SUMMARY
In Saccharomyces cerevisiae, chemical or genetic inhibition of proteasome activity induces new proteasome synthesis promoted by the transcription factor RPN4. This ensures that proteasome activity is matched to demand. This transcriptional feedback loop is conserved in mammals, but its molecular basis is not understood. Here we report that Nuclear factor erythroid derived 2-related factor 1 (Nrf1), a transcription factor of the cap ‘n’ collar basic leucine zipper family, but not the related Nrf2, is necessary for induced proteasome gene transcription in mouse embryonic fibroblasts (MEFs). Promoter-reporter assays revealed the importance of antioxidant response elements in Nrf1-mediated upregulation of proteasome subunit genes. Nrf1-/- MEFs were impaired in the recovery of proteasome activity after transient treatment with the covalent proteasome inhibitor YU101 and knockdown of Nrf1 in human cancer cells enhanced cell killing by YU101. Taken together, our results suggest that Nrf1-mediated proteasome homeostasis could be an attractive target for therapeutic intervention in cancer.
INTRODUCTION
The 26S proteasome, a multicatalytic protease complex of over 2.5 MDa, is responsible for degradation of a variety of proteins in cells, and thus helps maintain intracellular protein homeostasis (Ciechanover, 2005; Finley, 2009; Pickart and Cohen, 2004). The actual proteolysis occurs in the 20S catalytic core subunit of the proteasome and is mediated through the chymotrypsin-like, trypsin-like, and caspase-like activities located therein (Kisselev and Goldberg, 2005). Structurally, the 20S core is composed of four stacked rings each of which is made of seven distinct subunits. Whereas the α-subunits encoded by genes PSMA1-7 make up the outer rings, the two inner rings are composed of the β-subunits which are encoded by genes PSMB1-7 (Unno et al., 2002). The 20S core is usually capped at one or both ends by the 19S regulatory subunit which is composed of a base and a lid (Glickman et al., 1998). The base in turn is made of 6 ATPase (encoded by genes PSMC1-6) and 3 non-ATPase (encoded by genes PSMD1, 2, and 4) subunits. At least 9 non-ATPase subunits (encoded by genes PSMD3, 6-9, and 11-14) form the lid subcomplex (Finley, 2009).
There exists a wealth of information on the structure (Groll et al., 1997; Unno et al., 2002) and assembly pathways (Kusmierczyk and Hochstrasser, 2008; Murata et al., 2009; Park et al., 2009; Roelofs et al., 2009) of the proteasome, but our understanding of the regulation of proteasome subunit (PSM) genes at the transcriptional level is rather limited. In the yeast Saccharomyces cerevisiae, it has been established that the transcription factor RPN4 mediates expression of PSM genes by binding to their promoters via a conserved motif (5′-GGTGGCAAA-3′) dubbed the PACE (Proteasome Associated Control Element) sequence (Mannhaupt et al., 1999). Interestingly, RPN4 is a short-lived protein that is ubiquitinated by Ubr2–Ubc2 (Wang et al., 2004) and then rapidly degraded by the proteasome (Xie and Varshavsky, 2001). Thus, when the proteasome levels in a cell are sufficient, RPN4 is constitutively degraded, and conversely, when proteasome is inhibited, RPN4 accumulates leading to increased synthesis of PSM genes resulting in accelerated recovery of proteasome activity (Dohmen et al., 2007).
Recent studies have indicated that elevated proteasome synthesis upon proteasome inhibition (which we refer to here as ‘bounce-back’) is well conserved in higher eukaryotes (Lundgren et al., 2005; Meiners et al., 2003; Mitsiades et al., 2002). In the case of Drosophila, either RNA interference (RNAi)-mediated knockdown of one of the proteasome subunit genes, PSMD4, or outright inhibition of the proteasome’s proteinase activity by MG132 results in transcriptional upregulation of PSM genes. Although a PACE-like motif remains to be identified, the 5′ UTR region of Drosophila PSM genes is necessary to mediate the bounce-back response (Lundgren et al., 2005). This bounce-back response is clearly operative in mammals too, because chemical inhibition of the proteasome in rat and human cells results in concerted upregulation of PSM mRNA levels followed by de novo formation of proteasomes (Meiners et al., 2003). Also, a microarray study aimed at understanding the mechanism of action of the proteasome inhibitor bortezomib (approved as a therapy for multiple myeloma and marketed under the trade name Velcade) revealed that several PSM genes are upregulated in multiple myeloma cells in response to the drug (Mitsiades et al., 2002).
Despite the pervasiveness of a bounce-back response in metazoans, the molecular mechanism behind this phenomenon and the identity of the transcription factor that mediates this response remains uncertain. Nrf2 has been implicated in mediating proteasome bounce-back in human fibroblasts (Kraft et al., 2006) but this result remains unconfirmed and we were unable to validate it in other cell types (our unpublished data). The unambiguous identification of this factor in mammalian cells is of keen interest because transcriptional upregulation of proteasome synthesis might limit the duration and intensity of proteasome inhibition and thereby attenuate the response of cancer patients to proteasome inhibitor therapy. In this study, using cell lines derived from gene knock-out mice in concert with knockdown and overexpression strategies, we identify Nuclear factor erythroid derived 2-related factor 1 (Nrf1) as a mediator of the mammalian proteasome bounce-back response.
