Abstract
The genomic termini of RNA viruses contain essential cis-acting signals for such diverse functions as packaging, genome translation, mRNA transcription, and RNA replication, and thus preservation of their sequence integrity is critical for virus viability. Sequence alteration can arise due to cellular mechanisms that add or remove nucleotides from terminal regions, or, alternatively, from introduction of sequence errors through nucleotide misincorporation by the error-prone viral RNA-dependent RNA polymerase (RdRp). To preserve template function, many RNA viruses utilize repair mechanisms to prevent accumulation of terminal alterations. Here we show that Bunyamwera virus (BUNV), the prototype of the Bunyaviridae family of segmented negative-sense RNA viruses, also can repair its genomic termini. When an intact nontranslated region (NTR) was added to the anti-genomic 3′ end, it was precisely removed, to restore both length and RNA synthesis function of the wild-type template. Furthermore, when nucleotides were removed from the anti-genome 3′ end, and replaced with a duplicate and intact NTR, both the external NTR were removed, and the missing nucleotides were restored, thus, indicating that the BUNV RdRp can both remove and add nucleotides to the template. We show that the mechanism for repair of terminal extensions is likely that of internal entry of the viral RdRp during genome synthesis. Possible mechanisms for repair of terminal deletions are discussed.
Keywords: Bunyamwera, RNA, polymerase, repair, virus
INTRODUCTION
Bunyamwera virus (BUNV) is the prototype of the Bunyaviridae family of segmented negative-sense RNA viruses. As with all bunyaviruses, the BUNV genome comprises three strands of RNA named small (S), medium (M), and large (L), and each BUNV genomic strand acts as template for generation of a single mRNA. The S mRNA encodes the N and NSs proteins from overlapping open reading frames (ORFs) (Fuller and Bishop 1982; Fuller et al. 1983; Elliott 1989b). The M mRNA encodes a polyprotein that is cleaved into Gn, NSm, and Gc proteins (Gentsch and Bishop 1979; Fuller and Bishop 1982; Elliott 1985), and the L mRNA encodes the RNA-dependent RNA polymerase (RdRp) (Elliott 1989a). This polymerase is responsible for two RNA synthesis activities: Replication to generate a replication intermediate known as the anti-genome (Fig. 1A), and transcription to generate mRNAs that are primed using capped oligonucleotides cleaved from host cell mRNAs (Fig. 2A). The signals responsible for these activities are located within terminal nontranslated regions (NTRs) of each functional template (Dunn et al. 1995; Bridgen and Elliott 1996; Barr et al. 2003) and recent work suggests that the separate promoters for replication and transcription involve some degree of cooperation between nucleotides from both 3′ and 5′ NTRs, implying that the functional templates for bunyavirus RNA synthesis are circular (Barr and Wertz 2004, 2005; Kohl et al. 2004; Barr et al. 2005). The genomic NTRs supply signals for transcription and replication, whereas the anti-genomic NTRs supply signals only for replication (Figs. 1A, 2A).
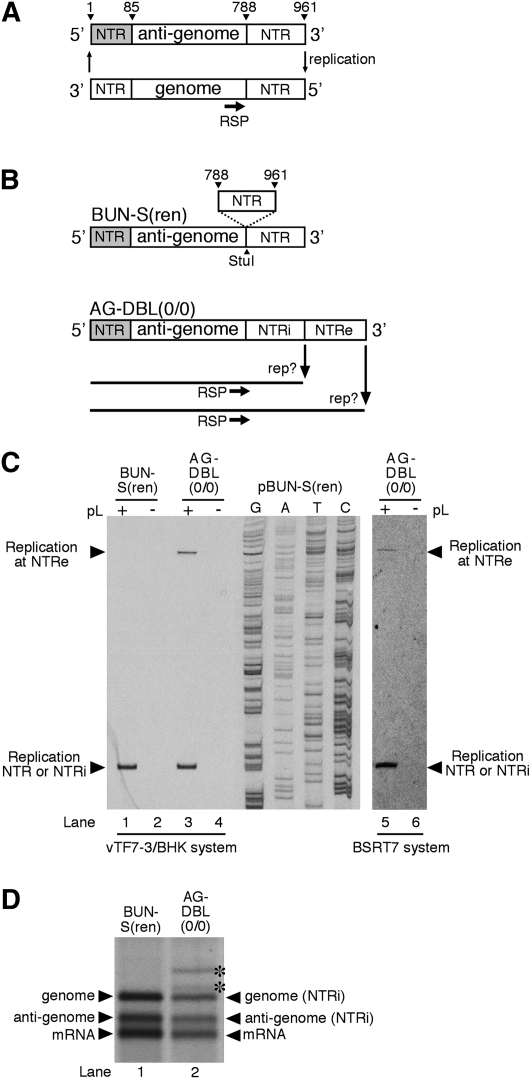
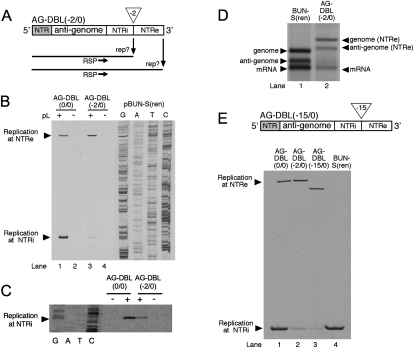
FIGURE 1.
