Abstract
Human papillomavirus type 16 (HPV) E1 protein provides helper function for the adeno-associated virus type 2 (AAV) life cycle. E1 is the replication protein of HPV, analogous to AAV Rep78, but without the endonuclease/covalent attachment activity of Rep78. Previously we have shown that E1 and Rep78 interact in vitro. Here we investigated E1’s effects on Rep78 interaction with AAV’s inverted terminal repeat (ITR) DNA in vitro, using purified Rep78 and E1 proteins from bacteria. E1 enhanced Rep78-ITR binding, ATPase activity, Rep78-ITR covalent linkage and Rep78-ITR-endonuclease activity (central to AAV replication). These enhancements occurred in a dose-dependent manner whenever assayed. However, overall Rep78-plus-E1 helicase activity was lower than Rep78’s helicase activity. These data suggest that E1’s broad-based helper function for the AAV life cycle (AAV DNA, mRNA, and protein levels are up-regulated by E1) is likely through its ability to enhance Rep78’s critical replication-required biochemistries on ITR DNA.
Introduction
Adeno-associated virus type 2 is a nonpathogenic human parvovirus with a single-stranded 4.68 kb genome. A unique feature of AAV biology is that productive infection usually requires simultaneous infection of a second virus, helper virus, such as adenovirus (Ad) (Atchison et al, 1965; Melnick et al, 1965; Hoggan et al, 1966), herpes virus (Buller et al, 1981; McPherson et al, 1985) or human papillomavirus (HPV) (Walz et al, 1997; Ogston et al, 2000; Meyers et al, 2001; You et al, 2006). However AAV can replicate autonomously under specific situations (Schlehofer et al, 1986; Yalkinoglu et al.. 1988; Yakobson et al., 1987, 1989; Meyers et al, 2000), and can establish chromosomal latency by integrating site-specifically on human chromosome 19 (termed AAVS1)(Kotin et al, 1992). The AAV genome consists of two large open reading frames (ORFs) including the rep and cap genes flanked by two inverted terminal repeats (ITRs). The main rep encoded proteins are designated as Rep78 and Rep68 both of which possess origin binding, helicase, ATPase, strand and site specific nicking, and covalent linkage required for AAV replication. The ends of the AAV genome, the ITRs, contain identical origins of DNA replication.
The molecular mechanism (s) through which the helper viruses exert helper effect on AAV DNA replication is an important topic in AAV biology and its nature seems to vary with the different types of helper viruses. The Ad and herpes virus helper genes have been studied extensively, however the specific biochemical functions of the HPV helper genes are yet to be determined. HPV-AAV interaction is also of interest as HPV is the main risk factor for the development of cervical cancer and AAV has been shown to inhibit papillomavirus-associated oncogenicity in epidemiologic, animal, and laboratory settings (Mayor et al, 1976; Georg-Fries et al., 1984; Hermonat, 1989; 1991; Su and Wu, 1996; Hermonat et al., 1997; Horer et al., 1995; Coker et al., 2001; Walz et al, 2002). The replication protein of HPV, E1, has a degree of homology with the Rep78 protein (Castella et al., 2006). Like Rep78, E1 is involved in HPV DNA replication initiation: origin (ori) recognition, ATP-dependent DNA-melting and unwinding of DNA (Wilson et al., 2001). This protein is highly conserved among papillomaviruses and consists of DNA –binding domain and an ATPase/helicase domain. We have shown previously that E1 could serve as a helper gene for productive AAV replication (You et al., 2006) and E1 protein can interact with E1 in vitro, modulating their overall ATPase activity (Bandyopadhyay et al., 2008). In this study, we examined E1’s effects on the biochemical activities of Rep78 with DNA, in particular with AAV ITR DNA. Our results demonstrate that E1 stimulates Rep78’s binding, ATPase, covalent linkage to ITR DNA and endonuclease activity. AAV also holds interest for its increasing use as a human gene-therapy vector (Hermonat and Muzyczka, 1984; Tratschin et al., 1984; Worgall et al, 2008; Maguire et al., 2008). An understanding of the mechanism by which HPV E1 modulates Rep78’s role in AAV replication may be helpful in optimizing the efficiency of vector production, suggest new approaches of combining E1 helper activities with other characterized helper proteins of Ad and HSV, and help us to understand the AAV-HPV interaction and how it leads to low cervical cancer rates.
Materials and methods
Purification of proteins
MBP-Rep78 and GST-E1 fusions were expressed and purified as previously described (Batchu et al, 1995; Bandyopadhyay et al., 2008). The quality and the predicted molecular size of these proteins were confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and Commassie blue staining (data not shown) following the standard protocol.
DNA substrates for in vitro biochemical assays
The DNA helicase substrate was generated by annealing a 32P-end labeled 17-mer complementary primer to M13mp18 ssDNA as described previously (Takashi et al., 2004) with some modifications. The DNA primer was 5’-end labeled by T4 polynucleotide kinase and [γ-32P) ATP and free ATP was removed by G-25 column (Amersham Pharmacia Biotech). This labeled oligomer was annealed to M13mp18 ssDNA in annealing buffer, heated to 95°C for 5 min followed by slow cooling to room temperature. The partial duplex substrate was purified using a MicroSpin S-400 HR column (Amersham Biosciences). Radiolabled AAV hairpin DNA in flop orientation for mobility shift assay and endonuclease assay was prepared by digestion of psub201 (Samulski et al., 1987) with XbaI and PvuII as previously described (Wu et al., 1999) and labeled at the 5’ end with T4 polynucleotide kinase. The substrate was boiled and snap cooled to form a hairpin with a double stranded trs. The ITR-stem, used in Figure 2B, is purchased commercially and the sequences are as follows: TCCAGGAACCCCTAGTGATGGAGTTGGCCACTCCCTCTCTGCGCGCTCGCTCGCTCACTGAGGC and Phos-GCCTCAGTGAGCGAGCGAGCGCGCAGAGAGGGAGTGGCCAA-CTCCATCACTAGGGGTTCCTGGA. The stem sequences contain both the Rep binding element (RBE) and trs nicking site. The oligos were annealed following the manufacturer’s instructions. Briefly, the oligos were annealed together in 50µl of reaction buffer which was heated to 65°C for 10 min and then allowed to cool slowly to room temperature. The annealed oligos were labeled with [γ-32P) ATP and T4 polynucleotide kinase and purified over a G25 column. The sequences of the oligo used to prepare the ITR-stem and ITR-hairpin substrate for the ATPase assay are as follows: TR1-TGAGGCCGCCCGGGCAAAGCCCGGGCGT-CGGGCGACC-TTTGGTCGCCC-GGCCTCAGTGAGC (61 mer); TR2- AGGAACCCC-TAGTGATGGAGTTGGCCACTCCCTCTCTG-CGCGCTCGCTCGCTCAC (55 mer); TR4-GAGCGAGCGCGCA-GAGAGGGAGTGGCC-AACTCCATCACTA (40 mer). ITR stem substrate was prepared by annealing TR4 and TR2 following manufacturer’s instruction. ITR – hairpin substrate was prepared by ligating TR1 to ITR-stem. Hairpin conformation of the ITR substrate was achieved by boiling and snap cooling the complete substrate. The 3’-labeled wt ITR for covalent attachment assay was prepared by boiling and quickly chilling an unlabeled XbaI-PvuII fragment and then filling in the 5’ overhang with Klenow fragment and [α-32P] dCTP (Chiorini 1994). The p5 substrate (AAV2 nucleotides 151–289) for electrophoretic mobility assays (EMSAs) was generated by PCR amplification using plasmid PSM620 (Samulski et al, 1982) as the template for amplification and the following primers AAV-p5-1: 5’ AGGGGTTCCTGGAGGGGTG and AAV-p5-2: 5’ CAAACCTCCCGCTTCAAAATG. Reaction conditions for p5 PCR was performed as described previously (McCarty et al. 1994).
