Abstract
Luteinizing hormone (LH) is a key regulator of male fertility through its effects on testosterone secretion by Leydig cells. Transcriptional control of this is, however, currently poorly understood. Mice in which the LH receptor is knocked out (LuRKO) show reduced testicular size, reduced testosterone, elevated serum LH, and a spermatogenic arrest that can be rescued by the administration of testosterone. Using genome-wide transcription profiling of LuRKO and control testes during postnatal development and following testosterone treatment, we show that the transcriptional effects of LH insensitivity are biphasic, with an early testosterone-independent phase and a subsequent testosterone-dependent phase. Testosterone rescue reenables the second, testosterone-dependent phase of the normal prepubertal transcription program and permits the continuation of spermatogenesis. Examination of the earliest responses to testosterone highlights six genes that respond rapidly in a dose-dependent fashion to the androgen and that are therefore candidate regulatory genes associated with the testosterone-driven progression of spermatogenesis. In addition, our transcriptional data suggest a model for the replacement of fetal-type Leydig cells by adult-type cells during testicular development in which a testosterone feedback switch is necessary for adult Leydig cell production. LH signaling affects the timing of the switch but is not a strict requirement for Leydig cell differentiation.
Keywords: early development, Leydig cells, luteinizing hormone, spermatid, spermatogenesis, testis, testosterone
Testosterone rescue of the spermatogenic arrest in luteinizing hormone receptor-deficient mice partially reenables the normal developmental transcriptional program and assists Leydig cell maturation.
INTRODUCTION
Prepubertal growth and development of the testis is controlled through a balance of genetic and hormonal factors operating in a microenvironment of somatic and gametic cells, involving extensive cell-cell interactions and hormonal signaling. Endocrine control of this process is exerted via the hypothalamo-pituitary-gonadal axis in which gonadotropin-releasing hormone from the hypothalamus triggers secretion of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) from the pituitary. These two hormones in turn support testis growth and development [1].
Deeper understanding the mode of action of these hormones can be achieved through the study of animal models in which the hormones themselves, or their receptors, are absent or nonfunctional. Such studies indicate that while FSH is important for the control of Sertoli cell proliferation and overall testis size, it is not required for fertility because male mice lacking FSH, or its receptor, are fertile despite reduced testicular volume [2–4]. In contrast, LH is strictly necessary for male function. There are two mutant mouse model systems in which the LH pathway is ablated [5, 6], and both are cryptorchid and infertile. Of these, the best characterized is the LuRKO (LH receptor knocked out) model in which exon 11 of the Lhcgr gene encoding the LH receptor is deleted. This exon encodes the intracellular and transmembrane domains, and its loss leads to a total lack of receptor function. The comparison of the prepubertal testis development of LuRKO mice with normal controls provides a unique insight into the contribution of LH to this process. In male LuRKO mice, testicular size is significantly reduced from around Day 13 postpartum, and normal germ cell differentiation is disrupted, arresting at the early round spermatid stage around Days 19–20 postpartum [5].
Evidence from several prior studies suggests that the effects of LH are largely mediated via the stimulation of testosterone production by the Leydig cells. Spermatogenesis in the hypogonadal (hpg) mouse, which lacks both FSH and LH, can be restored by treatment either with exogenous human chorionic gonadotropin (hCG), an alternate ligand for the LH receptor, or with testosterone, although the mice remain infertile due to impaired sexual behavior [7, 8]. In LuRKO mice, testosterone treatment restores spermatogenesis and partially restores fertility, although there is still a reduction in accessory gland function and impaired sexual behavior [9]. Sertoli cells are a key testosterone target in the developing postnatal testis, and the SCARKO (Sertoli cell androgen receptor knockout) mouse model, which has a selective loss of androgen receptor function in Sertoli cells, shows a similar phenotype to the LuRKO mouse [10]. Nevertheless, the presence of androgen receptors on other cell types in the testis, including peritubular myoid cells [11] and the Leydig cells themselves [12], indicates that Sertoli cells are not the only target for testosterone.
Luteinizing hormone levels in normal mice are low throughout pregnancy and after birth. Luteinizing hormone levels rise sharply with the onset of puberty between Days 20 and 25 of postnatal life, peaking around Day 30 and then declining slowly thereafter [13]. Testosterone levels, however, follow a more complex profile. Androgen support in fetal and perinatal life is provided by the fetal-type Leydig cells, which secrete androgen independently of LH. Testosterone levels rise throughout pregnancy, peaking around the time of birth, and then dropping rapidly thereafter [14]. Adult Leydig cells replace the fetal cells from around Day 7 postpartum and subsequently proliferate in an LH-dependent manner [15, 16]. Testosterone secretion in adult Leydig cells is LH dependent, and thus they are believed to remain largely quiescent until the pubertal LH surge. This then triggers a corresponding rise in testosterone from around Day 25 postpartum, reaching full adult levels by about Day 40 postpartum. In LuRKO males, where Leydig cells are markedly reduced in size and number, there is no pubertal surge in testosterone, and adult testosterone levels in the serum are very significantly reduced, indicating that pubertal androgen activity requires LH stimulation. Serum LH levels in adult LuRKO males are elevated over tenfold due to loss of negative feedback regulation by testosterone at the level of the pituitary [9]. Testicular descent, prostate and seminal vesicle growth, and maturation of the external genitalia are all dependent on this pubertal androgen surge and are abrogated in LuRKO male mice [5].
Given the hormonal profiles outlined above, the spermatogenic arrest in LuRKO presents an apparent paradox: the onset of arrest is at or before Day 19 postpartum, when testosterone levels are their lowest during normal development [17], and yet the arrest is clearly androgen dependent, as shown by hormone replacement experiments. A second puzzling feature is that older LuRKO males show breakthrough spermatogenesis, with all the germ cell stages being present in the testis at 12 mo of age despite continuing lack of LH activity, low number of Leydig cells, and sustained low testosterone levels [18]. The spermatogenic arrest is thus best characterized as an extreme delay in progression rather than a complete block.
The molecular mechanisms through which LH achieves its effects remains largely underexplored, and the transcriptional effects underlying the promotion of germ cell differentiation by LH are unknown. Through RT-PCR analysis, O'Shaughnessy et al. [17] identified five groups of genes based on their expression in normal fetal and adult Leydig cells, and Zhang et al. [19] subsequently examined the expression of some of these genes in prepubertal LuRKO testes. These studies however addressed only a few selected candidate genes, and a global overview of the transcriptional changes in LuRKO mice testes compared to normal controls has yet to be presented.
This study aims to investigate the role of LH and testosterone during prepubertal testicular development by performing transcriptional profiling of RNA from LuRKO mice testes at a range of prepubertal developmental time points compared to normal controls and at various time points following the administration of varying doses of testosterone. Because the delay/arrest seen in LuRKO mice occurs during this first wave, developmental profiling during the window surrounding the onset of arrest is a valid strategy for this model; this avoids the artifacts that can be introduced using an approach based on enzymatic digestion of the testis and cell separation. We chose a whole transcriptome approach to produce a global overview of gene expression changes as well as a detailed exposition of the individual genes involved, the data of which might be used for further studies. In particular, we sought:
To identify genes deregulated in the testes of LuRKO mice prior to the replacement of the fetal Leydig cell population
To determine whether the fetal Leydig cell androgen production is LH dependent in vivo and identify potential non-testosterone-mediated effects of LH
To identify gene changes associated with the onset of spermatogenic arrest in LuRKO mice
To determine whether the transcriptional changes were detectable in advance of the observed histological changes and whether the genes concerned included known LH/androgen response genes, which will also help resolve the issues surrounding the timing of the arrest in LuRKO germ cell progression
To determine which of the changes identified in 3 were corrected by testosterone replacement
To distinguish the upstream regulatory genes from their downstream targets by following the time course and dose-dependent responses to testosterone therapy
To determine which of the changes identified in 3 could be induced by low-dose testosterone therapy, which relates to the reinitiation of spermatogenesis in aging LuRKO males with constitutively low testosterone
In so doing, we have highlighted candidate regulatory genes responsible for the action of LH and testosterone that may well be applicable to human fertility.