RESULTS
Proteasome inhibitors induce the bounce-back response in human cancer cells
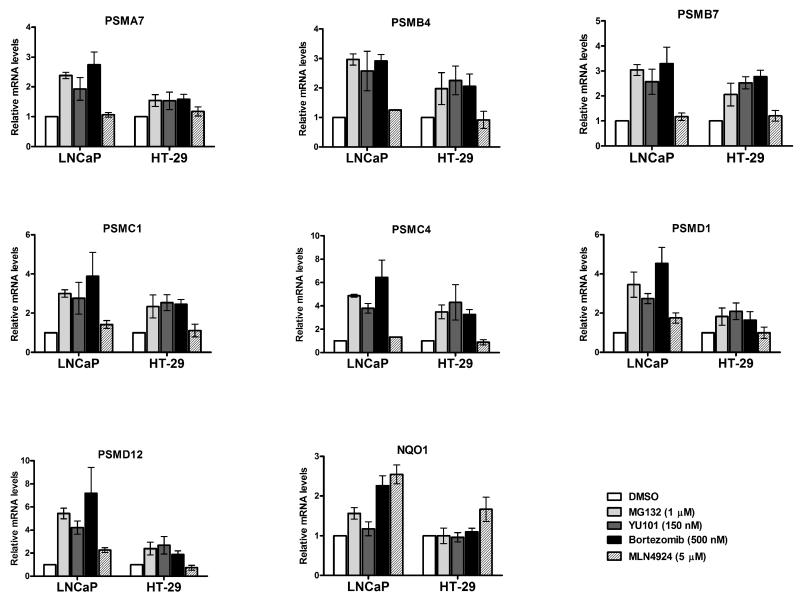
As a first step towards understanding the proteasome bounce-back response in mammals, we treated human prostate cancer LNCaP and colon cancer HT29 cell lines with different proteasome inhibitors (MG132, YU101, and Bortezomib) or the Nedd8 pathway inhibitor MLN4924 (Soucy et al., 2009). As expected, the proteasome inhibitors were able to robustly induce mRNA levels of several PSM genes that encode members of both the 20S (PSMA7, PSMB4, and PSMB7) and 19S (PSMC1, PSMC4, PSMD1, and PSMD12) complexes, albeit to varying degrees in the two cell lines that were surveyed (Fig 1). MLN4924 works by inhibiting the Nedd8-activating enzyme, the result of which is the accumulation of cullin-RING ligase (CRL) substrates (Soucy et al., 2009). Treatment of cells with MLN4924 stabilizes the transcription factor Nrf2 (Soucy et al., 2009), which should lead to activation of its downstream target genes. Indeed, we found this to be true for NQO1, a prototypical target gene of Nrf2 (Fig 1). In contrast, under the same treatment conditions, MLN4924 failed to appreciably induce the PSM genes in these cell lines (Fig 1), suggesting that inhibition of Nedd8 pathway alone is insufficient to elicit the bounce-back response.
Fig 1. Proteasome inhibitors, but not a Nedd8 pathway inhibitor, upregulate RNA levels of PSM genes in cancer cells.
Prostate cancer LNCaP and colon cancer HT-29 cells were treated with the indicated concentrations of proteasome inhibitors (MG132, YU101, and Bortezomib) or the Nedd8 pathway inhibitor (MLN4924) for 10 hrs, and mRNA levels of representative PSM genes were analyzed by quantitative RT-PCR. The values were normalized to GAPDH and for each cell line the DMSO treated sample was set to 1. Error bars denote SD (n=3).
Nrf1 is necessary to sustain the proteasome inhibitor-mediated bounce-back response
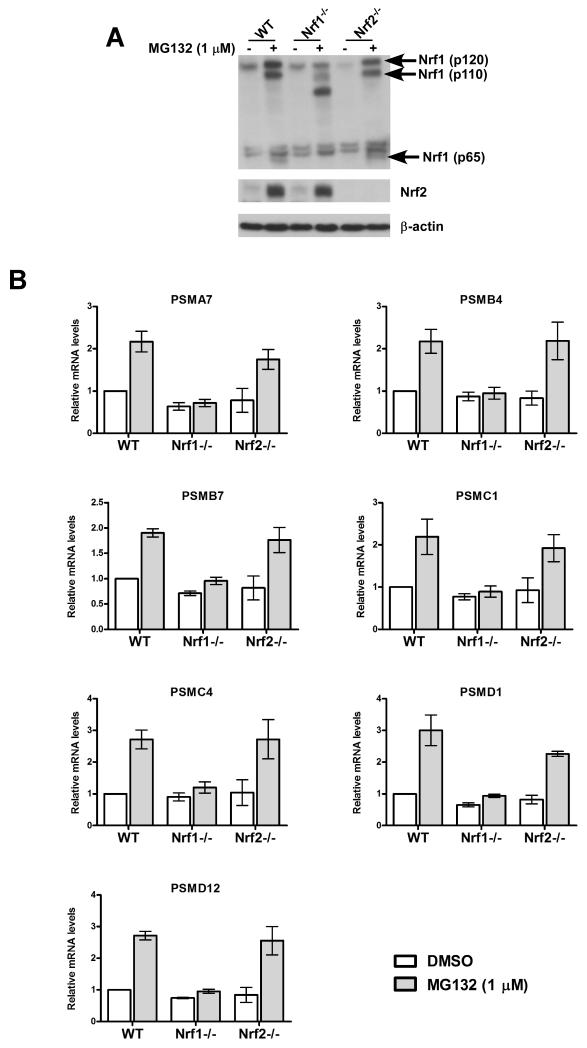
As noted in the Introduction, a recent study implicated Nrf2 in inducing proteasome activity in response to MG132 (Kraft et al., 2006), suggesting that Nrf2 mediates the bounce-back response. To test this hypothesis, we made use of mouse embryonic fibroblasts (MEFs) derived from Nrf2-/- mice (Chan et al., 1996). Whereas the wild-type (WT) MEFs accumulated Nrf2 protein after MG132 treatment, Nrf2-/- cells, as expected, did not show any detectable levels of the protein under the same conditions (Fig 2A), thereby confirming the identity of these cells. Importantly, MG132 induced mRNA levels of PSM genes in both WT and Nrf2-/- MEFs to a similar extent, thus ruling out an essential role for Nrf2 in eliciting the bounce-back response in these cells (Fig 2B). Interestingly, however, when we tested MEFs that are functionally deficient in the related transcription factor Nrf1 (Chan et al., 1998), we found that these cells were severely blunted in their ability to upregulate PSM genes in response to MG132 treatment (Fig 2B). Taken together, our data suggest that Nrf1, but not Nrf2 enables enhanced proteasome mRNA accumulation in MEFs treated with proteasome inhibitor.
Fig 2. Nrf1 but not Nrf2 is required for MG132-mediated upregulation of RNA levels of PSM genes.
(A) MEFs of different genotypes (WT, Nrf1-/-, and Nrf2-/-) were treated for 10 hrs with MG132 as indicated and the cell lysates were used for immunoblotting to detect protein levels of Nrf1 (with the antibody raised against the N-terminus) and Nrf2. β-actin protein levels were used as loading control. (B) RNA from MEFs under the same treatment conditions as above was used for quantitative RT-PCR to assess the mRNA levels of representative PSM genes. The values were normalized to GAPDH and the DMSO treated WT sample was set to 1. Error bars denote SD (n=3).
