Abstract
The signaling apparatus that controls bacterial chemotaxis is composed of a core complex containing chemoreceptors, the histidine auto-kinase CheA, and the coupling protein CheW. Site-specific spin labeling and pulsed-dipolar ESR spectroscopy (PDS) have been applied to investigate the structure of a soluble ternary complex formed by T. maritima CheA (TmCheA), CheW, and receptor signaling domains. Thirty-five symmetric spin-labels sites (SLSs) were engineered into the five domains of the CheA dimer and CheW to provide distance restraints within the CheA:CheW complex in the absence and presence of a soluble receptor that inhibits kinase activity (Tm14). Additional PDS restraints between spin-labeled CheA, CheW and an engineered single-chain receptor labeled at six different sites allows docking of the receptor structure relative to the CheA:CheW complex. Disulfide cross-linking between selectively incorporated Cys residues finds two pairs of positions that provide further constraints within the ternary complex: one involving Tm14 and CheW, and another involving Tm14 and CheA. The derived structure of the ternary complex indicates a primary site of interaction between CheW and Tm14 that agrees well with previous biochemical and genetic data on transmembrane chemoreceptors. The PDS distance distributions are most consistent with only one CheW directly engaging one dimeric Tm14. The CheA dimerization domain (P3) aligns roughly antiparallel to the receptor conserved signaling tip, but does not interact strongly with it. The angle of the receptor axis with respect to P3 and the CheW-binding P5 domains is bound by two limits differing by ~20°. In one limit, Tm14 aligns roughly along P3 and may interact to some extent with the hinge region near the P3 hairpin loop. In the other limit, Tm14 tilts to interact with the P5 domain of the opposite subunit in an interface that mimics that observed with the P5 homolog CheW. The time-domain ESR data can be simulated from the model only if orientational variability is introduced for the P5, and especially P3 domains. The Tm14 tip also binds beside one of the CheA kinase domains (P4); however, in both bound and unbound states, P4 samples a broad range of distributions that are only minimally affected by Tm14 binding. The CheA P1 domains that contain the substrate histidine are also broadly distributed in space under all conditions. In the context of the hexagonal lattice formed by trimeric transmembrane chemoreceptors, the PDS structure is best accommodated with the P3 domain in the center of a honeycomb edge.
Keywords: Histidine kinase, phosphotransfer, transmembrane signaling, coiled-coils, ESR/EPR spectroscopy, spin-labeling, simulation, ternary complex, structure
Bacterial chemotaxis, the ability of cells to adapt their motion to external stimuli, has long stood as a model system for understanding transmembrane signal transduction (1, 2). Three core proteins associate in an extended transmembrane complex, and along with accessory enzymes, compose a molecular device that can detect and amplify signals with remarkable sensitivity, dynamic range and gain (3, 4). The core signaling complex is composed of long, helical, chemotaxis receptors (MCPs, for methyl-accepting chemotaxis proteins), the multidomain histidine autokinase, CheA, and the receptor-coupling protein CheW. CheA, under the influence of MCPs, phosphorylates the response regulator protein CheY, which diffuses from the core complex and binds to the flagellar motor to switch its sense of rotation. Receptor modification enzymes, and phosphatases also associate with the core complex to tune net output. Despite intense study, much still remains to be understood about the structure of the signaling complex and how the enzymatic activity of CheA is regulated in response to extracellular ligand binding events.
Although the ligand binding domains differ among MCPs, they all have a similar construction and are exemplified by the four E. coli chemoreceptors, Tar, Tsr, Trg, and Tap (3, 5). Dimeric MCPs span the membrane with four helices (TM1, TM2, TM1′, and TM2′), bind ligands through a variable amino-terminal extracellular domain (MCPL) and bind cellular components through a well conserved carboxy-terminal cytoplasmic domain (MCPC). MCPC is linked to TM2 by a short cytoplasmic domain (the HAMP domain (6)) that transduces signals coming through the membrane into the large C-terminal regions of the receptor (3, 5). Each MCP subunit folds as two long anti-parallel helices that then dimerize into a four-helix bundle. The region most distal to the membrane (the tip of the bundle) interacts with CheA and/or the adaptor protein CheW. At sites ~140–195 Å away from the receptor tip, in the so-called “adaptation region” specific glutamate residues undergo reversible methylation/demethylation (by CheR and CheB or/and CheD (7), respectively) and specific glutamine residues undergo deamidation (by CheB or CheD) to tune receptor activation of CheA.
The histidine kinase CheA is the key enzymatic component for transducing ligand binding events into chemical changes (8–11). CheA is a dimer with each subunit containing five separate functional units (P1 to P5), strung together as distinct domains over the length of the polypeptide (12). P1 contains the substrate histidine autophosphorylated by the kinase domain (P4). P2 docks CheY for phosphotransfer from P1. The last three domains P3-P4-P5, comprise dimerization, kinase (ATP binding) and receptor-coupling modules, respectively, and their structures have been determined together for the T. maritima enzyme (CheAΔ289) (12). The attachments of P1 and P2 to CheAΔ289 by long linker regions (typically 25–45 residues) increase the local concentrations of these modules in the vicinity of P3-P4-P5 domains and may serve other functions (13–17) Signal transduction derives from the ability of receptors to modulate initial phospho-transfer within CheA. Trans autophosphorylation of P1 (i.e. one subunit phosphorylates the other) (18) is the rate-limiting step in CheA activation (19–22). CheW is a small protein composed of two intertwined β-barrels (subdomains 1 and 2) and a homolog of CheA P5 (23, 24). Interactions among MCPs and CheA are mediated by the adaptor protein CheW, which interacts with CheA P5 in a pseudosymmetric contact involving subdomain 1 of P5 and subdomain 2 of CheW (i.e. one CheW per CheA subunit) (23, 25–27). Receptor binding to E. coli CheA (EcCheA) generates a >100 fold increase in kinase activity (19–22, 28). This effect can be replicated by supplying the MCP kinase binding regions alone in different contexts (20, 29–32). In E. coli, attractant binding to the extracellular domain inhibits CheA activity (5, 19), but in other bacteria, such as B. subtilis, attractant activates the kinase, in accordance with the reverse effect of CheY-P on output (33, 34).
Biochemical studies provide strong evidence that MCPs dimers associate into higher order assemblies (3). Hill coefficients for the Tar receptor derived from membrane preparations are < 3 at all modification states (35), but much larger numbers are found for the serine receptor Tsr in its methylated form (36). Measurements of in vivo activity indicate that the cooperative responses of the system are well fit by a unit of 20–40 coupled receptors (37), but that the degree of cooperativity decreases as the receptors become demethylated in the adaptation region (36, 38). Semi-synthetic systems based on the assembly of receptor cytoplasmic domains on liposome surfaces give Hill coefficients consistent with >20 coupled receptors (31, 32). Despite the variation in size of the cooperative unit in these different contexts, the trimer-of-dimers structure formed by the structure of the Tsr cytoplasmic domain has been taken as a basic building block and requirement for CheA activation (39). Second-site suppressor genetic studies and multivalent cross-linking experiments demonstrate that the receptors tips are closely associated in a pattern consistent with the Tsr trimer-of-dimers structure (40–42). Incorporation of Tsr into soluble nanodiscs show that the ability to activate kinase requires > 2 receptors per nano-disc (43).
In many different bacteria, MCPs form mainly polar clusters in the membrane containing 1000s of receptors (44–49). Stoichiometry of components within the clusters is somewhat controversial with estimates ranging from roughly 9-3 receptor dimers for every CheA dimer plus two CheWs (28, 30, 44, 50, 51). Clustering is dependent on CheW and somewhat dependent on CheA (4, 52). Highly active soluble complexes of CheA, CheW, and truncated MCPs also show a clustered state of higher receptor stoichiometry that is consistent with cell membrane measurements (28, 30, 53). Cross-linking receptors with multivalent ligands potentiates these responses, as well as the responses of other MCPs not targeted by the ligands (54). EM tomography images of wild type as well as cells over-expressing chemoreceptors revealed the MCP clusters to have hexagonal symmetry and lattice spacing (70–78 nm per hexagon edge) that are remarkably conserved across many species of bacteria (46–49). The hexagonal lattices are most ordered at the “base plate” which constitutes the CheA:CheW binding layer and the symmetry is progressively lost toward the membrane (46–49). One study interpreted the images such that one trimer of receptor dimers occupies each vertex of the hexagon, while CheA and CheW only occur beneath alternating vertices (48). In another study, a continuous density for CheA and CheW has been observed under the hexagonal receptor arrays (46). The specific molecular interactions that produce these arrangements are largely uncharacterized. One of the initial models of this assembly predicted that the trimer-of-dimers to sit at the corners of vertices of the hexagon, separated from each other by a CheA dimer (55). The predicted hexagonal symmetry of this model agrees with the EM hexagonal patterns in cells, but not the lattice dimensions. Based on the solution structure of CheA:CheW and the arrangement of T. maritima receptor Tm1143 in the crystal lattice, we suggested a lattice based on a “hedgerow of dimers” (25). The receptor dimers were proposed to fit into the cleft formed between two symmetrically disposed CheWs and self association of the CheA P5 domains, as also observed in crystals structures, participated in the overall assembly. The size of the CheW cleft in the solution CheA:CheW structure could accommodate a receptor dimer, but not a trimer-of-dimers. Another study based on perturbation of protein-interactions-by cysteine modification identified a large receptor interaction surface on S. typhimurum CheA and proposed several association modes between CheA and MCPs based on the protection patterns of surface sites (27). CheA may engage receptors with multiple, coupled interfaces because the sites of modification that alter receptor interactions are broadly distributed over the surface of CheA (27).
A number of studies suggest that CheA activation requires an association with MCP dimers at high stoichiometry with respect to the CheA dimer (i.e. 3:1, 6:1, 12:1). Isolated receptor signaling domains activate CheA when in high concentration or templated in some fashion (20, 56). Receptor inactivation, however, likely involves physical separation of the receptors, either of the trimers within the lattice and/or of the dimers within the trimers (57–60). Also, the demethylated, less active receptors show reduced cooperativity (36, 38), which suggests less physical coupling. CheA does not dissociate from the clusters when MCPs are inactivated by attractant (61) and the purified core complexes themselves are extremely stable (50). Indeed EM studies of the clusters found evidence for both hexagonally ordered and disordered regions, which may represent different activity states of the signaling particles (48); however, it is not clear how CheA activity correlates with these structural states.
We have previously applied X-ray crystallography to define the structures of the core signaling components and site-specific spin labeling (62) with pulsed-dipolar ESR spectroscopy (PDS) (63–65) to investigate their association modes (63). We have focused on chemotaxis proteins from the thermophile T. maritima due to their ease of purification, stability, tendency to crystallize and lack of natural Cys residues. PDS has revealed the relative positioning of CheA domains in solution and the association mode between CheA and CheW, which was confirmed by crystallographic studies (25). We have now extended this PDS approach to probe the conformational changes that CheA:CheW undergoes when engaged by receptor signaling domains and the direct mode of interaction between receptors and CheA:CheW in soluble complexes of T. maritima proteins.
MATERIALS AND METHODS
Cloning, mutagenesis and spin labeling of proteins
Coding regions for T. maritima proteins CheAΔ289 (290–671), CheW (1–151) full-length Tm14, and Tm14C (40–213) were cloned and purified as previously described (24, 25, 66). In the cysteine-less background of CheAΔ289, six residues in the P5 domains (Q545, N553, S568, E646, D634 and S639), seven in the P4 domain (D371, E387, E401, K458, K496, D508, and S522) three in the P3 domain (E301, S318 and E331) and eleven on CheW (9, 15, 28, 31, 35, 72, 80, 101, 102, 137, 139) were separately changed to cysteines by Quickchange mutagenesis (Stratagene). Full length CheA has two native cysteines at site 63 and 208 which are in P1 and P2 domain respectively. Cysteine-less CheA was prepared by selectively substituting each of these cysteines to serine residues. This template was used for introducing five more cysteine substitutions in P1 (12, 14, 53, 76, 83) and one more in P2 domain (178). Proteins were labeled for 4 hours at room temperature with 5–10 mM MTSSL (1-oxyl-2,2,5,5-tetramethylpyrolinyl-3-methyl)-methanethiosulfonate, Toronto Research, Toronto) in gel filtration (GF) buffer (50 mM Tris, pH 7.5 150 mM NaCl) followed by overnight labeling at 4°C while the His-tagged proteins remained bound to Nickel-NTA agarose beads. The proteins were eluted by thrombin cleavage after 6–12 hours of treatment.