(A) Schematic representation of the BUNV S segment and its RNA replication ability. The annealing position of oligonucleotide RSP used to detect the 5′ end of the genome is shown. The anti-genomic 5′ NTR encompasses S segment nucleotides 1–85 and is shaded. The anti-genomic 3′ NTR encompasses S segment nucleotides 788–961 and is shown as an open box. (B) Schematic representation of model BUNV segment AG-DBL(0/0). This template was derived from BUN-S(ren) by the insertion of an additional anti-genomic 3′ NTR, resulting in two identical 3′ NTRs where replication could potentially initiate, designated NTRi (internal) and NTRe (external). (C) Identification of the position where replication initiated on the anti-genomic strands of AG-DBL(0/0). Model segment AG-DBL(0/0) was expressed in BHK-21 cells along with BUNV S and L ORFs (+), or the S ORF alone (−). The resulting BUNV RdRp-specific RNAs were analyzed by primer extension using end-labeled oligonucleotide RSP (lanes 1–4). Labeled DNA fragments were subjected to denaturing PAGE on a sequencing gel. Bands corresponding to either corrected or uncorrected negative-sense replication products are indicated with arrowheads. To act as size markers, RNAs generated by parental template BUN-S(ren) were analyzed alongside, and the corresponding cDNA was sequenced using oligonucleotide RSP. To eliminate a role of vaccinia virus-mediated effects in the repair process, the template AG-DBL(0/0) was also generated in BSRT7 cells, and its replication ability analyzed by primer extension as described above (lanes 5,6). (D) Metabolically labeled RNAs generated by AG-DBL(0/0) were also subjected to agarose-urea gel electrophoresis. Replication and transcription products are marked with adjacent arrowheads, and uncorrected RNA products are marked with an asterisk. RNAs generated by parental template BUN-S(ren) are included as size markers.
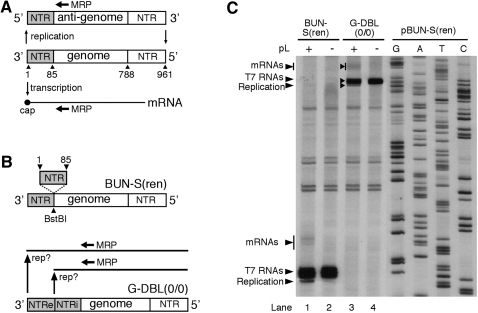
FIGURE 2.
(A) Schematic representation of the BUNV S segment genome and its RNA synthesis activity, which is both transcription and replication. The annealing position of oligonucleotide MRP used to detect the 5′ end of these RNAs is shown. (B) Schematic representation of model BUNV segment G-DBL(0/0). This template was derived from BUN-S(ren) by the insertion of an additional genomic 3′ NTR, resulting in two identical 3′ NTRs, where RNA synthesis could initiate, designated NTRi (internal) and NTRe (external). (C) Identification of the position where replication and transcription initiated on the genomic strand of G-DBL(0/0). Model segment G-DBL(0/0) was expressed in BHK-21 cells along with BUNV S and L ORFs (+), or the S ORF alone (−). The resulting BUNV RdRp-specific RNAs were analyzed by primer extension using end-labeled oligonucleotide MRP. Labeled DNA fragments were subjected to PAGE on a sequencing gel, and bands corresponding to BUNV RNAs are marked with arrowheads. To act as size markers, RNAs generated by parental template BUN-S(ren) were analyzed alongside, and the corresponding cDNA was sequenced using MRP.