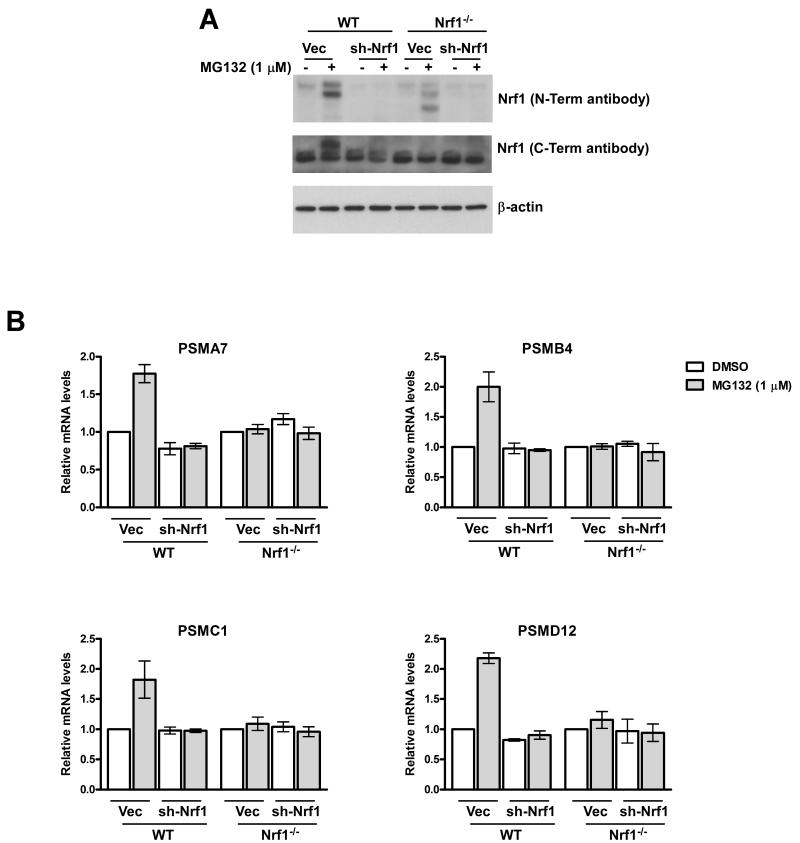
The Nrf1-/- MEFs that we used in this study retain the expression of a truncated form of the protein (Fig 2A), as was described originally (Chan et al., 1998). One concern was that Nrf1 might not actually be required for the proteasome recovery response, but that this truncated species could act in a dominant-negative or neomorphic fashion, thereby confounding our results. To test this possibility, we performed RNAi experiments with a retrovirus that expressed an shRNA targeted to the 5′ end of the Nrf1 mRNA. Knock-down of Nrf1 in WT MEFs (Fig. 3A) mimicked the Nrf1-/- phenotype, in that MG132 did not induce accumulation of proteasome mRNAs in depleted cells (Fig. 3B). Moreover, depletion of the truncated Nrf1 species in Nrf1-/- cells (Fig. 3A) did not revert the proteasome expression defect of these cells (Fig. 3B), ruling out the possibility that the truncated polypeptide could be acting as a dominant-negative. Taken together, these data establish an independent line of evidence for the requirement of Nrf1 in the bounce-back response and suggest that the lack of this response in Nrf1-/- cells was not an indirect consequence arising from a developmental adaptation to the continuous absence of Nrf1.
Fig 3. Knock-down of Nrf1 in WT MEFs abolishes MG132-mediated upregulation of RNA levels of PSM genes.
(A) WT and Nrf1-/- MEFs were transduced with retrovirus expressing sh-Nrf1 or vector control and 72 hrs later treated with MG132 for 10 hrs as indicated. The cell lysates were then used for immunoblotting to analyze protein levels of Nrf1 with either a rabbit polyconal antibody specific for the N-terminus or a mouse polyclonal antibody specific for the C-terminal region of Nrf1. β-actin protein levels were used as loading control. (B) RNA from MEFs under the same viral transduction and treatment conditions as above was used for quantitative RT-PCR to assess the mRNA levels of representative PSM genes. The values were normalized to GAPDH and for each cell line the vector-transduced DMSO-treated sample was set to 1. Error bars denote SD (n=3).
Exogenous expression of Nrf1 restores the proteasome inhibitor-mediated bounce-back response in Nrf1-/- MEFs
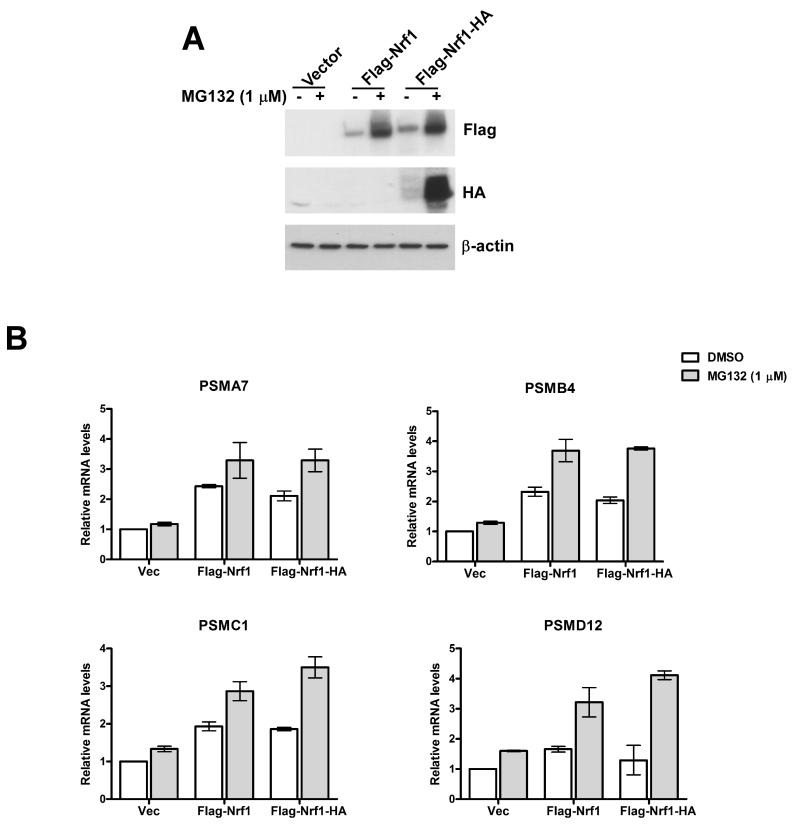
Next, we asked if reinstating Nrf1 expression in Nrf1-/- MEFs could rescue the defect in proteasome inhibitor-induced bounce-back response. To this end, we engineered singly tagged (Flag-Nrf1) and doubly tagged (Flag-Nrf1-HA) retroviral expression constructs and transiently overproduced these tagged proteins by infection of Nrf1-/- MEFs (Fig 4A). Nrf1-/- MEFs overexpressing tagged Nrf1, but not the vector control cells, upregulated mRNA levels of multiple PSM genes upon MG132 treatment (Fig 4B). Also, overexpression of tagged-Nrf1 induced PSM mRNA levels in untreated Nrf1-/- cells by ~1.5-2.0 fold compared to vector control (Fig 4B).