MATERIALS AND METHODS
LuRKO Testis Samples
LuRKO (official allele symhol Lhcgrtm1Hht) mice were maintained at the University of Oxford Department of Human Anatomy and Genetics, and all procedures were carried out in accordance with the Animals (Scientific Procedures) Act 1986 and with the approval of the Local Research and Ethics Committee. Testes were collected by dissection following cervical dislocation of the animal. Whole testes were snap frozen in liquid nitrogen and stored at −80°C prior to RNA extraction.
Experimental Design
Three experimental series were obtained. Series (a): a developmental time course examining LuRKO and wild-type littermate testes at Days 3, 8, 13, 19, and 27 postpartum, taking birth as Day 1. Series (b): a rescue time course following the testicular response to testosterone propionate treatment (2.5 mg) at 2, 4, 8, 12, 24, and 48 h postinjection. Series (c): a second rescue time course to examine the dose sensitivity of the androgen response at 24 h using varying doses of testosterone propionate (vehicle-only control, 250 ng, 2.5 μg, 25 μg, and 2.5 mg). Supplemental Data S1 (this and all supplemental data are available online at www.biolreprod.org) is a schematic showing the relationship between the three experimental series. For the two rescue series, Day 19 LuRKO males were given a single subcutaneous injection of the appropriate dose of testosterone propionate in 0.1 ml arachis oil. The controls for the two rescue series were the Day 19 sample from the developmental time course (constituting a time zero untreated sample) and a vehicle-only control harvested at 24 h postinjection. The 2.5 mg/24 h sample appears in both rescue series, as the penultimate time point of series (b) and the high-dose endpoint of series (c). Supplemental Data S2 shows the serum testosterone levels obtained in each experimental group. For all the treated samples, the time points and dose levels examined are before any overt histological change is apparent (Supplemental Data S3); thus, the observed expression changes are due to transcriptional regulation rather than changing cell numbers in the testis.
RNA Extraction
For each experimental sample, two biological replicates were obtained, each replicate being itself a pool of two animals to ensure sufficient material for hybridization. RNA from whole testis tissue was prepared using TRI reagent (Sigma) according to the manufacturer's protocol. Briefly, tissue samples were homogenized in TRI reagent, followed by chloroform extraction. The aqueous supernatant containing the RNA was retained, and the RNA precipitated with isopropanol. RNA pellets were washed with 70% ethanol, air dried, and resuspended in RNase-free water (Milli-Q). RNA concentration and integrity was assayed with a Nanodrop 1000 spectrophotometer and an Agilent 2100 Bioanalyzer.
Microarray Hybridization and Scanning
Array profiling was performed using the MouseWG-6 v2.0 oligonucleotide platform (Illumina). This comprises 45 281 probes from 30 789 different genes and splice variants, essentially covering the whole murine transcriptome. Probe labeling, hybridization, washing, and scanning were performed according to the manufacturer's protocols using the Illumina Total Prep kit (Applied Biosystems). Briefly, first-strand cDNA was synthesized in a total volume of 20 μl with the supplied reagents. The complete first-strand product was used for second-strand synthesis, followed by column purification. The purified product was then used for in vitro transcription using T7 polymerase. Biotin-16-dUTP was incorporated during this step, resulting in a biotinylated cRNA probe suitable for hybridization. Probe integrity was verified using the Nanodrop 100 and Agilent 2100, as was done for the initial RNA samples. Labeled cRNA (1.5 μg) was hybridized to the array overnight at 58°C in a total volume of 30 μl of the manufacturer's hybridization buffer, followed by posthybridization stringency washing and scanning using Illumina standard protocols.
Data Analysis
Scanned array expression data were imported into BeadStudio (Illumina) for normalization, preliminary analysis, and filtering. Quantile normalization without background subtraction was used, and the Illumina custom error model was used to generate present/absent calls for each probe (present is defined as P < 0.01 for signal detection) on each array and to call differentially expressed genes at each of the developmental stages (defined as P < 0.05 after false discovery rate correction). Two samples with low labeling efficiency and weak signal from housekeeping genes were judged as failed hybridizations and excluded from further analysis. This affected one replicate from the 24-h testosterone-treated time point and one replicate from the 27-h LuRKO developmental sample. Normalized data from BeadStudio was filtered to exclude genes not expressed in testis (i.e., data from probes that were classed as absent in all samples). This yielded a final data set covering 30 389 probes from 21 305 different genes. ANOVA analysis and figure generation were subsequently carried out using InforSense Discovery Edition (InforSense). The full array data set has been submitted to GEO, accession number GSE19453. Selected gene lists were used for gene ontology analysis using GOEAST (gene ontology enrichment analysis) and KEGG (Kyoto encyclopedia of genes and genomes) pathways using Pathway-Express. Array results for selected genes and samples were confirmed using quantitative RT-PCR (Supplemental Data S4; supplementary methods available as Supplemental Data S5).
RESULTS
LH Insensitivity Has Transcriptional Consequences Throughout Prepubertal Testis Development, Affecting over 10% of the Testis Transcriptome
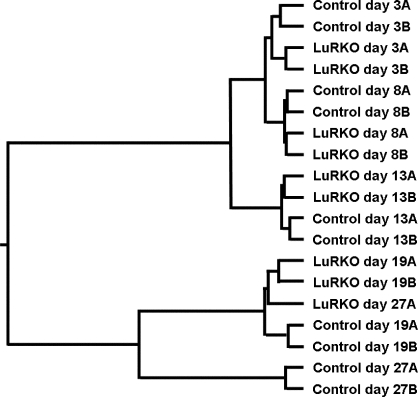
Hierarchical clustering of the LuRKO and normal time course samples according to the similarity of their global expression profiles resulted in a tree (Fig. 1) that was in good agreement with known histological data. That is, the Day 27 LuRKO samples clustered together with the Day 19 LuRKO samples rather than with the Day 27 normal samples, indicative of spermatogenic arrest at this stage. Significantly, however, the analysis also revealed differences between LuRKO and normal testes at Days 3, 8, and 13, with the knockout samples being more similar to each other than to the age-matched control samples. This signified that LH insensitivity had consistent effects on the testis transcriptome at all ages, even before hypoandrogenism set in.
FIG. 1.
Hierarchical cluster analysis of the complete expression profile for the LuRKO and normal developmental series. Biological replicates A and B for each time point have been grouped together in pairs, and LuRKO sample pairs have subsequently been grouped with the age-matched control pairs in most cases. The sole exception is the Day 27 LuRKO sample, which groups together with the Day 19 LuRKO samples, demonstrating the arrested testicular development at this age in LuRKO males.
In a paired t-test comparing the transcriptional profiles of the complete LuRKO and normal time courses, 2738 probes (2508 genes) were significantly up- or down-regulated with a 5% false discovery rate (FDR), and 833 probes (787 genes) of these were significant when a 1% FDR was used (see Supplemental Data S6). No fold change cutoff was used in this comparison, allowing even subtle changes to be detected so long as the changes were consistent throughout the time course. Analysis of the down-regulated genes using Pathway-Express [20] did not detect any known KEGG pathways [21] as significant (see Supplemental Data S7). In contrast, several KEGG pathways were found to be significantly overrepresented in the up-regulated genes. These pathways included axon guidance, focal adhesion, Wnt signaling, and ECM (extracellular matrix)-receptor interaction. Gene ontology analysis using GOEAST [22] also failed to detect significant patterns among the down-regulated genes (see Supplemental Data S7). Among the up-regulated genes, gene ontology terms for transcription and protein phosphorylation were significantly overrepresented as well as related parent nodes in the gene ontology tree.
The Effects of LH Insensitivity on Somatic and Germ Cell Transcriptomes Are Biphasic
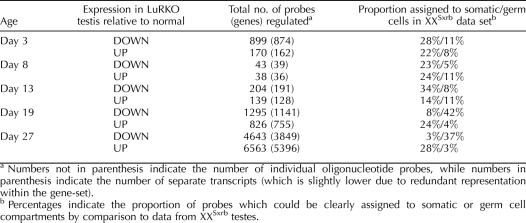
Since testicular cell composition varies widely throughout development, it is likely that there are many genes that are only regulated by LH within a given developmental window and are unchanged at other stages. Table 1 gives the number of genes found to be significantly up- and down-regulated at each individual time point postpartum. There proved to be a biphasic distribution in the numbers of genes deregulated at each time point. Days 3, 19, and 27 displayed the highest numbers of changes, with fewer changes being apparent at Days 8 and 13. This indicated that there were two successive waves of transcriptional response to LH deficiency. The switch between the two response phases occurred around Day 8 in our experiment, concurrent with the appearance of adult Leydig cells in the control testes.