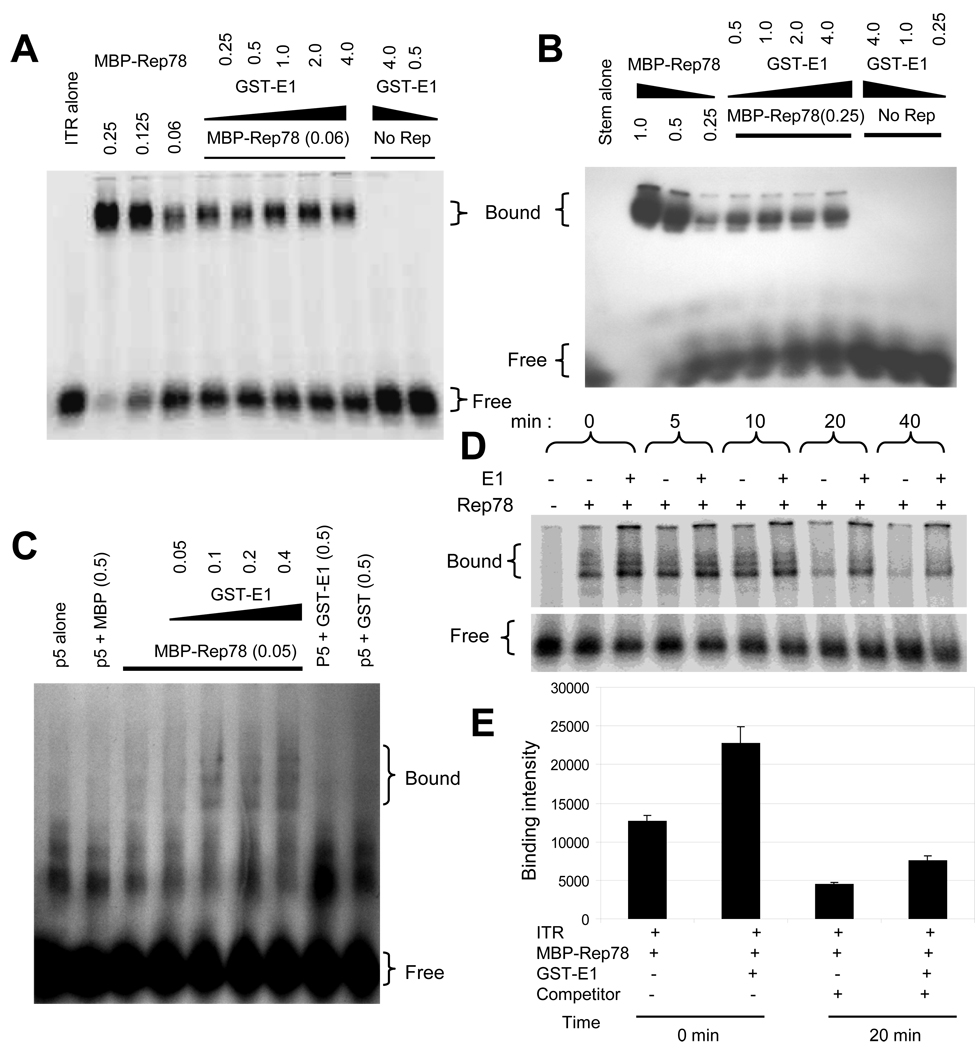
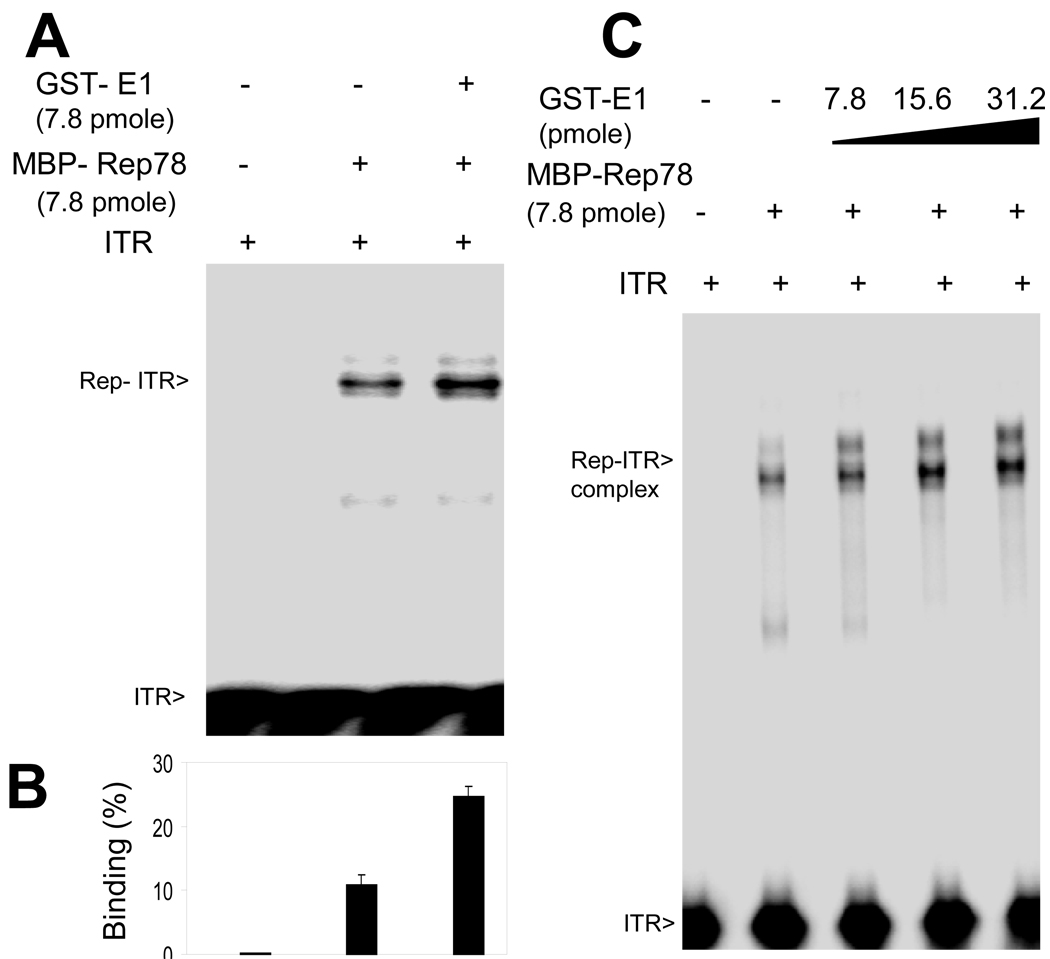
Figure 2. Enhanced binding of Rep78 to multiple AAV DNA substrates in the presence of E1.
32P-labeled DNA substrates, as indicated, were incubated with the indicated amounts of MBP-Rep78 in the absence or presence of GST-E1 and analyzed by EMSA. Numbers indicate the amount of the added proteins in micrograms. The positions of free probe and probe bound in a Rep complex are indicted as “free” and “bound”. (A) Binding of MBP-Rep78 to complete hairpin ITR DNA was enhanced by GST-E1 in a dose-dependent manner. (B) Binding of MBP-Rep78 to ITR-stem was also enhanced by GST-E1 in a dose-dependent manner. (C) Binding of MBP-Rep78 to p5 promoter DNA was also enhanced by GST-E1 in a dose-dependent manner. (D) Survival of the MBP-Rep78-ITR complex over time was enhanced by GST-E1. (E) Graphical representation of enhanced stability of Rep78-ITR complex in presence of E1 after challenging preformed complex with unlabeled ITR substrate for 20 min.