The critical information contained within the NTRs dictates that they are highly sensitive to nucleotide change. This is particularly important at the extreme ends of the NTRs, the functions of which are particularly sensitive to nucleotide alteration. Possible sources of such alteration are from cellular mechanisms that can add or remove nucleotides from exposed termini, or alternatively by action of the error-prone viral RdRp during RNA replication. To combat these possible sources of error, many RNA viruses possess mechanisms that are able to perform genome repair. These mechanisms are diverse in their mode of action: For example some plant RNA viruses have subverted the activity of cellular tRNA nucleotidyl transferase to maintain a 5′-CCA-3′ motif required at their 3′ termini (Rao et al. 1989), whereas several mammalian viruses utilize either host-cell-encoded or virus-encoded terminal adenylyl transferases to maintain 3′ poly(A) tails required for priming genome replication (Kusov et al. 2005; van Leeuwen et al. 2006; Rubach et al. 2009). Other viruses make use of terminal redundancy within their complement of genome strands to supply missing nucleotides from one genome strand to another. A particularly well-studied example is Turnip Crinkle virus (TCV), which possesses a genome comprising a single positive sense RNA strand, but is associated with satellite RNAs that share terminal sequences with the TCV genome. It has been shown that terminal deletions engineered into the satC satellite RNA can be repaired using abortive transcripts generated from the intact termini of the TCV genome (Nagy et al. 1997). In addition, repair can also occur through recombination between satC and TCV genomes, by the viral RdRp jumping strands during replication. Finally, the TCV RdRp can also perform nontemplated polymerization, to create a primer that is incorporated at the 5′ end of the newly synthesized replication product. Subsequently, repaired templates that possess permissible terminal sequence are then amplified, and predominate in the cellular pool (Guan and Simon 2000).
In the case of negative strand RNA viruses, several mechanisms have been identified that can eliminate additional terminal nucleotides, and these rely on the ability of the viral RdRp to access internally located replication promoters. Previous reports have shown that duplicated anti-genomic 3′ NTRs are eliminated during RdRp replication of vesicular stomatitis virus (VSV) (Whelan and Wertz 1999). In the case of Sendai virus, the viral RdRp was proposed to access the replication promoter by direct RdRp entry (Vulliemoz and Roux 2002), whereas in the case of human respiratory syncytial virus (hRSV), access to an internal promoter was thought to rely on prior scanning of upstream sequences (Cowton and Fearns 2005). It is also noteworthy that many negative stranded RNA viruses rescued from cDNA have additional nonviral nucleotides at their 5′ ends, and these are removed during replication. Although the mechanism of nucleotide removal is unknown, it hints that terminal repair may be a common ability of these viruses. Interestingly, it has been reported for the negative sense RNA virus Borna disease virus (BDV), that terminal changes are not repaired (Schneider et al. 2005, 2007), and furthermore are postulated to offer selective advantages during the virus life cycle, particularly during persistent BDV infections of neuronal cells.
Finally, there is evidence to suggest that the many segmented negative stranded RNA viruses may use a “prime and realign” mechanism to initiate both mRNA transcription and RNA replication activities, which may also have a role in terminal repair (Jin and Elliott 1993; Garcin et al. 1995). In the case of Hantaan virus, initiation of RNA synthesis is thought to occur at an internal location at the 3′ NTR within a repeated trinucleotide sequence (3′-AUCAUCAU-5′). Following limited polymerization, the nascent strand slips backward to realign with the template by virtue of the 3′ proximal triplet repeat. Subsequent RdRp extension results in both trinucleotides being represented in the nascent strand, despite only one of them actually acting as a replication template. We wanted to investigate whether the BUNV RdRp was capable of correcting sequence changes at the termini of model RNA segments, and if so, to examine the mechanism responsible.
RESULTS AND DISCUSSION
Additional NTR sequences at the anti-genomic 3′ end are removed
To examine terminal repair we inserted additional intact NTRs at the 3′ ends of the genomic or anti-genomic strands of previously described model BUNV segment BUN-S(ren) (Barr et al. 2003), which comprised wild-type S segment NTRs flanking the Renilla luciferase ORF (Lorenz et al. 1991). If the BUNV RdRp was capable of repairing the termini by eliminating the additional NTR, then the resulting replication product would be correspondingly shorter, which could be detected using primer extension analysis.
We first analyzed whether a duplicated anti-genomic 3′ NTR could be eliminated. Plasmid pBUN-S(ren) was altered by inserting a PCR-generated fragment representing the entire S segment anti-genomic 3′ NTR (nucleotides 788–961) at an existing Stu I site located at the NTR/ORF boundary (Fig. 1B). The resulting template AG-DBL(0/0) contained duplicate anti-genomic 3′ NTRs designated internal (NTRi) or external (NTRe) (Fig. 1B). The numbers in parentheses within the template name indicate whether nucleotides have been added or removed from the internal and external NTRs, respectively. The RNA synthesis ability of AG-DBL(0/0) was tested in the previously described RNA synthesis assay, based in BHK-21 cells (Barr et al. 2003). To allow reliable identification of BUNV RdRp-specific products, duplicate transfections were also performed with the omission of the L (RdRp) ORF expressing cDNA. The RNA synthesis products of the AG-DBL(0/0) anti-genome were identified by primer extension analysis using oligonucleotide RSP (Fig. 1A,B), as previously described (Barr et al. 2003). For comparison, RNA synthesis from wild-type template BUN-S(ren) was also analyzed.