Fig 4. Forced expression of Nrf1 in Nrf1-/- MEFs reinstates MG132-mediated upregulation of RNA levels of PSM genes.
(A) Nrf1-/- MEFs were transduced with retrovirus expressing one of Flag-Nrf1, Flag-Nrf1-HA or vector control and 72 hrs later treated with MG132 for 10 hrs as indicated. The cell lysates were used for immunoblotting to detect the levels of exogenous Nrf1 by using tag-specific antibodies. β-actin protein levels were used as loading control. (B) RNA from Nrf1-/- MEFs under the same viral transduction and treatment conditions as above was used for quantitative RT-PCR to assess the mRNA levels of representative PSM genes. The values were normalized to GAPDH and the vector-transduced DMSO-treated sample was set to 1. Error bars denote SD (n=3).
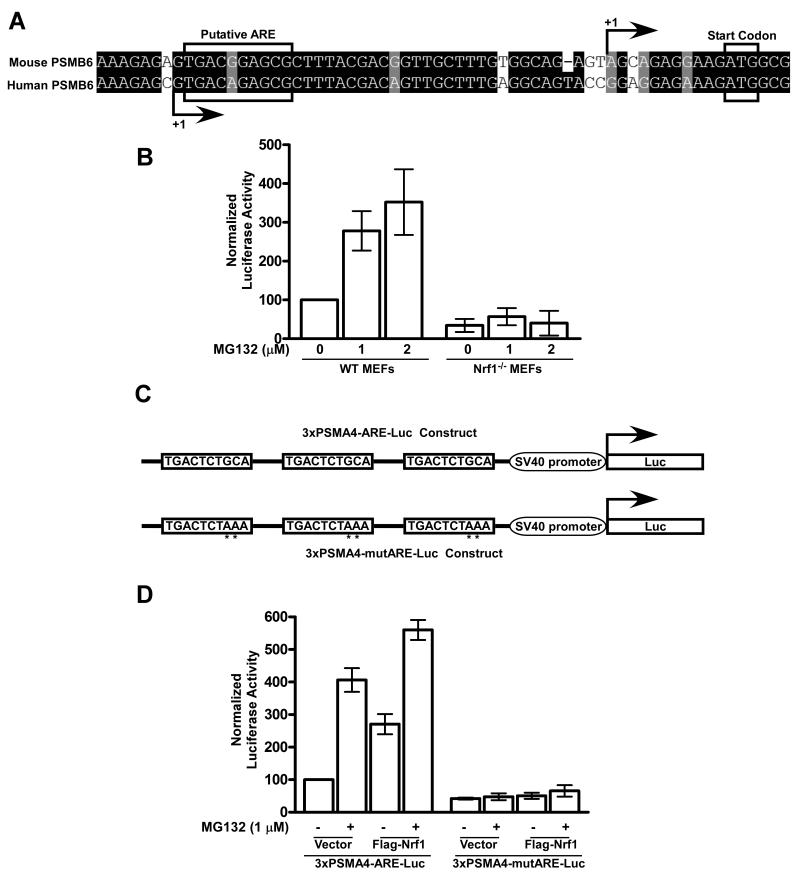
Nrf1 is necessary and sufficient to activate anti-oxidant response elements (AREs) from PSM genes
To further explore the link between Nrf1 and PSM gene expression in the context of the proteasome bounce-back response, we generated a construct with a ~3kb promoter region from murine PSMB6 fused to a firefly luciferase reporter. It has been well established that Nrf1 and the related transcription factors Nrf2 and Nrf3 bind to ARE, a cis-acting enhancer sequence found in the promoter regions of their target genes, thereby regulating their transcription (Biswas and Chan, 2009; Johnsen et al., 1998; Sankaranarayanan and Jaiswal, 2004; Venugopal and Jaiswal, 1996). Using a position weight matrix that has been derived from functional AREs (Wang et al., 2007), we computationally predicted a putative ARE close to the transcription start site of both the human and mouse PSMB6 genes (Fig 5A). When we performed luciferase assays with the murine PSMB6 promoter-reporter construct, we observed a dose dependent increase in luciferase activity after MG132 treatment of WT but not Nrf1-/- MEFs (Fig 5B), implying that Nrf1 was necessary to induce PSM gene promoters in response to proteasome inhibition. To confirm the importance of AREs in this context, we generated a synthetic promoter construct in which three copies of a putative ARE from the first intron of the human PSMA4 gene were inserted upstream of a minimal SV40 promoter driving the expression of a firefly luciferase reporter (Fig 5C). When LNCaP cells transfected with this construct were treated with MG132, we saw an increase in luciferase activity when compared to DMSO-treated control (Fig 5D). In the same experiment, we also found that overexpression of Flag-Nrf1 activated this synthetic promoter construct, and this activation was further enhanced when these cells were treated with MG132. In stark contrast, we found that neither MG132 nor overexpressed Flag-Nrf1 induced the synthetic promoter construct when the AREs were mutated (Fig 5D). Taken together, our data are consistent with a model where proteasome inhibition leads to Nrf1-dependent activation of PSM gene promoters specifically through AREs.
Fig 5. Nrf1 is necessary and sufficient to activate AREs from PSM genes.
(A) Sequence alignment of the genomic region close to the transcription start site (indicated by +1) of the PSMB6 gene in mouse and human. Putative ARE and the start codon are marked. (B) WT and Nrf1-/- MEFs were transfected with the PSMB6-Luc construct and 24 hrs post-transfection, the cells were treated with the indicated concentration of MG132 for 12 hrs after which luciferase assays were performed as described in Materials and Methods. Error bars denote SD (n=3). (C) Schematic representation of the luciferase reporter constructs – 3xPSMA4-ARE-Luc which has three copies of ARE derived from the first intron of the human PSMA4 gene and 3xPSMA4-mutARE-Luc which is identical to the above except that it has the AREs mutated in key positions indicated by asterisks. (D) LNCaP cells were transiently transfected with the indicated reporter constructs along with either a vector control or Flag-Nrf1 expression construct. Forty-eight hrs after transfection, the cells were further incubated with the indicated concentration of MG132 for 12 hrs after which luciferase assays were performed as described in Materials and Methods. Error bars denote SD (n=3).