Helicase assay
The standard helicase assay was performed under the conditions described previously (Wu et al., 1999) with some modifications. 32-P labeled helicase substrate (25,000) cpm was incubated with GST or MBP fusion proteins in a 20 µl reaction mixture containing 25 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 1mM DTT, 0.4mM ATP and 10µg/ml BSA. The amount of the proteins is indicated in the figure. The reaction mixture was incubated for 30 min at 37°C and the reaction was stopped by the addition of 10 µl gel loading buffer (0.5%SDS, 50 mM EDTA, 40% [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue, 0.1% Xylene Cyanol). The substrate and product were separated by nondenaturing 6% polyacrylamide gel electrophoresis (PAGE) in 1 X TAE buffer. The gels were dried and exposed to X-ray film for autoradiography. Densitometric scanning was done by the Molecular Imager (Bio Rad) and quantification was done by Quantity One software.
Electrophorectic mobility shift assay
Electrophorectic mobility shift assay (EMSA) was carried out as described previously (Wu et al., 1999). The amount of purified proteins is indicated in the figure. Briefly, radiolabeled AAV hairpin DNA was incubated at 24° for 30 min in a final reaction volume of 20 µl containing 25 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 1mM DTT, 0.5 µg BSA, 50mM NaCl, 2% glycerol, 0.01% NP-40, 1 µg of poly dI:dC. The DNA-protein complexes were resolved by 4.5% nondenaturing PAGE in 0.25X Tris-borate-EDTA buffer. The gels were dried and data were analyzed by PhosphorImager and Quantity One software.For ITR-stem substrate, the radiolabelled substrate was incubated in a 20 µl reaction in binding buffer (10 mM Tris pH 7.5, 50 mM NaCl, 4% (v/v) glycerol, 1mM MgCl2, 0.44mM EDTA, 0.5 mm DTT, 12.5 ug dI:dC per ml, 50 ug of BSA per ml) for 20 min at room temperature. The DNA-protein complexes were resolved by 4.5% nondenaturing PAGE in 0.25X Tris-borate-EDTA buffer. The gels were dried and data were analyzed by PhosphorImager and Quantity One software. The stability of Rep78-ITR complex in presence of E1 was determined by challenging preformed Rep78-ITR complex with an excess of unlabeled ITR substrate and measuring the decay of labeled ITR-protein complex . To determine the effect of adenosine triphosphate (ATP) on Rep78-E1 interaction on ITR, ATP, nonhydrolyzable ATP and ADP was added at 0.5mM to the reaction mixture.
Endonuclease assay
Endonuclease assay was performed as described by Wu et al., 1999, with minor modifications. Briefly, 5’-32P-end-labeled AAV hairpin DNA (25,000 cpm) was incubated with GST or MBP fusion proteins in a 20 µl reaction volume containing 25 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 1mM DTT, 0.4mM ATP and 10µg/ml BSA. The amounts of the proteins are indicated in the figure. The mixture was incubated for 30 min at 37°C, and the reactions were terminated by the addition of 10 µl gel loading buffer (0.5%SDS, 50 mM EDTA, 40% [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue, 0.1% Xylene Cyanol). Subsequently, the mixtures were boiled to release the nicked fragment. The products were resolved by 6% non-denaturing PAGE in 1 X TAE buffer. The gel was then dried and exposed to X-ray film for autoradiography. Densitometric scanning was done by Molecular Imager (Bio Rad) and quantification was done by Quantity One software.
Covalent attachment assay
Covalent attachment was assayed by performing trs endonuclease assay with 3’ labeled ITR probe. As described previously (Davis 2000), purified 3’-end labeled ITR DNA was incubated in the presence of MBP-Rep78 and/or GST-E1 in 20 µl reaction volume containing 25 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 1mM DTT, 0.4mM ATP and 10µg/ml BSA at 37°C for 30 min. Then, 5 µl of 5X SDS gel-loading buffer (125 mM TrisHCl [pH7.5], 5%[wt/vol] SDS, 50% [vol/vol] glycerol, 0.25% [wt/vol] bromophennol blue, 1% [vol/vol] β-mercaptoethanol) was added to the reaction mixture and boiled for 7 min. The 3’labeled probe covalently bound to MBP-Rep78 was separated from the free probe by electrophoresis on a SDS-6% polyacrylamide gel. The gel was then soaked for 30 min in a fixing solution containing 20% methanol, 10% acetic acid and 5% glycerol; dried and autoradiographed. Densitometric scanning was done by Molecular Imager (Bio Rad) and quantification was done by Quantity One software.
ATPase assays
ATPase assays were carried out as previously described (Bandyopadhyay et al., 2008) with some modifications. In a final volume of 10 µl containing 25 mM Tris-HCl (pH 7.4), 50 mM NaCl, 10mM MgCl2 ,1 mM DTT, 100 µg BSA/ml, 0.066 µM of [α-32P]ATP (3000 Ci/mmol), 100 ng of different DNA substrates (ssDNA/TR-stem/TR) were added. The reaction was carried out at 37°C for 30 min in the presence of either individual proteins or both the proteins together. The reaction was terminated by the addition of EDTA to a final concentration of 20 mM and the products were analyzed by thin layer chromatography (TLC). One microliter of the reaction mixture was taken out and spotted onto plastic-backed polyethyleneimine (PEI)-cellulose sheets (EM Sciences, Gibbstown, N.J). The sheets were developed by ascending chromatography in 1.0 M formic acid and 0.5M LiCl, dried and scanned by Molecular Imager (Bio Rad). Quantification was done by Quantity One software.
Results
The purpose of our study was to determine the mechanism of how AAV replication increases in presence of HPV E1 (You et al., 2006). In the course of AAV DNA replication, the Rep proteins perform a number of essential functions. These include binding to the ITR, nicking of the double stranded replication intermediate at the trs, concomitant covalent linkage to the ITR, and then strand displacement and synthesis from the 3’ end of the nick. Thus, we assessed whether these activities were affected by the addition of interacting HPV E1 protein. All experiments were performed using bacterially expressed Rep78 and E1 as fusions with MBP and GST, respectively. For ease of discussion, hereafter, we will use Rep78 and E1 instead of MBP-Rep78 and GST-E1, respectively.
