatory drug at the adenomatous polyp stage than at the adenocarcinoma stage.
- 154.Kokalj-Vokac N, Almeida A, Viegas-Pequignot E, Jeanpierre M, Malfoy B, Dutrillaux B. Specific induction of uncoiling and recombination by azacytidine in classical satellite-containing constitutive heterochromatin. Cytogenet Cell Genet. 1993;63:11–15. doi: 10.1159/000133492. [DOI] [PubMed] [Google Scholar]
- 155.Ji W, Hernandez R, Zhang XY, et al. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res. 1997;379:33–41. doi: 10.1016/s0027-5107(97)00088-2. [DOI] [PubMed] [Google Scholar]
- 156.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 157.Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001;159:465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 159.Jeanpierre M, Turleau C, Aurias A, et al. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 160.Tuck-Muller CM, Narayan A, Tsien F, et al. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 161.Vilain A, Bernardino J, Gerbault-Seureau M, et al. DNA methylation and chromosome instability in lymphoblastoid cell lines. Cytogenet Cell Genet. 2000;90:93–101. doi: 10.1159/000015641. [DOI] [PubMed] [Google Scholar]
- 162.Tsien F, Youn B, Fiala ES, et al. Prolonged culture of chorionic villus cells yields ICF syndrome-like chromatin decondensation and rearrangements. Cytogen Genome Res. 2002;98:13–21. doi: 10.1159/000068543. [DOI] [PubMed] [Google Scholar]
- 163.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15(Spec):417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 164.Ehrlich M. DNA methylation and cancer-associated genetic instability. In: Nigg E, editor. Genome Instability and Cancer. Kluwer; NY, USA: 2004. [Google Scholar]
- 165.Schulz WA, Elo JP, Florl AR, et al. Genome-wide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- 166.Ehrlich M, Ehrlich KC. Effect of DNA methylation on the binding of vertebrate and plant proteins to DNA. In: Jost JP, Saluz HP, editors. DNA Methylation: Biological Significance. Birkhauser Verlag; MA, USA: 1993. pp. 145–168. [DOI] [PubMed] [Google Scholar]
- 167.Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–3173. doi: 10.1038/sj.onc.1204340. [DOI] [PubMed] [Google Scholar]
- 168.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 169.Xu Y, Sengupta PK, Seto E, Smith BD. Regulatory factor for X-box family proteins differentially interact with histone deacetylases to repress collagen a2(I) gene (COL1A2) expression. J Biol Chem. 2006;281:9260–9270. doi: 10.1074/jbc.M511724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J. 2001;20:2812–2822. doi: 10.1093/emboj/20.11.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gibbons RJ, McDowell TL, Raman S, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 172.Denegri M, Moralli D, Rocchi M, et al. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol Biol Cell. 2002;13:2069–2079. doi: 10.1091/mbc.01-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 176.Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 177.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. Cell differentiation induces TIF1β association with centromeric heterochromatin via an HP1 interaction. J Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- 179.Jolly C, Metz A, Govin J, et al. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gasser SM. Positions of potential: nuclear organization and gene expression. Cell. 2001;104:639–642. doi: 10.1016/s0092-8674(01)00259-8. [DOI] [PubMed] [Google Scholar]
- 181.Kosak ST, Skok JA, Medina KL, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 182.Papait R, Pistore C, Grazini U, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol Biol Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Enukashvily NI, Malashicheva AB, Waisertreiger IS. Satellite DNA spatial localization and transcriptional activity in mouse embryonic E-14 and IOUD2 stem cells. Cytogenet Genome Res. 2009;124:277–287. doi: 10.1159/000218132. [DOI] [PubMed] [Google Scholar]
- 184.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 185.Rizzi N, Denegri M, Chiodi I, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 187.Lee KH, Lotterman C, Karikari C, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 189.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 190.Werbowetski-Ogilvie TE, Bhatia M. Pluripotent human stem cell lines: what we can learn about cancer initiation. Trends Mol Med. 2008;14:323–332. doi: 10.1016/j.molmed.2008.06.005. [DOI] [PubMed] [Google Scholar]; • The identification of cancer-initiating cells as normal stem cells is still controversial.