Construction of a single-chain receptor
The single chain-dimer was cloned using Tm14C as a template. PCR introduced oligonucleotides coding for the C-terminal linker GASGGTG into a Tm14C fragment along with an NdeI N-terminal restriction site and a BamHI C-terminal restriction following the linker. Overlap PCR then produced the following construct NdeI-Tm14-Linker-BamHI-Tm14-SalI, from the Tm14 template and Tm14-linker fragment. This PCR product was ligated into pet28a using the NdeI and SalI sites. Quickchange Mutagensis (Novagen) was used to introduce Cys residues into the single homodimer construct, which was then PCR amplified with primers that introduced a linker and BamH1 site at the 3′ end. The NdeI-Tm14C(mutant)-Linker-BamHI fragment was then cloned into the single chain construct, replacing the 5′ repeat. It was found that in E. coli BL21(DE3) the single chain dimer was subject to recombination, which resulted in a dimer of non-covalent subunits. Overexpression in BLR(DE3) cells, which have no homologous recombination system, produced a high degree of single change dimer; the protein could be expressed and purified with an Ni-NTA column as described above. The yields were only 25% of Tm14C, but the single-chain dimer was stable and easily purified.
Preparation of samples for PDS
All the spin labeled proteins used in our experiments were divided into small aliquots and stored at −80°C for future use. However, we observed that some of the proteins lost a significant amount (roughly 30%) of spin label over a period of 2–3 months. The loss in spin label is reflected in reduction in amplitude of the primary echo. For measuring signals from protein complexes, the proteins were mixed together and the sample incubated at room temperature for 30–60 min. before flash cooling in liquid N2 for ESR experiments. Protein concentrations within the range of 25–50 μM were used for DEER experiments.
Pulsed dipolar ESR measurements
Pulsed dipolar electron spin resonance spectroscopy (PDS ESR, or PDS for short) yields the distance, r between electron spins residing on a molecule of interest. PDS involves measuring magnetic dipolar couplings between two (or more) unpaired electrons. In our case, the spins are nitroxide spin-labels attached specifically to genetically engineered cysteine residues on a protein (67). The dipolar coupling A(r, θ) between two such spins (A and B) separated by r is given by:
| (1) |
where is the dipolar coupling constant in angular frequency units and γe is the gyromagnetic ratio of an electron spin, Ħ is Planck’s constant divided by 2π, r is the magnitude of the vector r, separating the two spins, and θ is the angle between this vector and the direction of the external magnetic field, B0.
Currently, the two most common methods for distance measurements from dipolar spin-couplings are pulsed double electron-electron resonance (DEER or PELDOR) (64, 68) and double-quantum coherence (DQC) (63–65, 69). The two methods provide similar information, but have different virtues and target somewhat different systems and situations. DQC resolves dipolar couplings over a wider distance range, minimizes orientational selection effects (in standard 1-D implementation) present in DEER (70) is less prone to the constant signal background, and yields stronger signals particularly in dilute samples. On the other hand DEER requires a less demanding experimental setup and better references the dipolar signal to the subtracted background, which is a desirable feature in the context of this work. In DQC, similar referencing would require more effort. The nature of the background in DQC is less understood, since it originates from the bath of nearby spins and is relatively small in dilute systems. Clustering of spins will reduce the DQC signal and produce a larger background of uncertain shape. Since CheA and its complexes do cluster, but in an unknown manner, DQC was not used for this study. For more details regarding the nature and analysis of the DEER signals measured in this study see Supporting Information.
Four-pulse DEER experiments were carried out at 17.3 GHz on a specially constructed 2D-FT ESR spectrometer modified to support PDS as described previously (25, 71). Distance distributions were reconstructed using Tikhonov regularization (72) and further refined by a maximum entropy regularization method (MEM) (73).
Detection and analysis of intermolecular dipolar signals in the ternary complex
In the context of the ternary complex, the addition of spin labeled single chain receptor dimer to spin labeled CheA/CheW results in a total of three spin label sites: two on the CheA dimer, or on two CheW molecules bound to CheA, and one on the receptor dimer. Thus, the inter-domain signal (i.e. from receptor to CheA/CheW complex) is accompanied by an intra-dimer CheA or CheW-CheW signal, which significantly complicates the data analysis. Additionally, if the single-chain receptor binds to CheA/CheW with both of its symmetric surfaces to CheA/CheW, it will produce two distances with their maximum separation ~30 Å given by the width of the receptor dimer and the length of nitroxide tethers. This yields a total of up to five characteristic distances in the ternary complex in a distinct conformation (c.f. Supplemental Figure S1). However, the conformational range of spin labels, and especially the subunit mobility of CheA, gives rise to rather broad distance distributions. In some cases, when the inter-subunit distances in the CheA/CheW complex lie outside the range of where they can be reliably processed by regularization algorithms, the inter-domain distance can be detected as a distinct change in the dipolar signal time domain envelope, and can be isolated using a suitable subtraction. For instance, some of the sites on the P5 sub-unit have separations of 50–60 Å in the dimer. If the probing site on the receptor is located at a distance of 15–40 Å from either of these sites, then the new distance is visible in the time domain signal, as well as in the distance distributions. However, when that is not the case, and two distances are close, the isolation of the inter-domain signal becomes problematic and requires signal deconvolution, e.g. as described below in the Signal Modeling section below.
Furthermore, the addition of the third spin creates a situation when there could be pairs and triads of coupled spins, with their stoichiometries dependent on the binding constant, (Kd) and spin labeling efficiency, x.
Because of all the real and potential difficulties associated with isolation of the inter-domain signal in the complex, each distance measurement between spin labeled receptor and CheA/CheW entailed two control experiments: 1) spin labeled receptor with wild type CheAΔ289/CheW and 2) unlabeled receptor with spin labeled CheAΔ289/CheW. Samples of receptor dimers with a single spin label in complex with wild type CheAΔ289/CheW developed over a period of time weak dipolar signals amounting to as much as 0.02–0.08 of the total spin-echo amplitude, i.e. 6–12% of the nominal dipolar signal amplitude employed in this work. We believe that over long periods (> hours), receptors have a tendency to associate in a non-specific manner. However, freshly prepared receptor proteins with wild type CheAΔ289/CheW showed almost no unwanted dipolar signals and provided good referencing as well as receptor binding properties.
Rigid body refinement
Molecular models of the CheA:CheW complex in the presence of unlabeled receptor were refined against distance restraints obtained from PDS measurements by applying the conjugate gradient minimization algorithms of CNS as previously described (74). For the refinement, the initial model of CheAΔ289:CheW was developed from combining the coordinates from the crystal structures of CheAΔ289 and CheW in complex with CheAΔ354. In most cases, Ravg (probability weighted average spin separation) was extracted from the P(r) for modeling, as opposed to Rmax (spin separation of highest probability). For each measurement, we assigned the uncertainty in Ravg to be dminus=5Å and dplus=1Å, as defined (74). In our refinement procedure, we assumed that in each subunit, CheW and P5 together move as a rigid body. In the final refined structure, the change in conformation of domains was evaluated by superimposing the P3 domains of the initial and final structures of CheA/CheW.
Modeling of dipolar signals in the ternary complex
Whereas the combination of signal analysis by the Tikhonov regularization and maximum entropy methods (72, 73) followed by rigid-body refinement can be an efficient means to yield the ternary structure, at the current stage of development, rigid-body modeling does not rigorously treat nitroxide side-chains. Accordingly, it seeks the most probable structure based on the simple approach given above for the uncertainty in the distance between the spin label and the backbone. The inability to account for the approximate orientations of the nitroxide side-chains with respect to the backbone limits accuracy (74).
In order to validate and further refine the structure provided by rigid-body modeling, but also evaluate the protein flexibility underlying its function we have reproduced the time-domain dipolar signals and distance distributions based on the proposed structure of the complex to compare with the experimental data. These simulations require reasonable assumptions regarding the orientation of the nitroxide side-chains and the range of conformations sampled by mobile domains and subunits of the protein components. Importantly, this procedure also provides further insight into the dynamics (or more precisely the spatial distributions of subunits) within the ternary complex. To simulate the experimental data, a nitroxide moiety, modeled as an extension of its native residue (i.e. at the distal residue atoms), was confined inside a sphere with the radius RN of 3.0–6.0 Å. Although more elaborate modeling, (69, 75, 76) can be used, this simple model was quite sufficient for present purposes. The exact locations of the spheres can be adjusted by applying small shifts of their centers (Δr ≤ 3.5 Å). For spin labels attached to the CheA P3 domain, this simple modeling yielded dipolar signals in virtually perfect agreement with the experiment, but for the more mobile P4 and P5 the average distances and especially their distributions showed deviations in some cases which could be remedied by fine adjustments in the structural modeling. The very wide distance distributions observed between certain pairs of sites residing on mobile subunits P4, P5 and on CheW originate primarily from the range of orientations the CheA domains sample due to the flexibility provided by the short interconnecting linkers between domains (~30 Å or more in some cases; cf. PDB ID: 2CH4). Again we note that a more detailed modeling of the domain flexibility ultimately should result in even better agreement with the experimental data. In the absence of such domain flexibility, all five distances in the ternary complex would typically appear well-resolved. Given the fact that this is a complicated case of three coupled spins, as noted above, we simulated the time-domain dipolar signals, using at the outset the ternary structures A and B (see below) produced by the rigid-body modeling and assuming a “linear” (on log scale) background. Further details appear in the Supporting Information.
The examples of simulations for (100/331, 100/545, 111/331, 149/301, 149/80, 149/9, 160/545, 167/80) are shown in Supplemental Figures S1–S6 of the Supporting Information.
Most of the simulations were based on static models in one of two orientations (A or B) defined by the bounds of the ESR constraints (see below), but in two cases the receptor together with its CheW and P5 domain were allowed to “sweep” over distance of about 10 Å away and 25 Å along P3 symmetry axis. We found better agreement with the model for these cases in our present study. More detailed modeling of mobile domains would be useful, as noted above, but it was not necessary for purposes of this work.
Disulfide Cross linking
The stock solution of the initiator Cu(II)(1,10 phenanthroline)3 was prepared according to the procedure of Bass and Falke(77). For each reaction, the final reaction volume was kept constant at 15 μl, which included 5 μl of NuPAGE LDS sample dye. All the proteins were solubilized in GF buffer. The final concentration of cysteine-substituted CheAΔ289 proteins varied between 1–2μM in the final reaction mixture, whereas the initiator concentration was fixed to 0.1mM in all cases. 10 μl of the reaction mixture was loaded on SDS PAGE gel for analysis by coomassie staining.
Production of heterodimers of CheAΔ289
Equimolar amounts of CheAΔ289 E301C, E331C and S318C/Q545C dimers with the His-tag removed and CheAΔ289 WT dimer with His-tag intact were mixed at 65 °C for 10 minutes to enable subunit exchange (78). His-tagged heterodimers and remaining wild-type homodimers were purified from non-His tag containing dimers by Ni-NTA affinity chromatography at 4°C to minimize further subunit exchange and then reacted with MTSSL.
RESULTS
Experimental Strategy
Soluble complexes between TmCheA, CheW and soluble cytoplasmic domains (MCPC) of TmMCPs (Tm1143, Tm14, Tm14C, and Tm1428) were reconstituted from protein purified from recombinant expression in E. coli. All three MCPCs bound with similar affinities, caused inhibition of CheA activity and produced similar changes in spin-label signals; thus, the receptor domain that bound CheA with highest affinity, Tm14C (Tm14: residues 40–213), was used for most PDS experiments. As described previously, Tm14 forms a low stoichiometry complex with CheA and CheW, with one dimeric receptor binding a dimer of CheA that has each subunit bound to one CheW (CheA:CheW) (66). The affinity of CheW for CheA is relatively high (~100 nM; (78)), but much lower between CheA:CheW and Tm14C (20–50 μM see below). When bound, Tm14 inhibits CheA activity (66). We attribute the low binding affinity to the extraction of the system from its cellular environment where the ternary complex could be organized in higher order structures (47). To investigate the protein association modes in this complex, spin labels (SLs) were distributed over all of TmCheAs five domains, CheW (35 spin label sites (SLSs) and Tm14C (6 SLSs) (Figure 1). Pulsed dipolar ESR was employed to extract distance restraints from the dipolar interactions between spins. Thus, a single label on CheA will generate one distance across the dimer interface, as will a single site on CheW, as one CheW binds each CheA subunit. The P(r) function, which expresses the relative probability of all spin-spin separations in the sample < 80 Å was then calculated in the presence and absence of the receptor fragment. We then measured interactions between SLSs on Tm14C or either CheW or CheA. These measurements are more complicated due to inherent symmetries that generate multiple spin-spin separations. To limit signal overlap, single chain receptor fragments that combine two receptor sequences in one polypeptide were generated to present only one free Cys for SL incorporation (Figure 2).