Primer extension analysis showed that the additional 3′ NTR markedly affected RNA synthesis. Instead of generating the customary single RNA replication product (Fig. 1C, lane 1), the AG-DBL(0/0) anti-genome was replicated to generate two RNAs having different 5′ ends (Fig. 1C, lane 3). The position of these ends relative to the RSP-binding site was mapped by comparison to the BUN-S(ren) genome band (Fig. 1C, lane 1) and the adjacent sequence ladder. This showed that the slow-migrating product corresponded precisely to initiation at the external 3′ NTR (NTRe). Interestingly, the fast-migrating product corresponded precisely to initiation at the first nucleotide of the internal 3′ NTR (NTRi), generating an RNA having a 5′ end indistinguishable from the BUN-S(ren) genome. This suggested that a proportion of the AG-DBL(0/0) replication products had been shortened by elimination of the external NTRe. To provide independent confirmation that the change in 5′ end positions also corresponded to shortened overall length of the replication products, we visualized metabolically labeled RNAs generated by template AG-DBL(0/0) using agarose-urea gel electrophoresis and fluorography. As predicted, this analysis showed that AG-DBL(0/0) generated a replication product that precisely comigrated with the BUN-S(ren) genome (Fig. 1D, cf. genome band, lane 1, and genome band, lane 2). This gel also showed that AG-DBL(0/0) generated RNAs that comigrate with the BUN-S(ren) anti-genome and mRNAs, suggesting that the newly truncated genome performed RNA synthesis that was indistinguishable from parental template BUN-S(ren). The uncorrected full-length genome and anti-genome replication products generated by AG-DBL(0/0) were also visible, and with a reduced abundance that correlated with that observed in the primer extension analysis. The greater abundance of the corrected BUN-S(ren)-specific RNAs compared to those of AG-DBL(0/0) suggested that the shortened template was a better substrate for replication by the RdRp.
We also considered alternatives for how NTR elimination may have occurred, including elimination through DNA recombination of the corresponding plasmids, either during plasmid amplification in bacterial cells or through activities encoded by the recombinant vaccinia virus vTF7-3. Thus, we have performed controls to eliminate these possibilities: Plasmid stocks showed no alterations, and PCR analysis of plasmids transfected into BHK-21 cells showed no sequence elimination. However, to completely remove all possibility that vTF7-3 was involved in NTR elimination, we also tested the replication activity of templates BUN-S(ren) and AG-DBL(0/0) in BSRT7 cells, which express T7 RNA polymerase without the need for vTF7-3 infection. Although the abundance of BUNV-specific RNAs was ∼10-fold reduced compared to the vTF7-3 system, primer extension analysis of resulting RNAs using oligonucleotide RSP identified RNA products corresponding to precise and abundant elimination of NTRe from AG-DBL(0/0) (Fig. 1C, lane 5). This confirms that activities of vTF7-3 played no role in NTRe elimination, and showed that this terminal repair was a function of the BUNV RdRp. Because the repair ability of the BUNV RdRp was equivalent in both RNA synthesis systems, we chose to perform subsequent experiments using the more active vTF7-3 assay.
Additional NTR sequences at the genomic 3′ end are not removed
We next analyzed whether an additional NTR could be maintained at the 3′ end of the genomic strand. Plasmid pBUN-S(ren) was modified by inserting a PCR-generated NTR (S segment nucleotides 1–85) at a BstBI site located immediately downstream of the existing 3′ NTR (Fig. 2B). The resulting model segment G-DBL(0/0) possessed duplicate 3′ NTRs (NTRe and NTRi) and its RNA synthesis characteristics were tested in the RNA synthesis assay, and analyzed by primer extension using oligonucleotide MRP (Fig. 2A).
This analysis showed the G-DBL(0/0) genome generated a single replication product (Fig. 2C, lane 3). By comparing this product to those generated by BUN-S(ren) (Fig. 2C, lane 1) and the adjacent sequence ladder, we determined that it had initiated precisely at NTRe. If replication had initiated at NTRi, the products would comigrate with the BUN-S(ren) anti-genome, but these were not detected. These results suggested that the BUNV RdRp was unable to initiate RNA replication at NTRi, and thus the duplicate NTRs were maintained.
