Abstract
Two obligatory DNA strand transfers take place during reverse transcription of a retroviral RNA genome. The first strand transfer involves a jump from the 5′ to the 3′ terminal repeat (R) region positioned at each end of the viral genome. The process depends on base pairing between the cDNA synthesized from the 5′ R region and the 3′ R RNA. The tertiary conformation of the viral RNA genome may facilitate strand transfer by juxtaposing the 5′ R and 3′ R sequences that are 9 kb apart in the linear sequence. In this study, RNA sequences involved in an interaction between the 5′ and 3′ ends of the HIV-1 genome were mapped by mutational analysis. This interaction appears to be mediated mainly by a sequence in the extreme 3′ end of the viral genome and in the gag open reading frame. Mutation of 3′ R sequences was found to inhibit the 5′–3′ interaction, which could be restored by a complementary mutation in the 5′ gag region. Furthermore, we find that circularization of the HIV-1 genome does not affect the initiation of reverse transcription, but stimulates the first strand transfer during reverse transcription in vitro, underscoring the functional importance of the interaction.
Keywords: HIV-1, RNA–RNA interaction, reverse transcription, strand transfer
INTRODUCTION
Embedded in the RNA genomes of retroviruses are sequence motifs and structural elements that play a regulatory role in virus replication. Many of these signals are clustered in the untranslated leader regions (UTR) that are present at both ends of the retroviral genome (Fig. 1A; Berkhout 1996; Damgaard et al. 2004; Wilkinson et al. 2008). The 5′ UTR contains the repeat (R) region (present at both ends of the genome), the U5 region (unique at the 5′ terminus) and the leader sequence. The 3′ UTR is subdivided into the U3 region (unique at the 3′ terminus) and the R region. The R region of the human immunodeficiency virus type 1 (HIV-1) is relatively extended (97 nucleotides [nt]) compared to that of other retroviruses, and encodes two well-conserved stem–loop structures, the TAR and poly(A) hairpins. The TAR hairpin binds the viral Tat protein and is essential for Tat-mediated activation of transcription (Berkhout et al. 1989; Baudin et al. 1993; Brady and Kashanchi 2005). The adjacent poly(A) hairpin encompasses the polyadenylation signal and is important for the regulation of polyadenylation (Ashe et al. 1995; Berkhout 1996; Das et al. 1999). Polyadenylation of the HIV-1 RNA occurs within the 3′ R region, resulting in a truncated poly(A) hairpin. The R region also plays a pivotal role in the process of reverse transcription, which is initiated near the 5′ end of the HIV-1 genome using a tRNAlys3 primer. Two obligatory DNA strand transfers take place during reverse transcription of the HIV-1 genome (Basu et al. 2008). The first strand transfer depends on base pairing between the de novo synthesized cDNA of the 5′ R and the 3′ R RNA (Gilboa et al. 1979). Several studies suggest that the first strand transfer is facilitated by the higher-order RNA structure of the HIV-1 genome. The dimeric state of the RNA genome was found to be required for efficient strand transfer (Berkhout et al. 1998; Balakrishnan et al. 2003). Dimerization is mediated through base pairing of a palindromic sequence motif in the exposed loop of the DIS hairpin located in the 5′ leader RNA, an interaction referred to as loop–loop kissing (Moore and Hu 2009). Furthermore, a loop kissing interaction between the TAR hairpin in the de novo synthesized cDNA and the TAR hairpin in the 3′ R RNA was proposed to play a role in strand transfer (Berkhout et al. 2001). Finally, the tertiary conformation of the viral RNA genome may stimulate strand transfer by juxtaposing the 5′ R and 3′ R sequences that are 9 kb apart in the linear sequence.
FIGURE 1.
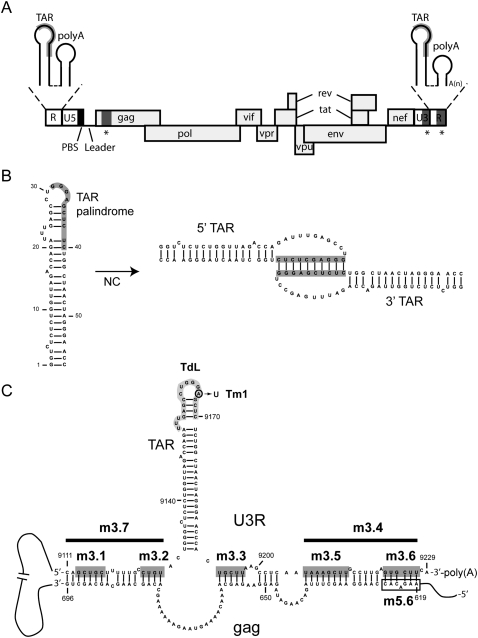
(A) Schematic representation of the HIV-1 viral RNA genome. The positions of the open reading frames are indicated. The coding region is flanked at the 5′ and 3′ ends of the genome by untranslated leader regions (UTR). The 5′ UTR contains the repeat (R) region, the U5 region, and the leader sequence (L). The primer binding site (PBS) is marked by a black box and the TAR and poly(A) hairpins in the R region are shown as insets above. The 3′ UTR contains the U3 region and the R region. The sequences involved in the proposed TAR–TAR interaction are marked in gray, and the sequences involved in the proposed gag–U3R interaction are marked by asterisks. (B) Model for the TAR–TAR interaction (Andersen et al. 2004). The palindromic sequence that was found to induce dimerization, dependent on the nucleocapsid (NC) protein, is marked by a gray box. (C) Model for the gag–U3R interaction (Ooms et al. 2007). Sequences in the gag open reading frame (positions 619–696) were proposed to interact with sequences flanking the TAR hairpin in the U3R region. Mutations introduced in the U3R region are indicated. The sequences that were substituted in mutants m3.1, m3.2, m3.3, m3.5, and m3.6 are marked by dark gray boxes. The deletions introduced in mutants m3.4 and m3.7 are marked by a black line. Mutant Tm1 contains one substitution, A9166U, in the TAR hairpin. The sequence in the TAR hairpin marked in light gray was deleted and replaced by three A residues in mutant TdL. An open box marks the gag sequence substituted in mutant m5.6.