FIGURE 1. Spin label positions on CheA, CheW and Tm14.
Ribbon representation of CheA (P1 – red, P2 – green, P3 – dark purple and grey, P4 – light pink and blue, P5 – magenta and light blue) and CheW (cyan) and Tm14 (yellow and orange) showing positions of residues mutated to Cys (yellow balls) and labeled with MTSSL for dipolar ESR or applied in disulfide cross-linking experiments. P1 and P2 are connected to P3, P4 and P5 by long unstructured linkers (dotted lines). For clarity, only one P1 and P2 domain is shown and spin-label sites are marked on only one CheA, CheW, and Tm14 subunit. CheW and P5 have related folds composed of two pseudosymmetric β-barrels known as subdomain 1 (SD1) and subdomain 2 (SD2).
FIGURE 2. Possible spin-spin interactions in a ternary complex of Tm14:CheAΔ289:CheW.
Schematic diagram for spin label separations containing a CheAΔ289 dimer, bound to two CheW proteins and single chain receptor. In CheAΔ289, P4 domains are not shown for clarity. Intermolecular and intramolecular distances are represented by solid and dashed double headed arrow respectively.
Heterodimers of CheA were also produced that were labeled on only one subunit. For this purpose, we selected two sites on the P3 domain (301 and 331). However, the results of distance measurements between these sites and spin-labeled single chain receptor were inconclusive because the dipolar signals produced very weak amplitude due to the low population of SL-hererodimers in solution and the low affinity of these hetereodimers for the single chained receptor. (The procedure for preparing heterodimers produces unlabeled CheAΔ289, which cannot be easily separated from spin labeled heterodimers (25).) For these reasons, and the difficulty in preparing these samples, heterodimers were not utilized in most measurements.
In all cases, the PDS time-domain data, which directly reflects the dipolar interaction energy of the spins, was processed to give distance distributions (P(r)). The inter-subunit separation between symmetrical sites on CheAΔ289 dimer was recorded in the absence and presence of unlabeled receptor. The changes in distances are a direct measure of conformational adjustments in CheA as it binds to the receptor. It should be noted that the receptor binding can cause the domains in the two subunits to move in a manner that does not change the separation between the spin label sites. Where no changes in dipolar signals were observed upon addition of unlabeled receptor, binding interactions were confirmed by pull-down assays.
The possibility of higher association states in solution was evaluated by consideration of Vinter and Vintra contributions to the dipolar signal (see Methods), the details of these studies will be reported separately (Bhatnagar et al. unpublished). In short, CheA was found to form tetramers through interactions with its P5 domains, however, these larger species constituted less than 4% of the molecules at the concentrations where measurements were made, and hence do not interfere with the strongest dipolar signals, which in all cases represent intra-dimer spin separations. Below we first describe the effects of unlabeled Tm14C on each domain of CheA:CheW; which are structural changes associated with inactivation of CheA autophosphorylation activity. We then describe interactions between spin-labeled receptor fragments and spin-labeled CheA:CheW. In combination, the two studies allow determination of a model for the ternary complex. Aspects of this model are then confirmed by cross-linking experiments. Finally, we discuss the implications of the Tm14:CheA:CheW structure for interactions with a trimer of receptor dimers and incorporation into the hexagonal transmembrane receptor arrays that have been visualized in cells.
P1
The P1 domain of CheA is a five helix bundle connected to the P2 domain by a 42 residue linker; P1contains the phosphorylatable histidine residue, His46, on the face of its 2nd α-helix (79, 80). We recorded dipolar signals between the two P1 domains in the CheA dimer by modifying six residues (A12, A14, A53, A63, A76 and A83) with spin-labels (SLs) (Figure 1). All sites reported that the P1 domains are far apart from each other and broadly distributed (Figure 3 and Table 1). The long separation and spatial extent of the SLSs (especially A12, A14, A53 and A83) produced weak amplitudes that made Tikhonkov regularization unreliable and prevented detection of obvious changes to the signals in the presence of Tm14C, however; some minor changes could be noted with A63 and A76. For example, with site A76, Rmax of the distance distribution increased by 4 Å, which indicates that Tm14 causes the P1 domains to move further apart, perhaps biasing their localization toward the adjacent subunit of the dimer for transphosphorylation (Figure 3A). However, this effect was not as clearly evident from other P1 SLSs. Overall, the P1 domains sample wide regions of space and on average are widely separated from each other in the absence and presence of Tm14C; nonetheless, Tm14C can modestly influence their spatial distribution.
FIGURE 3. Dipolar ESR data for symmetric SLSs on P1, P2 and P4.
Time domain signal (left) and corresponding distance distributions (right) for P1 sites (A) A76 (B) A178 and (C) P4 A496. For P1 and P2, signals are compared in the absence (dotted line) and presence of unlabeled receptor (solid line), and for P4 in the absence (dotted line) and presence (solid line) of ATP. All time domain signals and distance distributions are scaled to a common value for ease in comparison. In (A) the concentrations of CheA, CheW and Tm14 were 25 μM, 125 μM and 225 μM, respectively.
In (B), CheA, CheW and unlabeled Tm14C concentrations were 25 μM, 125 μM and 300 μM, respectively. In (C), concentrations of spin labeled CheAΔ289, CheW and ATP were 50 μM, 100 μM and 500 μM, respectively. MgCl2 was added at (500 μM).
Table 1.
CheA/CheW intersubunit SL- distances in the absence and presence of unlabeled receptor.
| Ravg (Å) between symmetric Cys-SL sites on CheA/CheW | ||
|---|---|---|
| Free CheA/CheW | CheA/CheW/Receptor | |
| CheW | ||
| W9 | *41, 51 (28) | 41, 51 (20) |
| W31 | 50 (22) | 49 (14) |
| W80 | 56 (6) | 56 (6) |
| W139 | 63 (19) | 67 (24) |
| P1 domain | ||
| A63 | 38 (32) | 39 (36) |
| A76 | 65 (38) | 72 (38) |
| P2 domain | ||
| A178 | 34(23) | 34 (23) |
| A208 | 39 (28) | 38 (26) |
| P3 domain | ||
| A301 | 28 (6) | 28 (6) |
| A318 | 28 (6) | 28 (6) |
| A331 | 24 (9) | 24 (9) |
| P4 domain | ||
| A371 | 50 (23) | 50 (23) |
| A387 | 50 (18) | 48 (20) |
| A401C | 50 (25) | 51(29) |
| A508 | 60 (20) | 62 (21) |
| P5 domain | ||
| A545 | 44 (23) | 40 (8) |
| A553 | 63 (25) | 58 (32) |
| A568 | 62 (22) | 63 (17) |
| A634 | 40 (15) | 38 (13) |
| A639 | 45 (13) | 45 (13) |
| A646 | 58 (14) | 61 (23) |
Indicates the Rmax values for bimodal distribution with almost equal population of peaks. The addition of receptor leads to increase in population of one peak which is indicated in bold. Full width at half maxima for each distance distribution is reported in parenthesis. Cysteine substitutions on P3, P4 and P5 domain were made on CheAΔ289. ESR measurements on spin labeled CheW were made in presence of wild type CheAΔ289.
P2
The P2 domains, which function as docking sites for CheY and are separated from their respective P1 domains by a 25 residue linker, show closer separation to each other than do the P1 domains (Figure 3B). The two SL sites, residues A178 and A208, produced similar broad distance distributions with Ravg between 34–40 Å. If the linker between P2 and the P3 dimerization domain were fully extended, it could measure ~100 Å. Thus, the close proximity of the P2 domains to each other suggests some form of structural constraint. However, the unusually wide width of the P(r) distribution does indicate substantial mobility of the domains as would be expected by the flexibility of their linker connections. In the context of full length CheA from E. coli, the flexible nature of the P2 domain has been previously revealed by NMR studies (81), which also concluded that the P2 domains shared no stable interactions with the rest of the protein. However, crystallographic structures of CheY in complex with the P2 domain show tight interactions between P2 domains in the crystal lattice that relate the molecules by two-fold crystallographic (82) and non-crystallographic (83) symmetry. No such interactions were found in crystal structures from T. maritima proteins (84). The close proximity of the P2 SLs provide some evidence for P2 self interactions in solution; however, the two linkers connecting the P2 domains to the more rigid structure of CheAΔ289 may interact in a manner that restricts their separation.
In the presence of CheY, we noticed only minor changes in the dipolar signals from P2 spin-labeled sites (data not shown). A possible explanation for the lack of signal change on CheY binding is that the sites A208 and A178 are removed from the interaction surface between CheY and P2 and hence are not ideal reporter sites to sense changes on binding of CheY. Nonetheless CheY induces no major differences in the positioning of the P2 domains. Likewise, unlabeled receptor produced no noticeable effects on the P2 distance distributions (Table 1).
Domains P3-P4-P5 (CheAΔ289)
P3
SL site separations on P3 (A301, A318 and A331) agree well with its anti-parallel 4-helix bundle structure. No changes in dipolar signals were observed with unlabeled receptor (Table 1). CheAΔ289 was confirmed to bind the spin-labeled proteins with similar affinity as unlabeled protein by pull-down assays with affinity-tagged components containing SLs (data not shown).
P4
In our previous work (25), we reported that P4 domains sample a range of orientations based on broad distance distributions between P4-P4 as well as between P4-P3 and P4-P5. Our conclusions were drawn from a limited number of SLSs on P4 that behaved similarly (A387, A496 and A508) and did not make use of the more recently developed techniques for extracting P(r) distributions. For a more thorough investigation, we introduced four new cysteine substitutions at A371, A401, A458 and A522 which, along with the previously studied positions, uniformly cover the surface of domain (Figure 1). Out of the seven sites, A496, A458 and A522 produced weak dipolar signals with only 40% of the expected full amplitude based on labeling efficiency, which may indicate that these sites are sometimes separated by a distance which exceeds the maximum limit of distance detection by DEER (~80 Å). Not surprisingly these sites cluster near the ATP binding region, at the periphery of the P4 domain and Δ289 module.
In the ATP-free form of P4, dipolar signals from sites A371, A401, A387 and A508 did not change significantly in the presence of receptor (Table 1) and continue to sample a wide range of orientations in solution (25). The lack of any stabilization effect may indicate that receptor does not interact strongly with the P4 domains.
ATP Binding
Addition of ATP produces substantial changes in symmetric SL separations on P4 that indicate the P4 domains are closer to each other than predicted from the Δ289 crystal structure (24). In particular, a ~38 Å separation between the 401 sites located on the P3-facing helices dominates the P(r) in the presence of ATP. Furthermore, the 496 position reports a dramatic shift from ~70 to ~50 Å when ATP binds (Figure 3C). The A496 positions reside on the ATP-lid and are separated by ~90 Å in the CheAΔ289 crystal structure (24). Crystallographic data also shows that the binding of substrate restructures the ATP-lid, which closes down over the nucleotide in structures of the isolated P4 domain (85). Relaxation properties from continuous wave ESR experiments indicate that the spin-label on A496 is highly ordered in the absence of ATP, but then experiences increased dynamics characteristic of a freely exposed SL, when ATP binds (25). This would be consistent with the SL first binding in the ATP pocket and then becoming displaced by ATP. The shift to closer separations implies that the loops of the ATP-lids are closer to each other across the dimer interface than the nucleotide binding pocket. However, similar to the ATP-free form, addition of unlabeled Tm14C produces only minimal changes to these distributions and they remain broad, which rules against a well defined orientation for the P4 domain in the presence of Tm14C.