- 191.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gowher H, Stuhlmann H, Felsenfeld G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev. 2008;22:2075–2084. doi: 10.1101/gad.1658408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chapman V, Forrester L, Sanford J, Hastie N, Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- 194.Sanford JP, Clark HJ, Chapman VM, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- 195.Jost JP, Oakeley EJ, Zhu B, et al. 5-methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29:4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 197.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 198.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 199.Park JS, Jeong YS, Shin ST, Lee KK, Kang YK. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev Dyn. 2007;236:2523–2533. doi: 10.1002/dvdy.21278. [DOI] [PubMed] [Google Scholar]
- 200.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 201.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 202.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:E1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 204.Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J Biol Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- 205.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 206.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- 207.Orend G, Kuhlmann I, Doerfler W. Spreading of DNA methylation across integrated foreign (adenovirus type 12) genomes in mammalian cells. J Virol. 1991;65:4301–4308. doi: 10.1128/jvi.65.8.4301-4308.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci USA. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lacey M, Ehrlich M. Modeling dependence in methylation patterns with application to ovarian carcinomas. Stat Appl Genet Mol Biol. 2009;8:40. doi: 10.2202/1544-6115.1489. [DOI] [PubMed] [Google Scholar]; • Evidence for a neighboring sites model governing DNA methylation changes from a statistical analysis.
- 210.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 211.Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 213.Ostler KR, Davis EM, Payne SL, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nimura K, Ishida C, Koriyama H, et al. Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation. Genes Cells. 2006;11:1225–1237. doi: 10.1111/j.1365-2443.2006.01012.x. [DOI] [PubMed] [Google Scholar]
- 215.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 216.Jiang YL, Rigolet M, Bourc'his D, et al. DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum Mutat. 2005;25:56–63. doi: 10.1002/humu.20113. [DOI] [PubMed] [Google Scholar]
- 217.Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 2009;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- 218.Minarovits J. Microbe-induced epigenetic alterations in host cells: the coming era of patho-epigenetics of microbial infections. A review Acta Microbiol Immunol Hung. 2009;56:1–19. doi: 10.1556/AMicr.56.2009.1.1. [DOI] [PubMed] [Google Scholar]
- 219.Piyathilake CJ, Frost AR, Bell WC, et al. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol. 2001;32:856–862. doi: 10.1053/hupa.2001.26471. [DOI] [PubMed] [Google Scholar]
- 220.Hernandez-Blazquez FJ, Habib M, Dumollard JM, et al. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–693. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Galusca B, Dumollard JM, Lassandre S, et al. Global DNA methylation evaluation: potential complementary marker in differential diagnosis of thyroid neoplasia. Virchows Arch. 2005;447:18–23. doi: 10.1007/s00428-005-1268-5. [DOI] [PubMed] [Google Scholar]
- 222.Lubbert M, Wijermans P, Kunzmann R, et al. Cytogenetic responses in high-risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5-aza-2′- deoxycytidine. Br J Haematol. 2001;114:349–357. doi: 10.1046/j.1365-2141.2001.02933.x. [DOI] [PubMed] [Google Scholar]
- 223.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 224.Mack GS. Epigenetic cancer therapy makes headway. J Natl Cancer Inst. 2006;98:1443–1444. doi: 10.1093/jnci/djj447. [DOI] [PubMed] [Google Scholar]
- 225.Sessa C, ten Bokkel Huinink W, Stoter G, Renard J, Cavalli F. Phase II study of 5-aza-2′-deoxycytidine in advanced ovarian carcinoma. The EORTC Early Clinical Trials Group. Eur J Cancer. 1990;26:137–138. doi: 10.1016/0277-5379(90)90295-5. [DOI] [PubMed] [Google Scholar]