Most cellular mRNAs and many viral RNAs adopt a circular conformation (Mazumder et al. 2003; Edgil and Harris 2006). The biological importance of 5′–3′ interactions of protein coding RNA is underscored by the diversity of mechanisms of transcript closure that have evolved in organisms ranging from viruses to humans. Circularization can be mediated by the recruitment of protein factors that simultaneously interact with both ends of the mRNA, facilitating the bridging between the 5′ and 3′ ends (Sachs et al. 1997). Alternatively, some viruses have developed RNA motifs located at both ends of the genome with complementary sequences that promote protein-independent circularization of the viral RNA. This is the case for flaviviruses (Hahn et al. 1987; Khromykh et al. 2001; Alvarez et al. 2005; Polacek et al. 2009), hepatitis C virus (Romero-Lopez and Berzal-Herranz 2009), and certain picornaviruses, such as FMDV (Serrano et al. 2006), among others. Genome circularization is essential for replication of these positive-stranded RNA viruses, and plays an important role in processes such as replication, transcription, and translation of the viral RNA genome. A 5′–3′ interaction was also reported for the LTR-retrotransposon Ty1 (Cristofari et al. 2002). Ty1 shares several features with HIV-1, including its genome organization and the process of reverse transcription. In this retrotransposon, RNA circularization was shown to play a role in the efficient initiation of reverse transcription. Genome circularization has also been suggested for retroviral genomes (Collett and Faras 1976; Darlix 1986a,b).
Several interactions were described that could mediate circularization of the HIV-1 genome. The TAR hairpin was found to harbor a palindromic sequence that can mediate dimerization, independently of the DIS sequence, in the presence of the viral nucleocapsid (NC) protein (Fig. 1B; Andersen et al. 2004). The HIV-1 NC protein is a nucleic acid chaperone that facilitates nucleic acid remodeling events, and is critical for virus replication (Bampi et al. 2004; Thomas and Gorelick 2008). Since the TAR hairpin is present at both ends of the genome, this interaction could mediate circularization. Another interaction described involves sequences in the gag open reading frame at the 5′ end of the genome and the 3′ terminal U3R region (Fig. 1C; Ooms et al. 2007). In vitro transcripts encompassing these regions were found to interact, and detailed RNA structure probing indicates that sequences flanking the 3′ TAR hairpin base pair with gag sequences. A similar gag–U3R interaction can be proposed for all HIV-1 subtypes (Ooms et al. 2007), suggesting this interaction has been conserved in evolution.
In this study, the suggested RNA sequences involved in an interaction between the 5′ and 3′ ends of the HIV-1 genome were mapped by mutational analysis. The interaction appears to be mediated by sequences in the U3R region and gag open reading frame, and depends on the presence of the NC protein. In particular, sequences in the R region at the extreme 3′ end of the viral genome were found to play a crucial role in circularization. Mutation of these sequences was found to inhibit the interaction, which could be restored by a complementary mutation in the 5′ gag region. We show that the interaction of the two termini of the HIV-1 genome does not affect the initiation of reverse transcription, but stimulates the first strand transfer during reverse transcription in vitro.
RESULTS
Mutation of the U3R region
To determine whether circularization of the HIV-1 genome can occur via the gag–U3R or TAR–TAR interaction, several mutations were introduced in the 3′ end of the genome (Fig. 1C; Table 1). Mutants m3.1–m3.7 contain substitutions or deletions designed to disrupt the proposed gag–U3R interaction. In mutants m3.1, m3.2, m3.3, m3.5, and m3.6 a multiple nucleotide substitution was introduced, that will result in loss of base pairing in each part of the extended gag–U3R interaction. In mutants m3.7 and m3.4 nucleotides 9111–9128 and 9207–9229 were deleted, respectively. To disrupt the proposed TAR–TAR interaction, mutant TdL was generated in which the top part of the TAR hairpin (positions 9154–9170) was deleted and replaced by three A residues. In addition, mutant Tm1 was generated that contains a single substitution in the loop of the TAR hairpin (A9166U). This mutant was described previously to disrupt the TAR–TAR interaction (Andersen et al. 2004).
TABLE 1.
Mutations introduced in the U3R and gag domain
Effect of U3R mutations on the 5′–3′ interaction
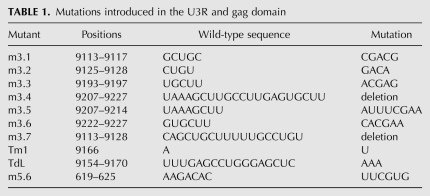
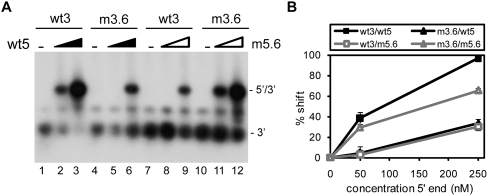
The ability of the U3R mutants to interact with the 5′ end of the HIV-1 genome was analyzed using band shift assays. Radiolabeled RNAs encompassing the 3′ terminal 219 nt of the HIV-1 genome (9011/9229) were generated by in vitro transcription. To prevent homodimerization, the 5′ transcript (1/699) contains a mutation in the DIS sequence, replacing the palindromic GCGCGC sequence with GCGgGC. This mutation was found to disrupt dimerization of the 5′ transcript (data not shown). Equal amounts of wild-type or mutant 3′ transcripts were incubated with the unlabeled 5′ transcript in the presence of NC protein. Subsequently, the NC protein was removed by phenol extraction, and the samples were analyzed on native TBM gels. Representative gels are shown in Figure 2, A–C. A prominent shift of the radiolabeled wild-type 3′ RNA (wt3) was observed upon incubation with the 5′ transcript (wt5) (Fig. 2A, lanes 1–3), demonstrating formation of a 5′–3′ complex. Quantification by phosphorimaging was used to calculate the percentage of complex formation (Fig. 2). Since the efficiency of complex formation varied considerably from experiment to experiment, the wt3 template was included in each individual experiment. Mutations m3.1, m3.2, m3.3, m3.5, and m3.7 did not significantly affect complex formation (Fig. 2A,B). In contrast, complex formation was significantly affected by mutations m3.6 and m3.4 (Fig. 2B). In this experiment, incubation of wt3 RNA with 50 and 250 nM of wt5 RNA resulted in 13% and 40% shifts, respectively. For mutant m3.6 we measured 0% and 23% shifts, whereas complex formation with the deletion mutant m3.4 was completely abolished. Mutations in the TAR hairpin did not significantly affect the 5′–3′ interaction (Fig. 2C). Sequences at the extreme 3′ end of the genome, thus, appear to play a crucial role in establishing the interaction between the two RNAs. These results indicate that the 5′–3′ interaction is mediated by U3R and gag sequences, and is not dependent on the TAR–TAR interaction.