P5
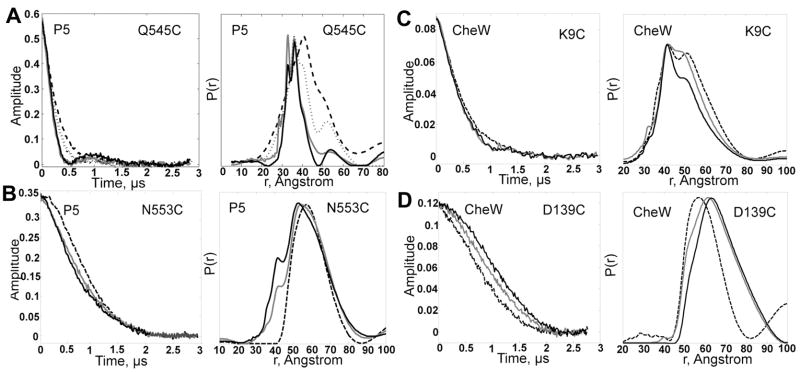
Unlike the other CheA domains, distance distributions from symmetric SLSs on P5 report moderate to substantial changes on addition of unlabeled receptor (Table 1). These effects were particularly striking for P5 sites A545 (on β7) and A553 (on the loop connecting β7 and β8, Figure 1). For A545, a widely distributed P(r) for CheA:CheW becomes narrow and bimodal and Ravg decreases from 44 to 40 Å on addition of Tm14C (Figure 4). The position of A545 may allow the SL to contact the surface of P3 when receptor is bound, which would explain the narrow width of the P(r). The bimodal peaks in the presence of receptor probably correspond to either two different orientations of the SL or two related domain orientations. In support of the latter, CheA still binds Tm14C in the absence of CheW, but the 545 P(r) does not contain the shortest-distance peak of the bimodal distribution (Figure 4A). Another site, A634 (on the β14 strand), which is close to A545 in CheA:CheW, responded in a similar manner to A545 when receptor binds (86). With site A553, the addition of receptor results in the appearance of a new separation peak at 42 Å whose amplitude increases and then appears to saturate with increasing receptor concentration (Figure 4B). This is a clear indication of ternary complex formation. These three P5 SLSs (A545, A553, and A634) all show similar changes over the same concentration range of the receptor, indicating that they reflect the same complex being formed. Unfortunately, due to the solubility limits of Tm14C and the low receptor binding affinity, a full saturation binding analysis could not be carried out.
FIGURE 4. Dipolar ESR data for symmetric SLSs on P5 and CheW.
Time domain signal (left) and corresponding distance distributions (right) for P5 sites (A) A545, (B) A553, (C) CheW W9, and (D) CheW W139. Signals are compared in the absence (dashed line) and presence of unlabeled receptor (solid line). In the case of A545, an additional data set is shown for Tm14 binding in the absence of CheW (dotted line). Darker shades of solid lines indicate increasing receptor concentrations (see below). All time domain signals and distance distributions are scaled to a common value for ease of comparison. CheA concentrations and CheW concentrations were constant at 25 μM, and 125 μM. (A) Tm14 is shown at 75 and 225 μM. (B) Tm14 shown 75 and 225 μM. (C) Tm14 shown at 150 and 300 μM (D) Tm14 shown at 100 and 200 uM.
The binding interaction between CheW and the P5 domain likely stabilizes the associated ends of P5 subdomain 1 and CheW subdomain 2 (Figure 1). Consistent with this, dipolar signals from sites A639 and A646, which localize to the interaction surface of P5 subdomain 1, produced relatively tight distance distributions (full width at half maximum is within 12–14 Å). Addition of receptor did not further shorten distances for A639 but did broaden the P(r) and increase the average separation by 3 Å for site A646 (Table 1). Site A568 which lies on subdomain 2, reported only a minor change in dipolar signal in the presence of receptor.
CheW
It is well established that CheW mediates interactions between CheA and MCPs (26, 28, 51, 87–90), and in keeping with this role, SLS sites on CheW also respond to Tm14C binding (Table 1). Previous DEER experiments with spin labeled CheW at sites W15, W72, W80 and W139 (in complex with wild type CheAΔ289) produced long distances, most of them within 60–70 Å (25). Because the accuracy of distance measurement in DEER in protonated solvents is limited to about 65 Å (63), we searched for new sites on CheW that would produce shorter CheW-CheW separations. We selected CheW sites W9, W28, W31, W35, W101, W102 and W137 all of which lie on the surface of CheW that faces inside the cleft formed by the two CheWs in the model of the CheAΔ289/CheW complex (25). The C β separations at these sites between the two CheWs are within 45 Å. However, within the subset of sites mentioned above, we were able to record and analyze dipolar signals only from sites W9 and W31. SLs on site W28 spontaneously oxidized, detached and accelerated the formation of disulphide linked CheW dimers. Cysteine substitution and subsequent spin-labeling at site W35 caused oligomerization of the protein. CheW proteins spin labeled at W101, W102 and W137 suffered from poor spin-labeling efficiency, possibly due to partial burial of these sites. Nonetheless, the other CheW positions provided useful reporters of Tm14 binding.
In free CheA:CheW, the P(r) from SLS W9 on CheW produced a distinctly bimodal P(r), which indicates two conformations of CheW or the SL itself (40 Å and 55 Å; Table 1) (Figure 4C). The broad distance distribution at this site (Full width at half maximum is 28 Å) may derive in part from the overlapping distributions and also from the flexibility of the CheW N-terminus (23). The interaction with receptor clearly favors the closer 40 Å separation and narrowing of the distance distribution by 8 Å (Figure 4), which is also reflected in a reduction of Ravg from 53 to 51 Å. Full-length CheA and CheAΔ289 produced identical results with CheW SLS W9. Even at the highest concentrations of receptor, where the change in signal saturates, both close and far separations contribute to the overall distribution, which indicates that the receptor favors the close position, but still allows occupation of the far position when bound in the complex. A reduction in distribution width was also seen for SLS W31 on receptor binding, even though the average separation remained the same (Table 1). Site W139 is located at the C-terminal helix of CheW. The CheW-CheW separations at this site are longer in the ternary complex than in binary interaction with only CheA (Figure 4C). Dipolar signals from site W80 did not change on addition of receptor. Thus, CheW reorients in the CheA:CheW complex when receptor binds, with the ends of subdomain 1 contracting towards each other in at least one configuration of the complex. However, the dipolar signals do not support a large scale translation of the entire protein.
Unlike most of the dipolar signals from CheAΔ289, the spin labeled sites on CheW consistently produced signals which were relatively weak in amplitude (only 50% of the expected full signal amplitude). It is unlikely that this derives from some of the CheA population only associating with one CheW, because isothermal calorimetry experiments demonstrated sub-micromolar binding of two molecules of CheW to a single CheAΔ289 dimer (78). Rather, while bound to the P5 domains, the two CheWs sometimes have separations that exceed the maximum distance detected with DEER in protonated systems (~70 Å) (63). Thus, the receptor does not greatly restrict motion of the two CheWs relative to each other, and hence may not bind them both simultaneously.
Modeling the overall conformation of CheA:CheW in the presence of receptor
We assumed that SL separations observed in the CheA/CheW complex on addition of Tm14C largely reflect rearrangements of individual domains, which move as rigid bodies. Furthermore, due to their extensive binding interface, we assumed that P5 and CheW move together. Previously, we developed a method that performs rigid body refinement on protein domains or modules under constraints of long-range distance restraints (74). Application of this method to the new SL distance restraints produced a new model for the CheAΔ289CheW complex in the presence of receptor (Figure 5). P1 and P2 were not modeled because of their positional uncertainty.
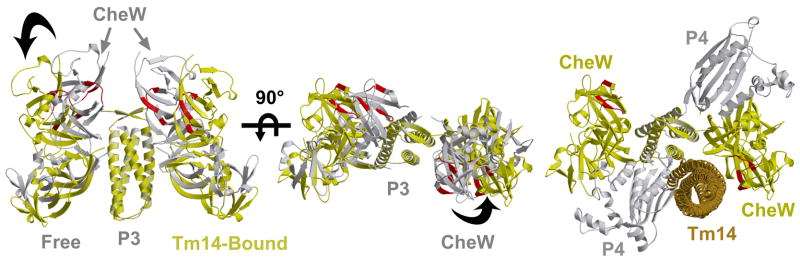
FIGURE 5. Conformational changes in CheA:CheW on binding Tm14.
Comparison of initial structure of CheAΔ289/CheW (grey) to the final structure (yellow) in the presence of Tm14 after rigid body refinement against PDS distance restraints of Table 1. P4 domains are omitted from the left and center structures for clarity. Regions of CheW important for binding MCPs are in red. Right - A dimeric MCP (brown) is docked in the pocket created by the rotation of CheW.
When compared with the model of CheAΔ289:CheW complex (based on crystal structures of CheAΔ289(24), CheAΔ354/CheW and ESR structure of the receptor free complex (25), the most pronounced differences in the new model involve the positions and orientations of the P5 domains and the CheW molecules. Residues important for receptor binding cluster on a lateral surface of the molecule at the fusion of the two subdomains in CheW (88, 91) (Figure 5). In the initial free conformation, the surface formed by these residues line a cleft at the top of CheA formed by both CheWs. The receptor-bound conformation of CheAΔ289:CheW indicates that at least one of the CheWs rotates outward to display the receptor binding surface on the side of the complex (Figure 5). This more exposed surface of CheW is oriented to interact with receptors located along side the P3 domain. Given the many multiple configurations of CheW are indicated by the PDS data, we applied Ravg values instead of Rmax values in the refinement so as not to bias the model toward one conformation of the many. Although, the ends of subdomain 1 of CheW do reside closer to each other in the Tm14C-bound complex compared to free CheA:CheW, refining against Ravg does not achieve the close separation indicated by the bimodal P(r) for site W9 alone (Figure 4). This may indicate that CheW does not respond as a rigid body when Tm14C binds, but rather undergoes some internal conformational changes, as well.
Certain SLSs on the P5 domains come closer together (e.g. A545, A553), but others (A634, A646, A639) move farther apart. This can be achieved by a slight rotation of the domains down toward P3 such that the subdomain 2 β-barrel approaches more closely to its symmetry mate. It should be noted that the receptor-free structure of CheA:CheW is assumed to be quite dynamic based on the observed conformationally heterogeneity of the domain positions. Furthermore, the domains are more widely spaced than would be indicated by the crystal structure. Although the receptor-bound structure is more rigid and compact than the receptor-free structure, it is still more open than indicated by the crystal structures.
Direct distance restraints between Tm14 and CheA:CheW
Mapping association modes in the ternary complex formed by the cytoplasmic fragment of Tm14C, CheAΔ289 and CheW with PDS is based on obtaining pair-wise intermolecular distances in the complex. To confirm that Tm14C primarily forms dimers in isolation and in complex with CheA:CheW, the receptor spin labeled at position 125 was shown to generate a symmetric separation indicative of a dimer under all conditions (Rmax at 28 Å; data not shown). The amplitude of the P(r) remained low at longer distances, which indicates the absence of higher order assemblies. The dimeric nature of both receptor and CheA complicates data analysis, since intermolecular distances (e.g. between CheA and Tm14) are accompanied by intramolecular separations. To reduce the number of spin-spin separations we prepared single-chain receptors, where the receptor subunits are covalently linked C-terminus-to-N-terminus such that a single Cys labeling site can be introduced into the receptor dimer to remove the strong inter-subunit distance of ~30 Å.
In order to test if the single chain construct has the same properties as the receptor homodimer, we studied its effect on distance distributions from CheA SL at position A545 and wild type CheW. We found that the distance distributions changed in the same manner, and in the same concentration ranges as was observed on addition of receptor homo-dimer (data not shown). Thus, the single chain version of Tm14C retains similar affinity and specificity in its interaction with CheA and CheW. Although single chain Tm14C harbors only one spin, it can still presumably bind to CheA:CheW through both of its covalently connected subunits, and hence will produce two possible intermolecular distances between receptor and a single site on CheA or CheW (Figure 2).