TABLE 1.
Number of probes found to be abnormally expressed in LuRKO testes relative to control at each postpartum age analyzed.
In order to identify which cellular compartment was predominantly affected in each wave of transcriptional regulation, we compared the expression data for LuRKO versus normal testes to data from testes of XXSxrb mice (Supplemental Data S8). XXSxrb mice bear a translocation on the X chromosome, which leads to sex reversal, the resulting males being germ cell deficient and sterile. Germ cell genes are underexpressed in XXSxrb testes, while somatic cell genes are relatively overexpressed, thus allowing any given transcript to be presumptively assigned as being of germ cell or somatic cell origin. Use of the mutant model is especially useful in that it allows an independent assessment of somatic and germ cell origin for any given transcript, in addition to the data obtained from developmental profiling. We found that approximately one-third of the probes altered in LuRKO testes could be assigned to the germ cell or somatic cell compartments, with the remainder either giving a mixed profile in the XXSxrb comparison or not being present in the MEEBO oligonucleotide set (http://www.microarray.org/sfgf/meebo.do) used for the XXSxrb profiling experiment. The final column in Table 1 summarizes the number of array elements found to be altered at each time point and their likely cellular origin. At early ages (Days 3–13), the regulated genes were preferentially identified as somatic cell genes irrespective of the direction of the regulation. At later ages (Days 19 and 27), a clear split became apparent. Genes that were underexpressed in LuRKO testes, and thus dependent on LH/testosterone activity, were predominantly identified as germ cell genes. Conversely, genes that were expressed at higher levels in LuRKO testes were predominantly identified as somatic cell genes. This is likely to be due to dilution effects, as a failure to activate germ cell transcription means that somatic transcripts form a larger proportion of total RNA in LuRKO testes and germ cell transcripts a lower proportion. We note that at Day 19 there was no clear histological difference between LuRKO and normal testes; thus, for this time point, the dilution may be the result of lower transcriptional activity per spermatid. At Day 27, there was a marked deficit in later stage spermatids in LuRKO testes, and the observed changes were therefore likely to be a result of the changing testicular composition.
Genes Affected by LH Insensitivity at Varying Ages Postpartum Show Differential Responses to Testosterone Treatment
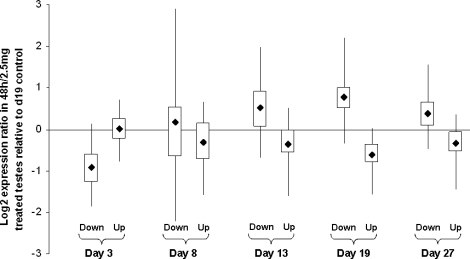
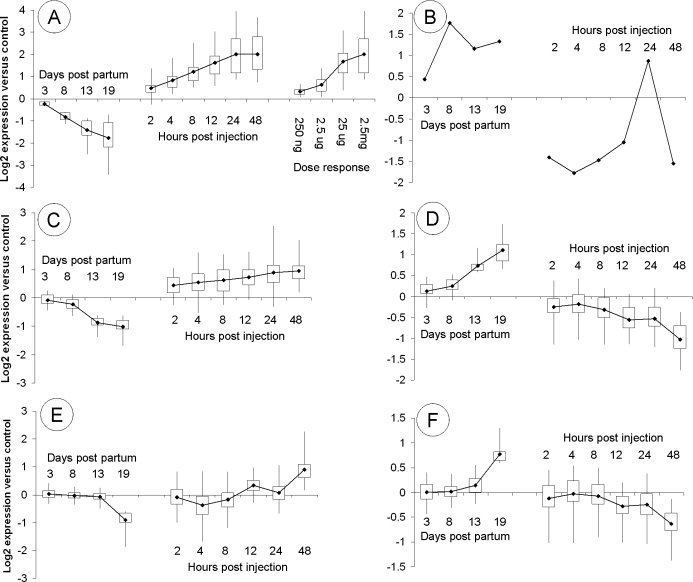
For each of the sets of genes identified as up- or down-regulated at each age, we examined the log2 expression ratio in RNA prepared from the maximally testosterone-treated animals (2.5 mg of testosterone for 48 h) relative to untreated controls in order to identify whether the genes concerned were androgen responsive. Figure 2 shows box and whisker plots indicating the net response of each group of genes to testosterone treatment. For Days 13–27, genes identified as down-regulated in LuRKO are on average induced by testosterone treatment, while genes identified as up-regulated in LuRKO are on average repressed by testosterone treatment. No such pattern was apparent at Days 3 and 8, although genes that were down-regulated in LuRKO testes at Day 3 surprisingly showed a down-regulation in response to testosterone treatment at Day 19.
FIG. 2.
Box and whisker plots indicating the response to testosterone treatment for the genes found to be up- or down-regulated in LuRKO testes at each developmental stage. X axis: various groups of developmentally regulated genes; y axis: log2 expression ratio of the 48-h treated sample relative to untreated control. Dark square indicates the mean, boxed area indicates 50% limit, and whisker indicates 95% limit of the expression ratio for each category.
First-Phase Effects of LH Insensitivity Affect Cell Adhesion, Cytoskeletal Organization, and Protein Turnover
We used Pathway-Express to analyze the genes deregulated at Day 3 in order to search for downstream pathways potentially affected in fetal Leydig cells. In addition to the WNT/focal adhesion pathway up-regulation, which was a general feature at all ages (see above), there was up-regulation of the KEGG pathways for the regulation of the actin cytoskeleton, leukocyte transendothelial migration, and adherens junction function. Down-regulated pathways were ubiquitin-mediated proteolysis, proteasome function, and basal transcription factors.
Consistent with the pathway analysis, gene ontology analysis of down-regulated genes showed an enrichment for ontology terms related to protein folding, ubiquitination, the mitochondrial electron transport chain, and the mitochondrial ribosome large subunit, while up-regulated genes showed an enrichment for ontology terms related to the actin cytoskeleton and histidine catabolism. Significantly, steroid synthesis was not identified as significantly changed by either the pathway analysis or the gene ontology analysis, while known androgen-responsive genes (see below) showed no change in expression between LuRKO and normal testes at this age. This, therefore, confirms the prior finding that androgen production by fetal Leydig cells is not LH dependent and there is therefore no disturbance of androgen pathway activity in fetal and early neonatal LuRKO testes.
Surprisingly, Day 3 LuRKO testes also showed down-regulation of several genes relating to synaptonemal complex formation, namely Sycp1, Sycp2, Syce, and Rad51. The protein products of these genes play a key role in meiosis. Their mRNAs are expressed in spermatogonia, with transcript levels peaking around Day 10 of postnatal life. At Day 3 postpartum, very few spermatogonia are present in the testis, and most germ cells are in the gonocyte stage. Absolute expression levels for all four genes were very low in both genotypes at Day 3, and thus the difference between LuRKO and normal at this age may therefore represent random noise or may represent a delay in differentiation of the first few spermatogonia in LuRKO testes.
At Day 8, comparatively few genes were regulated in either direction, and pathway analysis did not indicate any KEGG pathways to be significant. While the WNT/focal adhesion pathway did show up-regulation at Day 8, this was not significant. GOEAST analysis of the down-regulated genes, however, showed an enrichment for ontology terms related to C21 steroid synthesis, particularly steroid hydroxylase activity. This is consistent with the results of the previous candidate gene studies (see also below). Ontology analysis of the up-regulated genes indicated enrichment for rRNA transcription, ribosome biogenesis, and DNA-dependent transcription. The ontology term for steroid dehydrogenase activity was significant among the up-regulated genes, which proved to be due to the significant up-regulation of Hsd11b1 and Hsd17b3 (exon 8 probe) at this time point. Up-regulation of Hsd17b3 was observed by Zhang et al. [19] at Day 10 using primers that detect exon 8, suggesting this is a genuine biological result, although it was not statistically significant in their study.