Nrf1 accelerates recovery of proteasome activity in MEFs and restrains apoptosis in cancer cells treated with covalent proteasome inhibitors
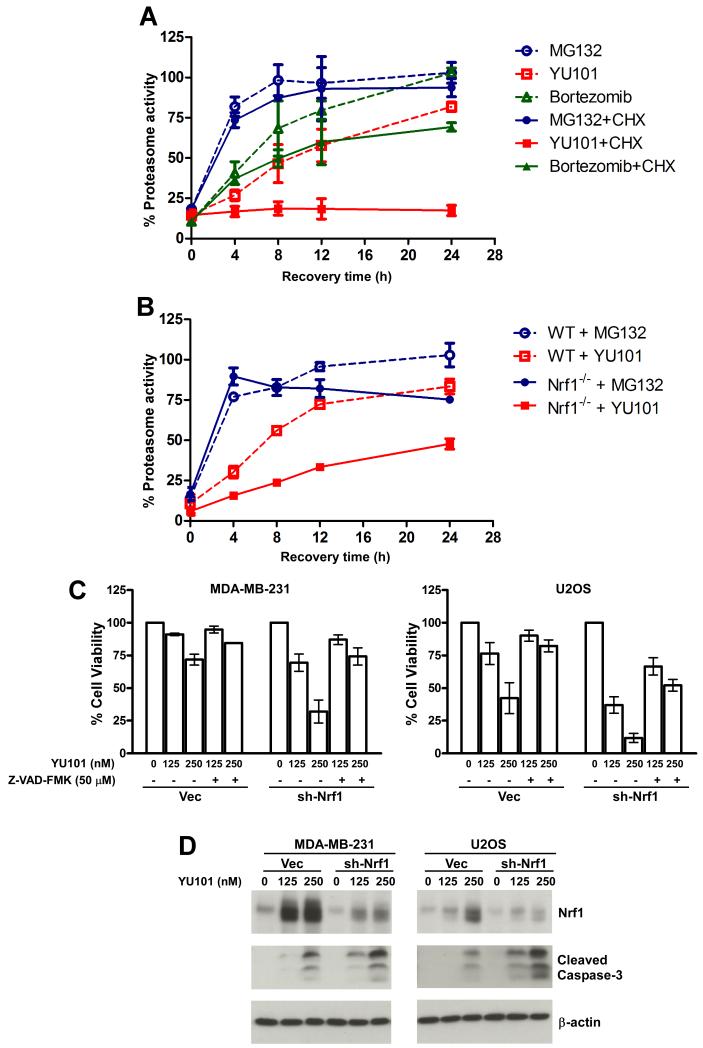
Upon inhibition, proteasome activity can be recovered in one of two ways: either the inhibitor can dissociate from the active site or the cell can produce new proteasomes. A reversible inhibitor such as MG132 is expected to dissociate rapidly from the active site upon clearance of the inhibitor, whereas in the case of a slowly-dissociating inhibitor like bortezomib, it is unclear whether the slow recovery of whole blood proteasome activity observed in human patients is due to dissociation, elevated proteasome synthesis, or both. For a covalent, non-dissociable inhibitor like YU101 or carfilzomib, it is anticipated that recovery of proteasome activity in dosed patients should depend entirely on synthesis of new proteasomes. This is a critical distinction because the nature of the recovery mechanism could have major therapeutic implications.
To test these ideas and to set the stage for investigating the importance of Nrf1 in this context, we performed a preliminary experiment in which HT29 colon cancer cells were briefly exposed to three different proteasome inhibitors – MG132, YU101, and bortezomib – each dosed to achieve 80-90% inhibition of the chymotryptic site. We found that with time, the proteasome activity recovered after drug wash-out in all of these cases as expected, although with different kinetics (Fig 6A, hatched lines), presumably dictated by the speed of dissociation of the agent from the active site of the proteasome. However, when the same experiment was performed in the presence of the protein synthesis inhibitor cycloheximide, we observed that whereas MG132- and bortezomib-treated cells were still able to recover (albeit with modestly slower kinetics for the bortezomib-treated cells), YU101-treated cells failed to reinstate their proteasome activity (Fig 6A, solid lines). Thus, our data suggest that new protein synthesis is absolutely required for the recovery of proteasome activity that follows proteasome inhibition elicited by irreversible agents. Next, we asked if this recovery of proteasome activity proceeded in an Nrf1-dependent fashion. To this end, we compared WT and Nrf1-/- MEFs in their ability to recover after a transient exposure to MG132 or YU101. We found that whereas the WT MEFs were able to recover from both insults, the Nrf1-/- MEFs were impaired in their ability to recover when treated with YU101 (Fig 6B). Specifically, for the YU101 treatments, a single-phase exponential curve fit estimates that the time taken to recover half of the inhibited activity was 6.92 ± 1.64 hrs and 15.96 ± 5.49 hrs respectively for the WT and Nrf1-/- MEFs. These results underscore the importance of the Nrf1-dependent proteasome synthesis pathway for the cells to recover rapidly from covalent inhibition of the proteasome.
Fig 6. Loss of Nrf1 influences recovery of proteasome activity and cancer cell apoptosis after proteasome inhibition.
(A) HT29 cells were treated with either MG132 (2 μM), YU101 (100 nM) or Bortezomib (40 nM) for 1 hr after which the drugs were washed off and the cells were allowed to recover in the absence or presence of 50 μg/ml cycloheximide (CHX). Cells were frozen at different time points as indicated and were used for proteasome activity assays as described in Materials and Methods. For each time point, the results are normalized to DMSO-treated control in the case of hatched lines and to CHX-treated control in the case of solid lines. Error bars denote SD (n=3). (B) WT and Nrf1-/- MEFs were treated with either MG132 (50 μM) or YU101 (300 nM) for an hour after which the drugs were washed off and proteasome activity was measured for cells collected at different time points as indicated. The results were normalized to DMSO-treated control for each cell type. Error bars denote S.E.M. (n=2). (C) MDA-MB-231 and U2OS cells were transduced with retrovirus expressing either sh-Nrf1 or vector control, and 48 hrs later were seeded in to 96-well plates. The next day, the cells were treated with indicated concentrations of YU101 and the pan-caspase inhibitor Z-VAD-FMK, and further incubated for 72 hrs. The cell viability was then assessed using the CellTiter-Glo method. The results were normalized to DMSO-treated control which was set to 100% in each case. Error bars denote S.E.M. (n=2). (D) MDAMB-231 and U2OS cells were transduced with retrovirus expressing either sh-Nrf1 or vector control, and 72 hrs later were treated with indicated concentrations of YU101 for 48 hrs (MDA-MB-231) or 24 hrs (U2OS). The cell lysates were used for immunoblotting to detect the levels of Nrf1 (with the antibody raised against N-terminus) and cleaved caspase-3. β-actin protein levels were used as loading control.