We introduced five SLs on the single chain receptor (R100, R111, R149, R160 and R167) and one on the homodimer (R125) (Figure 1). With the exception of R125 which has two labels, all of the Cys substitutions belong to the same receptor subunit. R149 is located directly at the tip, site R100 is located 75 Å away from the tip, and the other sites span the region in between R100 and R149 (Figure 1).
We collected PDS data between Tm14 SLSs (R100, R111, R125, R160, R167 and R149) and SLSs of the CheA:CheW complex (A545, A634, A639, A568 and A646 on P5; A371 and A387 on P4; A301, A318 and A331 on P3; W9, W31, W80 and W139 on CheW), which are summarized in Table 2 and Supplemental Table 1 of the Supporting Information. Of these, eleven measurements produced intermolecular distances in the P(r) distributions that were distinct and did not overlap with the intramolecular distances. This set of distance restraints (Table 2) was then used to construct a model of the CheA/CheW/Tm14C complex. In addition, we simulated the time domain PDS data that yielded the dipolar couplings represented in the model, as well as dipolar couplings that were held independent from the model. The simulations provided insights into the precise domain arrangements and dynamics of the complex and helped resolved situations where signals overlap (See below).
Table 2.
Comparison of intermolecular ESR distances with Cβ separations in the model.
| ESR Distances (Å) | Model Distances (Å) | ||
|---|---|---|---|
| Orientation A | Orientation B | ||
| CheW-R | |||
| W9-R149 | 28, 20–40 | 22, 20 | 17, 20 |
| W80-R167 | 21 (6); 31 (6) | 24, 17 | 29, 25 |
| P5-R | |||
| A545-R100 | 56 (16); 20–30 | 47, 47 | 56, 49, 21, 35 |
| A545-R111 | 42, 51, 40–65; 20–30 | 40, 37, 28, 25 | 48, 39, 11, 27 |
| A634-R100 | 59 (19) | 46, 49 | 52, 45 |
| A634-R111 | 55 (22) | - | 43 |
| A639-R111 | 49, 45–75; 20–30 | 37, 41, 49, 45 | 40, 28, 46, 52 |
| P3-R | |||
| A318-R100 | 55, 45–70 | 46, 42, 32 | 49, 36, 40 |
| A318-R111 | 49, 45–65 | 37, 31 | 42 |
| A331-R100 | 50, 35–70 | 44, 31, 25 | 38, 36, 30, 22 |
| A331-R111 | 42, 55, 35–70 | 39, 27, 22 | 35, 32, 24 |
| A331-R125 | 39, 52, 35–70 | 42, 33, 26, 35 | 43, 31, 29, 40 |
ESR distances in bold represent the Rmax values from the distance distributions. The corresponding half width at full maximum (FWHM) is indicated in parenthesis. In cases where there are closely separated multiple peaks, a distance range is provided along with the Rmax of the major peak. Semicolons offset multiple groups of distances within the same distribution, with the dominant distance indicated in bold. Because ESR distances are typically longer than the C β separations, the model distances that satisfy the following criteria are reported: Rmax − 14 Å < Model distance < Rmax +5 Å for cases where FWHM is indicated. For the rest, the Rmax is replaced by the lower limit of distance range.
Distances between P3 and Tm14
The experiments described above with unlabeled receptor confirmed that P3 maintains its anti-parallel 4-helix bundle structure in the ternary complex. Due to the rigid structure of this domain in the CheAΔ289 dimer, any SLS on P3 produces a signal corresponding to a distance of 30 Å across the helical bundle. If the intermolecular distance between Tm14C and the P3 domain exceeds this, it can be detected within the P(r). In order to determine the orientation of the P3 domain relative to the receptor tip, we measured dipolar signals between SLSs on Tm14C and three different P3 SLSs: A301 at the N-terminal end, A331 at the hairpin tip and A318 at the middle of the bundle (Figure 1; Table 2).
In spite of the considerable width of the distance distributions (which are well matched by the simulations in Supplemental Figure S1 of the Supporting Information), SLSs from the N-terminal end, middle and hairpin of the P3 bundle show a trend that is most consistent with an anti-parallel arrangement of P3 with the Tm14C 4-helix bundle. For example, R111 is located 58.5 Å from the tip of the receptor whereas R100 is further away by 16.5 Å. For A318, in the middle of P3, Rmax of the spin-spin separation increased from 48 to 55 Å as the receptor site changes from R111 to R100; i.e. A318 is closer to R111 than to R100 (Figure 6). All of the P3 to Tm14 distances are shorter than would be expected if P3 were positioned beneath the receptor in an orientation that aligned their symmetry axes. In contrast, it is difficult to distinguish among intermolecular distances between site A331 at the tip of P3 and either R100, R111 and R125. Nonetheless, some of the separations are much closer (35 Å) than any of the separations involving A318. This supports an antiparallel alignment of the respective bundles, with P3 oriented roughly alongside Tm14C. However, the similarity of the signals from receptor SLSs and A331 indicate that the P3 domain is not rigidly fixed to the side of the receptor, but is rather sampling a breadth of orientations offset from the receptor stalk. This assessment is further confirmed below by the simulations.
FIGURE 6. Dipolar signals and distance distributions between SL-CheA:CheW and SL-receptor.
Time domain signal (left) and corresponding distance distributions (right) for Tm14C:CheA:CheW complexes. PDS data are compared for SL-CheA:CheW in the presence of unlabeled receptor (dotted lines) to the presence of spin-labeled receptor (solid lines). (A) P3 A318 to R100 and R111: Tm14C concentrations were 300 μM and CheAΔ289/CheW were at 25 μM and 125 μM, respectively. (B) P5 A634 to R100 and R111. Tm14C concentrations were 300 μM and CheAΔ289/CheW were at 25 μM and 125 μM respectively. (C) CheW 9 to R149: CheAΔ289: spin labeled CheW were at 50 μM and 100 μM respectively, whereas R149 was at 300 μM. (D) CheW 80 to R167: CheAΔ289: spin labeled CheW was at 50 μM and 100 μM, respectively whereas R167 was at 400 μM.
Given the configuration indicated by the other P3-Tm14 restraints, we would expect to observe distances in the 30 Å – 40 Å range for separations between A301 and R167 or R149. Unfortunately, the predicted signals will overlap with those from the inter-subunit P3 SLS distances and thus not be obvious in the distance distributions. However, simulations of the time-dependent data for A301 and R167 provide support for the anti-parallel arrangement of P3 and Tm14C from these SLSs (Supporting Information and see below).
Distances between P4 and receptor
No clear inter-protein distances were observed between SLSs on P4 and Tm14C; although the interaction of CheA:CheW with SL-receptors increased the width of the distance distributions from P4 sites A371 and A387. This could be due to a number of factors, which could include the introduction of a receptor-based spin in the proximity of these positions, or an increase in the mobility of the P4 sites induced by receptor binding.
Distances between P5 domains and receptor
We detected four distinct intermolecular distances between sites A545 and A634 and two receptor SLSs: R100 and R111 (Figure 6). All were within the 40–70 Å range as was observed with P3 domain SLSs. Hence, this data further reaffirms that the P3 domain and the receptor do not stack with their dimer axes aligned. Taken together, these restraints also indicate that the receptor sits alongside P3 and that both CheW proteins have not rotated to the same side of P3 to engage the receptor. If this were the case, the two P5 domains would be too far removed to produce distances in this range. The two ranges of distances in the bimodal distribution of site A639 to R111 (20–30 Å and 40–70 Å; Table 2) indicate that the receptor is closer to one of the P5 subunits than the other and points to asymmetry in the ternary complex.
P3 and P5 residues that are close to each other in the crystal structure of CheAΔ289 give similar distance distributions with SLSs on Tm14C. For example, P5 sites A545 and A634 and P3 site A318 are within 10 Å of each other in the structure and produce very similar dipolar interactions with R100. Thus, the juxtaposition of the P5 and P3 domains is not greatly different than that observed in the crystal structure of the receptor-free kinase. Furthermore, a comparison of distances from R111 and R100 to A545 or A634 reveals that R111 is closer to both P5 sites (on average) than R100. This places at least one P5 domain close to the stalk of the receptor, centered ~50 Å from the tip.
Distances between CheW and receptor
Intermolecular distances between CheW and receptor were difficult to detect, in part because, CheW-CheW dipolar signals were just 50% of the maximum expected amplitude for reasons discussed above. Contrary to SLSs on P3 and P5, most separations between CheWs were greater than 30 Å and distance distributions were broad. Nonetheless, we successfully detected two intermolecular distances within 20–40 Å between CheW and receptor: W9-to- R149 and W80-to-R167 (Figure 6). R149 is at the tip of the receptor and R167 is well within the signaling domain region. These two restraints demonstrate that the receptor signaling domain is close to at least one molecule of CheW and provide a good guideline for aligning the receptor in the CheA/CheW complex.
Restraints from disulphide crosslinking
In order to confirm the conformation of receptor in the ternary complex, we tested the proximity of cysteine residues between receptor and CheA/CheW complex by performing oxidative disulphide crosslinking experiments (77). All of the single cysteine mutations were engineered individually into CheA devoid of Cys residues, CheW and receptor. We conducted sixty-three crosslinking experiments with eight, thirteen and six cysteine substituted CheWs, CheAΔ289 and receptor signaling domain (Supplemental Table S1). In the Tm14C receptor, all of the Cys substitutions were implemented on a single-chain construct with the exception of site R125. Each experiment involved Cys-substituted receptor with either CheAΔ289 or CheW. In some cases, we tested the ability of crosslinking between two partners in the presence of a third Cys-less protein component (Supplemental Table S2).
These experiments revealed only two pairs of sites that cross-linked in the presence of an oxidative initiator, and only one that cross-linked efficiently. In the first case, site E149C at the tip of the single chain receptor selectively bonded with the N-terminal residue K9C of CheW (Figure 7A). This crosslinked pair confirms the PDS results that detected short distances between SLs at these positions (Table 2). The cross-linking efficiency increased in the presence of wild type CheAΔ289, which indicates that the proximity of W9 with R149 increases in the ternary complex.
FIGURE 7. Disulfide cross-linking confirms points of interaction in the ternary complex.
(A) SDS-Page gel showing crosslinking between site CheW K9C and N149C on single chain Tm14C receptor. (B) Crosslinking between Tm14C homodimer N125C and CheΔ289K496C. All protein concentrations were 2μM and Cu(II)(phenanthroline)3 was used as the crosslinking initiator.
The second cross-link occurred between A496C on the P4 domain and R125C on the Tm14C homodimer (Figure 7). The efficiency of cross-linking was much less than in the R149-W9 case, but control reactions of A496 with other Tm14 sites and R125 with other CheA sites produced no similar products (Figure 7B). The presence or absence of wild type CheW did not change the cross-linking efficiency. Interestingly, no cross-linking was observed with the single-chain construct of R125, which was one of the few instances where the single chain receptor behaved differently than the homodimer.
Structural Model of the Ternary Complex formed by CheA, CheW and Tm14
Distance restraints provided by PDS and crosslinking experiments were used to orient the receptor with respect to the CheA/CheW complex. The conformation of CheA:CheW refined from the PDS-derived distance restraints in the presence of unlabeled receptor and the conformation of Tm14C from its crystal structure were taken for modeling the complex (66). We note that in the structure of the three known receptor signaling domains, all have very similar structures for the kinase interacting tips, despite different bends and distortions in the bundle stalks (25, 39, 66). Taking the fixed conformation of CheA:CheW in the presence of unlabeled receptor, the receptor dimer was placed such that the intermolecular constraints of Table 2 were satisfied. Due to the length of the spin label itself, ESR distances are typically longer than Cβ separations. To account for this, we considered the model distance (Cβ-Cβ separations) to agree with the ESR data if: Rmax −14 Å < Rmodel < Rmax + 5 Å. We tested the validity of each orientation limit by comparing the range of intermolecular distances with the Cβ separations from the corresponding sites (Table 2). In each case, there are four possible distances: two between symmetric sites on the CheAΔ289 dimer and the single site on the receptor dimer, and two more if the receptor binds to CheA:CheW with the symmetric surface (Figure 2). We also excluded model distances that would produce SL-SL separations overlapping with signals from the spin-labeled CheA:CheW complex with unlabeled receptor.