- 226.Vermorken JB, Tumolo S, Roozendaal KJ, Guastalla JP, Splinter TA, Renard J. 5-aza-2′-deoxycytidine in advanced or recurrent cancer of the uterine cervix. Eur J Cancer. 1991;27:216–217. doi: 10.1016/0277-5379(91)90493-w. [DOI] [PubMed] [Google Scholar]
- 227.Aparicio A, Weber JS. Review of the clinical experience with 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumors. Curr Opin Investig Drugs. 2002;3:627–633. [PubMed] [Google Scholar]
- 228.Silverman LR. Targeting hypomethylation of DNA to achieve cellular differentiation in myelodysplastic syndromes (MDS) Oncologist. 2001;6:8–14. doi: 10.1634/theoncologist.6-suppl_5-8. [DOI] [PubMed] [Google Scholar]
- 229.Kornblith AB, Herndon JE, 2nd, Silverman LR, et al. Impact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized Phase III trial: a Cancer and Leukemia Group B study. J Clin Oncol. 2002;20:2441–2452. doi: 10.1200/JCO.2002.04.044. [DOI] [PubMed] [Google Scholar]
- 230.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 231.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Issa JP, Gharibyan V, Cortes J, et al. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol. 2005;23:3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 233.Reu FJ, Bae SI, Cherkassky L, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 234.Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 235.Koschmieder S, Agrawal S, Radomska HS, et al. Decitabine and vitamin D3 differentially affect hematopoietic transcription factors to induce monocytic differentiation. Int J Oncol. 2007;30:349–355. [PubMed] [Google Scholar]
- 236.Watanabe Y, LePage S, Elliott M, et al. Characterization of preexisting humoral immunity specific for two cancer-testis antigens overexpressed at the mRNA level in non-small cell lung cancer. Cancer Immun. 2006;6:3. [PubMed] [Google Scholar]
- 237.Hoeppner LH, Dubovsky JA, Dunphy EJ, McNeel DG. Humoral immune responses to testis antigens in sera from patients with prostate cancer. Cancer Immun. 2006;6:1. [PubMed] [Google Scholar]
- 238.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 239.Dubovsky JA, McNeel DG, Powers JJ, Gordon J, Sotomayor EM, Pinilla-Ibarz JA. Treatment of chronic lymphocytic leukemia with a hypomethylating agent induces expression of NXF2, an immunogenic cancer testis antigen. Clin Cancer Res. 2009;15:3406–3415. doi: 10.1158/1078-0432.CCR-08-2099. [DOI] [PubMed] [Google Scholar]
- 240.Gjerstorff MF, Johansen LE, Nielsen O, Kock K, Ditzel HJ. Restriction of GAGE protein expression to subpopulations of cancer cells is independent of genotype and may limit the use of GAGE proteins as targets for cancer immunotherapy. Br J Cancer. 2006;94:1864–1873. doi: 10.1038/sj.bjc.6603163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Mund C, Hackanson B, Stresemann C, Lubbert M, Lyko F. Characterization of DNA demethylation effects induced by 5-aza-2′-deoxycytidine in patients with myelodysplastic syndrome. Cancer Res. 2005;65:7086–7090. doi: 10.1158/0008-5472.CAN-05-0695. [DOI] [PubMed] [Google Scholar]
- 242.Garcia-Manero G. Demethylating agents in myeloid malignancies. Curr Opin Oncol. 2008;20:705–710. doi: 10.1097/CCO.0b013e328313699c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Daskalakis M, Nguyen TT, Nguyen C, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 244.Yang AS, Doshi KD, Choi SW, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 245.Davidson S, Crowther P, Radley J, Woodcock D. Cytotoxicity of 5-aza-2′-deoxycytidine in a mammalian cell system. Eur J Cancer. 1992;28:362–368. doi: 10.1016/s0959-8049(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 246.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Scott SA, Dong WF, Ichinohasama R, et al. 5-aza-2′-deoxycytidine (decitabine) can relieve p21WAF1 repression in human acute myeloid leukemia by a mechanism involving release of histone deacetylase 1 (HDAC1) without requiring p21WAF1 promoter demethylation. Leuk Res. 2006;30:69–76. doi: 10.1016/j.leukres.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 248.Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–2085. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- 249.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci USA. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ehrlich M, Jiang G. DNA Hypo- vs hypermethylation in cancer: tumor specificity, tumor progression, and therapeutic implications. In: Szyf M, editor. DNA Methylation and Cancer. Kluwer Academic/Plenum Publishers and Landes Bioscience/Eurekah.com; Washington, DC, USA: 2003. [Google Scholar]
- 301.Gene cards website www.genecards.org