Taken together, these results showed that an internal duplicated 3′ NTR was maintained in the context of the genome, but was eliminated in the context of the anti-genome. We are unsure why the genomic and anti-genomic 3′ NTRs should behave in such different ways. One possible explanation is that there is a specific sequence or structure within the anti-genomic 3′ NTR that allows direct RdRp entry, which is missing from the genomic 3′ end. There are many sequence differences between these NTRs, and previous work with BUNV has indicated that these differences affect RNA synthesis (Lowen and Elliott 2005). Furthermore, work with influenza virus has shown that initiation strategies on its vRNA and cRNA promoters are different, and are specified by their precise nucleotide sequence (Deng et al. 2006). Alternatively, it is possible that repair of the genomic construct may occur at low frequency, and we are unable to detect the repaired product due to low abundance of the BUNV RdRp-generated genomic strand compared to that of the T7 RNA polymerase generated anti-genomic RNA.
Evidence that NTR removal is by direct RdRp entry to the replication promoter
We next wanted to determine how the BUNV RdRp had eliminated the external 3′ NTR during replication of AG-DBL(0/0). We identified two possibilities, which have been previously described for other negative strand RNA viruses: First, that the RdRp entered the template directly at the NTRi, as is thought to occur for Sendai virus (Vulliemoz and Roux 2002). Second, that the RdRp may attach to NTRe and scan to the NTRi, at which point replication begins, which is postulated to occur for HRSV (Cowton and Fearns 2005). In both cases, the result would be elimination of NTRe. However these two possibilities can be distinguished, because in the first example of direct internal RdRp entry, the NTRe is redundant, whereas in the latter case, the NTRe must be active.
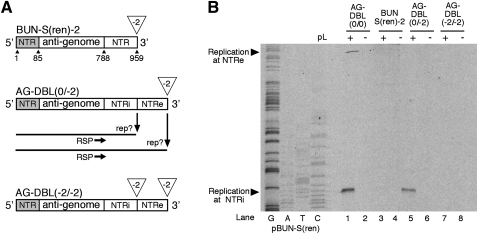
To test this possibility, we first needed to make NTRe inactive, and previous work by others has shown this can be achieved for BUNV by deleting nucleotides from the extreme end of the NTRs (Dunn et al. 1995). To confirm this strategy, we removed the terminal 2 nucleotides (nt) from the anti-genomic 3′ NTRe of BUN-S(ren) to create BUN-S(ren)-2 (Fig. 3A). The replication ability of this template was tested in our RNA synthesis assay, and primer extension using oligonucleotide RSP failed to detect replication products even after long exposure of autoradiographs (Fig. 3B, lane 3), confirming that a 2-nt deletion rendered NTRe inactive. A similar template AG-DBL(−2/−2) was also generated in which 2 nt were removed from both NTRi and NTRe (Fig. 3A), and primer extension analysis revealed that this template was also inactive (Fig. 3B, lane 7).
FIGURE 3.
(A) Schematic representation of the anti-genomic strand of model templates BUN-S(ren)-2, AG-DBL(0/−2) and AG-DBL(−2/−2). These templates were derived from BUN-S(ren) and AG-DBL(0/0) by making 2-nt deletions to either NTRi (internal) or NTRe (external) as shown. Deletions are shown by an open triangle. (B) Identification of the position where replication initiated on the anti-genomic strand of AG-DBL(0/−2). Model segments were expressed in BHK-21 cells along with BUNV S and L ORFs (+), or the S ORF alone (−). The resulting BUNV RdRp-specific RNAs were analyzed by primer extension using end-labeled oligonucleotide RSP. Labeled DNA fragments were subjected to PAGE on a sequencing gel, and bands corresponding to BUNV RNAs are marked with arrowheads. To act as size markers, RNAs generated by template AG-DBL(0/0) were analyzed alongside, and the BUN-S(ren) cDNA was sequenced using RSP.
The inactive NTRe was then engineered into AG-DBL(0/0) to create template AG-DBL(0/−2), and its replication activity was tested in the RNA synthesis assay. Primer extension analysis using RSP (Fig. 3A) detected a strong signal corresponding to genome replication that initiated at the NTRi (Fig. 3B, lane 5). As NTRe was inactive, this suggested that the only way the BUNV RdRp could access NTRi was not by scanning or diffusion from NTRe, but instead was by internal entry direct to NTRi.
