Abstract
While many RNA-binding proteins (RBPs) bind RNA in a sequence-specific manner, their sequence preferences alone do not distinguish known target RNAs from other potential targets that are coexpressed and contain the same sequence motifs. Recently, the mRNA targets of dozens of RNA-binding proteins have been identified, facilitating a systematic study of the features of target transcripts. Using these data, we demonstrate that calculating the predicted structural accessibility of a putative RBP binding site allows one to significantly improve the accuracy of predicting in vivo binding for the majority of sequence-specific RBPs. In our new in silico approach, accessibility is predicted based solely on the mRNA sequence without consideration of the locations of bound trans-factors; as such, our results suggest a greater than previously anticipated role for intrinsic mRNA secondary structure in determining RBP binding target preference. Target site accessibility aids in predicting target transcripts and the binding sites for RBPs with a range of RNA-binding domains and subcellular functions. Based on this work, we introduce a new motif-finding algorithm that identifies accessible sequence-specific RBP motifs from in vivo binding data.
Keywords: RNA-binding protein, accessibility, secondary structure, post-transcriptional regulation, gene regulation, cis-element
INTRODUCTION
In eukaryotic cells, post-transcriptional regulation of mRNA stability (Grigull et al. 2004; Tadros et al. 2007), translation (Wharton et al. 1998), localization (Lecuyer et al. 2007), and splicing (Blencowe 2006) involve the targeting of transcripts by various RNA-binding proteins (RBPs) that recognize cis-elements in the transcript sequence. To map out post-transcriptional networks (Keene 2007), transcripts associated with RBPs have been identified in genome-wide assays (Overall et al. 2004; Keene et al. 2006). In many cases, these target sets are enriched for short RNA sequence motifs (Gerber et al. 2004, 2006; Hogan et al. 2008; Ray et al. 2009) that reflect the sequence-binding preferences of the assayed RBPs. However, these sequence preferences do not provide sufficient specificity to distinguish the RBP-associated transcripts from unbound transcripts containing the same short sequence motifs. While some RBPs recognize their binding sites within a hairpin loop (e.g., Vts1p [Aviv et al. 2006]), most mRNA-binding RBPs bind unstructured single-stranded RNA (ssRNA) (Ellis et al. 2007), so specific RNA secondary structures are unlikely to provide the required specificity for many RBPs, thus limiting the applicability of recent algorithms developed to identify secondary structures bound by RBPs (Rabani et al. 2008; Foat and Stormo 2009).
mRNA secondary structure may instead provide specificity by sequestering potential RBP target sites within regions of double-stranded RNA (dsRNA), thus rendering them nonfunctional. This is an extension of the long-held view that sequence-specific RBPs, unlike DNA-binding proteins, require at least some of their binding site to be single-stranded (Mattaj and Nagai 1994). This belief stems from the fact that the A-form helical structure typically adopted by dsRNA has a major groove that is narrower than that of the B-form helix of dsDNA, thus preventing amino acid side chains from accessing and recognizing the bases within dsRNA. This belief is supported by recent surveys (Draper 1999; Allers and Shamoo 2001; Jones et al. 2001) of structures of RBP–RNA complexes deposited in the PDB (Berman et al. 2000) that report that base-specific interactions between RBPs and RNA only occur in regions of ssRNA or near irregularities in the RNA helix. These irregularities result from unpaired bases (i.e., bulges and internal loops) and have the effect of widening the major groove.
This bias toward structurally accessible binding sites can be exploited to improve in vitro predictions of binding of RBPs to RNAs. A role for accessibility in RBP binding has long been supported by in vitro selection (Levine et al. 1993; Gao et al. 1994) and measurements of in vitro binding affinity (Hackermuller et al. 2005). More recently, motif-finding algorithms have been developed that use measures of RNA single-strandedness to more accurately recover some RBP motifs from in vitro selection binding data (Hiller et al. 2006). In all cases, computational models of RNA folding were used to predict RNA secondary structure.
However, despite its predictive value in vitro, structural accessibility has not been used to aid in the prediction of in vivo binding of RBPs to mRNA, in part because of the perceived difficulty of accurately predicting mRNA secondary structure computationally. Popular RNA secondary structure prediction methods use simplified energy models and largely ignore the effect of cotranscriptional folding of mRNA on its secondary structure (see Geis et al. 2008, Kinwalker program). These approximations are thought to have a large impact on the accuracy of their predictions for longer RNAs such as mRNAs. However, despite these deficiencies, structural accessibility calculated by these programs does predict the in vivo binding sites of microRNAs (miRNAs) (Robins et al. 2005; Kertesz et al. 2007; Long et al. 2007) and small interfering RNAs (siRNAs) (Tafer et al. 2008), thus demonstrating that these methods do predict single-strandedness with some degree of accuracy.
Nonetheless, using target-site accessibility to predict in vivo RBP binding has remained largely untested because of obvious differences in RNA binding by RBPs versus miRNAs/siRNAs. Unlike these noncoding RNAs (ncRNAs), many RBPs function in the nucleus where mRNA secondary structure may be much more constrained by large heterogeneous ribonucleoprotein complexes (hnRNPs) associated with the transcript that are displaced during the export of the mRNA to the cytoplasm or during the first round of translation. It has also been suggested, based on the presence of RNA helicases and potential RNA chaperones within hnRNPs, that mRNA secondary structure undergoes extensive remodeling to facilitate RBP binding (Rajkowitsch et al. 2007). Furthermore, although miRNAs and siRNAs compete for the same binding interface as mRNA secondary structure, RBPs can bind RNA through a variety of interfaces, some of which may require only small disruptions in A-form helical structure to expand the major groove, thereby permitting recognition of bases flanking the disrupted region. Under this circumstance, only a subset of the bases within the sequence-specific binding site need be unpaired.
The recent availability of mRNA target sets for a large number of RBPs from yeast, flies, and humans has allowed us to assess the impact of structural accessibility on sequence-specific binding of a diverse set of RBPs that carry a variety of RNA-binding domains and participate in a number of subcellular functions. Through a systematic analysis, we demonstrate that target site accessibility, predicted based on intrinsic mRNA secondary structure, plays a general role in RBP binding. Incorporating target site accessibility into computational models of sequence-specific RBP binding yields a statistically significant overall improvement in their ability to predict the outcome of large-scale assays of in vivo RBP–mRNA interactions for the majority of sequence-specific RBPs.