Minimal change in overall helicase activity in the presence of E1
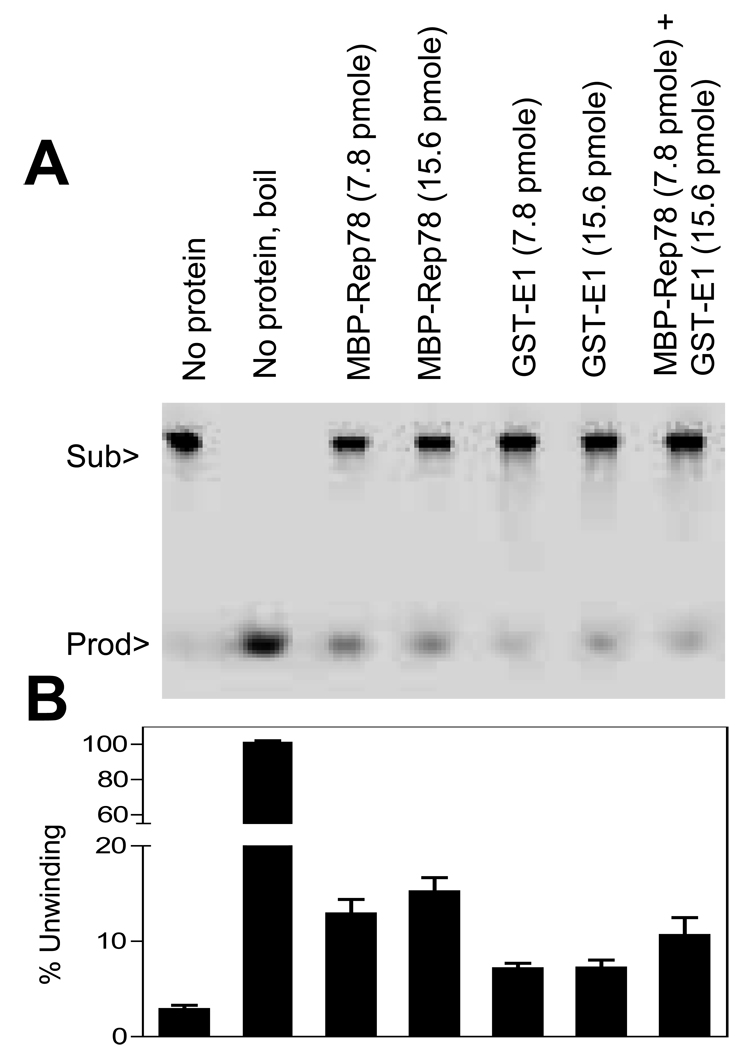
Both E1 and Rep78 have DNA helicase activity and possibly both their ATPase activities might be affected by E1-Rep78 interaction. To determine this, standard helicase assay was performed in the presence of Rep78 and E1 using radiolabeled M13-derived partial duplex substrate as described in Materials and Methods. As shown in Figure 1, the helicase activity of Rep78 plus E1 was not much changed from either protein by itself. This result suggests that the helicase activity is likely not the direct mechanism by which E1 can induce higher replication of AAV.
Figure 1. Minimal change in helicase activity of Rep78 in the presence of E1.
Standard helicase assay was performed in absence or presence of indicted amount of MBP-Rep78 and/or GST-E1 proteins. The helicase substrate consisted of a radiolabeled 17-mer annealed to M13mp18 ssDNA. (A) is one representative gel of helicase assay. The reaction mixture for the lane marked “No protein, boil” was heated to 100°C for 5 min before loading. (B) Quantitative estimation of helicase activity of the proteins. Each column represents the mean of three experiments and error bars indicate the standard error of each mean. Figure 1B is provided with a split X axis to allow easier reading of the experimental activity.
Increased binding of Rep78 to DNA substrates in the presence of E1
Rep78 recognizes DNA in multiple ways, however the strongest interaction is Rep78 binding to the “RBE” within the ITR DNA (McCarty et al., 1994). Because of an expected link between Rep78’s biochemical activity and DNA binding activities, we tried to assess E1’s effect on DNA binding by including it an ITR-Rep78 EMSA. Full length AAV ITR DNA in its hairpin configuration was purified and radiolabeled as described in Materials and Methods. As shown in Figure 2A, we observed an overall enhancement of the Rep78-ITR complex band in the presence of HPV E1, while E1 did not itself demonstrate the ability to bind the ITR. Similar assays using unrelated BSA protein failed to show binding to the ITR or affects upon Rep78-ITR binding (data not shown). We also examined the effect of E1 on the binding of Rep78 to the ITR stem [linear double stranded, minus ITR B and C sequences (McCarty et al., 1994) which contains both the RBE and nicking site]. As shown in Figure 2B, binding of Rep78 to the linear stem fragment was also enhanced by E1, and again, E1 alone did not bind the ITR stem. Though, the increase in binding intensity of Rep-ITR complex and the Rep-ITR-stem complex were only modest, these increases are real as confirmed by repeated experiments. The Rep-ITR complex formed in the presence of E1 could not be captured by anti-GST antibody (data not shown) indicating absence of E1 in the complex. Lastly, as shown in Figure 2C, binding of Rep78 to the p5 promoter fragment was also enhanced by E1, and again, E1 alone did not bind p5.
One possibility for enhanced binding is that the helicase activity of E1 protein helps to unwind the double-stranded DNA substrate in the region of RBE, allowing more Rep molecules to bind to the exposed single-stranded DNA resulting in an increase in Rep-ITR complex formation. Two other possibilities are that E1 enhances Rep’s binding by promoting multimerization of the protein or by bridging Rep into contact with its substrate. The increase in DNA binding could result from either promotion of complex formation or enhanced stability of the complex. Competition EMSA was used to approach this question. Initially, Rep78 and ITR complex formation was allowed in the presence or absence of E1 and the dissociation of the complex was assessed by the addition of an excess amount of unlabeled competitor ITR DNA to the assembled complex, as described in the Materials and Methods section. As shown in Figure 2D, the level of Rep78–ITR complexes increased in the presence of E1 (as in panels 2A–C). Moreover, complex band intensity remained higher in the presence of E1 over time. Figure 2E shows a quantification of complex survival at 20 minutes post-competitor addition and indicate higher amounts ITR-Rep78 complex remain in the presence of E1. However, this higher amount does not appear due to an effect on complex stability as an equal proportion of Rep78 remains bound in presence or absence of E1, but due to the enhanced higher initial binding of Rep78 to ITR.
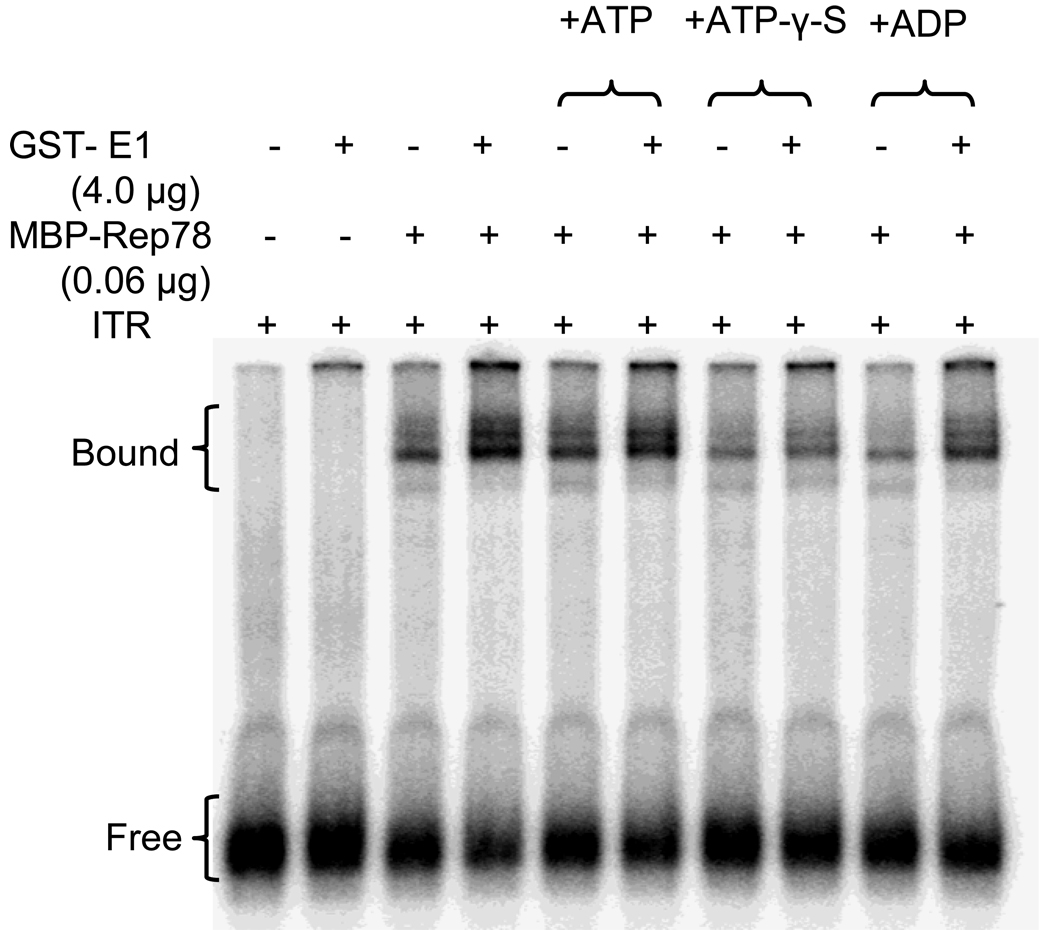
We also examined the effect of ATP on Rep-ITR complex formation in the presence or absence of E1. No significant difference had been observed in the Rep78-ITR complex formation in the presence of E1 in regards to the addition of ATP (Figure 3). However, the addition of ATPγS to the reaction, reduced Rep78’s binding to ITR in the presence or absence of E1 implicating ATP stimulates Rep binding to the ITR irrespective of the presence E1. Since ATPγS is a non-hydrolyzable analogue, it would presumably remain bound to Rep, unlike ATP, which would be hydrolyzed. These data suggest that ATP hydrolysis may be involved in Rep78-ITR complex formation or stabilization regardless of E1. The addition of ADP, non-hydrolyzable by Rep78, also seems to support this issue.