Motivated by these results, we sought to investigate if our findings could have any bearing on cancer treatment strategies, especially in regimens involving proteasome inhibitors. Proteasome inhibition has recently emerged as a viable mode of anti-cancer therapy (Orlowski and Kuhn, 2008), and given the important role of Nrf1 in the accelerated recovery of proteasome activity, we reasoned that Nrf1 depletion could have a synergistic effect when employed in combination with a covalent proteasome inhibitor. In order to test this hypothesis, we knocked down Nrf1 in MDA-MB-231 breast cancer and U2OS osteosarcoma cell lines and subjected these to YU101 treatments. We found that depletion of Nrf1 sensitized both cell types to killing by YU101, and that this effect could be blunted by co-treatment with the pan-caspase inhibitor Z-VAD-FMK (Fig 6C), thereby implicating a role for caspases in this context. Consistent with this notion, we observed that YU101 treatment elicited enhanced level of cleaved caspase-3 in Nrf1-depleted cells (Fig 6D). Thus, our results point to Nrf1 as an intriguing target for development of anti-cancer drugs to be used in conjunction with covalent proteasome inhibitors.
DISCUSSION
In this study, we have identified the transcription factor Nrf1 as a mediator of the proteasome recovery (‘bounce-back’) response that is observed upon proteasome inhibition in mammalian cells. Nrf1 belongs to the cap ‘n’ collar basic leucine zipper (CNC-bZIP) family of proteins which also includes p45 NF-E2, Nrf2, Nrf3, Bach1, and Bach2 (Andrews et al., 1993; Kobayashi et al., 1999; Moi et al., 1994; Oyake et al., 1996). Although some of these factors, particularly Nrf2, have been studied in detail, relatively little is known about the biological functions of Nrf1. Nrf2 is a master transcription factor that is activated in response to oxidative stress, and helps maintain cellular redox homeostasis (Kensler et al., 2007). In the absence of oxidative stress, the transcriptional activity of Nrf2 is kept in check by the Keap1-Cul3-Rbx1 E3 cullin-RING ligase complex CRL3Keap1, which sequesters Nrf2 in the cytoplasm and subsequently targets it for degradation by the proteasome. In contrast, Keap1 affects neither the localization nor the transcriptional function of Nrf1, although both of these proteins can still interact with each other (Wang and Chan, 2006; Zhang et al., 2006).
Based on the high degree of sequence similarity with Nrf2 it is tempting to ascribe a redundant role for Nrf1 in antioxidant response. However, gene knockout experiments in mice paint a different picture. Whereas Nrf2 appears to be dispensable for vertebrate growth and development (Chan et al., 1996), deletion of Nrf1 in mice results in embryonic lethality at midgestation (Chan et al., 1998; Farmer et al., 1997). It is possible that the additional role that we have uncovered here for Nrf1 in PSM gene expression could explain the difference in outcome of the gene knockout studies. However, further experiments are necessary to test this hypothesis.
The yeast RPN4 has been implicated in both basal as well as stress induced synthesis of PSM genes (Dohmen et al., 2007). In contrast, while Nrf1 is required to mediate the bounce-back response after proteasome inhibition, its role in basal expression of PSM genes in unstressed mammalian cells appears to be rather modest. We were able to detect only a ~20% reduction in the mRNA levels of some PSM genes in Nrf1-/- MEFs when compared to the WT control (Fig 2B). Thus, our results point to the existence of other transcription factor(s) in maintaining basal levels of PSM gene expression in MEFs. It remains possible that the contribution of Nrf1 to basal expression of proteasome genes varies between different cell types.
Although both Nrf1 and Nrf2 can bind AREs, it is presently unclear why Nrf1 is the preferred mediator of PSM gene expression in response to proteasome inhibition. It could be that proteasome inhibitors, in addition to stabilizing both factors, mobilize additional signal(s) that result in selective deployment of Nrf1. Or it could simply be a cell-type dependent effect. In fact, in some cells, Nrf2 has been reportedly linked to proteasome subunit gene expression and/or proteasome activity (Arlt et al., 2009; Kraft et al., 2006; Kwak and Kensler, 2006; Kwak et al., 2003). For instance, it was shown that administration of antioxidant 3H-1,2-dithiole-3-thione (D3T) enhances the expression of several PSM genes in the liver of WT but not Nrf2-/- mice (Kwak et al., 2003). Also, another recent study, using samples obtained from colon cancer patients, attributed increased proteasome subunit protein expression and proteasome activity to higher nuclear levels of Nrf2 (Arlt et al., 2009). Furthermore, in human skin fibroblasts, Nrf2 was demonstrated to be responsible for inducing proteasome activity in response to a low dose of MG132 (Kraft et al., 2006). However, in our hands, we were unable to implicate Nrf2 in the bounce-back response. First, MLN4924, an inhibitor of the Nedd8 pathway, which is known to stabilize Nrf2 (Soucy et al., 2009), failed to induce PSM gene expression in two different human cancer cell lines, whereas it upregulated NQO1, a downstream target gene of Nrf2 (Fig 1). Second, and more importantly, Nrf2-/- MEFs were as proficient as their WT counterparts in inducing PSM gene expression in response to MG132 treatment (Fig 2). Nevertheless, it remains possible that the bounce-back response is mediated by Nrf1 in some cell types and Nrf2 in others.