Second-Phase Effects of LH Insensitivity Can Be Further Subdivided into Early, Medium, and Late Categories
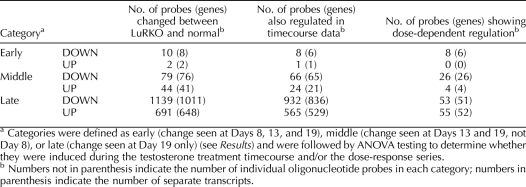
In order to analyze the second phase of LH-driven transcriptional changes, we filtered the data set to select genes found to be significant (P < 0.05 and ≥1.5-fold change) on at least Day 8, 13, or 19, but which were unchanged (P > 0.05 and <1.5-fold change) at Day 3. Day 27 was not included in this comparison as most changes seen at Day 27 are due to changes in the gross cellular composition of the testis rather than changes in transcriptional activity of given genes. This yielded an initial set of 2163 probes (1942 genes). These up- and down-regulated genes were then subcategorized as early/mid/late regulated, depending on whether they showed a change on all three days (early), on Days 13 and 19 only (middle), or on Day 19 only (late), giving six categories in total. In all, 1965 probes (1769 genes) fell into one of these six categories (see Supplemental Data S9 for gene lists and heat maps). The remainder showed more noisy mixed patterns of regulation, being up-regulated at some ages and down-regulated at others. These mixed patterns were not analyzed further. Table 2 shows summary data for the number of probes falling into each category.
TABLE 2.
Genes from the second phase of responses to LH insensitivity, defined as a significant change at Day 8, Day 13, or Day 19 and no significant change at Day 3.
Early Second-Phase Effects Include Known Sertoli Cell Androgen-Responsive Genes, while Late Second-Phase Effects Herald the Onset of Spermatogenic Arrest
Very few of the second-phase transcriptional changes were detected at Day 8, with only 10 genes falling into the early category. Eight of these were down-regulated in LuRKO (Defb45, Drd4, Insl3, Kcnk3, Lmcd1, Pla1a, Rhox5, and Spinlw1), and two were up-regulated (Npm3 and Rock2). Importantly, of these, Drd4, Rhox5, and Spinlw1 are known to be androgen-responsive Sertoli cell genes [23]. This strongly suggests that hypoandrogenism in LuRKO testes sets in as early as Day 8, concurrent with the first appearance of adult-type Leydig cells in the control testes. Pathway analysis and gene ontology analysis was not possible owing to the small number of genes in the early category. In the middle second-phase category, once again the number of regulated genes was too low for conclusive gene ontology results.
GOEAST analysis of down-regulated transcripts in the late second-phase category showed highly significant enrichment for multiple terms related to germ cell development and function, including the ontologies for sexual reproduction, spermatogenesis, male gamete generation, spermatid development, flagellum, outer dense fiber, acrosomal vesicle, acrosomal membrane, sperm motility, sperm-egg recognition, and fertilization. Despite this, no known KEGG pathways were found to be significant in the late-phase gene list, indicating the very poor state of knowledge concerning the genetic pathways underlying spermatid maturation. Once again, components of the WNT pathway related to tight junctions and focal adhesion were significant among the genes up-regulated in LuRKO; however, in this case the up-regulated genes appeared to relate to the tight junction pathway rather than the focal adhesion pathway seen up-regulated at earlier ages, including Day 3. The late second-phase category included several further genes previously identified as Sertoli cell androgen response genes, specifically Gpd1, Serpina5, Tpd52l1, and Tsx [23]. These may be downstream or secondary responses occurring after the upstream responses in the early category.
Testosterone Treatment in LuRKO Reinitiates the Normal Prepubertal Transcriptional Program
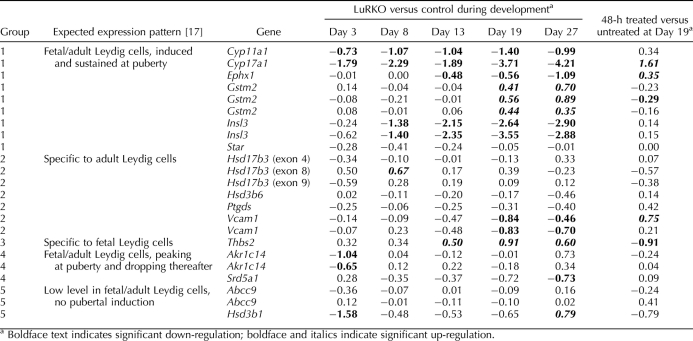
Our initial analysis (see above) indicated that the second-phase genes were testosterone responsive. We therefore performed a close analysis of the time course response of these second-phase genes to treatment with 2.5 mg testosterone. First, we filtered the second-phase gene list for each category to select for probes showing significant changes in expression across the testosterone treatment time course (ANOVA P < 0.05 after FDR correction) and where the direction of change was consistent between the developmental series and the treatment time course, that is, a down-regulation in LuRKO being corrected by testosterone administration or vice versa. The large majority (1596 of 1965) of oligonucleotide probes matching second-phase genes passed this filter, indicating that most of the second-phase responses to LH insensitivity were corrected to some extent by testosterone treatment. Table 2 shows the breakdown of these genes according to our early/mid/late categorization, and Figure 3 shows box and whisker plots for the testosterone-responsive genes in each of the categories.
FIG. 3.
Box and whisker plots indicating the response to testosterone treatment for the various subcategories of testosterone-responsive genes. A) Early down-regulated genes, (B) early up-regulated genes, (C) middle down-regulated genes, (D) middle up-regulated genes, (E) late down-regulated genes, and (F) late up-regulated genes. Early, medium, and late categories are defined in the Results and shown in Table 2. X and y axes, boxes, and whiskers are as in Figure 2. Panel B contains only a single probe, and thus there are no boxes for this panel. Developmental time course data and testosterone treatment time course data are shown for all six categories. Panel A additionally shows the dose-response data for early down-regulated genes because this category was responsive to low-dose testosterone.
Early genes, where there is a difference between LuRKO and normal from Day 8 onward, responded to testosterone within 12 h of treatment, while middle and late genes, where there is no difference between LuRKO and normal until Days 13 and 19, only responded to testosterone after 48 h of treatment if at all. Thus, the temporal sequence of gene changes in response to testosterone rescue of the LuRKO phenotype mirrored the events seen in normal prepubertal development.
Dose-Dependency of Early Testosterone-Responsive Genes
We wished to establish whether any of our candidate testosterone-responsive genes showed a dose-dependent response to testosterone stimulation. We therefore filtered the list of second-phase genes to select for probes showing significant change in expression across the dose response sample series time course (ANOVA P < 0.05 after FDR correction) and where the direction of change was consistent with the existing data. Table 2 indicates the number of genes in each category that passed this final filter. Most of the genes in the middle and late categories did not show any significant dose sensitivity. This is almost certainly due to the fact that most of these genes only change in expression after 48 h of testosterone stimulation, and the dose response sample series was taken at 24 h after the treatment. However, all of the remaining six genes in the early down-regulated category passed this filter. These genes—Defb45, Drd4, Lmcd1, Pla1a, Rhox5, and Spinlw1—were activated in the normal testis relative to LuRKO from Day 8 onward, they responded to androgen treatment within 12 h, and they were sensitive to low dosages of androgen. They are thus key candidates for a regulatory role in the androgen-dependent progression of spermatogenesis. It is interesting to note that these changes occurred well before the spermatogenic block became evident. This suggests that these genes have an upstream role in preparing the somatic cell compartment to support the subsequent germ cell development.
Known Leydig Cell Genes: Developmental Profile and Testosterone Sensitivity
Finally, we examined the expression of the known Leydig cell genes identified by O'Shaughnessy et al. [17]. Table 3 shows the log2 ratio expression data in LuRKO mice relative to control testes across the time course and the log2 expression ratio for the sample from the maximally treated animals (2.5 mg of testosterone for 48 h) relative to the untreated control. Of the genes they identified, the Lhcgr data is not shown here because the expression is negligible at all ages in LuRKO. Estrogen sulfotransferase (Sult1e1) was not present in our data set. Multiple measurements for the same gene indicated the presence of multiple probes on the array. In the case of Hsd17b3, probes directed at different exons showed differing expression patterns, potentially indicating differential regulation splice variants for this transcript.