The PDS distance restraints greatly limit the possible orientations that the receptor can take with respect to CheA:CheW and thus provides a solid foundation for docking the receptor into the kinase complex (Table 2 and Figure 8). There are short distances (< 25 Å) observed between SLS W9 and R149 and W80 and R167. These two positions bracket the receptor tip along the CheW molecule and match well with the receptor-interacting surface of CheW that has been identified through biochemical and genetic studies (87, 88). The specific close restraint of R149 to W9 and R167 to W80 establishes the general orientation of CheW with respect to receptor. The disulphide crosslink between the Tm14C tip and CheW N-terminus places a further constraint on the proximity between the receptor tip and CheW subdomain 1. These conditions are generally satisfied if the long axis of the receptor, (while still being at the side of the P3 domain) aligns anti-parallel to the P3 long axis and the height of the receptor is adjusted such that the tip region lies close to the N-terminus of CheW. Interestingly, this orientation places the signaling domain of the receptor directly facing the proposed receptor-interaction-region surface of CheW in the new CheA:CheW conformation (88, 91). The low amplitudes of the CheW-to-CheW dipolar signals in the ternary complex suggest that the second CheW is not rigidly disposed with respect to the first, which would be the case if both CheW molecules were binding the same receptor dimer. Thus, it is most likely that only one CheW binds the receptor dimer in this ternary complex. Nonetheless, receptor binding does favor a conformation where the peripheral subdomains of the CheW proteins are close (Rmax ~40 Å). This is consistent with the receptor binding between CheW and P3 and in doing so drawing one CheW toward the CheA dimer axis (Figure 5).
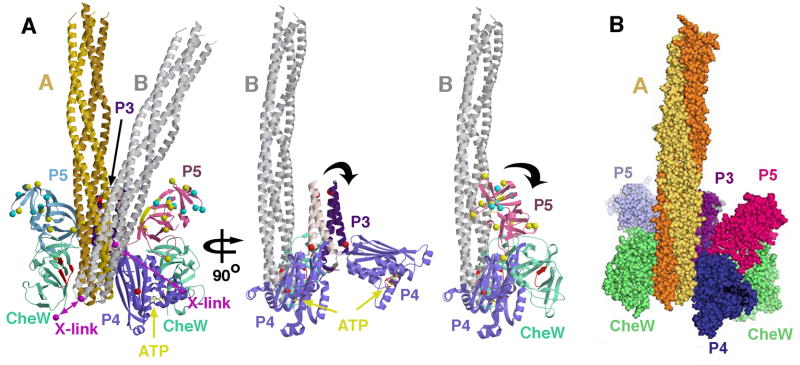
FIGURE 8. PDS structure of the CheA:CheW:Tm14 ternary complex.
PDS data bounds Tm14 within two orientations (A; yellow) and (B; grey). Tm14 resides between CheW and P4 of an adjacent subunit. The P3 domain aligns roughly antiparallel to the the tip of Tm14, which sits close to the N-terminus of CheW. Secondary structure elements on CheW important for binding with MCPs (88, 91) are colored in red, with the analogous regions on P5 in yellow. Positions on P3 and P4 predicted to interact from protection studies on S. typhimurium CheA (27) are shown as red spheres. Residues on P5 have been implicated in receptor-mediated activation of E. coli CheA (yellow spheres), and also ligand-mediated deactivation (cyan spheres). Magenta balls and lines designate positions that undergo disulfide cross linking. In middle view, P5 and CheW domains are removed for clarity, in right view, P5 and CheW are shown without P3. Black arrows denote directions of domain motions indicated from simulation of the ESR time-domain data. (B) Space filling representation of (A), left panel, with Tm14 orientation A shown.
Distance restraints among P3 A318 and A331 with R100, R111, R125 and R167 in the 30–50 Å range confirm that P3 aligns with the side of the receptor in an antiparallel fashion, fully consistent with its position set by the CheW interactions described above. For example, A331 is closer to R100 than is A318 and much closer than is A301. The distances between the P5/P3 domains and the receptor are more widely distributed than those between CheW and receptor. Hence, the signaling domain of the receptor is positioned close to CheW, but the receptor orientation with respect to P3 and P5 is determined with less certainty. We considered orientations of the receptor which had the tip fixed with respect to CheW but the angle between the stalk axis and the P3 domain variable. The receptor orientations that satisfied the distance restraints to P3/P5 fall within two limits: the first (A) localizes the receptor axis completely antiparallel to P3, and the second (B), produces an angle of ~20° between the receptor and P3 symmetry axes that directs the receptor stalk toward the P5 domain of the adjacent subunit that binds the receptor-engaged CheW (Figure 8).
The P5 domain also shows restraints consistent with the docking position imposed by P3- and CheW interactions. A634 and A545 are both closer to R111 than to R100, and both are closer to R111 than is A639 (Table 2). Interestingly, A634 appears to have a shorter interaction with R111 than A545, despite these two sites projecting from residues that are beside each other on two adjacent β–strands. As the receptor extends away from one CheW, it could run along the P5 domain of the same subunit that the interacting CheW is bound (orientation A), or with a small deviation in angle, cross over to interact with the P5 domain of the adjacent subunit (orientation B). Orientation B would predict that A634 would be closer on average to R111 than A545; and thus, in this case, is more consistent with the data (Figure 8).
The position of the P4 domain has the greatest uncertainty in the ternary complex. In the presence of ATP the A496-A496 separations (~50 Å) and the A401-A401 separations (~40 Å), indicate that P4 is much closer to its symmetry mate than indicated by the CheAΔ289 crystal structure. This placement does not change on binding Tm14C. The only way such restraints could be satisfied is if the P4 domains rotate down toward one another relative to the receptor tip. However, this moves A496 away from the receptor stalk, where a modest cross-linking interaction is seen between this position and R125. By-in-large, the data indicates that the P4 domain is quite mobile when Tm14C is bound, and thus it may indeed sample a large space that includes regions close to the receptor stalk as well as down below the tip.
Simulations of time-dependent data based on the ternary complex model
Simulation of PDS time dependent data was carried out based on the SLSs in the modeled ternary complex (Table 3 and Supplemental Figures S1–S6 of the Supporting Information). The simulations indicate that inclusion of the multi-spin coupling terms of Eqn 3 (Supporting Information) only provide a modest improvement in data agreement (Figure S1). However, a number of additional model attributes were necessary in order to properly fit the experimental time domain signals. These features largely introduced conformational variability (presumably due to molecular dynamics) in CheA:CheW and the spin-label itself. Also important, was refinement of individual T2 values for spins on CheA:CheW or Tm14, labeling efficiencies, and binding constants (Figure S1). Inclusion of label mobility over a spherical volume of radius of 3.5 Å, T2 (CheA or W) = ~2–2.5 μs; T2(Tm14) = ~1.5 μs, and Kd = 20–70 μM (usually < 50 μM) consistently produced good agreement. In the case of CheW-to-Tm14C restraints reported by W9:R149, W80:R167 and W31:R167, orientations A and B provide equally reasonable fits without the necessity of conformational variability in the domains (Figure S2); this is not surprising because at the contact of CheW with the receptor tip the A and B conformations are quite similar. In contrast, conformational variability of the P3 and P5 domains must be accounted for in the modeling (Table 3; Figure S3–S4). This was achieved by assuming a rocking displacement about the P4-P5 hinge so as to move CheW:P5:Tm14 roughly in a vertical plane relative to P3 (Figure S2–S6). This accommodation was necessary to accurately fit the time domain data of A331:R100; A318:R100, A318:R111, where a 10–20 Å range of separations provide a reasonable fit to the data (Figure S3–S4). One exception was the separation between A301:R149 (Figure S5), which could be fit with a narrow range and small offset. This restraint fixes the top of the P3 dimerization domain relative to the tip of the receptor bundle and confirms the anti-parallel arrangement of these helix bundles. Both orientations limits, A and B, were tested, with B generally producing better agreement with the experimental data for these SLS interactions.
Table 3.
Distance restraints verified through time-domain simulations.
| Model | Favored Orientation | |
|---|---|---|
| CheW-R | ||
| W9-R149 | VSP | A or B |
| W80-R167 | VSP | A or B |
| W31-R167* | VSP | A or B |
| P5-R | ||
| A545-R100 | VSP/VDP | B |
| A545-R111 | VSP/VDP | B |
| A545-R160 | VSP/VDP | A or B |
| P3-R | ||
| A318-R100 | VSP/VDP | B |
| A318-R111 | VSP/VDP | B |
| A331-R100 | VSP/VDP | B |
| A331-R111 | VSP/VDP | B |
| A301-R149* | VSP/VDP | B |
Model: In modeling, parameters were included for labeling efficiencies, variable spin dephasing times (T2) and CheA:CheW to Tm14 dissociation constants (KD). Variable spin-label positions (VSP), and variable domain positions (VDP) also implemented where noted.
Agreement between modeled and experimental time-domain data was bench marked by comparing residuals to those from well determined cases (See Supporting Information).
Spin-spin interactions that did not give reliable P(r) distributions due to substantial overlap of signals, but whose time-domain data could be simulated from the model.
Simulation of P5 SLS to receptor interactions also agreed with the overall ternary complex model. When positional heterogeneity was introduced into P5, orientation B provided better agreement for sites A545:R111 and A545:R100 but A545:R160 agreed equally well with both A and B orientations. Dipolar interactions between A646 and receptor sites were difficult to model which suggests perhaps that an isotropic treatment of SLSs distributions is not valid in this case, and/or displacements associated with this region of P5 are complex.
In sum, the simulations strongly emphasize the dynamic nature of CheA:CheW when bound to Tm14C. The orientational variability of the domains likely reflects the relative binding contribution that each domain makes to the total interaction with Tm14C. For domains where Tm14C distance restraints could be measured, CheW is relatively well fixed in the complex, P5 less so, and P3 quite dynamic and the least constrained by receptor interactions. P4, P1 and P2 are the most mobile domains, in that they provide no well-resolved distance restraints to Tm14C.
DISCUSSION
Implications of the PDS Structure
The overall structure of the ternary complex reveals that Tm14 makes contact with CheW and P5 and transiently with P3 and P4. The P4 and P5 domains that bind the single receptor dimer belong to the adjacent subunit that harbors the receptor-engaged CheW. Although CheW interactions with receptors are likely the most extensive, CheA contacts are also important, as evidenced by the ability of Tm14C to bind and perturb CheA in the absence of CheW. The receptor affinity is less without CheW, but changes in SL distributions (where measured) indicate that CheA has a similar, but not identical, conformation when bound to the receptor in the absence CheW. Thus, interactions between Tm14C and CheA are only partially modulated by CheW binding. Orientation B places the receptor stalk close to the P5 analogous to the CheW surface that interfaces with the conserved signaling tip. A minor adjustment of the refined model that is within the range of movements modeled for P5 would allow for P5 to interact with the receptor stalk in the same manner as homologous CheW does. Such a contact would explain the competitive binding of CheA and CheW for receptors (92); both proteins may use very similar surfaces to recognize the helical bundle of the receptor. Nonetheless, the interaction of Tm14C for P5 is weaker than for CheW; some conformational variability must be introduced into the P5 position to model the dipolar interactions between this domain and the receptor bundle.
Based on the solution ESR structure of free CheA:CheW, and knowledge of interfaces involved in ternary complex formation, we previously proposed a model of the ternary complex where the receptor binds into the cleft formed by two CheWs and the tip of the receptor sits above the N-terminal end of the P3 domain (25). This model assumed that symmetry would be maintained (i.e. each CheA, receptor and CheW subunit or molecule would be engaged in the same interactions as its symmetry mate) and that CheA:CheW would not significantly change conformation on binding the receptor. Data presented herein establishes that both of these assumptions are invalid. The intermolecular distances between the receptor and either P3 or P5 domains disagree with the Cβ separations predicted from the previous model (25). Instead, the 40–70 Å range of distances observed between SLSs on Tm14 and P3 and P5 are only consistent with the receptor symmetry axis residing roughly parallel to the P3 axis. However, P3 is not tightly associated with Tm14. The distance restraints between P3 and Tm14 indicate an offset of the domains (~10 Å) and furthermore, the dipolar interactions between P3 and Tm14C SLSs can only be simulated if the position of P3 is allowed to vary relative to the receptor stalk, i.e. P3 is not fixed against the receptor in this complex.