RESULTS
To investigate the role of mRNA secondary structure on RBP binding, we compiled data on the in vivo mRNA targets of a set of 30 eukaryotic RBPs from Saccharomyces cerevisiae, Drosophila melanogaster, and humans derived from RNP immunoprecipitation microarray (RIP-chip) copurification assays. Details of each data set are available in the Materials and Methods and Supplemental File 1. We assessed the impact of mRNA secondary structure on putative RBP binding sites by scoring their accessibility and determining whether more accessible target sites were more likely to be bound. We define “target site accessibility” as the probability that the entire target site is unpaired as estimated by a computational method, RNAplfold (Bernhart et al. 2006), which considers the relative stabilities of all possible secondary structures containing the site and its flanking sequence (see Materials and Methods for details).
Target site accessibility predicts mRNA targets of Pumilio and Puf3p
We began our investigation by examining RIP-chip-derived target sets for Drosophila Pumilio (Gerber et al. 2006), a well-studied protein with conserved RNA binding specificity, and S. cerevisiae Puf3p (Gerber et al. 2004), the likely yeast ortholog of Pumilio. These two proteins, along with the Fem-3-binding factor in Caenorhabditis elegans, share a conserved RNA-binding domain consisting of eight repeats of the Pum-homology domain (PumHD). To date, all known Puf proteins bind their targets through this domain and subsequently regulate the stability and/or translation of these targets (Wharton et al. 1998; Olivas and Parker 2000; Goldstrohm et al. 2006; Hook et al. 2007). Structural (Wang et al. 2002; Miller et al. 2008; Zhu et al. 2009), small-scale (Dalby and Glover 1993; Jackson et al. 2004), and large-scale (Gerber et al. 2004, 2006) studies are consistent with a conserved single-stranded consensus binding sequence, UGUAHAUA, for Pumilio and Puf3p (H indicates that A, C, or U is permitted).
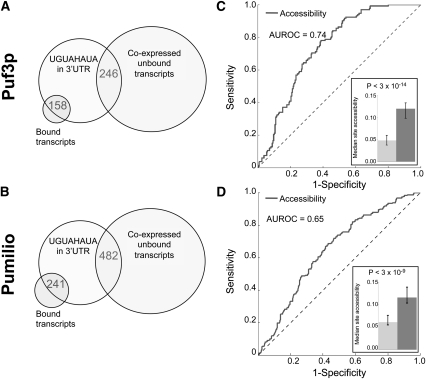
To evaluate the role of accessibility in Puf3p and Pumilio targeting in vivo, we defined a set of mRNAs likely to be bound by fly Pumilio and yeast Puf3p based on their relative enrichment in the bound fraction of mRNA using FDR cutoffs established in the original studies (Gerber et al. 2004, 2006). As a negative control, we also defined a set of mRNAs expressed under the queried conditions, and thus available for binding, but that were not enriched in the coimmunoprecipated fraction (see Supplemental File 1 for details). We called these transcripts “unbound.” All experimentally validated target sites to date for Pumilio and Puf3p occur in the 3′ UTR, so we scanned the 3′ UTRs of mRNAs in the bound and unbound sets and identified those that contained a match to the UGUAHAUA consensus. As expected, a larger proportion of bound transcripts contained a 3′ UTR match to UGUAHAUA (Puf3p 75% vs. 9%, P < 3.0 × 10−94, Pumilio 51% vs. 12%, P < 1.0 × 10−81, Fisher's Exact Test). However, there were more unbound transcripts with matches than bound ones (158 vs. 246 in yeast, 241 vs. 482 in fly) (Fig. 1A,B). As such, target-site recognition for Pumilio and Puf3p cannot be explained by RNA sequence preference alone. To determine whether target site accessibility could distinguish unbound transcripts from bound ones, we estimated the probability that each match to UGUAHAUA was single-stranded, using a computational method that predicts target site accessibility based only on RNA sequence flanking the target site (see Materials and Methods), and compared the accessibility of matches in the 3′ UTRs of bound and unbound transcripts. Multiple consensus sites within the same 3′ UTR may increase the affinity of the RBP for the mRNA; to control for this, we only compared bound and unbound transcripts with the same number of matches. Figure 1, C and D, contains the results for transcripts with a single match in their 3′ UTR, which constitute the vast majority of transcripts. The results for transcripts with multiple matches are similar (Supplemental Table 1).
FIGURE 1.
Puf3p and Pumilio consensus binding sites have higher accessibility in the 3′ UTRs of their bound mRNA targets. (A,B) While the consensus matches were significantly enriched in the set of bound transcripts for yeast Puf3p and fly Pumilio, more unbound transcripts contained consensus matches than bound ones (158 vs. 246 for yeast Puf3p [A], 241 vs. 482 for fly Pumilio [B]). (C,D) Comparison of site accessibility of transcripts coimmunoprecipitating (co-IPing) with Puf3p (C) and Pumilio (D) and those coexpressed but not co-IPing. All compared transcripts have only a single copy of the Puf3p/Pumilio consensus UGUAHAUA (H matches A, C, or U) in their 3′ UTRs (132 bound and 235 unbound transcripts for Puf3p; 201 bound and 414 unbound transcripts for Pumilio). The ROC curve (solid line) plots the sensitivity (i.e., the proportion of bound transcripts recovered; vertical axis) against [1 – specificity] (i.e., the proportion of unbound transcripts recovered; horizontal axis) as the accessibility threshold is adjusted from the highest to the lowest. (Inset) Median site accessibility for the bound set (dark gray bar) and the unbound set (light gray bar). Error bars represent the 95% confidence interval of the median calculated using 5000 bootstrap samples. P-values were calculated using the Wilcoxon–Mann–Whitney Rank Sum test.
As Figure 1 shows, the median accessibility of sites in bound mRNAs was almost twofold higher than in unbound mRNA in both yeast (Fig. 1C, inset) and fly (Fig. 1D, inset). Furthermore, receiver operating characteristic (ROC) analysis demonstrated that target-site accessibility is a statistically significant predictor of coimmunoprecipitation of a transcript with Pumilio or Puf3p (Fig. 1C,D; Puf3p area under ROC [AUROC] curve = 0.74, P = 3 × 10−14; Pumilio AUROC = 0.65, P = 3 × 10−9, Wilcoxon–Mann–Whitney test). These results demonstrate that target site accessibility plays a role in RBP binding, and thus target mRNA selection, by Pumilio and Puf3p.