TABLE 3.
Log2 ratio expression data for the known Leydig cell genes identified by O'Shaughnessy et al. [17] and grouped according to the expression patterns observed.
Of these known Leydig cell genes, Cyp11a1, Cyp17a1, and Hsd3b1 were down-regulated at Day 3, indicating LH-dependence in fetal Leydig cells. Insl3, Star, and Srd5a1 also showed moderate down-regulation at early ages, although they were not significant. The above six genes were all significantly down-regulated at later ages, indicating LH-dependence in adult-type Leydig cells. Ephx1 appeared to be LH dependent in adult but not in fetal Leydig cells, while Vcam1 was specific to adult cells and was LH dependent. Akr1c14 was down-regulated at Day 3 but not at later ages. The fetal cell-specific marker Thbs2 showed progressive up-regulation throughout the first wave in LuRKO relative to control, demonstrating the late retention of fetal-type Leydig cells in LuRKO testes. Gstm2 showed this pattern also. These results are broadly in agreement with those of Zhang et al. [19] except that their study did not detect the early down-regulation of Cyp11a1, Cyp17a1, and Hsd3b1. It is likely that the semiquantitative RT-PCR technique they used is less sensitive than the array method used here. It may be significant that their RT-PCR results for Hsd17b3 are consistent with our results for exon 8 of this gene because their study used a forward primer in exon 1 and a reverse primer at the exon 7/8 border, thus, selectively detecting transcripts containing these exons.
When we examined the testosterone sensitivity of the known Leydig cell genes, the results were mostly as expected. Those genes that were down-regulated in LuRKO were induced by testosterone treatment and vice versa, although this was not always statistically significant at 48 h. Insl3 showed little response to testosterone, indicating that its expression was directly dependent on LH and not on androgen activity. The same was true of Srd5a1. Star showed only a moderate nonsignificant reduction in expression at early ages with no difference between normal/LuRKO/treated LuRKO at Day 19, indicating that expression of this gene may be independent of the LH/androgen pathway in neonatal and juvenile testes. This is consistent with reports that the antiandrogen flutamide did not affect Star expression in neonatal rat testes [24]. The fetal Leydig cell marker Thbs2 was down-regulated by testosterone, despite the fact that its expression was not LH sensitive.
DISCUSSION
Microarray analysis of spermatogenesis is a rapidly developing field. Array analysis has been used to analyze various stages of germ cell development in mouse and human to look for transcriptional consequences of given infertility phenotypes, to identify disease genes affecting testis function, and to identify regulatory interactions at the single gene and whole chromosome level [25–35]. A number of microarray studies have attempted to identify androgen targets within the testis; however, there is considerable variation in the published findings that likely reflects the different mutants used, different background strains on which the mutations are maintained, different ages analyzed, and various array platforms. A particular difficulty in the field is the interpretation of data from later stages of testicular development where there are marked differences in cellular composition between the mutant and control samples. For example, in the Tfm mouse, where there is inactivation of the androgen receptor throughout the testis, O'Shaughnessy et al. [36] identified 20 genes of somatic tubular origin as significantly down-regulated at Day 20 postnatal and 6 genes as up-regulated, suggesting that the predominant action of testosterone is to activate gene transcription. In contrast, Eacker et al. [37] compared an androgen receptor hypomorph, in which receptor action is reduced in cells throughout the testis, to the same model with an additional Sertoli cell-specific deletion of the androgen receptor. This group found the opposite pattern, with more genes showing up-regulation than down-regulation and consequently concluded that testosterone has a predominantly repressive action in the adult testis. It is, however, likely that these conclusions are affected by the methodology used in the two studies. In both cases, at the age used for the study, spermatogenic arrest has already set in, and there is a significant increase in somatic cell proportion in the testis. O'Shaughnessy et al. [36] corrected for this by applying a more stringent fold-change filter to up-regulated than to down-regulated genes, consequently detecting comparatively few up-regulated genes. Eacker et al. [37] did not apply such a correction, therefore, preferentially detecting up-regulated genes and being less sensitive to down-regulated genes.
In the work presented here, we avoid this difficulty by using age-matched controls for the developmental time course analysis and by focusing on the early responses to LH insensitivity and testosterone rescue before there are gross changes in testicular histology. Our results are in good agreement with other studies using a similar methodology, for example, Denolet et al. [23], who examined SCARKO testes at Day 10 postnatal, prior to spermatogenic arrest. This group identified nine genes that were strongly down-regulated in SCARKO mice relative to control littermates and that were also repressed by the antiandrogen flutamide. Of these nine genes, namely, Rhox5, Spinlw1, Galgt1, Drd4, Tsx, Gpd1, Tubb3, Serpina5, and Tpd52l1, all except Galgt1 and Tubb3 were found to be androgen responsive in our study. Our study extends their results by separating androgen effects into early and late responses (see below), thus, giving further information about the regulation of Sertoli cell function by testosterone. Interestingly, several of these genes were also found to be regulated by FSH in the hpg mouse [38], suggesting that Sertoli cells potentiate Leydig cell function when stimulated by FSH. This latter study also used an experimental design focused on the early transcriptional responses to treatment, avoiding the confounding effect of changes in testicular composition. We believe this to be critically important to the correct interpretation of future studies in this field.
Our study is the most systematic investigation of in vivo testicular androgen responses to date. The key finding is that there is a biphasic response to LH insensitivity, that is, a first transient phase at Day 3 that is largely self-correcting by Day 8 and a more sustained second phase with onset from Day 8 onward. The next question is which of the changes seen in LuRKO are direct effects of LH insensitivity and which are due to reduced androgen secretion. For the first-phase genes, although we observed down-regulation of Cyp11a1 and Cyp17a1 at Days 3 and 8 in LuRKO relative to control, these being the rate-limiting enzymes for steroid synthesis, the majority of genes changing at these ages did not appear to be testosterone responsive. The first phase thus appears to be predominantly an androgen-independent response to the lack of LH stimulation. A significant caveat here is that the testosterone response was measured in Day 19 testes. To formally establish whether the first-phase genes in Day 3 testes are testosterone responsive would require experimental (and unphysiological) treatment of neonatal animals with testosterone. It is however anticipated that perinatal transcriptional changes in LuRKO should be androgen independent because the normal masculinization of LuRKO male internal genitalia indicates that fetal androgen activity is unaffected by the receptor knockout [5]. Androgen secretion by fetal Leydig cells is thus presumed not to depend on LH, or at least to be redundantly controlled by other factors [38]. Fetal Leydig cells are responsive to LH in culture [12]; however, the physiological import of this is unclear. Direct measurements of perinatal testosterone in LuRKO are contradictory, with Zhang et al. [19] finding no change in testosterone at birth and Abel et al. [38] finding a marked reduction. A final factor to consider here is that the androgen receptor is not immunologically detectable on Sertoli cells until Day 5 of postnatal development [39], and thus even if perinatal testosterone production is affected in LuRKO, this may not have any consequence in the testis. Overall, our results in Day 3 testis should be taken as supporting the findings of those of Zhang et al. [19] and indicative of no major changes in androgen pathway activity in neonatal LuRKO males.
