Abstract
OBJECTIVE
The mammalian target of rapamycin (mTOR)/p70 S6 kinase 1 (S6K1) pathway is a critical signaling component in the development of obesity-linked insulin resistance and operates a nutrient-sensing negative feedback loop toward the phosphatidylinositol 3-kinase (PI 3-kinase)/Akt pathway. Whereas acute treatment of insulin target cells with the mTOR complex 1 (mTORC1) inhibitor rapamycin prevents nutrient-induced insulin resistance, the chronic effect of rapamycin on insulin sensitivity and glucose metabolism in vivo remains elusive.
RESEARCH DESIGN AND METHODS
To assess the metabolic effects of chronic inhibition of the mTORC1/S6K1 pathway, rats were treated with rapamycin (2 mg/kg/day) or vehicle for 15 days before metabolic phenotyping.
RESULTS
Chronic rapamycin treatment reduced adiposity and fat cell number, which was associated with a coordinated downregulation of genes involved in both lipid uptake and output. Rapamycin treatment also promoted insulin resistance, severe glucose intolerance, and increased gluconeogenesis. The latter was associated with elevated expression of hepatic gluconeogenic master genes, PEPCK and G6Pase, and increased expression of the transcriptional coactivator peroxisome proliferator–activated receptor-γ coactivator-1α (PGC-1α) as well as enhanced nuclear recruitment of FoxO1, CRTC2, and CREB. These changes were observed despite normal activation of the insulin receptor substrate/PI 3-kinase/Akt axis in liver of rapamycin-treated rats, as expected from the blockade of the mTORC1/S6K1 negative feedback loop.
CONCLUSIONS
These findings unravel a novel mechanism by which mTORC1/S6K1 controls gluconeogenesis through modulation of several key transcriptional factors. The robust induction of the gluconeogenic program in liver of rapamycin-treated rats underlies the development of severe glucose intolerance even in the face of preserved hepatic insulin signaling to Akt and despite a modest reduction in adiposity.
The mammalian target of rapamycin (mTOR) is a Ser/Thr kinase that belongs to the phosphatidylinositol (PI) kinase-related protein kinase family (1). mTOR integrates signals from growth factors, hormones, nutrients, and cellular energy level to regulate protein translation and cell growth, proliferation, and survival (1). mTOR exists in two complexes. The mTOR complex 1 (mTORC1) is composed of mTOR, GβL, PRAS40, deptor, and raptor and regulates cell growth, whereas the complex 2 (mTORC2) consists of mTOR, GβL, protor-1, mSIN1, deptor, and rictor and controls cytoskeleton regulation and cell survival (1,2). mTORC1 activates cellular processes by phosphorylating two downstream effectors, p70 S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (3). Both mTORC1 and S6K1 are activated by insulin through the insulin receptor/insulin receptor substrate (IRS)/phosphatidylinositol 3-kinase (PI 3-kinase)/Akt pathway (4). We and others have shown that chronic activation of the mTORC1/S6K1 pathway by nutrients or prolonged insulin treatment promotes insulin resistance through increased IRS-1 serine phosphorylation, leading to a reduction in IRS-1 function and impaired activation of the PI 3-kinase/Akt pathway, thereby creating a negative feedback loop on insulin action. Accordingly, acute inhibition of mTORC1 by rapamycin was found to restore insulin action on the PI 3-kinase/Akt pathway and to prevent the insulin-resistant effects of excess nutrients on insulin-mediated glucose transport in muscle and adipose cells (3,5–7).
Because rapamycin protects from insulin resistance caused by excess nutrients in vitro, it was proposed that it may hold promise as a drug for treatment of insulin resistance in obesity. In fact, rapamycin is already used in the clinic as an effective immunosuppressant in human transplant recipients to prevent graft rejection (8). However, somewhat paradoxically it appears that a significant side effect of chronic inhibition of mTORC1/S6K1 signaling by rapamycin treatment is deranged lipid and glucose metabolism (9). Indeed chronic rapamycin administration was found to cause hyperlipidemia and to reduce fat mass, as well as to promote glucose intolerance and a diabetes-like syndrome (8,10,11). However, the underlying mechanisms behind these metabolic derangements remain unknown and particularly intriguing when considering that rapamycin exerts positive metabolic actions when used in vitro in insulin target cells.
In the present study, we have evaluated the effects of chronic rapamycin treatment in rats and explored potential mechanisms by which this mTORC1 inhibitor impacts on lipid and glucose metabolism. We show that rapamycin treatment reduces fat deposition in white adipose tissue through altered expression of key enzymes required for fatty acid uptake and triglyceride synthesis. Despite such loss in fat mass, rapamycin treatment also causes insulin resistance and glucose intolerance, in part through inducing hepatic gluconeogenesis by enhancement of the expression of gluconeogenic enzymes and nuclear recruitment of important gluconeogenic transcriptional regulators. This hepatic phenotype was observed despite preservation of insulin signaling through the IRS/PI 3-kinase/Akt pathway as expected from the ability of rapamycin to block the mTORC1/S6K1-dependent phosphorylation of IRS-1 on serine residues. Our data thus provide a mechanism to explain the higher prevalence of glucose intolerance and a diabetes-like syndrome in rapamycin-treated patients who underwent a transplant and further highlight the importance of the mTORC1/S6K1 pathway in modulating glucose homeostasis.
RESEARCH DESIGN AND METHODS
Animals.