Figure 3. Modulation of Rep78-ITR complex formation by E1 and ATP analogues.
32P-labeled ITR DNA was incubated with MBP-Rep78 in absence or presence of GST-E1 plus the presence of ATP, ADP or ATPγS in an EMSA analysis. ATPγS is a non-hydrolyzable ATP analogue.
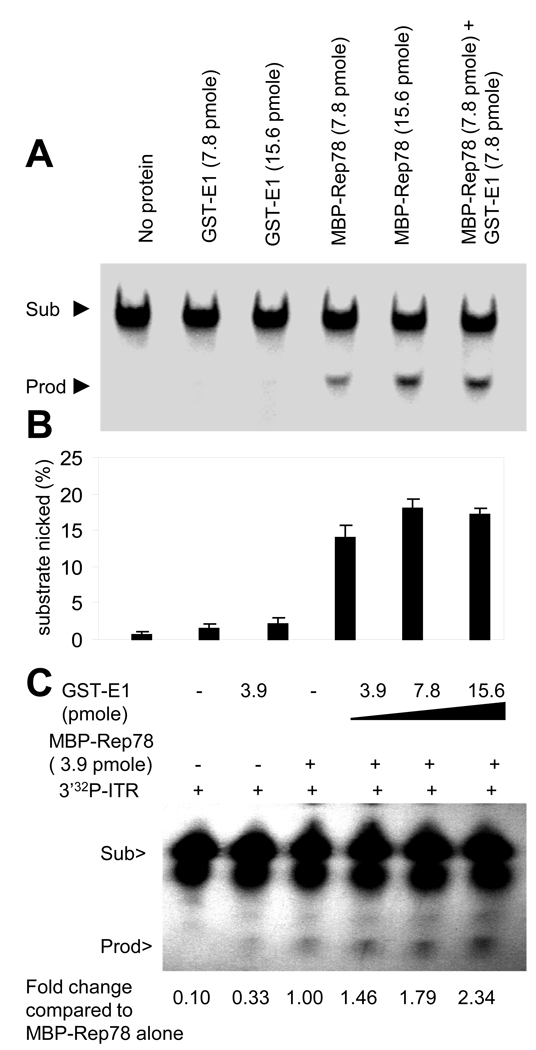
Enhanced Rep78 endonuclease activity on the ITR in the presence of E1
One critical biochemical activity of Rep78 for AAV DNA replication is endonuclease (nickase) activity. E1, which has no endonuclease activity, was assayed for the ability to modulate Rep78’s endonuclease activity using ITR as a substrate with resulting site-specific cleavage at the trs site. Endonuclease activity of Rep78 was measured by the formation of a specific sized small cleavage product, a higher mobility band (lower band) on a polyacrylamide gel upon incubation of Rep78 with 5’radiolabaled ITR in a trs endonuclease assay. As shown in Figure 4A the amount of the endonuclease product was enhanced in the presence of E1 and the overall activity of the two proteins together was greater than Rep activity alone. Figure 4B represents average quantification of the results from 3 experiments. This result strongly suggests that E1 is able to enhance Rep78’s endonuclease activity on the AAV ITR. Lastly, as shown in Figures 4C the endonuclease activity of Rep78 on the ITR was enhanced by the addition of E1 in a dose-dependent manner. The activity was quantified by measuring the product intensities over the substrate intensities and then normalized to Rep78 alone activity. This result suggests that the enhanced binding of Rep78 to ITR in the presence of E1 likely results in increased Rep78 endonuclease activity.
Figure 4. Enhanced endonuclease activity of Rep78 on the ITR hairpin substrate in presence of E1.
Representative Standard trs endonuclease activity of Rep78-ITR (hairpin) in presence or absence of E1 (A) as described in Materials and Methods. Quantitative measurement of the nicked product is shown graphically (B). Each column represents the mean of four experiments. Error bars indicate the standard error of each mean. (C) Analysis of Rep78-ITR nicking in the presence of increasing amounts of E1. The number indicates % of nicking normalized to MBP-Rep78 alone.
Enhanced Rep78-covalent linkage to the ITR in the presence of E1
During AAV DNA replication, concomitant with Rep78’s endonuclease activity is covalent attachment of Rep78 to the 5’ nicked DNA end. The amount of 5’covalently attached Rep78-DNA that results from the endonuclease assay was measured by the formation of the lower mobility band (higher band) on an SDS-polyacrylamide gel upon incubation of Rep78 with 3’radiolabaled ITR in a trs endonuclease assay (Figure 5A). The substrate was 3’ end labeled so that the label would be present on the fragment which was covalently linked to Rep78. As shown in Figure 5B, Rep78 could be covalently linked to hairpin ITR more efficiently in the presence of E1, and Figure 5C shows that covalent linkage occurs in a dosage-dependent manner with increasing addition of E1. Again, it must be mentioned that HPV E1 protein has never been described to have any ITR- or generalized DNA-covalent linkage activity of any type. In turn, these results are fully consistent with and confirm the increase in Rep78’s endonuclease activity on hairpin ITR DNA in the presence of E1 (Figure 3A and B). This result also suggests that the enhanced binding of Rep78 to ITR in the presence of E1 results in increased in endonuclease and covalent attachment activity.
Figure 5. Enhanced Rep78-ITR hairpin-covalent linkage in the presence of E1.
The covalent attachment assay was performed similar to the standard trs endonuclease assay as described in materials and methods. 1 µg of MBP-Rep78 and/or 1 µg GST-E1 was incubated with 3’-labeled ITR hairpin probe, SDS sample buffer was added, the sample was boiled and the probe covalently attached to MBP-Rep78 was separated from the free probe by electrophotresis on a SDS-6% polyacrylamide gel. (A) is a representative gel of covalent attachment assay. (B) is a graphical representation of the comparison of the amounts of covalently linked probe to MBP-Rep78 in absence or presence of GST-E1. (C) Analysis of Rep78-ITR covalent attachment in presence of increasing amounts of E1.