Even with the limited knowledge that we now possess of the Nrf1-mediated bounce-back response, it seems reasonable to predict that this pathway could be dysregulated in a few scenarios. One such example is those cancers in which the proteasome is found to be overproduced. As early as 1990, it was observed that hematopoietic malignant tumor cells had higher levels of proteasome as well as PSM mRNA levels when compared to peripheral blood mononuclear cells (Kumatori et al., 1990). More recently, ovarian and breast cancer tissues were shown to exhibit higher levels of proteasome content and activity (Bazzaro et al., 2006; Chen and Madura, 2005). Also, a compilation of microarray data from different solid tumors revealed that up to 10 of the PSM genes are overexpressed when compared to normal tissues (Pilarsky et al., 2004). It could be that in this subset of cancers, the bounce-back response is constitutively active as opposed to its regular role as a “molecular rheostat” for sensing and upregulating proteasome levels only when the need arises. A subtle variation on this theme could be that there is a sustained greater demand for proteasome activity in cancer cells and this actually triggers the chronic activation of the bounce-back response. A recent study in yeast illustrates this point. A substantial fraction of yeast mutants that are aneuploid for single chromosomes display heightened sensitivity to MG132 (Torres et al., 2007). Although the RPN4 feedback loop was not monitored in these cells, a logical proposal is that chromosome aneuploidy (which is rampant in solid tumors) leads to an imbalance in the production of subunits for multi-protein complexes (Torres et al., 2007). These excess subunits are metabolized via the ubiquitin system, placing a greater load on the proteasome and leading to chronic dependence on elevated proteasome levels.
Regardless of the mechanism by which some cancer cells overproduce proteasome, if the Nrf1 pathway is found to be activated in some cancers, it would provide a strong rationale for targeting this node in anticancer therapeutic strategies. Compounds that antagonize Nrf1 activation or function could be particularly efficacious in combination therapies with irreversible proteasome inhibitors. One common problem that has been noted with both bortezomib (an FDA approved drug for the treatment of Multiple Myeloma) and carfilzomib (an experimental therapeutic) is that they are rapidly cleared from the patients’ blood within the first hour of their administration (Papandreou et al., 2004; Schwartz and Davidson, 2004). Thus, treatment with these compounds amounts to an in vivo pulse-chase experiment. Regardless of the extent of proteasome inhibition that occurs during this first hour, the inhibited state is transient because once the drug is cleared proteasome activity recovers through a combination of drug dissociating from the active site and new proteasome synthesis. Whereas the extent and duration of proteasome inhibition that is currently achieved may be good enough to kill multiple myeloma tumor cells, it may be insufficient to be effective in other cancers. This could explain the limited efficacy of bortezomib in a number of other cancer types. Although this problem can not be readily rectified for bortezomib because it dissociates from the chymotryptic site even if new synthesis is blocked (Fig. 6A), our in vitro data suggest that it may be possible to sustain profound inhibition of the proteasome for a longer duration with the irreversible inhibitor carfilzomib by attenuating the proteasome recovery response mediated by Nrf1 (Fig. 6B). This could expand the repertoire of cancer types that can be successfully treated with proteasome inhibitors.
One other potentially relevant observation that has been made in the clinic is the emergence of bortezomib resistance (Orlowski and Kuhn, 2008). Although this could be explained by a number of other mechanisms such as mutation of the drug binding site (Oerlemans et al., 2008) and amplification of selected proteasome subunit genes (Altavilla et al., 2009), one possibility that has been overlooked so far is the dysregulation of the bounce-back response. Further understanding of this pathway, in terms of the precise mechanism and elucidating the other molecular players involved may facilitate development of new therapeutics that target the ubiquitin proteasome system.
EXPERIMENTAL PROCEDURES
Constructs
The coding region of human Nrf1 was amplified from a full-length cDNA-containing plasmid (Open Biosystems) using primers 5′-CAC TCA CTG CGG CCG CT C TTT CTC TGA AGA AAT ACT TAA CGG AA-3′ (forward) and 5′- TCA CTT TCT CCG GTC CTT TGG C-3′ (reverse), digested with NotI and cloned in-frame in to the NotI-HpaI site of the pMSCV-hyg retroviral vector (Clontech) that was previously modified to encode an N-terminal 3xFLAG tag, resulting in the construct Flag-Nrf1 (RDB-2411). The construct Flag-Nrf1-HA (RDB-2412) was obtained as above except that the reverse primer (5′-TCA GGC GTA GTC GGG CAC GTC GTA GGG GTA CTT TCT CCG GTC CTT TGG C-3′) encoded an HA tag sequence.
The shRNA expression construct shNrf1 (RDB-2413) targeting Nrf1 was based on a 19-mer sequence (GGGATTCGGTGAAGATTTG) present in the coding region of both human and mouse genes and was cloned in to pSUPER.retro.puro (Oligoengine).
To obtain 3xPSMA4-ARE-Luc (RDB-2415), an oligo (5′-cgagccgtgggcacga TGACTCTGCA ccgcctcctctgagccgtgggcacga TGACTCTGCA ccgcctcctctgagccgtgggcacga TGACTCTGCA ccgcctcctctg-3′) containing three copies of a putative ARE (shown in upper-case) derived from the first intron of the human PSMA4 gene was annealed to its corresponding reverse-complement oligo and cloned into pGL3-promoter vector (Promega). The construct 3xPSMA4-mutARE-Luc (RDB-2416) was obtained as above except that the putative AREs were modified to TGACTCTAAA, where the mutation is shown underlined. To obtain the PSMB6-Luc construct, the ~3kb promoter region of the mouse PSMB6 gene was amplified using primers 5′-TGA TGG CTC ATC GCC ATC CAT-3′ (forward) and 5′-GGC CGC CAT CTT CCT CTG CTA-3′ (reverse) from mouse genomic DNA and cloned in to pGL3-Basic vector (Promega).
Cell Culture and Retroviral Transductions
Prostate cancer LNCaP, colon cancer HT29, breast cancer MDA-MB-231, osteosarcoma U2OS, and 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Atlanta Biologicals), penicillin and streptomycin (Invitrogen) at 37°C in 5% CO2. Mouse embryonic fibroblasts (MEFs) were grown as above except that the medium was additionally supplemented with β-mercaptoethanol and non-essential amino acids (Invitrogen). Primary MEFs derived from knock-out animals were used in Fig-2 and for all subsequent experiments, MEFs immortalized with an shRNA construct (RDB-2418; a kind gift from Dr. Mei-Ling Kuo, City of Hope, Duarte, CA) targeting p19ARF and selected for puromycin resistance were used.
For retroviral production, 293T cells were transfected with the required retroviral construct along with helper plasmids. Forty-eight hours after transfection, media supernatant containing the retrovirus was collected every 4-5 hrs for two days. This retrovirus-containing medium, supplemented with polybrene (10 μg/ml), was used to transduce the target cells.