FIGURE 2.
5′–3′ complex formation for the U3R mutants. Equal amounts of the radiolabeled wild-type (wt3) and mutant 3′ transcripts m3.1, 3.2, m3.3, and m3.7 (A), m3.5, m3.6, and m3.4 (B), or TdL and Tm1 (C) were incubated with 0, 50, and 250 nM of the unlabeled 5′ transcript (wt5) in the presence of NC. Subsequently, the NC protein was removed and the samples were analyzed on agarose gels containing TBM. The positions of the radiolabeled 3′ transcript and the 5′–3′ complex are indicated. Quantification by phosphorimaging was used to calculate the percentage of complex formation and indicated below each lane. (D) Equal amounts of the radiolabeled wild-type and mutant 3′ transcripts m3.5, m3.6, and m3.4 were heated together with either 0, 10, 50, or 250 nM of the unlabeled 5′ transcript, followed by slow cooling to room temperature. The samples were analyzed on native TBM gels to measure 3′–5′ complex formation.
To test if 5′–3′ complex formation depends on the NC protein, similar assays were performed in the absence of NC. No complexes were formed upon incubation of the 3′ and 5′ transcripts at 37°C in the absence of NC (data not shown). However, heating the 3′ and 5′ transcripts to 85°C followed by slow cooling to room temperature did result in efficient complex formation. A representative gel is shown in Figure 2D. The 3′ RNAs were incubated with 10, 50, and 250 nM of wt5 RNA, which for the wt3 RNA resulted in 34%, 62% and 100% shifts, respectively. Complex formation was not affected by mutation m3.5, but was reduced for mutations m3.6 and m3.4. For mutant m3.6 we measured 0%, 28%, and 42% complex formations, and for mutant m3.4 only 20% complex formation was measured upon incubation with the highest amount of wt5 RNA. For the TAR and other U3R mutants, a 5′–3′ complex was formed with efficiencies similar to the wild-type (wt3) RNA (data not shown). Mutations m3.6 and m3.4 thus affect complex formation both by heat and by incubation with NC.
Stability of mutant 5′–3′ complexes
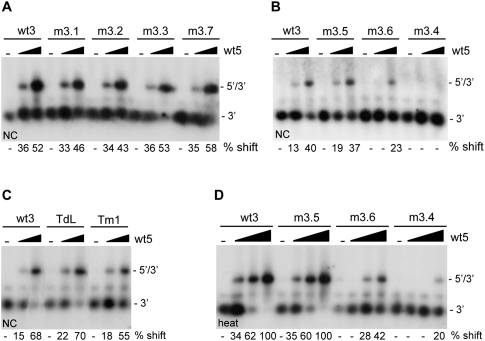
Although we measured no significant effects of mutations m3.1, m3.2, and m3.3 and mutations in the TAR hairpin on complex formation, these mutations could destabilize the 5′–3′ interaction. To analyze the effect of these mutations in further detail, we compared the melting temperature of the 5′–3′ complexes formed. Radiolabeled wild-type or mutant 3′ transcripts were heated together with the wt5 transcript, after which the samples were slowly cooled to room temperature. Samples were diluted 10-fold and subsequently incubated for 10 min in the 37–65°C temperature range. The dilution prevents reassociation of the melted 5′–3′ complexes. Reactions were analyzed on a nondenaturing TBE gel (Fig. 3), and the percentage of shift was determined. The melting temperature (Tm), defined as the temperature at which 50% of the 5′–3′ complex is melted, was determined for each complex. For the wild-type complex a Tm of 64°C was measured, whereas for the mutants m3.1, m3.2, and m3.3 and mutations in the TAR hairpin a Tm of around 57–59°C was measured. This shows that these mutations have a modest effect on the stability of the 5′–3′ interaction.
FIGURE 3.
Stability of the 5′–3′ complexes. The 3′ and 5′ transcripts were heated, followed by cooling to room temperature, to allow 5′–3′ complex formation. Subsequently, the samples were diluted and aliquots were incubated for 10 min at 37, 50, 55, 60, or 65°C. The melting temperature was calculated by PhosphorImager quantification of the gels. The positions of the radiolabeled 3′ transcript and the 5′–3′ complex are indicated.