Protection studies (27) in which site-directed Cys substitutions on CheA were evaluated for their ability to undergo chemical modification and perturb the ternary complex of CheA:CheW and transmembrane MCPs have identified four sites on the surface of S. typhimurium CheA that are involved in receptor interactions. Two of the sites are on the P3 domain (each at the ends of the domain) and one on each of the P4 and P5 domain. Orientation A allows the receptor to interact with both the sites on the P3 domain, but only one in orientation B (Figure 8). If the concave side of the P4 β-sheet were to bind directly to the receptor, an identified interaction site would lie at the interface (Figure 8). The ternary complex model suggests that the average P4 domain position approaches such an orientation and that some configurations within the broad positional population may achieve this mode of binding. The site identified in P5 by protection studies is buried within the P5 domain, hence it is unlikely to interact directly with the receptor and probably exerts its effect indirectly by influencing the conformation of the binding surface. The protection data lead to the proposal that P3 binds up against the receptor tip in an anti-parallel fashion (27). Whereas this general juxtaposition of P3 with receptor is recapitulated in the PDS model, the simulations rule against a strong contact between P3 and Tm14C in this soluble complex. It should be noted that the compared complexes in the protection and PDS studies come from different sources and are in different contexts. The S. typhimurium complex contained transmembrane receptors and represents an activated state of CheA, whereas the T. maritima complex is formed in vitro with a soluble receptor-like protein and represents of an inhibited form of CheA. It is possible that in the confines of the transmembrane receptor arrays, a stronger interaction is formed between P3 and receptors than we observe here.
In the case of CheW, the interaction surface between CheW and Tm14C displayed by the PDS model agrees well with previous genetic and biochemical data (88, 89, 91) as previously described (25). For P5, which is tightly associated with CheW, the PDS model also suggests possible contacts to Tm14C. Extensive random and directed mutagenesis studies have been carried out on the CheA P5 domain in E. coli (26, 90). That work, which also utilized chemical modification of site-directed Cys replacement, has identified P5 residues important for CheW interactions, receptor-mediated activation of CheA, and ligand induced deactivation of CheA activity in ternary complexes (26, 90). The P5 residues identified to interact with CheW agree well with P5:CheW contacts found by crystallography and ESR spin-labeling (25). Nonetheless, some P5 residues whose mutation or modification affect CheW binding reside far from the P5:CheW interface, which perhaps points to long-range structural coupling within the domain. In the PDS ternary complex, the interaction between P5 and Tm14 does not fix the domains to nearly the same extent as the CheW to Tm14 interface. Nonetheless, the model does predict that P5 can contact the receptor stalk. In orientation B, the P5 surface that sits closest to Tm14C is analogous to the surface of CheW that binds receptor. However, the P5 loop on subdomain 1 that would contact the receptor also contacts CheW on the other side (Figure 8). Thus, the importance of P5 subdomain 1 residues in receptor activation/deactivation may not be easily assigned because their substitution or modification also disrupts CheW binding, and hence ternary complex formation. Residues on subdomain 2 of P5 have also been implicated in receptor activation and deactivation by ligand (26, 90). A number of these sites map to positions that could contact the receptor in orientation B (Figure 8), although there does not appear to be a clear trend with respect to mutation/modification phenotype and location of the residue in the potential interface.
Implications for Activity
The binding of Tm14 (and other MCPC domains) to TmCheA:CheW results in kinase inhibition (66). At this stage we cannot provide a definitive answer for why inhibition occurs; however, several features of the PDS structure may be relevant for rationalizing the inhibitory state of CheA:CheW when bound to Tm14 (66). The close spatial proximity of the receptor tip with the P4 domain could allow the receptor to influence kinase activity. This could be achieved by either blocking access of P1 to P4 or by affecting the catalytic machinery of P4; for example, the conformation of the ATP-lid (85), which could influence phosphotransfer chemistry or P1 recognition. In order to satisfy the PDS restraints between A496-A496, the ATP-lids must be facing inward toward the CheA dimer axis; and in this orientation the P4 domains may be too close to allow one P1 domain to bind between them. Such simultaneous closing off of both active sites may explain how one receptor dimer can impact the activity of both CheA subunits. On the other hand, it has been recently reported that TmCheA displays a large degree of negative cooperativity in binding ATP (93), and thus, under the ATP concentrations used, only one subunit may be active. The cross-link formed between P4 A496 and R125 indicates that the ATP-lid and receptor stalk are sometimes in contact (Figure 8). This domain is clearly inconsistent with Rmax measurements from P4 SLS, but can be explained by the broadness of the distributions, which ultimately reflect substantial mobility of the P4 domains. Furthermore, Tm14 may influence the P4 domain indirectly through action on the P5 domain, which clearly responds to receptor binding and is closely associated with P4.
We were somewhat surprised to find only modest affects of Tm14 on the phospho-accepting P1 domain, which appeared very broadly distributed under all conditions. Recent disulfide cross-linking experiments on S. typhimurium CheA have also revealed that P1 samples a large region of space, but is directed toward the adjacent subunit for trans-phosphorylation (17). This indicates some influence of CheA P2-P5 on P1 localization. Although Tm14 does not grossly change the P1 distribution it could preclude a subset of conformations necessary for productive interaction with P4.
Considerations for a trimer of receptor dimers and a hexagonal lattice of transmembrane receptors
Numerous studies have implicated a trimer of receptor dimers as an essential feature of the active signaling particle (3). In particular, electron microscopy tomograms of polar clusters of signaling complexes from many organisms (including T. maritima) find honey-comb lattices of hexagonal symmetery at the CheA:CheW receptor layers (46–49). These structures have been interpreted as consisting of a trimer of receptor dimers at each of the hexagon vertices (46–49). However, EM studies have found that not all regions of the polar signaling clusters are ordered in hexagonal arrays (48); the “disordered regions” may also be necessary for the clusters to function (59). This heterogeneity could imply that different activity states of the system are realized by modulating its architecture. Several investigations of E. coli systems indicate that increasing the separation between receptors dimers is associated with kinase inhibition (57–60). The T. maritima complex that we have characterized could be indicative of such a state.
Although Tm14 is a naturally soluble protein void of a transmembrane region, its kinase interacting tip has high homology with those from transmembrane MCPs. We thus considered the implications of the inhibited PDS ternary complex to the hexagonal assemblies of the membrane arrays, which are presumably activating with respect to CheA. If we assume that the basic mode of association of CheA:CheW with transmembrane MCPs is similar to that observed with Tm14, the single dimer binding mode would also have to be compatible with a trimer-of-dimers. The range of Tm14 orientations constrained by the PDS data maps out an interaction surface on CheA:CheW large enough to accommodate a trimer-of-dimers (Figure 9). If we further assume that both subunits of CheA can bind receptors simultaneously the dimensions of the membrane lattices place considerable constraints on how CheA:CheW could be incorporated within them. Another important parameter in constructing the lattice is the stoichiometry of components. Both receptors and CheA are dimers, and one CheW binds to one CheA subunit (23, 78, 94). Whole cell analysis suggests a stoichiometry of ~3 receptor dimers, to 1 CheA dimer to 2 CheW proteins (or 3:1 in terms of receptor chains to CheA:CheW subunits) (44). However, reconstituted signaling particles, of both membrane bound receptors and soluble fragments possess stoichiometries of receptors that are considerably higher at 6–9 receptor dimers/1 CheA dimer/2–3 CheWs (28, 50). If a receptor trimer-of –dimers occupies each hexagon vertex (46, 47), then each hexagon can be thought of containing two net trimers-of-dimers (or 6 receptors). The 70–78 Å hexagonal edge spacing (~12 nm honeycomb spacing) in the EM images (27, 46, 47), which presumably indicate the distance between the trimer centers, is too long for the receptor trimers to be in direct contact. An appropriate edge distance results if trimers are separated by an additional four-helix bundle, such as the CheA P3 domain, which is a structural homolog of the receptor signaling tip (Figure 9). In fact, taking the separation of three aligned four helix bundles found in the hedgerows of the Tm14C (66) or Tm1143 (25) crystal structures along with the Tsr trimer crystal structure (39) adjusted to fit the dimensions of recent EM reconstructions (57), produces a honeycomb edge distance that agrees well with the EM edge spacing (Figure 9B). Placing a P3 domain in the center of alternating edges of the hexagon generates three CheA:CheW subunits per hexagon, each from a different CheA dimer (12 receptor subunits: 3 CheA:CheW; or 4:1). Three CheA:CheW subunits could fit within the hexagon (120 Å lattice spacing), but six could not. In order for CheA:CheW to bind a trimer of receptor dimers and for all of the receptor stalks to align perpendicular to the membrane, one CheW would contact a receptor dimer in an “A” orientation, whereas the P5 domain of the adjacent CheA subunit would contact a different receptor dimer of the same trimer in roughly a “B” orientation (Figure 9). If 2 CheA:CheW subunits were present per hexagon, with P3 domains in the center of adjacent edges, the stoichiometry would be 12 receptor subunits: 2 CheA subunits (6:1). One full CheA:CheW dimer in the interior of each hexagon would produce the same stoichiometry. In this latter arrangement, with the P3 domain interior to the receptor walls of the honeycomb network, one CheA:CheW dimer could contact up to four receptor trimers, but no other CheA:CheW dimers could be accommodated. With one interior CheA:CheW dimer, only one CheA:CheW subunit would engage the receptors, which would present a concave face, very different from the edge-on binding mode found in the PDS structures.
FIGURE 9. Incorporation of the PDS ternary complex into an MCP membrane array.
(A) Spacing of the transmembrane MCP trimers observed by cryo-EM (70–78 Å) taken with the PDS structure suggest that the P3 domain could bridge adjacent receptor trimers. If PDS orientation A is superimposed on a single dimer within the trimer, the receptor from orientation B overlaps with the position and an adjacent dimer within the trimer. CheW and P5 domains associated with the same CheA subunit are also spaced appropriately to bridge adjacent trimers with similar interaction surfaces (red and yellow). In left view, no P4 domains are shown, in right view, only one. Receptor trimers were generated from the coordinates of the Tsr cytoplasmic domain (39), adjusted to fit EM reconstruction envelopes (57). (B) Schematic diagram for how edge positioning of the CheA P3 domain could be elaborated into a hexagonal lattice of chemoreceptors. Domain sizes and edge spacings are roughly to scale and thus, only alternating edges could accommodate a P3 domain and still allow the associated P5 and CheW domains to fit within a hexagon.
We believe that the extrapolation of the Tm14:CheA:CheW PDS model to the constraints of the hexagonal membrane arrays is a useful exercise (Figure 9). However, it is important to recall that we detect only a single Tm14 dimer binding to the CheA:CheW dimer by PDS. Nonetheless, that single dimer binds in a range of orientations, and this orientation range provides a reasonable footprint on CheA:CheW for a receptor trimer-of-dimers. In principle, both subunits of the CheA:CheW dimer could bind such a trimer, and hence the extension to the bridging structure of Figure 9. The reason we do not observe the binding of trimers-of-dimers to one CheA:CheW subunit or CheA dimers bridging multiple receptor dimers or trimers-of-dimers in solution may derive from isolating these proteins from their cellular context. For transmembrane chemoreceptors, the membrane environment may facilitate trimer formation, which then provides the high affinity binding entity for CheA:CheW. The affinity of one Tm14 for CheA:CheW is already quite weak and without the membrane scaffold organizing the receptors at high effective concentration, soluble complexes containing additional receptor units may have very high dissociation constants. Even though Tm14 itself is not a transmembrane receptor, it may associate within the transmembrane receptor arrays or in some other higher order complex. On the other hand, there is also the possibility that Tm14 functionally binds CheA:CheW as a single receptor dimer. Nonetheless, the same T. martima CheA must interact with the transmembrane receptors, which appear to form hexagonal lattices based on trimers (47). Given the high sequence conservation at the CheA binding regions on the soluble and membrane receptor proteins, we would expect there to be similarities in the interactions they make with CheA:CheW.