We also performed a similar test for one of the human homologs of Pumilio, human Pum1, using RIP-chip-derived target sets from Morris et al. (2008) (Supplemental Fig. 1). Although in this case there are more bound than unbound transcripts with 3′ UTR copies of UGUAHAUA (Supplemental Fig. 1A), accessibility remained a statistically significant predictor in determining Pum1 binding upon comparison of bound versus unbound transcripts containing the same number of consensus sites in their 3′ UTRs (Supplemental Fig. 1B; Supplemental Table 1).
Accessibility improves motif-based mRNA target prediction for diverse RBPs
Having established proof-of-principle that target site accessibility, predicted on the basis of mRNA sequence alone, has a measureable impact on RBP binding in vivo, we sought to determine the generality of this observation by assessing the impact of target site accessibility for RBPs with a diverse range of RNA-binding domains, sequence-binding preferences, and subcellular functions. To do so, we compiled RIP-chip data and consensus sequence motifs for additional RBPs from yeast and human. Of 18 such RBPs (including Puf3p, Pumilio, and Pum1), we removed three (Pab1p, Nsr1p, Nrd1p) whose bound sets were not significantly enriched for the reported consensus sequence (Wilcoxon–Mann–Whitney P-value > 0.05) and one RBP (Ssd1p) for which only seven transcripts matched the consensus. Thus, we were left with 14 likely sequence-specific RBPs with a large enough number of putative target mRNAs for our analysis. Although the fact that these RBPs are sequence-specific suggests that at least some portion of their binding site needs to be structurally accessible, many of these RBPs lack crystal structures, so it is not clear how much of the site needs be accessible or whether the structural accessibility of the site can be predicted by computational folding of the mRNA sequence without consideration of the influence of trans-factors on the mRNA's secondary structure. For each of these RBPs, we again used the relative enrichment among mRNAs copurifying with the RBP, as measured using the RIP-chip assay, to define its bound mRNA transcripts and a set of unbound mRNA transcripts that were coexpressed with the RBP but showed no evidence of being bound (see Supplemental File 1 for details).
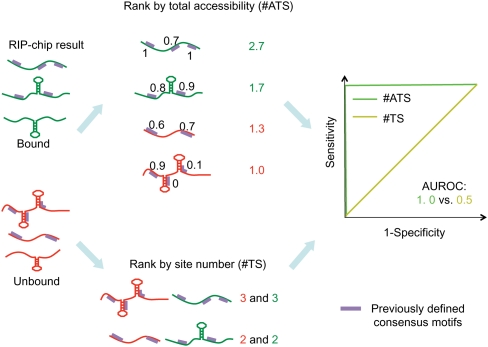
Some of the RBP consensus sites matched in a large number of positions, making it difficult to directly compare transcripts with the same number of potential target sites. We therefore adopted a new and more general analytical procedure (described schematically in Fig. 2) that compared all bound transcripts with all unbound ones and scanned the entire mature mRNA sequence for binding sites. To assess the predictive value of target site accessibility, we assigned each transcript a score equal to the sum of the accessibilities of sites in the transcript, and then evaluated how well that score distinguished bound and unbound transcripts. We call this score the “total accessibility” and abbreviate it by “#ATS” because it is equal to the expected number (#) of Accessible Target Sites per transcript. To control for the fact that transcripts with more RBP binding sites were more likely to be bound, we also assessed how well the number of target sites (“#TS”) in a transcript predicted whether or not it was bound. As before, we evaluated the predictive accuracy of both #ATS and #TS using AUROC. We tested for a significant difference between the two AUROCs by combining a permutation test with the Delong–Delong–Clarke–Pearson procedure (DeLong et al. 1988; see Materials and Methods).
FIGURE 2.
Schematic of the in silico assay for measuring the impact of target site accessibility on RBP binding. The flowchart displays the procedure for evaluating accuracy at distinguishing bound and unbound sets of mRNA using either #ATS- or #TS-based scoring of an RBP consensus sequence. For each RIP-chip data set, transcripts were sorted in decreasing order by their relative enrichment among mRNAs copurifying with the RBP. We defined those with relative enrichment larger than the “positive threshold” to be the “bound” set of transcripts and those with relative enrichment smaller than the “negative threshold” to be the “unbound” set of transcripts. In this way, it was guaranteed that the transcripts in the unbound set were coexpressed with the RBP. We then identified all consensus-sequence matches (which we call “target sites”) in each transcript and removed transcripts with no target sites. We then ranked transcripts in decreasing order of number of target sites and used this ranking to calculate the #TS AUROC. To calculate #ATS, we first calculated the accessibility of each target site. This calculation considered all possible secondary structures, weighted according to their stability, so even sites that were single-stranded or paired in the most probable secondary structure (as displayed) could have a value <1 or >0, respectively. We ranked transcripts in decreasing order by the sum of the accessibilities of their target sites (i.e., #ATS) and calculated the associated AUROC.
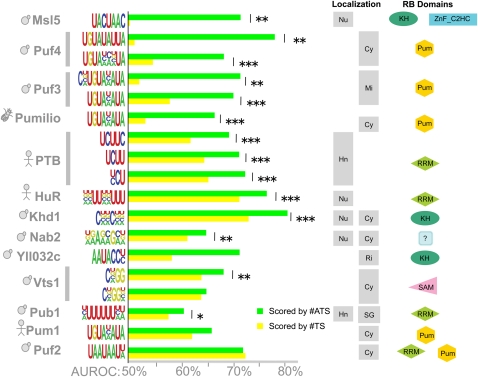
Considering only the transcripts that contained at least one copy of the consensus sequence, for 13 of the 14 RBPs there was a statistically significant increase in #TS or #ATS among the bound transcripts (P < 0.05, Wilcoxon–Mann–Whitney) compared with unbound. These 13 RBPs, along with their AUROCs, are displayed in Figure 3. For 10 of 13 RBPs, there was a statistically significant increase in AUROC when #ATS versus #TS was used to predict whether a transcript would be bound. In some cases the increase in AUROC was quite large, while in other cases it was more modest. However, it should be noted that because random performance for AUROC is 50%, absolute increases in AUROC translate into relative decreases in error that are at least twice as large (where error is measured as 100% − AUROC). For example, although the average absolute increase in AUROC for all motifs in Figure 3 is 9.3%, the average relative decrease in error is 22.1%. Notably, in no case did #ATS have an AUROC significantly smaller than the AUROC for #TS.