Compensatory mutations in the 5′ RNA
To confirm the essential role of the distal 3′ R sequences in formation of the 5′–3′ interaction, a compensatory mutation (m5.6) was introduced in the gag region of the 5′ RNA (Fig. 1C; Table 1). This mutation is complementary to mutation m3.6 in the 3′ RNA, restoring base-pairing possibilities between the 3′ and 5′ transcripts. Radiolabeled wt3 and m3.6 transcripts were incubated with either the unlabeled wt5 or m5.6 transcripts. The samples were heated followed by slow cooling to room temperature after which the samples were analyzed on a native TBM gel (Fig. 4). The wild-type 3′ and 5′ transcripts formed complexes with 97% efficiency. Combining wild-type and mutant transcripts (wt5 with m3.6 or wt3 with m5.6) both resulted in low levels of complex formation (30%–33%). Incubation of the two mutant RNAs (m3.6 and m5.6) with complementary mutations partially restored complex formation to 65%. Hence, mutation m5.6 in the 5′ RNA inhibits formation of the 5′–3′ complex with wt3 RNA, whereas it stimulates this interaction with the mutant m3.6 RNA. Similar results were obtained upon incubation of the 3′ and 5′ transcripts in the presence of the NC protein (data not shown). These results indicate that the 5′–3′ interaction can be restored, at least partially, by restoring the base-pairing possibilities between the 3′ terminal R sequences and gag coding region.
FIGURE 4.
Complex formation for mutant m5.6. (A) Radiolabeled wild-type (wt3) or mutant m3.6 3′ transcript was incubated with the unlabeled 5′ transcript wt5 or mutant m5.6. The samples were heated followed by slow cooling to room temperature after which the samples were analyzed on a native TBM gel. The position of the radiolabeled 3′ transcript and the 5′–3′ complex is indicated. (B) Two independent experiments were quantitated, and the percentage of complex formation is plotted as a function of the 5′ transcript concentration.
Role of the 5′–3′ interaction in initiation of reverse transcription
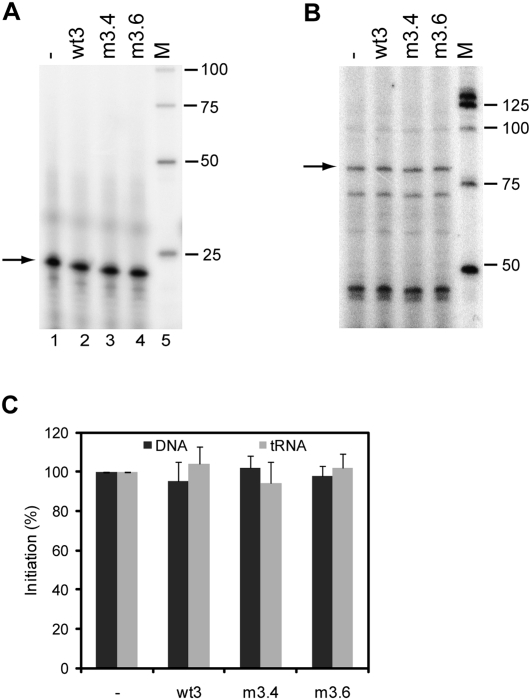
We next analyzed the potential role of the interaction between the two RNAs in the initiation of reverse transcription. In vitro reverse transcription reactions were performed using the DNA oligonucleotide Lys (complementary to positions 182–202 in the PBS) as a primer, or calf liver tRNA as a source of tRNAlys3 primer. The primer and the 5′ and 3′ transcripts were heated followed by slow cooling to room temperature and reverse transcription was initiated by addition of HIV-1 RT enzyme and (α-32P)-dCTP. This resulted in extension of the primer with 1 nt, which was analyzed on a denaturing sequencing gel (Fig. 5A,B). The yield of extended product was quantitated and is summarized in Figure 5C. The presence of the wt3 RNA did not stimulate reverse transcription on the wt5 template. In addition, no significant differences in initiation were measured between wt5/wt3 and wt5/m3.4 or m3.6 complexes. These results suggest that the 5′–3′ interaction does not affect DNA- or tRNAlys3-primed initiation of HIV-1 reverse transcription.
FIGURE 5.
Initiation of reverse transcription on the 5′ transcript in the presence or absence of the wt3, m3.4, or m3.6 transcript. Reverse transcription was initiated using a DNA primer complementary to the PBS (A) or tRNAlys3 (B), HIV-1 RT enzyme and (α-32P)-dCTP. This results in extension of the primer with a single C-nucleotide, and the 22-nt product of the DNA primer and the 77-nt product of the tRNA primer are marked by an arrow. M, end-labeled 25 bp marker. (C) Relative initiation efficiency. Two independent experiments were quantitated, and the activity of 5′ transcript alone was arbitrarily set at 100%.
Role of the 5′–3′ interaction in strand transfer
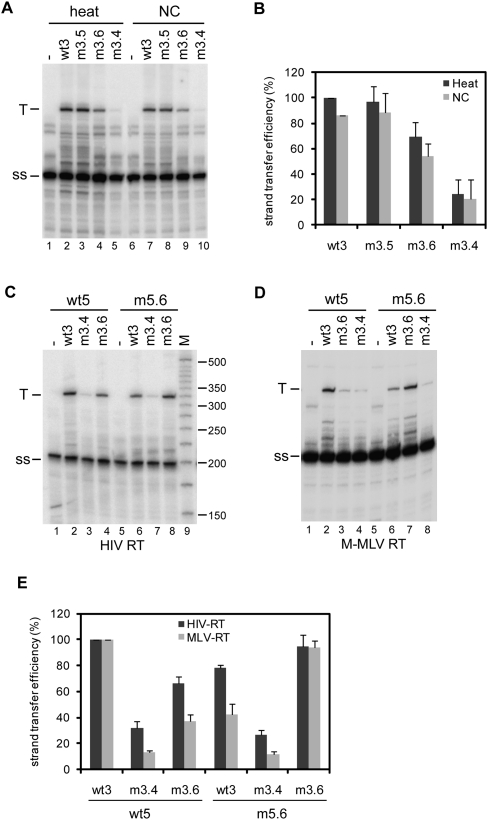
It is a generally accepted idea that the 5′ and 3′ genome ends should be in close proximity during reverse transcription to facilitate the first strand transfer. To study the role of the 5′–3′ interaction in strand transfer, in vitro reverse transcription reactions were performed in which the 5′ transcript acts as a donor template and the 3′ transcript as an acceptor template. The radiolabeled DNA oligonucleotide Lys was used as a primer. The 5′–3′ complexes were formed by heating the 5′ RNA and 3′ RNA together with the Lys primer, followed by slow cooling to room temperature. Or alternatively, the Lys primer was heat annealed onto the 5′ transcript, after which 5′–3′ complexes were allowed to form by incubation with the 3′ RNA in the presence of NC protein. After complex formation, a similar amount of NC was added to the heat-annealed reactions, since this protein was reported to stimulate strand transfer (Thomas and Gorelick 2008). Reverse transcription was performed by addition of HIV-1 RT enzyme and all four dNTPs, and the cDNA products were analyzed on denaturing sequencing gels (Fig. 6A). Reverse transcription of the donor template extended the Lys primer to a 202-nt strong-stop (ss) product. Strand transfer by means of the 97-nt R region results in the synthesis of an extended 324-nt transfer cDNA product (T). These products were quantified to calculate the strand transfer efficiency, and the results of three independent experiments are summarized in Figure 6B. Strand transfer for the wt5 donor and wt3 acceptor pair was set at 100%. The strand transfer efficiency using the m3.5 transcript as an acceptor was similar to that of the wt3 transcript. However, transfer efficiency was reduced to ∼60% using the m3.6 acceptor, and 25% using the m3.4 acceptor. Similar results were obtained using either heat or NC to generate 5′–3′ complexes (Fig. 6A). The effect was similar using the M-MLV RT enzyme, although in general the transfer efficiency is more severely affected by the mutations using the RT enzyme from M-MLV than from HIV-1 (Fig. 6D,E).