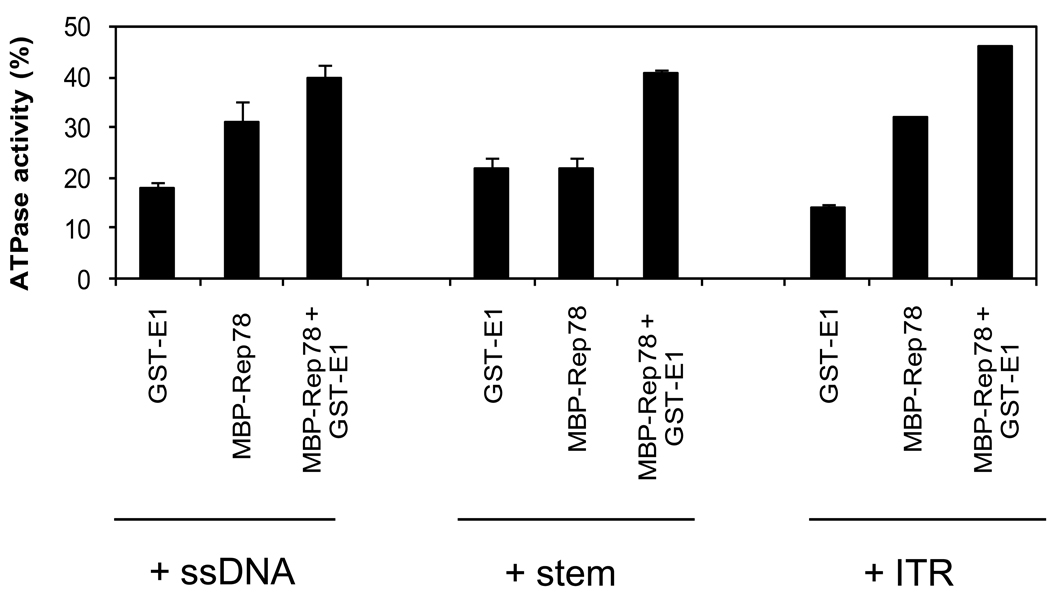
ATPase activity is enhanced by E1 specifically in the presence of hairpined ITR DNA
An important feature of Rep78 is its ATPase activity which is required for the helicase function, endonucleolytic cleavage in the ITR, and for covalent attachment of Rep78 to ITR DNA. E1 was previously shown (Bandyopadhyay et al., 2008) to reduce Rep78’s ATPase activity. In contrast, here we have shown that ATP dependent endonuclease activity of Rep78 is increasing in the presence of E1. It was suspected that the presence of DNA might exert some effect on ATPase activity of Rep78 in the presence of E1. To answer this question, we assayed the effect of E1 on the ATPase activity of Rep78 in the presence of three different DNA substrates (unrelated single-stranded DNA, double stranded ITR-stem, hairpin ITR). ATPase assays were performed in the presence of 100ng of the three different DNA substrates as described in Materials and Methods. As shown in Figure 6, maximum ATPase activity occurred with the addition of hairpin ITR DNA.
Figure 6. Modulation of ATPase activity of Rep78 in the presence of E1 and three DNA substrates.
Standard ATPase assays were performed as described in Materials and Methods. The ATPase reaction mixture contained 7.8 pmole of E1 or Rep78 or both the proteins together in presence of different DNA substrates as indicated. The percentage of ATP hydrolyzed was quantified in each case. Error bars represent one standard deviation from at least three trials.
Discussion
Rep78 is a critical protein driving AAV’s life cycle. Previous work has shown that human papillomavirus replication protein E1 has significant helper activity for AAV (You et al., 2006)) and that it could bind Rep78 in vitro (Bandyopadhyay et al., 2008). This study demonstrates that the E1 up-regulates multiple AAV Rep78-inverted terminal repeat DNA biochemistries related to replication. Here it is shown that E1 mildly enhances the binding of Rep78 to hairpin ITR as well as to the ITR-stem, similar to that seen Stracker et al. (2004) by the addition of HSV ICP8, Ad-DBP, and RPA. While the hairpin portions (B and C) of the ITR do not play a role in the Rep78 binding, its presence is required for efficient trs endonuclease activity. Most important, both trs endonuclease activity and coupled covalent attachment of Rep78 to hairpin ITR were more efficient in the presence of E1 compared to Rep78 alone. During lytic infection, Rep78 trs endonuclease activity/covalent attachment must be efficient to complete DNA replication, produce monomeric genomes, and prepare for their encapsidation into viral particles forming in the nucleus. Due to the critical nature of Rep78 trs endonuclease activity/ITR covalent linkage, these data suggest this is at least one mechanism through which E1 exerts its helper effect. Consistent with these elevated activities, ATPase activity is also enhanced by E1 in presence of hairpin ITR. Enhanced Rep78-ITR binding in the presence of E1 is also consistent with enhanced nickase and covalent attachment activity.
E1’s prime function for HPV is as an origin-binding helicase, thus, it is also possible that E1 is opening up the duplex ITR to facilitate Rep78’s binding to ITR and in turn the nicking activity. However, we didn’t observe any increase in helicase activity by Rep78 in the presence of E1. It is important to note that the limiting factor of the standard helicase assay is the use of the non-AAV substrate. HPV E1’s effect on Rep’s DNA binding and endonuclease activities are similar to the effect observed for the high mobility group chromosomal protein 1, which also directly interact with Rep proteins. Another Rep interacting protein ‘single stranded DNA–binding protein’ has been reported for having similar effect on ITR (Stracker et al., 2004). However, these studies did not assay for increases in ATPase activity and covalent attachment, all of which we observe to be enhanced by E1. It must also be mentioned that in most of the experiments we used higher concentrations of E1 than Rep78, that the in vitro concentrations of these proteins was arbitrary, without accurate knowledge as to whether these concentrations mimic the in vivo situation, and that in vivo there are many additional cellular proteins that are likely able to bind either Rep78 or E1 and that may alter or modify Rep78-E1 interactions that we observe here.
Rep78 protein is one of the members of SF3 helicases (Tuteja and Tuteja, 2004). Though the mechanism of these helicases is not entirely understood, they function as motor proteins coupling the energy from the hydrolysis of ATP to the unwinding of duplex DNA with 3’ to 5’ polarity (von Hippel and Delagoutte, 2001, Caruthers and McKay, 2002). In their function as motor proteins, their oligomeric structure is important. Rep78 forms oligomers when it binds to DNA, in some cases the association is promoted by ATP, though the exact oligomeric structure is unknown. A hexameric structure is proposed in one study (Smith et al., 1997) based on the behavior of Rep78-DNA complex by cross-linking experiment and on chromatography. Our EMSA analysis indicates that Rep78 forms oligomers on the target DNA and this could be enhanced by E1. As we have seen and also reported by others (Li et al., 2003), multiple species in mobility shift assay suggest multiple oligomeric states of Rep78 bound to ITR DNA (Weitzman et al., 1994) and some of these oligomeric forms are promoted by ATP. Similarly, Christensen et al. (1995) have reported that the binding of the homologous NS1 protein of minute virus of mice to the viral p38 promoter is ATP dependent. Furthermore, NTPs and/or non-hydrolyzable NTP analogs have been shown to increase and/or stabilize the oligomerization of certain prokaryotic helicases, including helicase II of E.Coli (Christensen et al., 1995) and the gp41 and gp4 helicases of bacteriphages T4 and T7, respectively (Dong et al., 1995).
Rep78’s ATPase activity is required both for the helicase function and endonuclease/covalent attachment activity. The ATPase activity is stimulated by DNA and is associated with the oligomerization of the protein (Dignam et al., 2007). Oligomerization promotes a conformational change of the protein activating its ATPase activity. Presence of target DNA helps to form more active oligomers. We have seen activation of Rep78’s ATPase activity in the presence of single stranded DNA, duplex stem and full hairpin structure of ITR.The ITR specific sequence requirement of Rep78’s ATPase activity has been reported by Dignam et al., 2007. In our experiments, maximum ATP hydrolysis is observed in the presence of ITR DNA in its hairpin conformation, consistent with Dignam et al. E1 enhanced Rep78 ATPase activity, binding to the ITR, ITR-endonuclease and ITR-Rep78 covalent attachment. These enhanced activities are consistent with the increase in AAV DNA replication observed previously in the presence of E1 (You et al., 2006). In future work we intend to map the interaction domain/s of these two proteins by in vivo analysis in which our laboratory is presently engaged.