CONCLUSIONS
Our PDS and cross-linking studies have probed the domain architecture of the T. maritima CheA kinase when bound to CheW and a soluble MCP-like protein that inhibits kinase activity (Tm14C). Signaling domains of transmembrane MCPs bind CheA and CheW in an analogous fashion. CheW engages the sequence conserved tip of Tm14 through conserved motifs that are consistent with regions identified through prior genetic and biochemical studies. CheA does not reside below the conserved tip, as we previously suggested, but rather alongside the tip, with the symmetry axes of the P3 domain and the receptor tip roughly anti-parallel, but not strongly interacting. The receptor tip binds between CheA-associated CheW and P4 of the adjacent CheA subunit. The receptor stalk runs up toward the P5 domains and likely crosses the dimer to the adjacent subunit to interact with the symmetry related P5. This would allow the related surfaces on CheW and homologous P5 to engage the receptor in similar ways. Variability in the positions of the P3 and P5 domains relative to Tm14 must be introduced to successfully model the ternary complex. P4 is very broadly distributed in the presence and absence of the receptor, with little change in its positioning or mobility on receptor binding. PDS-derived distances suggest P4 samples space below the receptor tip, whereas cross-linking data indicates that it can also interact with the receptor stalk above the tip. Given the proximity of the receptor to P4, the mechanism by which the receptor inhibits kinase activity could involve interference with P1 access to P4 or a direct effect on the catalytically competent conformations of the P4 domains. Incorporation of this structure into a hexagonal lattice of clustered transmembrane receptors could be achieved by placing the P3 domain of the CheA dimer in the center of a honeycomb edge; although other arrangements are also possible.
Supplementary Material
Abbreviations
- A
CheA residue
- CheY-P
phosphorylated CheY
- DEER
double electron electron resonance
- DQC
double quantum coherence
- EM
electron microscopy
- HAMP domain
domain found in Histidine kinases, Adenylyl cyclases, Methyl-accepting chemotaxis proteins, and Phosphatases
- GF
gel filtration
- MCP
methyl-accepting chemotaxis protein
- MCPL
MCP ligand binding domain
- MCPC
MCP cytoplasmic domain
- P1
CheA phosphotransferase domain
- P2
CheA CheY docking domain
- P3
CheA dimerization domain
- P4
CheA kinase domain
- P5
CheA regulatory domain
- PCR
polymerase chain reaction
- PDS
pulsed dipolar ESR spectroscopy
- Ravg
probability weighted average spin separation
- Rmax
spin separation of highest probability
- R
Tm14 residue
- SL
spin label
- SLS
spin label site
- Tm14
soluble MCP-like chemoreceptor from T. maritime
- Tm14C
shortened, more soluble fragment of Tm14
- W
CheW residue
Footnotes
This work supported by grants: NIGMS GM06675 (B.R.C), NIH/NCRR P41-RR016242 (J.H.F.) and NIH/NIBIB 2R01EB003150 (J.H.F.)
Details of PDS signal measurement and analysis. Table of Intermolecular distances between CheA:CheW and Tm14. Summary of disulfide cross-linking between Cys-engineered Tm14. Modeling of distance distributions from intermolecular spin-spin dipolar signals. Simulations of PDS data. This material is available free of charge via the Internet at http://pubs.acs.org.
LITERATURE CITED
- 1.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 3.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends In Biochemical Sciences. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 7.Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Hess JF, Bourret RB, Simon MI. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 9.Kofoid EC, Parkinson JS. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988;85:4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goudreau PN, Stock AM. Signal transduction in bacteria: molecular mechanisms of stimulus-response coupling. Curr Opin Microbiol. 1998;1:160–169. doi: 10.1016/s1369-5274(98)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.West AH, Stock AM. Histidine kinases and response regulators proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 12.Bilwes AM, Park SY, Quezada CM, Simon MI, Crane BR. Structure and Function of CheA, the Histidine Kinase Central to Bacterial Chemotaxis. In: Inouye M, Dutta R, editors. Histidine Kinases in Signal Transduction. Academic Press; San Diego, CA: 2003. pp. 48–74. [Google Scholar]
- 13.Stewart RC, Jahreis K, Parkinson JS. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 14.Stewart RC, VanBruggen R. Phosphorylation and binding interactions of CheY studied by use of badan-labeled protein. Biochemistry. 2004;43:8766–8777. doi: 10.1021/bi0495735. [DOI] [PubMed] [Google Scholar]
- 15.Stewart RC, Van Bruggen R. Association and dissociation kinetics for CheY interacting with the P2 domain of CheA. J Mol Biol. 2004;336:287–301. doi: 10.1016/j.jmb.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 16.Stewart RC. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry. 1997;36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- 17.Gloor SL, Falke JJ. Thermal Domain Motions of CheA Kinase in Solution: Disulfide Trapping Reveals the Motional Constraints Leading to Trans-autophosphorylation. Biochemistry. 2009;48:3631–3644. doi: 10.1021/bi900033r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson RV, Bourret RB, Simon MI. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 19.Wolanin PM, Stock JB. Transmembrane Signaling and the Regulation of Histidine Kinase Activity. In: Inouye M, RD, editors. Histidine Kinases in Signal Transduction. Academic Press; San Diego: 2003. pp. 74–123. [Google Scholar]
- 20.Ames P, Parkinson JS. Constitutively signaling fragments of Tsr, the E. coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-depenent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninfa EG, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 23.Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nature Struct Biol. 2002 doi: 10.1038/nsb753. in press. [DOI] [PubMed] [Google Scholar]
- 24.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 25.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor: kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 26.Zhao JH, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA kinase of bacterial chemotaxis: Chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levit MN, Grebe TW, Stock JB. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem. 2002;277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 29.Seeley SK, Wittrock G, Thompson LK, Weis RM. Oligomers of the cytoplasmic fragment from the Escherichia coli aspartate receptor dissociate through an unfolded transition state. Biochemistry. 1996;35:16336–16345. doi: 10.1021/bi961749i. [DOI] [PubMed] [Google Scholar]
- 30.Wolanin PM, Baker MD, Francis NR, Thomas DR, DeRosier DJ, Stock JB. Self-assembly of receptor/signaling complexes in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2006;103:14313–14318. doi: 10.1073/pnas.0606350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montefusco DJ, Shrout AL, Besschetnova TY, Weis RM. Formation and activity of template-assembled receptor signaling complexes. Langmuir. 2007;23:3280–3289. doi: 10.1021/la062717r. [DOI] [PubMed] [Google Scholar]
- 32.Shrout AL, Montefusco DJ, Weis RM. Template-directed assembly of receptor signaling complexes. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- 33.Bischoff DS, Ordal GW. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992;6:23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 34.Garrity LF, Ordal GW. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology. 1997;143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- 35.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: Limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 37.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 38.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 40.Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc Natl Acad Sci U S A. 2006;103:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boldog T, Grimme S, Li MS, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li MS, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 46.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Zhuo L, Chen S, Muller A, Iancu CV, Murphy G, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A Early Edition. 2009 doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khursigara CM, Wu XW, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang PJ, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erbse AH, Falke JJ. The Core Signaling Proteins of Bacterial Chemotaxis Assemble To Form an Ultrastable Complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and Kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 52.Lybarger SR, Nair U, Lilly AA, Hazelbauer GL, Maddock JR. Clustering requires modified methyl-accepting sites in low-abundance but not high-abundance chemoreceptors of Escherichia coli. Mol Microbiol. 2005;56:1078–1086. doi: 10.1111/j.1365-2958.2005.04593.x. [DOI] [PubMed] [Google Scholar]
- 53.Levit MN, Liu Y, Stock JB. Biochemistry. 1999;38:6651–6658. doi: 10.1021/bi982839l. [DOI] [PubMed] [Google Scholar]
- 54.Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 55.Shimizu TS, Le Novere N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signaling proteins involved in bacterial chemotaxis. Nature Cell Bio. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 56.Besschetnova TY, Montefusco DJ, Asinas AE, Shrout AL, Antommattei FM, Weis RM. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc Natl Acad Sci U S A. 2008;105:12289–12294. doi: 10.1073/pnas.0802868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khursigara CM, Wu XW, Zhang PJ, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamanna AC, Ordal GW, Kiessling LL. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol Microbiol. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- 60.Borrok MJ, Kolonko EM, Kiessling LL. Chemical probes of bacterial signal transduction reveal that repellents stabilize and attractants destabilize the chemoreceptor array. ACS Chem Biol. 2008;3:101–109. doi: 10.1021/cb700211s. [DOI] [PubMed] [Google Scholar]
- 61.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nature Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 63.Borbat PP, Freed JH. Measuring distances by pulsed dipolar ESR spectroscopy:spin-labeled histidine kinases. Meth Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 64.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Physical Chemistry Chemical Physics. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 65.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 66.Pollard AM, Bilwes AM, Crane BR. The Structure of a Soluble Chemoreceptor Suggests a Mechanism for Propagating Conformational Signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hubell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Op Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 68.Milov AD, Maryasov AG, Tsvetkov YD. Pulsed electron double resonance (PELDOR) and its applications in free-radicals research. Applied Magnetic Resonance. 1998;15:107–143. [Google Scholar]
- 69.Borbat PP, Mchaourab HS, Freed JH. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J Am Chem Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 70.Maryasov AG, Tsvetkov YD. ELDOR in ESE structure studies. J Appl Magn Reson. 1998;14:101–113. [Google Scholar]
- 71.Borbat PP, Crepeau RH, Freed JH. Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J Magn Reson. 1997;127:155–167. doi: 10.1006/jmre.1997.1201. [DOI] [PubMed] [Google Scholar]
- 72.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Chiang YW, Borbat PP, Freed JH. Maximum entropy: A complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. Journal Of Magnetic Resonance. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 74.Bhatnagar JaFJH, Crane BR. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Meth Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- 75.Hustedt EJ, Stein RA, Sethaphong L, Brandon S, Zhou Z, DeSensi SC. Dipolar coupling between nitroxide spin labels: The development and application of a tether-in-a-cone model. Biophys J. 2006;90:340–356. doi: 10.1529/biophysj.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sale K, Song LK, Liu YS, Perozo E, Fajer P. Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J Am Chem Soc. 2005;127:9334–9335. doi: 10.1021/ja051652w. [DOI] [PubMed] [Google Scholar]
- 77.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: Applications to soluble and transmembrane receptors of bacterial chemotaxis. Two-Component Signaling Systems. 2007;(Pt B):25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SY, Quezada CM, Bilwes AM, Crane BR. Subunit exchange by CheA histidine kinases from the mesophile Escherichia coli and the thermophile thermotoga maritima. Biochemistry. 2004;43:2228–2240. doi: 10.1021/bi0352419. [DOI] [PubMed] [Google Scholar]
- 79.Zhou H, Lowry DF, Swanson RV, Simon MI, Dahlquist FW. NMR studies of the phosphotransfer domain of the histidine kinase CheA from Escherichia coli: assignments, secondary structure, general fold, and backbone dynamics. Biochemistry. 1995;34:13858–13870. doi: 10.1021/bi00042a018. [DOI] [PubMed] [Google Scholar]
- 80.Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 81.McEvoy MM, Muhandiram DR, Kay LE, Dahlquist FW. Structure and dynamics of a CheY-binding domain of the chemotaxis kinase CheA determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:5633–5640. doi: 10.1021/bi952707h. [DOI] [PubMed] [Google Scholar]
- 82.Gouet P, Chinardet N, Welch M, Guillet V, Cabantous S, Birck C, Mourey L, Samama JP. Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY-CheA(124–257) complex. Acta Crystallogr D. 2001;57:44–51. doi: 10.1107/s090744490001492x. [DOI] [PubMed] [Google Scholar]
- 83.Welch M, Chinardet N, Mourey L, Birck C, Samama JP. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nature Struct Biol. 1998;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 84.Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci U S A. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nature Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 86.Bornhorst JA, Falke JJ. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: Development of a heterogeneous two-state model. J Mol Biol. 2003;326:1597–1614. doi: 10.1016/s0022-2836(03)00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boukhvalova M, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar - Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- 89.Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interactions on CheW. J Biol Chem. 2002;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 90.Zhao JS, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 93.Eaton AK, Stewart RC. The Two Active Sites of Thermotoga maritima CheA Dimers Bind ATP with Dramatically Different Affinities. Biochemistry. 2009;48:6412–6422. doi: 10.1021/bi900474g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gegner JA, Dahlquist FW. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci U S A. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.