FIGURE 3.
Target site accessibility predicts in vivo binding for a diverse range of RBPs. Bar graphs compare the accuracy of #ATS and #TS at predicting bound transcripts based on a given consensus. To the left of the bar graph, each row is labeled by the RBP, the associated consensus sequence used for classification, and a cartoon indicating the species of origin (yeast, fly, or human). Some RBPs have multiple reported consensus sequences; these are grouped and indicated by a vertical bar. To the right of the bar graph, for each RBP, we show its known subcellular localization and its known RNA-binding domains (using SMART domains). (Left localization column) Nuclear localization (if any) as indicated: (Hn) hnRNP, (Nu) nucleus; (right localization column) cytoplasmic localization as indicated: (Cy) cytoplasm, (Mi) mitochondrion, (Ri) ribosome, (SG) stress granule. Supplemental File 1 contains the evidence for the reported localization and domains. The statistical significance of differences between #ATS AUROC (green bars) and #TS AUROC (yellow bars) was calculated using the Delong–Delong–Clarke–Pearson procedure: (*) P < 0.05, (**) P < 0.01, (***) P < 10−4. Exact P-values are in Supplemental File 2.
Target site accessibility predicted not only the targets of proteins thought to bind unstructured ssRNA (e.g., the Puf family of proteins) but also the targets of Vts1p, a stem–loop-binding protein that recognizes loops containing CNGG(N)0-3 (Aviv et al. 2006). Indeed, our procedure was sensitive enough to detect the fact that Vts1p binds loops of length four bases or more: #ATS had a significant improvement in accuracy over #TS when used with the CNGG motif but not a CNGGN motif because the latter would score four-base loops bound by Vts1p as inaccessible.
In summary, target site accessibility is a statistically significant predictor of in vivo binding for 71% (10 of 14) of the sequence-specific RBPs tested.
Improvement due to accessibility is not explained by nucleotide composition biases
We performed a number of computational controls to confirm that the observed role of target site accessibility was not due to other potential properties of functional binding sites. One possible alternative explanation for the observed differences is that functional RBP binding sites are in regions of biased, low-order nucleotide composition. For example, many cis-regulatory mRNA elements are located in 3′ UTRs, which tend to be AU-rich. This AU-richness of flanking sequence has been suggested as an explanation for the predictive value of accessibility for miRNA binding (Grimson et al. 2007). However, if this were the case for RBPs, then one would expect either (1) that the effect would disappear if our analysis was restricted by only scanning sites in the 3′ UTR or (2) that the flanking regions around target sites in bound transcripts would be biased toward a single type of dinucleotide. However, target site accessibility remained a strong predictor of in vivo binding when we restricted our scans to 3′ UTRs (Fig. 4). Furthermore, although there were statistically significant differences in the dinucleotide composition of sequence flanking sites in bound transcripts, the enriched and depleted dinucleotides depended on the consensus sequence and tended to favor dinucleotides less likely to pair with the consensus (Fig. 5). Indeed, most dinucleotides were significantly enriched for some RBPs and significantly depleted for others. We also confirmed that the increase in predictive power was not due to the sequence composition of the consensus; when we repeated our analysis using the reverse consensus (e.g., CNGG became GGNC), we only saw a significant increase for #ATS when the reversed consensus was a strong match to the forward consensus (Supplemental Fig. 2).
FIGURE 4.
3′ UTR target site accessibility predicts in vivo binding. Results are presented as in Figure 3, but only target sites within the 3′ UTRs of transcripts were used to calculate #TS and #ATS. (+) AUROC is significantly different from random (Wilcoxon–Mann–Whitney, P < 0.05). Exact P-values are in Supplemental File 2.
FIGURE 5.
Differences in dinucleotide composition around putative RBP binding sites between bound and unbound transcripts. (A) Heat map showing the t-statistic of the difference in di- and single-nucleotide frequencies in 40 bases upstream and downstream of the target site. (Rows) The RBP and its consensus binding sequence motifs (in IUPAC representation) used to identify target sites. (Columns) Single nucleotide versus dinucleotide. Rows and columns were ordered based on two-dimensional hierarchical clustering. Those t-statistics with absolute value <2 are not statistically significant at α = 0.05 and are set to 0; those with an absolute value >4 remain statistically significant after a Bonferroni correction and are thresholded at 4 or −4, as appropriate. (B) As for A, but using 20 bases up- and downstream.
Many RBPs require the entire target site to be accessible
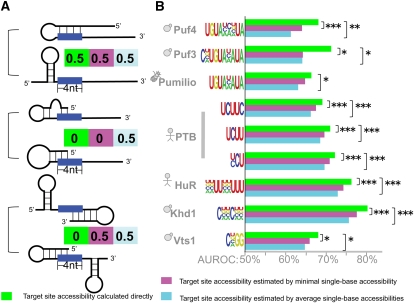
We next sought to determine whether different approaches to calculating target site accessibility had an impact on the performance of our assay. Different approaches make different assumptions about the role of target site accessibility in RBP binding. The method we employed, as proposed by Hackermuller and coworkers (Hackermuller et al. 2005), requires the whole site to be unpaired for the protein to bind. However, other methods, including the EF option of MEMERIS (Hiller et al. 2006), and some methods used to predict target site accessibility for miRNAs (Robins et al. 2005; Ellis et al. 2007; Kertesz et al. 2007; Long et al. 2007; Geis et al. 2008; Tafer et al. 2008), allow target sites to be partially paired. To determine which of these approaches most accurately predicts in vivo RBP binding, we compared transcripts scored by #ATS based on the accessibility of the whole target site and those scored by #ATS when target site accessibility was approximated by either the average or the minimum single-base accessibility of all bases in the target site.
The estimates of target site accessibility from these two methods (single-base and whole-site) diverge most when the target site was partially paired. For example, if in all stable mRNA secondary structures exactly half of the bases in the target site were paired, then the average single-base accessibility for that site would be 0.5 but the accessibility of the whole-target site would be 0. On the other hand, if the target site was completely unpaired in some structures and completely paired in others, then both the average single-base accessibility and the whole-site accessibility would be exactly the same (Fig. 6A).