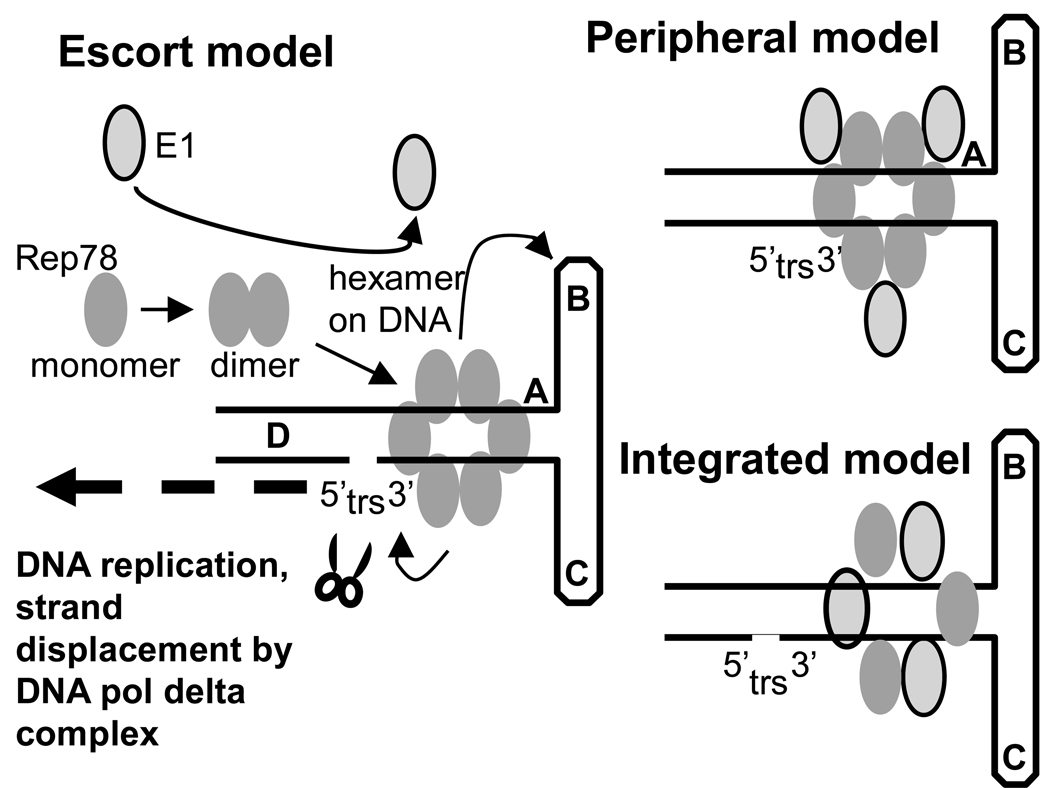
Figure 7 shows three potential models for E1’s effects on Rep78-ITR interaction. These include the “escort” model, in which E1 facilitates Rep78 multimerization and, thus, enhances multiple downstream Rep78 biochemistries on ITR DNA, including enhanced Rep78-DNA binding. However, direct E1 interaction with the pre-formed Rep78-ITR complex is low. Another model is the “peripheral” model in which E1 maintains a minimal association with the Rep78-ITR complex and facilitates Rep78-ITR biochemistries. Finally there is the “integrated” model in which E1 is hypothesized to substitute for Rep78 in the protein hexamer-DNA complex. The failure to identify super-shifted E1-Rep78-ITR complexes in EMSA assays by the addition of anti-GST antibodies suggests the “integrated” model is unlikely (data not shown). It must be noted however that MBP-Rep78 and GST-E1 are very similar in size and that if E1 were replacing some Rep78 molecules in the protein hexamers, the difference in size would be minimal and might not be observable. Both the “escort” and “peripheral” models would seem to be compatible with most of the data presented here. Weak E1 interaction with Rep78-ITR complexes might be present yet not strong enough to be visible by EMSA. Furthermore, preliminary experiments suggest that E1 may have multiple Rep78-binding domains (Bandyopadhyay and Hermonat, unpublished), which suggests that there may be multiple E1-binding sites within Rep78. Most Rep78-Rep78 interaction resides in the carboxy terminal of Rep78 and known to responsible for most Rep-Rep interactions (Hermonat and Batchu, 1997). It is possible that E1 monomer-dual Rep78 binding could facilitate Rep78 dimer formation and enhance all downstream Rep78 biochemistries.
Figure 7. Proposed models for HPV-E1 and AAV Rep78 interaction.
Shown are depictions of three models for E1-associated enhancement of Rep78-ITR biochemistry. The “escort” model depicts E1 promoting Rep78-multimerization. The “peripheral” model depicts the “escortd model plus a continued weak association of E1 with the Rep78-ITR complex, which is too weak to be observed by EMSA. The “integrated” model suggests substitution of E1 in place of some Rep78 molecules within the Rep78-ITR complex itself. This latter model is the least likely.
Thorough understanding of helper virus functions for AAV biology is important for its successful application as a gene therapy vector. In fact, it may be possible to improve rAAV production by combining different helper genes/proteins, such as E1 with those of other helper genes viruses, each with a defined and different mechanism of action. Finally, AAV-HPV interaction also regulates the risk of cervical cancer. HPV’s E1 helper functions for AAV constitutes an important component of this interaction.
Conclusions
Here we investigated HPV E1’s effects on AAV Rep78 interaction with its ITR DNA substrate in vitro. It was found that E1 enhanced Rep78-ITR binding, ATPase activity, Rep78-ITR-endonuclease activity and Rep78-ITR-covalent linkage (central to AAV replication). These data suggest that E1’s helper function for the AAV life cycle is likely through its ability to enhance Rep78’s critical biochemistries on ITR DNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work is supported a grant, R01 CA104873 from the NIH, a VA Merit grant, and intramural funding support from the UAMS College of Medicine Research Council to PLH .