FIGURE 6.
Strand transfer assays using the 5′ transcript as a donor template and the 3′ transcript as anacceptor template. (A) The formation of the 5′–3′ complex was mediated by heating the samples, or by incubation with NC as is indicated above the gel. Reverse transcription was performed using HIV-1 RT enzyme in the presence of all four dNTPs. Reverse transcription on the donor template results in a 202-nt strong-stop (ss) product and strand transfer results in an extended 324-nt transfer cDNA product (T). (B) Three independent experiments were quantitated, and the activity of wild-type 5′–3′ complex was arbitrarily set at 100%. (C) Strand transfer assay using the 5′ transcripts wt5 and m5.6 as donor, and the 3′ transcripts wt3, m3.4, and m3.6 transcripts as acceptor templates. Complexes were allowed to form in the presence of NC, and reverse transcription was performed using HIV-1 RT enzyme. (D) Strand transfer assay using the M-MLV RT enzyme. (E) Two independent experiments were quantitated for the RT enzymes of HIV-1 and M-MLV, and the strand transfer activity of the wild-type 5′–3′ complex was arbitrarily set at 100%.
To confirm the role of the 5′–3′ interaction in the strand transfer process, the mutant m5.6 transcript, containing a compensatory mutation to the m3.6 mutation. was used as a donor template. The 5′–3′ complexes were allowed to form by incubation in the presence of NC. After performing reverse transcription using the HIV-1 RT enzyme, the cDNA products were analyzed on a denaturing gel (Fig. 6C). The results of two independent experiments are summarized in Figure 6E. Strand transfer for the wt5 donor and wt3 acceptor pair was set at 100%. Strand transfer efficiency increased from ∼60% for the wt5/m3.6 donor–acceptor pair to 100% for the m5.6/m3.6 pair, whereas the strand transfer efficiency decreased from 100% for the wt5/wt3 donor–acceptor pair to ∼75% for the m5.6/wt3 pair. No significant differences were measured between the wt5 and m5.6 RNAs using the m3.4 acceptor template. Similar results were obtained using M-MLV RT (Fig. 6D,E). These results suggest that the 5′–3′ interaction plays an important role in HIV-1 reverse transcription by facilitating the strand transfer process.
DISCUSSION
Although often viewed as one- or two-dimensional structures, RNAs are three-dimensional molecules with complex tertiary interactions that may span long distances relative to the linear arrangement of nucleotides. Viral RNAs may even be considered four-dimensional, since conformational changes regulating their function occur over time during infection. In many cases, the communication required between distal RNA elements is provided by RNA–RNA interactions. Here, we analyzed the RNA elements involved in an interaction between the 5′ and 3′ ends of the HIV-1 genome and the putative role of this interaction in retrovirus replication. Two interactions were described that could potentially mediate circularization of the HIV-1 genome, an interaction between the TAR hairpins at both ends of the genome (Fig. 1B; Andersen et al. 2004), and an interaction between sequences in the gag open reading frame and the U3R region (Fig. 1C; Ooms et al. 2007). The proposed interactions do not involve the same sequences, and therefore, in principle, could occur at the same time. To analyze the role of these interactions in circularization, several mutations were introduced in the U3R region at the 3′ end of the HIV-1 genome (Fig. 1C; Table 1). The 5′ and 3′ transcripts were incubated and complex formation was analyzed using band shift assays (Fig. 2). Complex formation between the wild-type 5′ and 3′ RNAs during incubation at 37°C was dependent on the presence of NC. In the absence of NC, a heating step was required to mediate complex formation. Mutations introduced in the TAR region did not to affect formation of the 5′–3′ complex, although a modest effect on the stability of the complex was measured (Figs. 2, 3). In a previous study, short RNA transcripts (1/81) representing the TAR hairpin were demonstrated to dimerize efficiently in the presence of NC, whereas reduced TAR dimerization was measured for longer transcripts (1/744) (Andersen et al. 2004). In our study, using in vitro assays and longer 5′ and 3′ transcripts, complex formation did not seem to depend on a 5′-TAR/3′-TAR interaction. These results are consistent with recent studies demonstrating that the HIV-1 TAR hairpin can be deleted, when not required for Tat-mediated activation of transcription (Das et al. 2007). Although the TAR hairpin apparently is not essential for other processes besides transcription, the proposed TAR–TAR interaction may, for instance, play a role in facilitating genome dimerization, in addition to the DIS hairpin, to further stabilize the RNA dimer.