To add further weight to the argument that direct RdRp entry was responsible for the elimination of NTRe, we constructed an additional variant of AG-DBL(0/0) that had 2 nt removed from the beginning of NTRi, creating template AG-DBL(−2/0) (Fig. 4A). As this deletion renders the NTRi inactive, we would predict that the RdRp would not be able to enter NTRi directly, and thus replication would only initiate at NTRe. The replication activity of AG-DBL(−2/0) was examined in the RNA synthesis assay, and the resulting RNAs were analyzed by primer extension using oligonucleotide RSP (Fig. 4A). As we predicted, this analysis detected a strong signal corresponding to genome replication that initiated at the NTRe (Fig. 4B, lane 3, upper part). To confirm these findings, we also visualized the RNAs generated by template AG-DBL(−2/0) using agarose-urea gel electrophoresis and fluorography, which confirmed that the replication products were full length (Fig. 4D). Taken together, the analysis of the replication ability of AG-DBL(0/−2) and AG-DBL(−2/0) indicated that an active NTRe was not required for NTR elimination, which strongly supported direct entry of the BUNV RdRp as the predominant mechanism of its removal. This explanation of direct RdRp entry to an internal 3′ NTR remains compatible with our previous findings and those of others that the NTRs at opposite ends of a template must interact in order to promote RNA replication (Barr and Wertz 2004; Kohl et al. 2004). We propose that both NTRe and NTRi have the potential to interact with the single NTR at the opposite end of the template. When this interaction involves NTRe, the RdRp will contact 3′ and 5′ terminal nucleotides that comprise the replication promoter, and replication will proceed as normal. However, when the terminal interaction involves NTRi, the RdRp will still be able to contact 3′ and 5′ terminal residues of the base-paired replication promoter, but only if internal RdRp entry and subsequent removal of the NTRe sequence occurs.
FIGURE 4.
(A). Schematic representation of the construction of model BUNV segment AG-DBL(−2/0). This template was derived from BUN-S(ren) by the insertion of an additional NTR that had the extreme 2 nt deleted (shown by an open triangle), and resulted in the presence of two NTRs, where RNA replication could potentially initiate, designated NTRi (internal) and NTRe (external). (B) Identification of the position where replication initiated on the anti-genomic strand of AG-DBL(−2/0). Model segment AG-DBL(−2/0) was expressed in BHK-21 cells along with BUNV S and L ORFs (+), or the S ORF alone (−). The resulting BUNV RdRp-specific RNAs were analyzed by primer extension using end-labeled oligonucleotide RSP. Labeled DNA fragments were subjected to PAGE on a sequencing gel, and bands corresponding to BUNV RNAs are marked with arrowheads. To act as size markers, RNAs generated by template AG-DBL(0/0) were analyzed alongside, and the BUN-S(ren) cDNA was sequenced using RSP. (C) The position at which RNA replication was initiated on AG-DBL(0/0) and AG-DBL(−2/0) was also analyzed in BSRT7 cells using primer extension as described above. Bands corresponding to replication products initiated at NTRi are marked with an arrowhead. (D) Metabolically labeled RNAs generated by AG-DBL(−2/0) were also subjected to agarose-urea gel electrophoresis. Replication and transcription products are marked with adjacent arrowheads. RNAs generated by parental template BUN-S(ren) are included as size markers. (E) Schematic representation of template and AG-DBL(−15/0), and identification of the position where replication initiated. The replication ability of AG-DBL(−15/0) was tested as described above. Primer extension products from templates AG-DBL(0/0), AG-DBL(−2/0), and BUN-S(ren) are included for comparison.
Deleted NTR sequences can be precisely repaired
Interestingly, long exposure of the primer extension autoradiograph, shown in Figure 4B, showed a low abundance of a product that corresponded to initiation at the partially deleted NTRi of AG-DBL(−2/0). This same primer extension product is shown with longer exposure in Figure 4E, lane 2. Remarkably, the mobility of this primer extension product indicated that the deletion of 2 nt had been repaired. To remove any possibility that this observed repair could be mediated by any vTF7-3 activities, we also repeated this experiment using BSRT7 cells, and the results were the same: The AG-DBL(−2/0) replication product was repaired, such that its 5′ end was indistinguishable from the AG-DBL(0/0) replication product (Fig. 4C).
Although these primer extension data indicated that this repair was not a common event, we were intrigued by how it had occurred, in particular we were curious as to the identity of the template on which the missing nucleotides were copied. Based on our work described above, and the previous work of others, one possibility was that the RdRp directly entered at NTRi, and repaired the missing 2 nt using the prime and realign mechanism (Jin and Elliott 1993; Garcin et al. 1995), and the 3′ ends of the BUNV segments possess the necessary triplet repeat for this to be possible (3′-UCAUCACA-5′). However, if this mechanism was capable of repairing two missing nucleotides from AG-DBL(−2/0), we expected that the missing nucleotides of templates BUN-S(ren)-2 and AG-DBL(−2/−2) (Fig. 3B, lanes 3,7) would also be repaired, and our results show they are not.