In contrast, the second phase of responses to LH insensitivity is largely testosterone dependent in that replacement therapy acts to correct the transcriptional changes seen in LuRKO males. The majority of second-phase changes only set in at postnatal Day 13 or Day 19; however, six genes (see below) show an androgen-dependent change as early as Day 8. This stands in stark contrast to measurements of testicular and serum testosterone levels, which show low (and decreasing) androgen levels at this age [38]. It is, however, consistent with the down-regulation of androgen synthesis genes seen at this age by us and others [19], and with the growing body of results from the SCARKO model, indicating that there are some androgen-dependent changes in gene expression as early as postnatal Day 4 [39]. Strikingly, we found that these early genes were induced within 8–12 h of testosterone administration, with Rhox5 and Drd4 showing even earlier induction within 2–4 h of treatment. Early genes were also induced by lower doses of testosterone, showing almost full induction at 25 μg and some induction with as little 2.5 μg; this is 100-1000-fold lower than the dosage required to mimic pubertal testosterone levels. Serum testosterone measurements for the dose-response experiment surprisingly showed little difference between the doses at the low end of the range analyzed in our study (see Supplemental Data S2) despite the fact that significant transcriptional changes were seen between, for example, the 2.5 and 25 μg samples. It may be that serum levels do not directly reflect intratesticular levels of testosterone. Further experimentation will be required to pinpoint the critical range of intratesticular androgen levels required to activate the spermatogenic program. Nevertheless, taken together, our data indicate that there are indeed early acting androgen effects during prepubertal testis development mediated by basal levels of testosterone.
Efforts to date at identifying key androgen response genes have focused largely on the Sertoli cell because of the availability of a selective receptor knockout model. Subsequent use of the same Cre/lox technology has generated models with a meiotic/postmeiotic germ cell androgen receptor knockout [40], a peritubular myoid cell androgen receptor knockout [11], and a Leydig cell androgen receptor knockout [12]. Our analysis is complementary to and extends these other studies in that the time course analysis can separate early androgen responses from later responses. For example, of the Sertoli cell androgen responsive genes identified by Denolet et al. [23], Drd4, Rhox5, and Spinlw1 were early second-phase genes in our analysis, while Gpd1, Serpina5, Tpd52l1, and Tsx fell into the late second-phase category. Moreover, our analysis is not restricted by cell type and will detect androgen response genes irrespective of their site of expression. Defb45, Lmcd1, and Pla1a may well be responding in cell types other than Sertoli cells because they were not identified in the Denolet et al. SCARKO study [23].
The six genes we identified as showing an early change between LuRKO and control, an early response to testosterone treatment, and a response to low-dose testosterone are Defb45, Drd4, Lmcd1, Pla1a, Rhox5, and Spinlw1. Three of these—Drd4, Rhox5, and Spinlw1—have previously been identified as androgen-responsive Sertoli cell genes, while the other three are here identified for the first time as putative androgen targets. Of these genes, Rhox5 is the best categorized. It is a reproductive homeobox gene with two promoters, the proximal promoter being Sertoli cell-specific and androgen responsive [41]. It has four androgen response elements in the proximal promoter, and members of the GATA family of transcription factors are also necessary for successful transcription of Rhox5 [42]. In this light, it is interesting that another of the six genes in question, Lmcd1, is a transcriptional cofactor repressing the activity of GATA6 [43]. It may thus be playing a role in moderating Rhox5 expression or in restricting expression of Rhox5 to Sertoli cells by repressing GATA6 activity in other cell types. Spinlw1 (eppin) and Drd4 (dopamine receptor 4) are androgen regulated in SCARKO testes [23]. Nothing is known about the function of either during testis development, although Spinlw1 is also an epididymal protein that coats mature sperm and has been investigated for a potential role in contraceptive vaccination [44]. Defb45 has been identified by automatic genome annotation as a member of the defensin family of antimicrobial peptides. Nothing is yet known about its role in the testis. Pla1a is a phosphatidylserine-specific phospholipase that hydrolyzes cell surface lipids to generate lysophosphatidic acid [45], which in turn is a paracrine signal promoting germ cell survival [46].
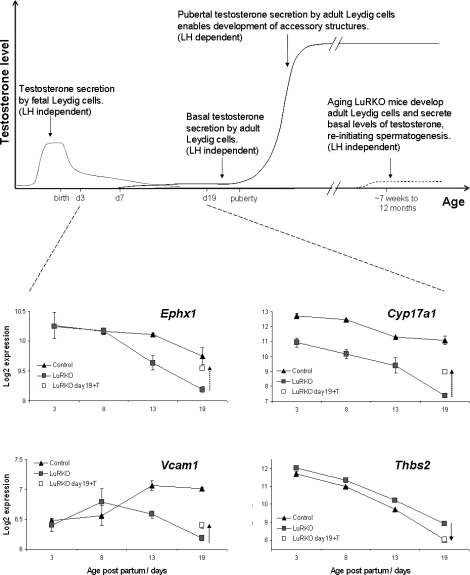
What then governs the onset of hypoandrogenism in LuRKO males? Does it relate to the appearance of the androgen receptor on Sertoli cells (Day 5 onward) or to the appearance of adult-type Leydig cells in the testis (Day 7 onward)? While our current data set does not address this directly, evidence from older LuRKO mice suggests that the critical factor is Leydig cell maturation. Old (12 mo) LuRKO males show breakthrough spermatogenesis, with every type of germ cells being present in the testis even though testosterone levels are still at basal levels (2% of controls). This spermatogenic breakthrough is abolished by treatment with the antiandrogen flutamide, indicating that the breakthrough is androgen dependent. Twelve-month-old LuRKO males remain cryptorchid, and accessory organs such as the epididymis, seminal vesicles, and external genitalia do not develop, indicating that the LH-driven pubertal testosterone surge is necessary for these structures, though not for spermatogenesis itself. Crucially, these aging males with breakthrough spermatogenesis also have adult-type Leydig cells [18]. This has two key implications: firstly, that adult Leydig cells (but not fetal cells) secrete a basal level of testosterone that is sufficient to reinitiate and support spermatogenesis, and secondly, that this basal level of testosterone production is independent of LH. In our data set, we see an overexpression of the fetal Leydig cell gene Thbs2 from Day 8 onward, corresponding to the retention of fetal-type cells. We also see an underexpression of several of the group 1 (adult Leydig cell specific) genes identified by O'Shaughnessy et al. [17]. Taken together, it appears that the critical factor behind the androgen-dependent spermatogenic delay in LuRKO is a failure to develop adult-type Leydig cells at the normal age.
So what drives Leydig cell maturation? Our data set shows that the fetal Leydig cell gene Thbs2 was down-regulated by testosterone replacement therapy, suggesting that testosterone treatment acts to reduce fetal Leydig cell activity. Conversely, several genes specific to adult Leydig cell genes were induced by testosterone in this experiment. Importantly, this included Cyp17a1 itself, the rate-limiting enzyme for androgen synthesis—a paradoxical result since this enzyme is well documented to be repressed by testosterone in adult Leydig cells via an autocrine feedback mechanism [12]. Our interpretation of this is that testosterone treatment triggers differentiation of adult-type Leydig cells, which are not present at all in the untreated Day 19 LuRKO testis. The appearance of these cells in the testis following treatment thus presents as an up-regulation of Cyp17a1 despite the fact that testosterone acts to repress Cyp17a1 on a per-cell basis. Alternatively, it may be that Cyp17a1 is regulated differently by testosterone in fetal and adult Leydig cells.
We therefore propose a model whereby the ability to produce adult Leydig cells is dependent on androgen activity. Once the first few adult Leydig cells have differentiated, they secrete androgens favoring further differentiation, thereby setting up a positive feedback switch leading to ongoing maintenance of an adult Leydig cell population. The 12-mo LuRKO phenotype shows that while LH stimulation is necessary for the correct timing of the switch from fetal- to adult-type cells during normal testis development and for establishing normal Leydig cell numbers, it is not an absolute necessity for adult Leydig cell differentiation. In aging LuRKO animals, the switch may be tripped by other endocrine factors, or it may even be entirely stochastic because in the suggested model the replacement is self-reinforcing once initiated. Testosterone levels in LuRKO at 7 wk were higher than in castrated mice, indicating that the adult Leydig cell population may have begun to appear by this point [5]. In support of our model, adult Leydig cells fail to develop in Tfm mice lacking androgen receptor function [47] but do develop in SCARKO mice [48], in mice lacking the androgen receptor in Leydig cells [12], and in mice lacking the androgen receptor in peritubular myoid cells [11]. Also consistent with our proposal, Hardy et al. [49] found that LH and androgens had a synergistic effect on the differentiation of adult Leydig cells from peritubular mesenchymal precursors in rat cell culture.