FIGURE 6.
Target site accessibility is a better predictor than average/minimal accessibility of single bases in the target site. (A) Diagrams represent examples of how secondary structure leads to differences in the calculated target site accessibility when the calculation is for the entire site (green), minimal single-base accessibility (magenta), or average single-base accessibility (light blue). In each case, two equally stable structures are shown, and the numbers represent accessibility calculated for a four-base site assuming each secondary structure is equally probable. (B) As per Figure 3 except that light blue and magenta bars show AUROCs for #ATS scoring when the target site accessibility is replaced with the average and minimal accessibility of all single bases in the target site. Exact P-values are in Supplemental File 2.
When we compared the predictive accuracy of #ATS calculated using whole-site, average single-base, and minimum single-base accessibilities (Supplemental Table 2), we found that for six proteins (Puf3p, Puf4p, PTB, HuR, Khd1p, Vts1p) both of the single-base approximations significantly decreased predictive accuracy (Fig. 6B). Four of these six—Puf3p (Zhu et al. 2009), Puf4p (Miller et al. 2008), PTB (Oberstrass et al. 2005), and Vts1p (Aviv et al. 2006)—have solved co-crystal structures showing that the RBP binds to a completely unpaired target site, and there is other evidence to suggest that the entire HuR binding site must be unpaired (Levine et al. 1993; Gao et al. 1994; Meisner et al. 2004). Our data thus suggest that Khd1p will also require its entire binding site to be unpaired. It should be noted that we found no instances where either of the single-base approximations significantly improved accuracy compared with the whole-site method.
The impact of other variations in the calculation of target site accessibility
We evaluated two other methods for calculation of target site accessibility. First, when transcripts were scored using the maximum accessibility of the target sites in the transcripts, AUROC scores almost always decreased relative to #ATS, often significantly (Supplemental Table 2); the only exceptions were Vts1p and Msl5p. These results suggest that multiple accessible sites in the same transcript contribute to binding. Second, we investigated whether also considering the accessibility of sequence flanking the target site helped predict binding. Unlike the case for miRNAs (Kertesz et al. 2007), we found that for RBPs there was little improvement in AUROC when a measure of flanking region accessibility was added to target site accessibility (Supplemental Table 2).
In vivo sequence motif finding using accessibility
We next attempted to recover the RBP sequence-binding preferences from the in vivo data using motif-finding methods that either did or did not incorporate target site accessibility. We carried out these analyses for two reasons: First, we wanted to ensure that the increased predictive accuracy of accessibility did not arise from how the RBP consensus sequences were originally defined; second, we wished to assess whether incorporation of accessibility as a feature improved either the accuracy or the statistical power of RBP motif finding based on in vivo copurification data. We performed this motif-finding analysis on all yeast RBPs in our collection: These included 14 with previously associated motifs and 12 with no associated sequence motifs (Bfr1p, Cbc2p, Cbf5p, Gbp2p, MRN1p [also known as Ypl184c], Nab3p, Nab6p, Nop56p, Npl3p, Puf1p, Scp160p, and Tdh3p).
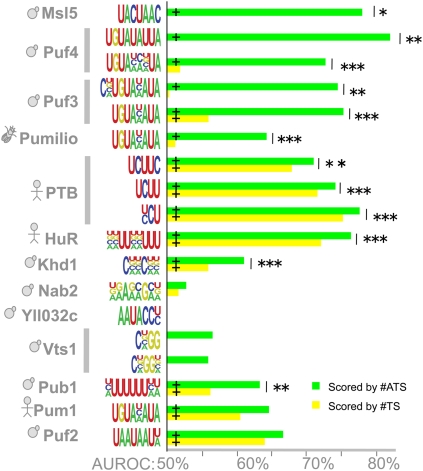
We used a discriminative motif-finding procedure that attempted to identify consensus sequences that, when scored with either #ATS or #TS, best distinguished bound and unbound transcripts by being more often present and/or having either more sites (for the #TS-derived motif) or more accessible sites (for the #ATS-derived motif) among the bound transcripts (see Materials and Methods). In Figure 7, we report two motifs for each RBP: one based on #TS and the other #ATS. These motifs were derived from the entire set of bound and unbound transcripts. However, the AUROCs that we report for #ATS and #TS were calculated on held-out data using a cross-validation training procedure that we employed to avoid over-fitting (see Materials and Methods). Motifs learned during cross-validation were similar to those reported in Figure 7. (Supplemental File 1 contains all learned motifs; note we do not display consensus sequence motifs for the nine RBPs for which our procedure was unable to find motifs whose predictive accuracy was significantly better than random.)
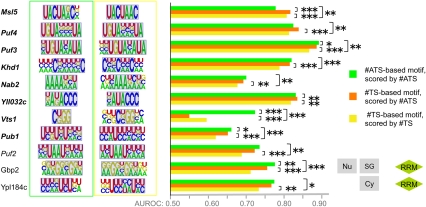
FIGURE 7.
RBP motifs optimized to distinguish bound versus unbound transcripts. Each RBP is shown associated with two motifs: the #ATS-derived motif with the highest AUROC (green box) and the #TS-derived motif with the highest AUROC (yellow box) after motif finding was performed on the complete set of bound and unbound transcripts. (Gray background) Overlap of manually aligned regions of the #ATS and #TS motifs for the same RBP. The bar graphs display median AUROC over 30 held-out test sets for motifs trained to maximize #ATS AUROC and scored with #ATS (green bar), trained to maximize #TS AUROC and scored with #TS (yellow bar), and trained to maximize #TS AUROC but scored with #ATS (orange bar). We assessed P-values for differences in distributions of 30 AUROCs on matched test sets using the Wilcoxon sign-rank test. (Italicized RBP names) The RBP has a previously defined consensus sequence, (bold italics) a significant increase in #ATS reported in Figure 3. The P-value threshold is indicated as for Figure 3, and exact P-values are available in Supplemental File 2. Subcellular location and RBD domains are displayed for RBPs not represented in Figure 3.