To further examine whether the prime and realign mechanism was involved in terminal repair, we designed a final template AG-DBL(−15/0), which had 15 nt removed from the 3′ end of NTRi (Fig. 4E). This deletion encompassed nucleotides that were not represented in the terminal repeats, and thus if repair occurred, it would seem to rule out involvement of the prime and realign mechanism. The replication ability of template AG-DBL(−15/0) was examined in the RNA synthesis assay, and the resulting RNAs were analyzed by primer extension using oligonucleotide RSP (Fig. 4E). The major replication product of AG-DBL(−15/0) corresponded to initiation at NTRe, with its 5′ end position reflecting the missing 15 nt of NTRi (Fig. 4E, lane 3, top). However, we also clearly detected a primer extension product corresponding to initiation at NTRi (Fig. 4E, lane 3, bottom), and remarkably, the comigration of this replication product relative to the genomic 5′ ends of AG-DBL(0/0), AG-DBL(−2/0) and BUN-S(ren) indicated that all 15 nt had been repaired (Fig. 4E, lanes 1,2,4). For reasons stated above, the repair of these nucleotides is unlikely to be mediated by the prime and realign mechanism, and consequently the template for these repaired nucleotides is not likely to be within NTRi. A more likely template that allows restoration of these nucleotides is the 3′ end of NTRe. One possible mechanism by which this may occur is by homologous recombination, in which the RdRp partially copies NTRe and then jumps down the template by virtue of common nucleotide sequence to continue copying NTRi. Further examination of this repair mechanism may provide important information of the RNA synthesis capabilities of the bunyavirus RdRp, and the possible role of recombination in sequence restoration. The possibility of recombination as a repair mechanism is particularly interesting in the case of segmented viruses, such as bunyaviruses, due to the increased terminal redundancy of their multistranded genomes, which allows multiple opportunities for repair to occur.
MATERIALS AND METHODS
Plasmids and model BUNV segments
All plasmids designed to express model BUNV segments having duplicated NTRs were derived from previously described plasmid pBUN-S(ren), which contained the entire 3′ and 5′ NTRs of the BUNV S segment surrounding the Renilla luciferase ORF (Barr et al. 2003). This plasmid contained BstBI and StuI restriction sites flanking the Renilla ORF (Figs. 1B, 2B), which were used to insert duplicated NTRs. The duplicated NTR sequences were generated by PCR, and were each flanked with the appropriate restriction sites, depending on which side of the Renilla ORF they were to be inserted. All nucleotide deletions to NTRs were made using Quikchange mutagenesis (Stratagene), and were confirmed as correct by sequence analysis. Plasmids stocks were amplified and maintained using Sure 2 cells (Stratagene) and showed no evidence of sequence rearrangements.
RNA synthesis assay
The RNA synthesis ability of model BUNV segments was examined by assembly of templates active for RNA synthesis in BHK-21 cells, as previously described (Barr et al. 2003). Briefly, cDNAs encoding model RNA segments and S and L segment ORFs were transfected into BHK-21 cells previously infected with vaccinia virus recombinant vTF7-3, which expressed T7 RNA polymerase. The anti-genomic sense primary T7 RNA polymerase transcript was assembled with S and L ORF protein products to form model segments active for RNA synthesis. Resulting BUNV RNAs were harvested using the RNeasy procedure (Qiagen), and used for further RNA analysis, as described below.
In addition, model BUNV segments were also generated in BSRT7 cells (a kind gift from Karl Klaus Conzelmann, Ludwig-Maximillians University), in which vaccinia virus recombinant vTF7-3 was absent. Briefly, plasmid cDNAs expressing model BUNV segments were transfected into BSRT7 cells using lipofectamine 2000 (Invitrogen) along with pTM1-based plasmids expressing S and L ORFs. Control transfections were also prepared, in which the L ORF plasmid was omitted. Resulting BUNV RNAs were harvested using the RNeasy procedure (Qiagen) and analyzed as described below.
Primer extension analysis
RNAs harvested from the RNA synthesis assay were used as template for primer extension analysis using 33P end-labeled PAGE-purified oligonucleotides (Invitrogen) and superscript II reverse transcriptase (Invitrogen). Negative sense RNAs were annealed to oligonucleotide RSP (5′-ATCAAATCGTTCGTTGAGCGAG-3′), and positive sense RNAs annealed to oligonucleotide MRP (5′-GTTATCATCCGTTTCCTTTGTTCTGG-3′). Primer extension reactions were performed as previously described (Barr et al. 2003), and the products were electrophoresed on standard 6% acrylamide denaturing sequencing gels, alongside appropriate sequence ladders to act as size markers.