Figure 4 shows a schematic representation of this model indicating androgen secretion by fetal and adult Leydig cells and the point at which the SCARKO and LuRKO models perturb the normal situation. Also included in this figure is the absolute expression data for LuRKO and control males for the fetal/adult Leydig cell antigens found to be testosterone regulated in our study, illustrating the switch over from fetal- to adult-type cells in controls and the failure of this process in LuRKO males. Ephx1 and Cyp17a1 are expressed in both fetal and adult cells. In control mice, there is a small drop in transcript levels as fetal cells are replaced by quiescent adult cells (quiescent due to low LH levels at this age). In LuRKO, there is a progressive reduction relative to control as the adult cells fail to develop. Testosterone treatment allows adult cell development and increases levels of these transcripts. Vcam1 is expressed specifically in adult Leydig cells. It becomes activated between Days 8 and 13 in control males but not in LuRKO. Testosterone treatment of LuRKO males increases the levels of this transcript. Thbs2 is expressed specifically in fetal Leydig cells. It decreases markedly in control males throughout development as fetal cells are replaced. In LuRKO, transcript levels are a little higher due to the retention of fetal-type cells. Testosterone treatment lowers the level of this transcript, implying that testosterone represses fetal Leydig cell activity and/or triggers their replacement by adult Leydig cells.
FIG. 4.
Schematic representation of the secretion of testosterone by fetal and adult Leydig cells during gestation, postnatally, and in the adult. The switch over from fetal- to adult-type Leydig cells occurs prepubertally, starting around Day 7. LuRKO males are hypoandrogenic from this point forward due to a failure to develop adult Leydig cells. Lower panels show transcriptional data for selected fetal/adult Leydig cell antigens during the period. The dotted arrows in these panels indicate the effect of 48 h of testosterone treatment at a dose of 2.5 mg.
The model makes three key predictions, which we aim to test in future experiments.
Low-dose testosterone therapy in LuRKO will allow resumption of spermatogenesis without being sufficient to trigger testicular descent and accessory gland development, that is, it will mimic the 12-mo-old LuRKO phenotype.
Testosterone therapy will drive Leydig cell maturation in the absence of LH; thus, in rescued LuRKO mice, the Leydig cells should show an adult phenotype.
Once the adult Leydig cells are present in the testis, they will be able to maintain the necessary basal testosterone level; thus, in rescued LuRKO mice, spermatogenesis will continue even if testosterone treatment is subsequently stopped.
A final question to pose is what is the biochemical basis of the delayed Leydig cell maturation in LuRKO? Clues to this can be found by looking to the transcriptional changes found in common between all the analyzed ages, in particular, the changes seen at Day 3. In this regard, a clear conclusion from the gene ontology pathway analysis is that genes affecting the WNT pathway and cell adhesion are up-regulated in LuRKO; however, the specific type of adhesion involved is age dependent. Focal adhesion genes and matrix adhesion genes, including integrins Itga6 and Itga11, collagens Col3a1 and Col5a2, laminins Lama1 and Lamc1, vitronectin, fibronectin, actinin Actn4, and beta actin, were up-regulated at all ages. Tight junction genes, including key junction components claudin Cldn1 and occludin Ocln, were only up-regulated from Day 19 onward.
The later effect on tight junctions is likely to be a downstream consequence of hypoandrogenism, although the interpretation is complex because flutamide has been shown to down-regulate tight junction components in rat testes [50], contrary to the up-regulation seen here. Since the focal/matrix adhesion effects are seen in the paired t-test analysis, which examines all the days including Day 3, these appear to be testosterone-independent effects of LH and may relate to the correct maturation of Leydig cells. The origin of mature Leydig cells is unclear in that they do not appear to differentiate directly from fetal cells. Peritubular mesenchymal cells and perivascular smooth muscle cells have both been proposed as candidate stem cells for this population, with the evidence largely favoring the former. These mesenchymal cells are also the precursors for the peritubular myoid cell lineage in the adult testis [51]. A fruitful avenue for future study will be the influence of testosterone on the precursor mesenchymal cells. Does testosterone affect the decision of whether to become an adult Leydig cell or a mature myoid cell? In particular, is it a necessary permissive factor allowing differentiation of adult Leydig cells? What junctional connections exist between peritubular mesenchymal cells and other cell types, and do alterations in these connections affect mesenchymal cell fate?
Acknowledgments
The luteinizing hormone receptor knockout model (LuRKO), which was developed by Dr. Fu-Ping Zhang, was a kind gift of Professor Ilpo Huhtaniemi. Measurements of serum testosterone levels (see Supplemental Data S1) were performed by Professor Peter O'Shaughnessy. We thank Dr. Oriane Chausiaux for assistance with RNA preparation, and Jo Bacon for assistance with qPCR validation (see Supplemental Data S4).
Footnotes
Supported by the Biotechnology and Biological Sciences Research Council (BB/F007434/1 to P.J.I.E. and N.A.A. BB/E024211/1 to D.K.G., and Career Development Fellowship to D.K.G.) and the Wellcome Trust (WT078137 to M.H.A. and Peter O'Shaughnessy).
These authors contributed equally to this work
REFERENCES
- Greep RO, Fevold H.The spermatogenic and secretory function of the gonads of hypophysectomised adult rats treated with pituitary FSH and LH. Endocrinology 1937; 21: 611–619. [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM.Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet 1997; 15: 201–204. [DOI] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, Sassone-Corsi P.Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci U S A 1998; 95: 13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel MH, Wootton AN, Wilkins V, Huhtaniemi I, Knight PG, Charlton HM.The effect of a null mutation in the follicle-stimulating hormone receptor gene on mouse reproduction. Endocrinology 2000; 141: 1795–1803. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Poutanen M, Wilbertz J, Huhtaniemi I.Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol Endocrinol 2001; 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Lei ZM, Mishra S, Zou W, Xu B, Foltz M, Li X, Rao CV.Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Mol Endocrinol 2001; 15: 184–200. [DOI] [PubMed] [Google Scholar]
- Singh J, O'Neill C, Handelsman DJ.Induction of spermatogenesis by androgens in gonadotropin-deficient (hpg) mice. Endocrinology 1995; 136: 5311–5321. [DOI] [PubMed] [Google Scholar]
- Spaliviero JA, Jimenez M, Allan CM, Handelsman DJ.Luteinizing hormone receptor-mediated effects on initiation of spermatogenesis in gonadotropin-deficient (hpg) mice are replicated by testosterone. Biol Reprod 2004; 70: 32–38. [DOI] [PubMed] [Google Scholar]
- Pakarainen T, Zhang FP, Mäkelä S, Poutanen M, Huhtaniemi I.Testosterone replacement therapy induces spermatogenesis and partially restores fertility in luteinizing hormone receptor knockout mice. Endocrinology 2005; 146: 596–606. [DOI] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lécureuil C, Heyns W, Carmeliet P, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A 2004; 101: 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yeh S, Chen YT, Wu CC, Chuang KH, Lin HY, Wang RS, Chang YJ, Mendis-Handagama C, Hu L, Lardy H, Chang C.Oligozoospermia with normal fertility in male mice lacking the androgen receptor in testis peritubular myoid cells. Proc Natl Acad Sci U S A 2006; 103: 17718–17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Lin HY, Yeh SD, Yu IC, Wang RS, Chen YT, Zhang C, Altuwaijri S, Chen LM, Chuang KH, Chiang HS, Yeh S, et al. Infertility with defective spermatogenesis and steroidogenesis in male mice lacking androgen receptor in Leydig cells. Endocrine 2007; 32: 96–106. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Baker P, Sohnius U, Haavisto AM, Charlton HM, Huhtaniemi I.Fetal development of Leydig cell activity in the mouse is independent of pituitary gonadotroph function. Endocrinology 1998; 139: 1141–1146. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Morris ID, Huhtaniemi I, Baker PJ, Abel MH.Role of androgen and gonadotrophins in the development and function of the Sertoli cells and Leydig cells: data from mutant and genetically modified mice. Mol Cell Endocrinol 2009; 306: 2–8. [DOI] [PubMed] [Google Scholar]