REFERENCES
- Atchison RW, Casto BC, Hammond WMcD. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Raney KD, Liu Y, Hermonat PL. AAV-2 Rep78 and HPV-16 E1 interact in vitro, modulating their ATPase activity. Biochemistry. 2008;47:845–856. doi: 10.1021/bi701579v. [DOI] [PubMed] [Google Scholar]
- Batchu RB, Miles DA, Rechtin TM, Drake RR, Hermonat PL. Cloning, expression and purification of full length Rep78 of adeno-associated virus as a fusion protein with maltose binding protein in Escherichia coli. Biochem. Biophy. Res. Comm. 1995;208:714–720. doi: 10.1006/bbrc.1995.1396. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Matson SW. A partially functional DNA helicase II mutant defective in forming stable binary complexes with ATP and DNA. A role for helicase motif III. J. Biol. Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- Buller RM, Jamik JE, Sebring ED, Rose JA. Herpes simplex virus types 1 and 2 completely help adeno-associated virus replication. J. Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers JM, McKay DB. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Castella S, Bingham G, Sanders CM. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Research. 2006;34:3008–3019. doi: 10.1093/nar/gkl384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorini JA, Wiener SM, Owens RA, Kyöstió SR, Kotin RM, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cotmore SF, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker AL, Russell RB, Bond SM, Pirisi L, Liu Y, Mane M, Kokorina N, Gerasimova T, Hermonat PL. Adeno-associated virus is associated with a lower risk of high-grade cervical neoplasia. Exp. Mol. Pathol. 2001;70:83–89. doi: 10.1006/exmp.2000.2347. [DOI] [PubMed] [Google Scholar]
- Davis MD, Wu J, Owens RA. Mutational analysis of adeno-associated virus type 2 Rep68 protein endonuclease activity on partially single-stranded substrates. J. Virol. 2000;74:2936–2942. doi: 10.1128/jvi.74.6.2936-2942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam SS, Correia JJ, Nada SE, Trempe JP, Dignam JD. Activation of the ATPase activity of adeno-associated virus Rep68 and Rep78. Biochemistry. 2007;46:6364–6374. doi: 10.1021/bi602412r. [DOI] [PubMed] [Google Scholar]
- Dong F, Gogol EP, von Hippel PH. The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J. Biol. Chem. 1995;270:7462–7473. doi: 10.1074/jbc.270.13.7462. [DOI] [PubMed] [Google Scholar]
- Georg-Fries B, Biederlack S, Wolf J, zur Hausen H. Analysis of proteins, helper dependence, and seroepidemiology of a new human parvovirus. Virology. 1984;134:64–71. doi: 10.1016/0042-6822(84)90272-1. [DOI] [PubMed] [Google Scholar]
- Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermonat PL. The adeno-associated virus Rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Hermonat PL. Inhibition of H-ras expression by the adeno associated virus Rep 78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- Hermonat PL, Batchu RB. The adeno-associated virus Rep78 major regulatory protein forms multimeric complexes and the domain for this activity is within the carboxy-half of the molecule. FEBS Letters. 1997;401:180–184. doi: 10.1016/s0014-5793(96)01469-x. [DOI] [PubMed] [Google Scholar]
- Hermonat PL, Plott RT, Santin AD, Parham GP, Flick JT. The adeno-associated virus Rep78 gene inhibits oncogenic transformation of primary keratinocytes by a human papillomavirus type 16-ras chimeric. Gynecologic Oncology. 1997;66:487–494. doi: 10.1006/gyno.1997.4789. [DOI] [PubMed] [Google Scholar]
- Hoggan MD, Blacklow NR, Rowe WP. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA. 1966;55:1457–1471. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt JA. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J. Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuma T, Ohtsu M, Utsugi T, Koga S, Okuhara K, Eki T, Fujimori F, Murakami Y. Dbp9p, a Member of the DEAD Box Protein Family, Exhibits DNA Helicase Activity. J. Biol. Chem. 2004;279:20692–20698. doi: 10.1074/jbc.M400231200. [DOI] [PubMed] [Google Scholar]
- Kotin RM, Linden RM, Berns KI. Characterization of a preferred site of human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Brister JR, Im DS, Muzyczka N. Characterization of the adenoassociated virus Rep protein complex formed on the viral origin of DNA replication. Virology. 2003;313:364–376. doi: 10.1016/s0042-6822(03)00340-4. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell'Osso L, Hertle R, Ma JX, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor HD, Drake S, Stahmann J, Mumford DM. Anti-bodies to adeno-associated virus and herpes simplex in sera from cancer patients and normal adults. Am. J. Obstet. Gynecol. 1976;126:100–104. doi: 10.1016/0002-9378(76)90472-5. [DOI] [PubMed] [Google Scholar]
- McCarty DM, Ryan JH, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J. Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson RA, Rosenthal LJ, Rose JA. Human cytomega;ovirus completely helps adeno-associated virus replication. Virology. 1985;147:217–222. doi: 10.1016/0042-6822(85)90243-0. [DOI] [PubMed] [Google Scholar]
- Melnick JL, Mayor HD, Smith KO, Rapp F. Association of 20 millimicron particles with adenovirusees. J. Bacteriol. 1965;90:271–274. doi: 10.1128/jb.90.1.271-274.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Mane M, Kokorina N, Alam S, Hermonat PL. Ubiquitous adeno-associated virus type 2 replicates in a model of normal skin. Virology. 2000;272:338–346. doi: 10.1006/viro.2000.0385. [DOI] [PubMed] [Google Scholar]
- Meyers C, Alam S, Mane M, Hermonat PL. Altered biology of adeno-associated virus type 2 and human papillomavirus during dual infection of natural host tissue. Virology. 2001;287:30–39. doi: 10.1006/viro.2001.0968. [DOI] [PubMed] [Google Scholar]
- Muzyczka N, Berns KI. Parvoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th ed. vol. 2. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001. pp. 2327–2346. [Google Scholar]
- Ogston P, Raj K, Beard P. Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J. Virol. 2000;74:3494–3504. doi: 10.1128/jvi.74.8.3494-3504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski RJ, Berns KI, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. U. S. A. 1982;79(6):2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski RJ, Chang LS, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlehofer JR, Ehrbar M, zur Hausen H. Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology. 1986;152:110–117. doi: 10.1016/0042-6822(86)90376-4. [DOI] [PubMed] [Google Scholar]
- Smith RH, Spano AJ, Kotin RM. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Cassell GD, Ward P, Loo YM, van Breukelen B, Carrington-Lawrence SD, Hamatake RK, van der Vliet PC, Weller SK, Melendy T, Weitzman MD. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 2004;78:441–453. doi: 10.1128/JVI.78.1.441-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PF, Wu FY. Differential suppression of the tumorigenicity of HeLa and SiHa cells by adeno-associated virus. Brit. J. Cancer. 1996;73:1533–1537. doi: 10.1038/bjc.1996.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratschin JD, West MH, Sandbank T, Carter BJ. A human parvovirus, adeno-associated virus, as a eukaryotic vector: transient expression and encapsidation of the prokaryotic gene for chloramphenicol acetyltransferase. Mol. Cell. Biol. 1984;4:2072–2081. doi: 10.1128/mcb.4.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur. J. Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their "coupling" within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- Walz C, Deprez A, Dupressoir T, Durst M, Rabreau M, Schlehofer JR. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J. Gen. Virol. 1997;78:1441–1452. doi: 10.1099/0022-1317-78-6-1441. [DOI] [PubMed] [Google Scholar]
- Walz CM, Correa-Ochoa MM, Muller M, Schlehofer JR. Adenoassociated virus type 2-induced inhibition of the human papillomavirus type 18 promoter in transgenic mice. Virology. 2002;293:172–181. doi: 10.1006/viro.2001.1256. [DOI] [PubMed] [Google Scholar]
- Weitzman MD, Kyöstiö SR, Kotin RM, Owens RA. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VG, West M, Woytek K, Rangasamy D. Papillomavirus E1 proteins: form, function, and features. Virus Genes. 2002;24:275–290. doi: 10.1023/a:1015336817836. [DOI] [PubMed] [Google Scholar]
- Worgall S, Sondhi D, Hackett NR, Kosofsky B, Kekatpure MV, Neyzi N, Dyke JP, Ballon D, Heier L, Greenwald BM, Christos P, Mazumdar M, Souweidane MM, Kaplitt MG, Crystal RG. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum. Gene Ther. 2008;19(5):463–474. doi: 10.1089/hum.2008.022. [DOI] [PubMed] [Google Scholar]
- Wu J, Davis MD, Owens RA. Factors affecting the terminal resolution site endonuclease, helicase, and ATPase activities of adeno-associated virus type 2 Rep proteins. J. Virol. 1999;73:8235–8244. doi: 10.1128/jvi.73.10.8235-8244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of helper virus. J. Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakobson B, Hrynko TA, Peak MJ, Winocour E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J. Virol. 1989;63:1023–1030. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalkinoglu AO, Heilbronn R, Burkle A, Schlehofer JR, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. J. Virol. 1988;65:3175–3184. [PubMed] [Google Scholar]
- You H, Liu Y, Prasad CK, Agrawal N, Zhang D, Bandyopadhyay S, Liu H, Kay HH, Mehta JL, Hermonat PL. Multiple human papillomavirus genes affect the adeno-associated virus life cycle. Virology. 2006;344:532–540. doi: 10.1016/j.virol.2005.08.039. [DOI] [PubMed] [Google Scholar]