The 5′–3′ interaction appears to be mediated primarily by sequences in the extreme 3′ end of the U3R region and in the gag open reading frame. Substitution of m3.6 and deletion of m3.4 at the 3′ genome terminus of the HIV-1 genome were found to inhibit formation of the 5′–3′ complex (Fig. 2). Surprisingly, mutations m3.3, m3.2, and m3.1 in the U3R region did not affect the efficiency of complex formation, and only had a modest effect on the stability of the complex (Fig. 3). Complex formation for mutant m3.6 was restored by the complementary mutation m5.6 in the gag region of the 5′ transcript (Fig. 4). These results suggest that the terminal 3′ R sequences play a crucial role in establishing the circular interaction in the HIV-1 genome. Since the R region is present at both ends of the genome, potentially, 5′ R sequences could also interact with the gag region. However, no dimerization was observed in band shift assays using a radiolabeled DIS mutant template (data not shown), demonstrating that intermolecular 5′ R-gag interactions do not occur in vitro. These results suggest that U3 sequences are required to stabilize this interaction. The 3′ R region contains the polyadenylation signal and was suggested to fold into a shortened form of the poly(A) hairpin after polyadenylation at the 3′ end of the genome (Das et al. 1999; Kanevsky et al. 2005). However, structure probing data obtained in two independent studies are inconsistent with formation of a shorter poly(A) hairpin. Whereas structure probing of the 3′ and 5′ R regions showed that the TAR hairpin is present at both ends, the poly(A) hairpin was not observed in the 3′ R region, possibly due to pairing with gag sequences (Kanevsky et al. 2005). Detailed RNA structure probing indicated that sequences in the poly(A) and U3 region base pair with gag sequences (Ooms et al. 2007). Furthermore, this gag–U3R interaction can be formed by all HIV-1 subtypes despite considerable sequence variation, thus providing phylogenetic support for the proposed interaction (Ooms et al. 2007). Computational folding predictions for multiple HIV-1 strains demonstrated that the 3′ poly(A) region preferentially interacts with sequences in the gag open reading frame (Gee et al. 2006). Mfold structure prediction (Zuker 2003) for the complete HIV-1 genome also supports the proposed 5′–3′ interaction (data not shown). The interaction was not observed by RNA structure probing of the whole HIV-1 genome (Watts et al. 2009), but in this study no structural information was provided for the 3′ poly(A) region due to the position of the primer at the terminal part of the genome. Finally, the tertiary conformation of the HIV-1 RNA genome may be more complex, and additional RNA–RNA interactions may bring the 5′ and 3′ ends of the HIV-1 genome in even closer proximity.
An interaction between gag sequences and the U3 region was also reported for the LTR-retrotransposon Ty1 (Cristofari et al. 2002). This interaction was found to be required for the efficient initiation of reverse transcription. In our study, the role of the 5′–3′ interaction in the HIV-1 genome was analyzed using in vitro reverse transcription assays. Mutations disrupting the circular interaction did not affect DNA- or tRNAlys3-primed initiation of HIV-1 reverse transcription (Fig. 5). However, the role of the 5′–3′ interaction in the initiation of reverse transcription by the in vivo assembled tRNA-viral RNA-RT complex remains to be determined. Production of a full-length copy of the retroviral genome during reverse transcription involves two DNA strand transfer steps (Basu et al. 2008). The first strand transfer depends on base pairing between the cDNA of the 5′ R and the 3′ R regions. Bringing the 5′ and 3′ ends of the genomic RNA together could facilitate this strand transfer step. In accordance with this we found that mutations disrupting the circular interaction affect the efficiency of the first strand transfer. Reduced strand transfer efficiency was measured for the mutant m3.4 and m3.6 acceptor templates (Fig. 6). Strand transfer efficiency was increased by restoring base-pairing possibilities using m5.6 as a donor template and m3.6 as the acceptor. Similar results were obtained with the HIV-1 and M-MLV RT enzyme, suggesting that stimulatory effect of the 5′–3′ interaction is not limited to the homologous RT enzyme (Fig. 6). Obviously, we cannot exclude effects caused by reduced complementarity between the 5′ R and mutant 3′ R regions. However, mutations m3.4 and m3.6 are both located in the 3′ poly(A) region, which was previously shown to have no significant effect on strand transfer efficiently in assays using a short donor (1/202) template lacking the gag sequences involved in the 5′–3′ interaction. Mutations in the poly(A) region did not significantly affect strand transfer, and even a deletion of the complete 3′ poly(A) region only had a minor effect (Berkhout et al. 2001). Mutation m5.6 does not restore complementarity with the 3′ R region of the m3.6 acceptor template, therefore this mutation likely stimulates strand transfer by restoring the 5′–3′ interaction. These results suggest that the proposed interaction between the 5′ and 3′ end of the HIV-1 genome facilitates the first strand transfer during reverse transcription.