Agarose-urea gel electrophoresis
BUNV-specific RNAs generated in the RNA synthesis assay were metabolically labeled with 3H uridine (Perkin Elmer) in the presence of actinomycin D (Sigma-Aldrich) and subjected to agarose urea electrophoresis. RNAs were directly visualized by subjecting the finished gels to fluorography followed by autoradiography, as described previously (Wertz and Davis 1981).
ACKNOWLEDGMENTS
We thank Professor R. M. Elliott (St. Andrews) for continued use of plasmids expressing BUNV S and L segment ORFs, and BUN-S(ren), and K. K. Conzelmann for the gift of BSRT7 cells. J.N.B. is supported by a Research Council UK academic fellowship and The Wellcome Trust.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1962010.
REFERENCES
- Barr JN, Wertz GW 2004. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J Virol 78: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Wertz GW 2005. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J Virol 79: 3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Elliott RM, Dunn EF, Wertz GW 2003. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 311: 326–338 [DOI] [PubMed] [Google Scholar]
- Barr JN, Rodgers JW, Wertz GW 2005. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J Virol 79: 12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen A, Elliott RM 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci 93: 15400–15404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowton VM, Fearns R 2005. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol 79: 11311–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Vreede FT, Brownlee GG 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol 80: 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Pritlove DC, Jin H, Elliott RM 1995. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology 211: 133–143 [DOI] [PubMed] [Google Scholar]
- Elliott RM 1985. Identification of nonstructural proteins encoded by viruses of the Bunyamwera serogroup (family Bunyaviridae). Virology 143: 119–126 [DOI] [PubMed] [Google Scholar]
- Elliott RM 1989a. Nucleotide sequence analysis of the large (L) genomic RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. Virology 173: 426–436 [DOI] [PubMed] [Google Scholar]
- Elliott RM 1989b. Nucleotide sequence analysis of the small (S) RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. J Gen Virol 70: 1281–1285 [DOI] [PubMed] [Google Scholar]
- Fuller F, Bishop DH 1982. Identification of virus-coded nonstructural polypeptides in bunyavirus-infected cells. J Virol 41: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller F, Bhown AS, Bishop DH 1983. Bunyavirus nucleoprotein, N, and a nonstructural protein, NSS, are coded by overlapping reading frames in the S RNA. J Gen Virol 64: 1705–1714 [DOI] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol 69: 5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Bishop DL 1979. M viral RNA segment of bunyaviruses codes for two glycoproteins, G1 and G2. J Virol 30: 767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Simon AE 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: An activity involved in 3′ end repair of viral RNAs. Proc Natl Acad Sci 97: 12451–12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Elliott RM 1993. Nonviral sequences at the 5′ ends of Dugbe nairovirus S mRNAs. J Gen Virol 74: 2293–2297 [DOI] [PubMed] [Google Scholar]
- Kohl A, Dunn EF, Lowen AC, Elliott RM 2004. Complementarity, sequence and structural elements within the 3′ and 5′ noncoding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J Gen Virol 85: 3269–3278 [DOI] [PubMed] [Google Scholar]
- Kusov YY, Gosert R, Gauss-Muller V 2005. Replication and in vivo repair of the hepatitis A virus genome lacking the poly(A) tail. J Gen Virol 86: 1363–1368 [DOI] [PubMed] [Google Scholar]
- Lorenz WW, McCann RO, Longiaru M, Cormier MJ 1991. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci 88: 4438–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Elliott RM 2005. Mutational analyses of the nonconserved sequences in the Bunyamwera orthobunyavirus S segment untranslated regions. J Virol 79: 12861–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PD, Carpenter CD, Simon AE 1997. A novel 3′-end repair mechanism in an RNA virus. Proc Natl Acad Sci 94: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AL, Dreher TW, Marsh LE, Hall TC 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc Natl Acad Sci 86: 5335–5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Schwemmle M, Staeheli P 2005. Genome trimming: A unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci 102: 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Martin A, Schwemmle M, Staeheli P 2007. Genome trimming by Borna disease viruses: Viral replication control or escape from cellular surveillance? Cell Mol Life Sci 64: 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen HC, Liefhebber JM, Spaan WJ 2006. Repair and polyadenylation of a naturally occurring hepatitis C virus 3′ nontranslated region-shorter variant in selectable replicon cell lines. J Virol 80: 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliemoz D, Roux L 2002. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J Virol 76: 7987–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz GW, Davis N 1981. Characterization and mapping of RNase III cleavage sites in VSV genome RNA. Nucleic Acids Res 9: 6487–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW 1999. The 5′ terminal trailer region of vesicular stomatitis virus contains a position-dependent cis-acting signal for assembly of RNA into infectious particles. J Virol 73: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]