Both #ATS-based and #TS-based motifs were consistent with previously determined binding preferences for all nine yeast RBPs displayed in Figure 3. Also consistent with Figure 3, the AUROC of the #ATS motif on held-out data was significantly higher than that of the #TS for seven of the nine RBPs reported therein. As expected, the #ATS-trained motif for Vts1p recovered the bound loop sequence and achieved a higher AUROC than the #TS-trained motif which attempted to model the stem sequence. For eight of the nine RBPs, there was also a significant increase in AUROC when we used #ATS to score the held-out data based on the #TS-trained motif (the only exception was Vts1p). Thus, even motifs trained to maximize the #TS-based score remained more predictive of RBP binding when scored with #ATS, demonstrating that target site accessibility increased accuracy regardless of how the sequence motif was derived.
Figure 7 also contains two RBPs not previously associated with motifs, Gbp2 and MRN1/Ypl184c. For these RBPs, the #ATS-derived and #TS-derived motifs were similar and there was a statistically significant increase in accuracy when scoring either of these motifs using #ATS, thus following the same pattern as most of the RBPs already associated with motifs. These results suggest that these two RBPs also bind unstructured ssRNA. Supplemental Figure 3 displays motifs for the six other RBPs: For three of these RBPs (Bfr1p, Scp160p, and Tdh3p), the #ATS-derived motif was contained within the motif discovered by #TS, and the AUROC for #ATS scoring based on the #TS-derived motif was smaller than the two other AUROCs. This pattern mirrors Vts1p, suggesting that the additional bases in the #TS-derived motif are likely to be inaccessible.
In summary, our motif analysis demonstrated two things. First, the ability of target site accessibility to improve accuracy (as shown in Fig. 3) is not an artifact of how the previously reported RBP sequence motifs were defined; even motifs trained to maximize #TS AUROC in our assay underwent a significant improvement in accuracy when scored using #ATS. Second, the #TS- and #ATS-derived motifs were very similar for RBPs that bind unstructured ssRNA and for these RBPs, we often observed no significant difference in AUROCs when the two motifs were scored using #ATS. This observation suggests that mRNA secondary structure functions primarily to sequester nonfunctional matches to an RBP's sequence preferences rather than to reveal binding sites for RBPs with highly degenerate sequence preferences.
DISCUSSION
We have demonstrated that binding-site accessibility has a significant impact on mRNA target selection for 12 of 14 RBPs (86%) with previously determined sequence-binding preferences. Also, using a novel discriminative in vivo motif finding approach that incorporates target site accessibility, we were able to identify two additional RBPs, Gbp2p and MRN1p, that are likely to bind unstructured ssRNA. Together, these 14 RBPs include five different classes of RNA-binding domains (RRM, KH, Pum-repeats, SAM, C2H2-Zn-finger), are not biased toward either nuclear or cytoplasmic function, and include examples of RBPs known to bind both unstructured ssRNA and loop sequences. Thus, accessibility predicts target selection by RBPs with a diverse set of RNA-binding domains that bind within different secondary structure contexts and that have different subcellular locations of binding.
We have also identified some differences between features of RBP binding sites and miRNA and siRNA binding sites. First, as previously reported for in vitro binding (Hiller et al. 2006), allowing partial pairing of putative RBP binding sites significantly reduces in vivo predictive accuracy for six RBPs. Five of these RBPs have previously been reported to require their entire binding site to be unpaired, strongly suggesting that the sixth, Khd1p, will have a similar requirement. It should be noted that we never observed an advantage to allowing partial pairing. Also, unlike miRNA binding (Kertesz et al. 2007), requiring flanking sequence also to be accessible never significantly improved predictive accuracy.
Our observations, taken together with similar observations on the role of mRNA secondary structure in small regulatory RNA targeting (Kertesz et al. 2007; Bompfunewerer et al. 2008; Long et al. 2008; Tafer et al. 2008), demonstrate that accessibility plays a role in target selection throughout the lifetime of an mRNA. Because in all cases structural accessibility was predicted using methods that consider only the mRNA sequence, these data suggest that intrinsic mRNA secondary structure forms prior to trans-factor binding and constrains subsequent binding events at all levels of post-transcriptional regulation. These data also provide a possible mechanism by which the clustering of target sites in the transcript increases the likelihood of RBP binding (Stadler et al. 2006; Ule et al. 2006; Akerman et al. 2009): Many of the RBP consensus motifs that we considered do not form stable RNA secondary structures when concatenated. Thus, site clustering, in addition to providing more target sites for the RBP, cooperatively enhances the accessibility of sites in the cluster. Preferential RBP binding at accessible target sites also provides a mechanism that explains why accessibility modulates the cis-regulatory impact of known splicing enhancer and suppressor elements on nearby splice sites (Hiller et al. 2007).
Our data predict that HuR, Puf3p, and Puf4p require their whole binding site to be accessible and are thus consistent with previous in vitro studies for HuR (Levine et al. 1993; Gao et al. 1994; Meisner et al. 2004; Ray et al. 2009) and solved co-crystal structures for Puf3p (Zhu et al. 2009) and Puf4p (Miller et al. 2008). However, our observations appear to conflict with recent reports for HuR (Lopez de Silanes et al. 2004) and Puf3p and Puf4p (Rabani et al. 2008), in which the binding sites we propose for these RBPs are predicted to be partially paired within a hairpin. There are a number of possible explanations for this disparity. First, it is possible that these proposed hairpins do not form because they are not energetically favored. The algorithms used to identify these stem–loops, COVE (Eddy and Durbin 1994) and RNApromo (Rabani et al. 2008), employ Covariance Models (CM) to predict RNA secondary structure. CMs consider only the existence of possible pairings but consider neither their thermodynamic stability nor the impact of flanking sequence on the predicted structure. Indeed, as stated in the user guide to COVE, “covariance models routinely overpredict [RNA secondary] structure, because a) they don't look for Watson–Crick complementarity and b) it is often statistically advantageous for the model to pair as many positions as possible” (Eddy 1993; Guide.tex accessed from ftp://selab.janelia.org/pub/software/cove/cove-2.4.4.tar.Z on February 7, 2010). Another explanation may that the hairpins do form but are comparatively less stable in bound than unbound mRNA. If true, this would be consistent with our observations because our model considers only the relative, not the absolute, accessibility of sites in bound mRNAs.
Our results have important consequences for large-scale analyses of RNA–protein interactions and RNA processing, both experimentally and computationally. We have shown that target-site accessibility almost always increases—and never significantly decreases—the ability to predict sequence-specific RBP binding to mRNAs. We have also found that rewarding a transcript for containing multiple target sites improves predictive accuracy. Thus, methods that attempt to identify RBP target sites in mRNAs or to infer regulatory networks should be augmented with target site accessibility data for all potential binding sites.