- Huhtaniemi I, Pelliniemi LJ.Fetal Leydig cells: cellular origin, morphology, life span, and special functional features. Proc Soc Exp Biol Med 1992; 201: 125–140. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Sha JA, McBride MW, Peng L, Payne AH, O'Shaughnessy PJ.Expression of 3beta-hydroxysteroid dehydrogenase type I and type VI isoforms in the mouse testis during development. Eur J Biochem 1999; 260: 911–917. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Willerton L, Baker PJ.Changes in Leydig cell gene expression during development in the mouse. Biol Reprod 2002; 66: 966–975. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Pakarainen T, Poutanen M, Toppari J, Huhtaniemi I.The low gonadotropin-independent constitutive production of testicular testosterone is sufficient to maintain spermatogenesis. Proc Natl Acad Sci U S A 2003; 100: 13692–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FP, Pakarainen T, Zhu F, Poutanen M, Huhtaniemi I.Molecular characterization of postnatal development of testicular steroidogenesis in luteinizing hormone receptor knockout mice. Endocrinology 2004; 145: 1453–1463. [DOI] [PubMed] [Google Scholar]
- Khatri P, Sellamuthu S, Malhotra P, Amin K, Done A, Draghici S.Recent additions and improvements to the onto-tools. Nucleic Acids Res 2005; 33(Web server issue):W762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M.KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 1999; 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ.GOEAST: a web-based software toolkit for gene ontology enrichment analysis. Nucleic Acids Res 2008; 36(Web server issue):W358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KA, Sharpe RM, Saunders PT, Swinnen JV, Verhoeven G.The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol 2006; 20: 321–334. [DOI] [PubMed] [Google Scholar]
- Mikkilä TF, Toppari J, Paranko J.Effects of neonatal exposure to 4-tert-octylphenol, diethylstilbestrol, and flutamide on steroidogenesis in infantile rat testis. Toxicol Sci 2006; 91: 456–466. [DOI] [PubMed] [Google Scholar]
- Sha J, Zhou Z, Li J, Yin L, Yang H, Hu G, Luo M, Chan HC, Zhou K.Identification of testis development and spermatogenesis-related genes in human and mouse testes using cDNA arrays. Mol Hum Reprod 2002; 8: 511–517. [DOI] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL.A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A 2003; 100: 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Yu Z, Guan J, Ge Y, Ma J, Li S, Wang S, Xue S, Han D.Stage-specific and tissue-specific expression characteristics of differentially expressed genes during mouse spermatogenesis. Mol Reprod Dev 2004; 67: 264–272. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Nielsen JE, Hansen MA, Tanaka M, Skakkebaek NE, Leffers H.Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol Reprod 2004; 70: 1751–1761. [DOI] [PubMed] [Google Scholar]
- Ellis PJ, Furlong RA, Wilson A, Morris S, Carter D, Oliver G, Print C, Burgoyne PS, Loveland KL, Affara NA.Modulation of the mouse testis transcriptome during postnatal development and in selected models of male infertility. Mol Hum Reprod 2004; 10: 271–281. [DOI] [PubMed] [Google Scholar]
- Maratou K, Forster T, Costa Y, Taggart M, Speed RM, Ireland J, Teague P, Roy D, Cooke HJ.Expression profiling of the developing testis in wild-type and Dazl knockout mice. Mol Reprod Dev 2004; 67: 26–54. [DOI] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD.The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 2004; 36: 642–646. [DOI] [PubMed] [Google Scholar]
- Fox MS, Ares VX, Turek PJ, Haqq C, Reijo Pera RA.Feasibility of global gene expression analysis in testicular biopsies from infertile men. Mol Reprod Dev 2003; 66: 403–421. [DOI] [PubMed] [Google Scholar]
- Feig C, Kirchhoff C, Ivell R, Naether O, Schulze W, Spiess AN.A new paradigm for profiling testicular gene expression during normal and disturbed human spermatogenesis. Mol Hum Reprod 2007; 13: 33–43. [DOI] [PubMed] [Google Scholar]
- Ellis PJ, Furlong RA, Conner SJ, Kirkman-Brown J, Afnan M, Barratt C, Griffin DK, Affara NA.Coordinated transcriptional regulation patterns associated with infertility phenotypes in men. J Med Genet 2007; 44: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess AN, Feig C, Schulze W, Chalmel F, Cappallo-Obermann H, Primig M, Kirchhoff C.Cross-platform gene expression signature of human spermatogenic failure reveals inflammatory-like response. Hum Reprod 2007; 22: 2936–2946. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H, Baker PJ.Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive Tfm mouse testis. Endocrinology 2007; 148: 2914–2924. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Shima JE, Connolly CM, Sharma M, Holdcraft RW, Griswold MD, Braun RE.Transcriptional profiling of androgen receptor (AR) mutants suggests instructive and permissive roles of AR signaling in germ cell development. Mol Endocrinol 2007; 21: 895–907. [DOI] [PubMed] [Google Scholar]
- Abel MH, Baban D, Lee S, Charlton HM, O'Shaughnessy PJ.Effects of FSH on testicular mRNA transcript levels in the hypogonadal mouse. J Mol Endocrinol 2009; 42: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A, De Gendt K, Allemeersch J, Smith LB, Welsh M, Swinnen JV, Verhoeven G.Early effects of Sertoli cell-selective androgen receptor ablation on testicular gene expression. Int J Androl 2010; (in press). Published online ahead of print 12 April 2009: DOI 10.1111/j.1365-2605.2009.00964.x. [DOI] [PubMed]
- Tsai MY, Yeh SD, Wang RS, Yeh S, Zhang C, Lin HY, Tzeng CR, Chang C.Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc Natl Acad Sci U S A 2006; 103: 18975–18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Wilkinson MF.Pem homeobox gene regulatory sequences that direct androgen-dependent developmentally regulated gene expression in different subregions of the epididymis. J Biol Chem 2002; 277: 48771–48778. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Rao MK, Kaur R, Buttigieg MR, Wilkinson MF.GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol Cell Biol 2008; 28: 2138–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath N, Wang Z, Lu MM, Morrisey EE.LMCD1/dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding. Mol Cell Biol 2005; 25: 8864–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Widgren EE, Richardson RT, Orand MG.Eppin: a molecular strategy for male contraception. Soc Reprod Fertil Suppl 2007; 65: 535–542. [PubMed] [Google Scholar]
- Hiramatsu T, Sonoda H, Takanezawa Y, Morikawa R, Ishida M, Kasahara K, Sanai Y, Taguchi R, Aoki J, Arai H.Biochemical and molecular characterization of two phosphatidic acid-selective phospholipase A1s, mPA-PLA1alpha and mPA-PLA1beta. J Biol Chem 2003; 278: 49438–49447. [DOI] [PubMed] [Google Scholar]
- Ye X.Lysophospholipid signaling in the function and pathology of the reproductive system. Hum Reprod Update 2008; 14: 519–536. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Johnston H, Willerton L, Baker PJ.Failure of normal adult Leydig cell development in androgen-receptor-deficient mice. J Cell Sci 2002; 115: 3491–3496. [DOI] [PubMed] [Google Scholar]
- De Gendt K, Atanassova N, Tan KA, de França LR, Parreira GG, McKinnell C, Sharpe RM, Saunders PT, Mason JI, Hartung S, Ivell R, Denolet E, et al. Development and function of the adult generation of Leydig cells in mice with Sertoli cell-selective or total ablation of the androgen receptor. Endocrinology 2005; 146: 4117–4126. [DOI] [PubMed] [Google Scholar]
- Hardy MP, Kelce WR, Klinefelter GR, Ewing LL.Differentiation of Leydig cell precursors in vitro: a role for androgen. Endocrinology 1990; 127: 488–490. [DOI] [PubMed] [Google Scholar]
- Gye MC, Ohsako S.Effects of flutamide in the rat testis on the expression of occludin, an integral member of the tight junctions. Toxicol Lett 2003; 143: 217–222. [DOI] [PubMed] [Google Scholar]
- Campagnolo L, Russo MA, Puglianiello A, Favale A, Siracusa G.Mesenchymal cell precursors of peritubular smooth muscle cells of the mouse testis can be identified by the presence of the p75 neurotrophin receptor. Biol Reprod 2001; 64: 464–472. [DOI] [PubMed] [Google Scholar]