Quantitative Reverse Transcription PCR (RT-PCR)
RNA was isolated using the RNeasy kit (Qiagen). cDNA was prepared using the Superscript III first strand synthesis kit (Invitrogen) according to the manufacturer’s recommendations. Quantitative PCR (qPCR) was performed using the SYBR GreenER supermix (Invitrogen). Primers used in these qPCRs are listed in Supplemental tables S1 and S2.
Immunoblot Analysis
Cells were lysed in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP40, 1% Na.Deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktail (Pierce). For detecting Nrf1, either a rabbit polyclonal antibody raised against the N-terminus (Chan et al., 1998) or a mouse polyclonal antibody specific for the C-terminus (Novus Biologicals) was used. Other immunoblots were performed with antibodies specific for Nrf2 (SantaCruz Biotechnology), Flag tag (Sigma-Aldrich), HA tag (Roche Diagnostics), cleaved caspase-3 (Cell Signaling), and β-actin (Sigma-Aldrich).
Luciferase Assays
Cells were transiently transfected with the firefly (promoter reporters) and renilla luciferase (pRL-TK; Promega) constructs along with effector plasmid as required. After harvesting the cells, luciferase assays were performed using the Dual Luciferase reporter assay system (E1910; Promega) according to the instructions from the manufacturer. The firefly luciferase activity was normalized to renilla luciferase activity for all experiments.
Proteasome Activity Recovery Assays
Cells seeded in 96-well plates were treated with different proteasome inhibitors for an hour at concentrations determined to inhibit proteasome activity by 80-90%. The cells were then washed with PBS thrice and allowed to recover in fresh medium. At definite time points, the cells were freeze-thawed in TE buffer (20 mM Tris pH 8, 5 mM EDTA) and subsequently used for measuring proteasome activity as described previously (Demo et al., 2007).
Cell Viability Assays
Cells in 96-well plates were treated with different agents as required and cell viability was assessed using the Cell-Titer Glo kit (G7572; Promega) according to the protocol recommended by the manufacturer. By employing a luminescence read-out, this kit quantitates the level of ATP which in turn is proportional to the number of viable cells.
HIGHLIGHTS.
Nrf1 mediates proteasome inhibitor-induced proteasome gene transcription in mammalian cells
Depletion of Nrf1 sensitizes cancer cells to covalent proteasome inhibitor treatments
Supplementary Material
ACKNOWLEDGEMENTS
We thank Millenium Pharmaceuticals for their generous gifts of bortezomib and MLN4924, Mark Smythe and Craig Crews for YU101, and Mei-Ling Kuo (City of Hope) for the p19ARF shRNA construct. We thank S. Materna for help with qPCR. We are grateful to A. Varshavsky and members of the Deshaies’ lab for critical reading of the manuscript. S.K.R. is supported by the Multidisciplinary postdoctoral award (W81XWH-07-1-0641) from the Department of Defense Breast Cancer Research Program. P.Y. is supported by the Swedish Research Council and the Swedish Cancer Society. J.Y.C. and C.S.L. are supported by grants (CA091907 and NS065223) from the National Cancer Institute and National Institute of Neurological Disorders and Stroke. R.J.D. is an HHMI Investigator, and this work was supported in part by HHMI and Weston Havens Foundation. Stemming from founder’s shares in Proteolix, R.J.D. retains rights to milestone payments from Onyx contingent upon FDA and EMEA approvals of carfilzomib.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altavilla G, Arrigo C, Marabello G, Galletti G, Santarpia M, Sauta M, Pitini V. Amplification and overexpression of the PSMB5 gene contributes to bortezomib resistance in retreatment of patients with multiple myeloma. J Clin Oncol (Meeting Abstracts) 2009;27:e19500. [Google Scholar]
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Arlt A, Bauer I, Schafmayer C, Tepel J, Muerkoster SS, Brosch M, Roder C, Kalthoff H, Hampe J, Moyer MP, et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009 doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- Bazzaro M, Lee MK, Zoso A, Stirling WL, Santillan A, Shih Ie M, Roden RB. Ubiquitin-proteasome system stress sensitizes ovarian cancer to proteasome inhibitor-induced apoptosis. Cancer Res. 2006;66:3754–3763. doi: 10.1158/0008-5472.CAN-05-2321. [DOI] [PubMed] [Google Scholar]
- Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2009 doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65:5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, Jiang J, Laidig GJ, Lewis ER, Parlati F, et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67:6383–6391. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Willers I, Marques AJ. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim Biophys Acta. 2007;1773:1599–1604. doi: 10.1016/j.bbamcr.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Farmer SC, Sun CW, Winnier GE, Hogan BL, Townes TM. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 1997;11:786–798. doi: 10.1101/gad.11.6.786. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap‘n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- Kraft DC, Deocaris CC, Wadhwa R, Rattan SI. Preincubation with the proteasome inhibitor MG-132 enhances proteasome activity via the Nrf2 transcription factor in aging human skin fibroblasts. Ann N Y Acad Sci. 2006;1067:420–424. doi: 10.1196/annals.1354.060. [DOI] [PubMed] [Google Scholar]
- Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, Tachikawa T, Shin S, Ichihara A. Abnormally high expression of proteasomes in human leukemic cells. Proc Natl Acad Sci U S A. 1990;87:7071–7075. doi: 10.1073/pnas.87.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Kensler TW. Induction of 26S proteasome subunit PSMB5 by the bifunctional inducer 3-methylcholanthrene through the Nrf2-ARE, but not the AhR/Arnt-XRE, pathway. Biochem Biophys Res Commun. 2006;345:1350–1357. doi: 10.1016/j.bbrc.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren J, Masson P, Mirzaei Z, Young P. Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol. 2005;25:4662–4675. doi: 10.1128/MCB.25.11.4662-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Oerlemans R, Franke NE, Assaraf YG, Cloos J, van Zantwijk I, Berkers CR, Scheffer GL, Debipersad K, Vojtekova K, Lemos C, et al. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112:2489–2499. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou CN, Daliani DD, Nix D, Yang H, Madden T, Wang X, Pien CS, Millikan RE, Tu SM, Pagliaro L, et al. Phase I trial of the proteasome inhibitor bortezomib in patients with advanced solid tumors with observations in androgen-independent prostate cancer. J Clin Oncol. 2004;22:2108–2121. doi: 10.1200/JCO.2004.02.106. [DOI] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Davidson T. Pharmacology, pharmacokinetics, and practical applications of bortezomib. Oncology (Williston Park) 2004;18:14–21. [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.