Our models do not perfectly reproduce the in vivo binding data, suggesting that there remains room for further improvement. Although some of the errors that our models make may be due to external factors, such as noise in the original experimental assay or our inability to precisely recover the mRNA transcript sequence targeted by the RBP, many errors are likely to be due to in vivo regulatory mechanisms that are not captured by our model. First, our model considers the accessibility of target sites in an mRNA but does not take into account whether or not that site is associated with a particular element of RNA secondary structure such as a hairpin loop. Indeed, we have recently found that searching for Vts1p binding sites only within hairpin loops improves the predictive power of our model (K Cook and Q Morris, unpubl.). Second, we did not model possible competition for binding sites by other trans-factors or cooperative binding with different trans-factors. These factors could, for example, be responsible for our poor performance at predicting Pum1 sites because Pum1 has similar binding preferences to Pum2 and there is an enrichment of microRNA binding sites around human Pum sites (Galgano et al. 2008). Third, the method that we used to predict target site accessibility considers neither the effects on RNA secondary structure of other bound trans-factors nor long-range interactions within the mRNA (e.g., Kreahling and Graveley 2005).
Despite these caveats, our analysis demonstrates that mRNA secondary structure has a significant impact on RBP binding in the absence of any of these other considerations, thus suggesting that internal mRNA secondary structure is an important determinant of RBP binding. Our results and evaluation framework also provide a means by which the universality and predictive power (or lack thereof) of these other possible regulatory mechanisms may be demonstrated. Python scripts to reproduce our analysis, calculate #ATS, assess AUROC, and perform our motif finding, as well as our detailed benchmark results, are available in the Supplemental Material.
MATERIALS AND METHODS
RBP copurification and sequence motif data collection
We used RBP copurification data from six different sources, though in compiling these data we made extensive use of matched collections of RBP binding data and consensus sequences compiled by Hogan et al. (2008) and Ray et al. (2009). The RBP copurification data we used were derived from the RNP immunoprecipitation–microarray (RIP-chip) assay. To ensure that we had sufficient statistical power for our analyses, we only used RBPs that copurified with at least 30 mature mRNA targets. Supplemental File 1 describes the source of each data set.
Source of transcript sequences
The Drosophila melanogaster (BDGP5.4), Saccharomyces cerevisiae (SGD1.01), and Homo sapiens (NCB136) transcript sequences were downloaded from Ensembl using BioMart (http://www.biomart.org/). For fly and human, we downloaded all cDNA sequences and defined 3′ UTRs as the portion of the cDNA downstream of the 3′ end of the coding sequence, as defined by Ensembl. Full-length cDNAs including 5′ and 3′ UTRs were not available for most yeast genes, so, like Hogan et al. (2008), we defined the yeast cDNA as the longest ORF corresponding to each gene plus 200 nucleotides (nt) upstream and 200 nt downstream of the start and stop codons, respectively, removing any ORF sequence from the upstream or downstream genes.
Defining bound and unbound sets of transcripts
Transcripts were classified into these two sets by comparing their relative enrichment in the RNA fraction copurifying with the RBP to two thresholds. The positive threshold defined the bound set and the negative threshold defined the unbound set (refer to Fig. 2 for more details and Supplemental File 1 for the thresholds). We defined relative enrichment using either FDR, Z-score, or LOD as reported in the original manuscript. Whenever possible, we used positive and negative thresholds established in the original study describing the data. However, in some cases we used a more permissive negative threshold to increase the statistical power of our analyses.
Quantifying target site accessibility
Target site accessibility was assessed using RNAplfold (Bernhart et al. 2006). RNAplfold models cotranscriptional folding by calculating base-pair probabilities using a small window of sequence around the site of interest based on a computational model of thermodynamic stability of RNA secondary structures. Specifically, it estimates the probability that either a binding site, or a single base, is unpaired by calculating local-pair probabilities for bases with a maximal span of L nucleotides, by sliding a moving window of size W nucleotides along the input RNA sequence. It computes the probability that a region of U consecutive nucleotides is unpaired by averaging the probability over all windows of size W that contain this region. In our experiments, we fixed W = 80 and L = 40 and set U to either the width of the consensus sequence or to 1 when calculating single-base accessibility. These parameter settings were previously optimized for predicting siRNA binding (Bompfunewerer et al. 2008; Tafer et al. 2008). When calculating target site accessibilities for a 3′ UTR site, we input the entire transcript into RNAplfold to ensure that the target site accessibility for sites immediately downstream of the stop codon incorporated coding sequence.
Scoring accessibility of target sites and their flanking regions
We scored the accessibility of a target site and the flanking region up to X bases upstream of the site and Y bases downstream by summing the single base accessibilities (calculated as described above) for the X upstream and Y downstream bases and then adding the accessibility of the target site times the length of the target site. We adopted this procedure, rather than calculating the accessibility of the site including its flank, because the latter value often dropped below machine precision (and became inaccurate) for larger values of X and Y.
Statistical tests for the significance of difference between two AUROCs
We used the Delong–Delong–Clarke–Pearson (DDCP) (DeLong et al. 1988) procedure to assess the significance of differences between the AUROCs on #TS and #ATS. However, because the UcR R package (Lindbäck 2009) implementation of DDCP that we used did not correct for tied ranks when assessing the significance of difference between two AUROCs, whenever there were tied ranks we reported the median DDCP P-value over 100 random permutations of the ordering of these tied ranks.