Genome circularization may be a common feature for single-stranded RNA viruses, and was found to play an important role in essential steps of the viral replication cycle, such as translation, transcription, and replication of the viral RNA genome (Edgil and Harris 2006). A variety of strategies is used for circularization, which in many cases involves long-distance RNA–RNA interactions. Circularization of flavivirus genomes was proposed to be mediated by direct RNA–RNA interactions. For the mosquito borne flaviviruses, the 3′ UTR was found to interact with sequences in the capsid coding region near the 5′ end of the genome (Hahn et al. 1987; Khromykh et al. 2001; Corver et al. 2003; Alvarez et al. 2005; Polacek et al. 2009). The hepatitis C virus genome was also reported to circularize by a long-range RNA–RNA interaction between the 5′ and 3′ ends of the genome (Romero-Lopez and Berzal-Herranz 2009). In addition, kissing interactions between stem–loop structures at the 5′ and 3′ genome ends of several plant viruses were demonstrated (Miller and White 2006). Although these viral genomes were found to circularize through protein-independent RNA–RNA interactions, in vivo these interactions may be facilitated or stabilized by viral and/or cellular factors. We found the 5′–3′ interaction in the HIV-1 genome to depend on the viral NC protein. The NC protein is known to act as a nucleic acid chaperone and facilitates nucleic acid rearrangements (Bampi et al. 2004; Thomas and Gorelick 2008). During virion assembly, or immediately after budding from the host cell, the NC protein is released and becomes closely associated with the RNA genome. The NC protein thus may regulate formation of the 5′–3′ interaction, while situated in the viral particle. The cellular poly(A)-binding protein (PABP) is involved in circularization of cellular mRNAs by an interaction with 5′ cap-binding proteins (Mazumder et al. 2003). PABP was also found to bind the gag domain involved in circularization of the HIV-1 genome, in addition to the 3′ poly(A) tail (Burd et al. 1991). Simultaneous interaction of the PABP protein with the gag domain and the 3′ poly(A) tail could further facilitate the bridging between the 5′ and 3′ ends of the HIV-1 genome. The role of circularization of the HIV-1 RNA genome during virus replication in the host cell remains to be established. The interaction is exclusive for unspliced genomic RNA, as all spliced subgenomic mRNAs lack the gag interaction domain. Besides a role of this interaction in strand transfer during reverse transcription, it could function in other genomic RNA-specific processes, such as packaging. Finally, circularization could stimulate translation, as was reported for many cellular and viral mRNAs.
MATERIALS AND METHODS
Generation of mutant RNA transcripts
Substitutions and deletions in the U3R region at the 3′ end of the HIV-1 genome were generated by standard site-directed PCR mutagenesis, and cloned using the TOPO TA cloning kit (Invitrogen) according to the manufacturer's instructions. The mutagenesis reactions were performed on the HIV-1 pLAI construct. Nucleotide numbers refer to positions on the HIV-1 genomic RNA, with position 1 being the capped G residue. The mutations were verified by sequencing analysis using the ABI-3100 system (Applied Biosystems). The constructs were used as a template for PCR amplification and subsequent in vitro transcription. The U3R region (9111/9229) was PCR amplified with the sense primer T7-9011 (nucleotides 9011–9030, with a 5′ flanking T7 RNA polymerase promoter sequence) and the antisense primer rev9229 (nucleotides 9211–9229) for mutants in the U3 and TAR region. Mutants in the 3′ terminal poly(A) region were amplified using the mutagenesis primers as antisense primers.
The 5′ end of the HIV-1 genome was PCR amplified from the plasmid pUC-DIS3b, which contains one substitution in the palindromic DIS sequence, replacing GCGCGC with GCGgGC. The PCR was performed using the sense primer T7-1 (nucleotides 1–20, with a 5′ flanking T7 RNA polymerase promoter sequence) and antisense primer rev699 (nucleotides 680–699). Mutation m5.6 in the gag coding sequence was introduced by standard site-directed PCR mutagenesis.
In vitro transcription was performed using the Ambion MegaShortscript T7 transcription kit according to the manufacturer's instructions. Radiolabeled transcripts were generated by transcription in the presence of (α-32P)-UTP (PerkinElmer). Transcripts were purified on 4% denaturing polyacrylamide gels and visualized either by UV shadowing or by autoradiography. Upon overnight elution in TE buffer, the RNA was ethanol precipitated, redissolved in renaturation buffer (10 mM Tris-HCl, 100 mM NaCl) and quantified by UV spectrometry or scintillation counting. The RNA was renatured by incubation at 85°C for 2 min, 65°C for 10 min, followed by slow cooling to room temperature, aliquots were stored at –20°C.
Bandshift assays
The wild-type and mutant 3′ transcripts (∼200 counts/s) were incubated in 10 μL dimerization buffer (5 mM MgCl2, 10 mM Tris–HCl at pH 7.5, 40 mM NaCl) with different amounts of the 5′ transcript (0, 10, 50, or 250 nM) for 30 min at 37°C in the presence of 150 pmol of HIV-1 NCp7-p1 (synthetic 71 aminoacid protein, generous gift of Jean Luc Darlix). After incubation, 0.2% of SDS was added, and the reactions were incubated at room temperature for 5 min. Subsequently, the NC protein was removed by two steps of phenol extraction with 0.1% SDS and 1 step of phenol/chloroform extraction. Alternatively, the 3′ and 5′ transcripts were incubated for 2 min at 85°C, 10 min at 65°C, followed by slow cooling to room temperature in the absence of NC. After addition of 5 μL nondenaturing gel loading buffer (30% glycerol with BFB dye), the samples were analyzed on 1.5% agarose gels containing TBM (89 mM Tris, 89 mM boric acid, 0.2 mM MgCl2). Gels were run at 100 V at room temperature, dried, and exposed by autoradiography or phosphorimaging (Bio-Rad Molecular Imager FX). The percentage of complex formation, was calculated by dividing the amount of shift by the total amount of labeled RNA in each lane.
Measurement of melting temperature
The 3′ (∼500 counts/s) and 5′ transcripts (2.5 pmol) were heated, followed by cooling to room temperature as described above. Subsequently, the RNA was placed on ice and diluted 10-fold in H2O and 10 μL aliquots were incubated for 10 min at 37, 50, 55, 60, or 65°C. Samples were loaded immediately on a 1.5% agarose gel containing TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA), and the melting temperature was calculated by PhosphorImager quantification of the gel.