Motif finding procedure and cross-validation
In general, we used a training set of bound and unbound transcripts to fit motifs which we then assessed using a held-out test set. We applied a two-step strategy to fit motifs. First, we calculated #TS and #ATS AUROCs for all possible 6-mers (including transcripts with no target sites when ranking transcripts, assigning them a score of 0) using the training set. We selected the 6-mers used to seed the next step of motif finding based on these AUROCs. We fit two separate consensus sequence motif models for each RBP, one seeded with the five 6-mers with the highest AUROCs when scored with #ATS, and the other seeded with the five 6-mers with the highest #TS-scored AUROCs. Starting from each seed, we employed an iterative motif-refinement procedure that shortened, lengthened, or introduced degeneracy a single base at a time. At each iteration, the motif that gave the largest AUROC on the training set was selected (measured using #ATS or #TS, as appropriate); the procedure was terminated when the AUROC failed to increase or the associated Bonferroni-corrected Wilcoxon–Mann–Whitney P-value failed to decrease. As with the 6-mers, we also ranked transcripts with no target sites when calculating the AUROC. Once the motif finding converged for all five seeds, we selected the motif with the highest AUROC on the training set. We then evaluated the AUROC of each model on the test set to assess its predictive accuracy. We generated 30 training/test set splits using a 3 × 10 fold cross-validation procedure whereby we randomly split the bound and unbound sets into 10 equally sized bins, trained the motif models on the sequences in nine of the bins, and evaluated them on the remaining bin. We repeated this random split three times and collected 30 test set AUROCs for each motif finding method and each transcript scoring method.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://morrislab.med.utoronto.ca/datamain and http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was funded by a CIHR Team Grant in mRNP Systems Biology (CTP-79838) (to H.D.L.), a CFI/ORF LOI grant (to Q.M.), and a CIHR operating grant (MOP-93671) (to Q.M). We thank Drs. Craig A. Smibert and Timothy R. Hughes (University of Toronto) for critical feedback on the manuscript. We also thank Dr. Debashish Ray, who compiled the HuR RIP-chip data set, and Sepand Mavandadi, who provided scripts to help format figures (both at the University of Toronto).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2017210.
REFERENCES
- Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y 2009. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol 10: R30 doi: 10.1186/gb-2009-10-3-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers J, Shamoo Y 2001. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol 311: 75–86 [DOI] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F 2006. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol 13: 168–176 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE 2000. The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Stadler PF 2006. Local RNA base pairing probabilities in large sequences. Bioinformatics 22: 614–615 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ 2006. Alternative splicing: New insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Bompfunewerer AF, Backofen R, Bernhart SH, Hertel J, Hofacker IL, Stadler PF, Will S 2008. Variations on RNA folding and alignment: Lessons from Benasque. J Math Biol 56: 129–144 [DOI] [PubMed] [Google Scholar]
- Dalby B, Glover DM 1993. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J 12: 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL 1988. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44: 837–845 [PubMed] [Google Scholar]
- Draper DE 1999. Themes in RNA-protein recognition. J Mol Biol 293: 255–270 [DOI] [PubMed] [Google Scholar]
- Eddy SR 1993. User's guide for COVE—covariance models of RNA sequence families MRC Laboratory of Molecular Biology, Cambridge, UK [Google Scholar]
- Eddy SR, Durbin R 1994. RNA sequence analysis using covariance models. Nucleic Acids Res 22: 2079–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JJ, Broom M, Jones S 2007. Protein–RNA interactions: Structural analysis and functional classes. Proteins 66: 903–911 [DOI] [PubMed] [Google Scholar]
- Foat BC, Stormo GD 2009. Discovering structural cis-regulatory elements by modeling the behaviors of mRNAs. Mol Syst Biol 5: 268 doi: 10.1038/msb.2009.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3: e3164 doi: 10.1371/journal.pone.003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD 1994. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci 91: 11207–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis M, Flamm C, Wolfinger MT, Tanzer A, Hofacker IL, Middendorf M, Mandl C, Stadler PF, Thurner C 2008. Folding kinetics of large RNAs. J Mol Biol 379: 160–173 [DOI] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79 doi: 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackermuller J, Meisner NC, Auer M, Jaritz M, Stadler PF 2005. The effect of RNA secondary structures on RNA-ligand binding and the modifier RNA mechanism: A quantitative model. Gene 345: 3–12 [DOI] [PubMed] [Google Scholar]
- Hiller M, Pudimat R, Busch A, Backofen R 2006. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res 34: e117 doi: 10.1093/nar/gkl544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S 2007. Pre-mRNA secondary structures influence exon recognition. PLoS Genet 3: e204 doi: 10.1371/journal.pgen.0030204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6: e255 doi: 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook BA, Goldstrohm AC, Seay DJ, Wickens M 2007. Two yeast PUF proteins negatively regulate a single mRNA. J Biol Chem 282: 15430–15438 [DOI] [PubMed] [Google Scholar]
- Jackson JS Jr, Houshmandi SS, Lopez Leban F, Olivas WM 2004. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 10: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM 2001. Protein–RNA interactions: A structural analysis. Nucleic Acids Res 29: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB 2006. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kreahling JM, Graveley BR 2005. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol Cell Biol 25: 10251–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD 1993. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol 13: 3494–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbäck JR 2009. ucR R package: Version ucR_0.3-0 Uppsala Clinical Research Centre, Uppsala, Sweden [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y 2007. Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14: 287–294 [DOI] [PubMed] [Google Scholar]
- Long D, Chan CY, Ding Y 2008. Analysis of microRNA-target interactions by a target structure based hybridization model. Pac Symp Biocomput 2008: 64–74 [PubMed] [Google Scholar]
- Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci 101: 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Nagai K 1994. RNA–protein interactions Oxford University Press, Oxford [Google Scholar]
- Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, Auer M 2004. mRNA openers and closers: Modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. ChemBioChem 5: 1432–1447 [DOI] [PubMed] [Google Scholar]
- Miller MT, Higgin JJ, Hall TM 2008. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol 15: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Olivas W, Parker R 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 19: 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall CM, Tam EM, Kappelhoff R, Connor A, Ewart T, Morrison CJ, Puente X, Lopez-Otin C, Seth A 2004. Protease degradomics: Mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol Chem 385: 493–504 [DOI] [PubMed] [Google Scholar]
- Rabani M, Kertesz M, Segal E 2008. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc Natl Acad Sci 105: 14885–14890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4: 118–130 [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Castillo LP, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR 2009. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 27: 667–670 [DOI] [PubMed] [Google Scholar]
- Robins H, Li Y, Padgett RW 2005. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci 102: 4006–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Shomron N, Yeo GW, Schneider A, Xiao X, Burge CB 2006. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet 2: e191 doi: 10.1371/journal.pgen.0020191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155 [DOI] [PubMed] [Google Scholar]
- Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL 2008. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol 26: 578–583 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM 2002. Modular recognition of RNA by a human pumilio-homology domain. Cell 110: 501–512 [DOI] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell 1: 863–872 [DOI] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM 2009. A unique binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci 106: 20192–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]