Reverse transcription assay
The in vitro synthesized 5′ transcript (0.3 pmol) was incubated with the DNA primer Lys (nucleotides 182–202, 3 pmol) or calf liver tRNA as a source of tRNAlys3 (5 μg, Novagen), with or without the 3′ transcript (1.5 pmol) in 10 μL renaturation buffer for 2 min at 85°C, 10 min at 65°C, followed by slow cooling to room temperature. Reverse transcription was initiated in a 25 μL volume, containing RT buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM DTT), 0.5 U of HIV-1 RT (heterodimer ARP631.2, Center for AIDS Reagents) and 0.5 μL (α-32P)-dCTP (PerkinElmer). The reactions were incubated for 30 min at 37°C and terminated by addition of 5 μL formamide loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). The samples were heated for 2 min at 85°C and analyzed on denaturing 8% polyacrylamide-7 M urea gels. The products were visualized by autoradiography, and quantitated by phosphorimaging.
Strand transfer assays
The 5′ transcript (1/699) is used as a donor template and the 3′ transcript (9111/9229) is used as an acceptor template. The oligonucleotide Lys was used as a primer to initiate reverse transcription. This primer was 5′ end labeled using T4 polynucleotide kinase (NEBiolabs) and (γ-32P)-ATP (PerkinElmer). The donor (0.3 pmol) and acceptor (1.5 pmol) template were incubated together with the labeled primer (3 pmol) in 10 μL dimerization buffer for 2 min at 85°C, 10 min at 65°C, followed by slow cooling to room temperature. Alternatively, only the donor template and Lys primer were heated as described above. Subsequently, the acceptor template was added and the reaction was incubated for 20 min at 37°C in the presence of 75 pmol HIV-1 NC protein. Reverse transcription reactions were performed in a 25 μL volume, containing RT buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 10 μM dNTPs), 0.5 U of HIV-1 RT or 100 U of M-MLV (Invitrogen), and 75 pmol of HIV-1 NC. The reactions were incubated for 60 min at 37°C, terminated by addition of 5 μL formamide loading buffer and analyzed on denaturing 6% polyacrylamide-7 M urea gels. The products were visualized by autoradiography, and the ssDNA (ss) and transfer (T) cDNA products were quantitated by phosphorimaging. Strand transfer efficiency was calculated as follows: [T/(ss + T)] × 100%
ACKNOWLEDGMENTS
We thank Jean Luc Darlix for the kind donation of NC protein and Dr. D. Stammers for the gift of purified HIV-1 RT enzyme (obtained through the MRC AIDS Reagent Project). We also thank Ebbe S. Andersen for the HIV-1 full-genome fold, and for critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2039610.
REFERENCES
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV 2005. Long-range RNA–RNA interactions circularize the dengue virus genome. J Virol 79: 6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J 2004. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem 279: 22243–22249 [DOI] [PubMed] [Google Scholar]
- Ashe MP, Griffin P, James W, Proudfoot NJ 1995. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev 9: 3008–3025 [DOI] [PubMed] [Google Scholar]
- Balakrishnan M, Roques BP, Fay PJ, Bambara RA 2003. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J Virol 77: 4710–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL 2004. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int J Biochem Cell Biol 36: 1668–1686 [DOI] [PubMed] [Google Scholar]
- Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA 2008. Strand transfer events during HIV-1 reverse transcription. Virus Res 134: 19–38 [DOI] [PubMed] [Google Scholar]
- Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C 1993. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol 229: 382–397 [DOI] [PubMed] [Google Scholar]
- Berkhout B 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 54: 1–34 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Silverman RH, Jeang KT 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59: 273–282 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Das AT, van Wamel JL 1998. The native structure of the human immunodeficiency virus type 1 RNA genome is required for the first strand transfer of reverse transcription. Virology 249: 211–218 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H 2001. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA 7: 1097–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J, Kashanchi F 2005. Tat gets the ‘green’ light on transcription initiation. Retrovirology 2: 69 doi: 10.1186/1742-4690-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G 1991. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol 11: 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett MS, Faras AJ 1976. Evidence for circularization of the avian oncornavirus RNA genome during proviral DNA synthesis from studies of reverse transcription in vitro. Proc Natl Acad Sci 73: 1329–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 77: 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL 2002. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J 21: 4368–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J 2004. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol 336: 369–379 [DOI] [PubMed] [Google Scholar]
- Darlix JL 1986a. Circularization of retroviral genomic RNA and the control of RNA translation, packaging and reverse transcription. Biochimie 68: 941–949 [DOI] [PubMed] [Google Scholar]
- Darlix JL 1986b. Control of Rous sarcoma virus RNA translation and packaging by the 5′ and 3′ untranslated sequences. J Mol Biol 189: 421–434 [DOI] [PubMed] [Google Scholar]
- Das AT, Klaver B, Berkhout B 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol 73: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Harwig A, Vrolijk MM, Berkhout B 2007. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol 81: 7742–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Harris E 2006. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res 119: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee AH, Kasprzak W, Shapiro BA 2006. Structural differentiation of the HIV-1 polyA signals. J Biomol Struct Dyn 23: 417–428 [DOI] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100 [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 198: 33–41 [DOI] [PubMed] [Google Scholar]
- Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fosse P 2005. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J Mol Biol 348: 1059–1077 [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75: 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL 2003. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem Sci 28: 91–98 [DOI] [PubMed] [Google Scholar]
- Miller WA, White KA 2006. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu Rev Phytopathol 44: 447–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Hu WS 2009. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev 11: 91–102 [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Abbink TE, Pham C, Berkhout B 2007. Circularization of the HIV-1 RNA genome. Nucleic Acids Res 35: 5253–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek C, Foley JE, Harris E 2009. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J Virol 83: 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Lopez C, Berzal-Herranz A 2009. A long-range RNA–RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 15: 1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW 1997. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell 89: 831–838 [DOI] [PubMed] [Google Scholar]
- Serrano P, Pulido MR, Saiz M, Martinez-Salas E 2006. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA–RNA interactions with the 5′ end region. J Gen Virol 87: 3013–3022 [DOI] [PubMed] [Google Scholar]
- Thomas JA, Gorelick RJ 2008. Nucleocapsid protein function in early infection processes. Virus Res 134: 39–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]