Animal handling and treatment were approved by the Animal Care and Handling Committee of Laval University. Male Sprague-Dawley rats (200 g) were purchased from Charles River Laboratories (St. Constant, QC, Canada) and housed individually in cages in a room kept at 23 ± 1°C with a 12-h light/12-h dark cycle. After 4-day adaptation, rats were matched by weight and divided into control and rapamycin-treated groups. Vehicle (0.1% Me2SO, 0.2% carboxymethylcellulose) or rapamycin (2 mg/kg/day; Biomol, Plymouth-Meeting, PA) was injected intraperitoneally once daily. Because the mTORC1 complex acts as a cellular nutrient sensor, we performed experiments (except where indicated) on rats that had been fasted for 12 h followed by 3 h of refeeding to achieve physiological activation of insulin signaling. After 15 days of treatment, control and rapamycin-treated rats were killed by decapitation. Tissues and blood were rapidly harvested and frozen in liquid nitrogen.
Intraperitoneal glucose, insulin, and pyruvate tolerance tests.
Rats were fasted for 6 h and injected intraperitoneally with glucose (2 g/kg) or insulin (2.5 units/kg; human insulin; Eli Lilly, Toronto, ON, Canada) diluted in saline 0.9%. For the pyruvate tolerance test, rats were fasted for 12 h and refed for 3 h before being injected with pyruvate (2 g/kg; Sigma-Aldrich, Oakville, ON, Canada) diluted in saline 0.9%. Blood was collected from the tail tip prior to and at various times after injection.
Plasma/tissue determinations.
Blood glucose levels were measured by a Precision PCx glucometer (MediSense; Abbott Laboratories, Mississauga, ON, Canada). Plasma insulin, C-peptide, and glucagon were determined by radioimmunoassay (Linco Research, St. Charles, MO). Plasma adiponectin and leptin were measured by ELISA following the suppliers' recommendations (ALPCO Diagnostics, Salem, NH; Millipore, Nepean, ON, Canada). Triglycerides in plasma and in muscle/liver lipid extracts (Roche Diagnostics, Montreal, QC, Canada) and nonesterified fatty acids (WakoChemicals, Richmond, VA), glycerol (Sigma-Aldrich), and lactate (Eton Bioscience, Hayward, CA) levels were measured by enzymatic methods according to the manufacturers' instructions.
Immunoblotting and immunoprecipitation.
Tissue samples were homogenized in buffer, subjected to SDS-PAGE, and transferred to nitrocellulose membranes as previously described (3). For immunoprecipitation of IRS-1 and IRS-2, liver samples were homogenized in buffer (20 mmol/l Tris, pH 7.5, 140 mmol/l NaCl, 1% Nonidet P-40, 2 mmol/l Na4P2O7, 10 mmol/l NaF, 2 mmol/l Na3VO4, and protease inhibitors). Equal amounts of protein (1 mg) were immunoprecipitated with protein A–Sepharose and subjected to SDS-PAGE as described above. Antibodies used for immunoblotting are listed in supplemental Table 1, available in an online appendix at http://diabetes.diabetesjournals.org/content/early/2010/03/10/db09-1324/suppl/DC1. Densitometric analysis was performed with ImageQuant TL software (GE Healthcare, Little Chalfont, U.K.).
Nuclear extracts.
Liver nuclear extracts were prepared as previously described (12) and submitted to immunoblotting as described above.
Kinase assays.
PI 3-kinase activity was measured in IRS-1 or IRS-2 immunoprecipitates, and Akt kinase activity was measured in Akt immunoprecipitates as previously described using a commercial peptide (crosstide) (3).
RNA extraction and quantitative PCR analysis.
RNA extraction and quantitative PCR analysis were performed as described previously (13). The primers used are listed in supplemental Table 2. Data are expressed as the ratio between the expression of the target gene and the housekeeping gene 36B4 (also known as ARBP), the expression of which was not significantly affected by treatment.
Adipose tissue DNA content and adipocyte diameter.
Adipocyte diameter was measured as described previously (14). Tissue DNA content was determined using the DNeasy tissue kit (QIAGEN, Mississauga, ON, Canada) following the manufacturer's instructions.
Morphometric analysis of islets.
Islet size and β-cell mass were measured as previously described (15).
MIN6 cells.
MIN6 cells at 70% confluence were seeded in 24-well plates 1 day before treatments. On the day of the experiment, cells were incubated for 2 h in Krebs-Ringer bicarbonate HEPES buffer containing 2.8 mmol/l glucose before incubation for 2.5–30 min at 10 mmol/l glucose. Culture medium was collected and assayed for insulin content by ELISA (ALPCO Diagnostics).
Statistical analysis.
ANOVA was performed using the general linear model procedure of Statistical Analysis Software (SAS, Version 9.2; SAS Institute, Cary, NC). When appropriate, initial body weight was included in the statistical model as a covariate.
RESULTS
Chronic rapamycin treatment reduces adiposity and causes hyperlipidemia.
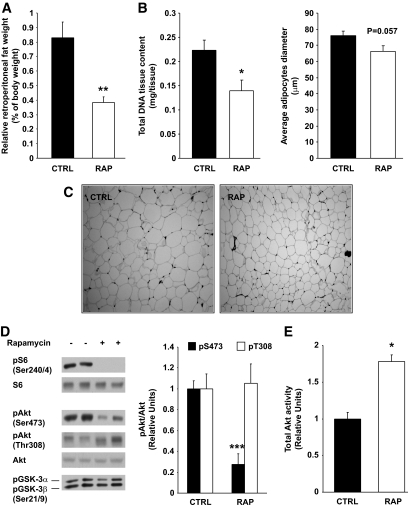
Rapamycin treatment significantly reduced (86%) body weight gain, which could be partly attributed to reduced (15%) food intake (Table 1). Rapamycin also decreased food efficiency by 84% and plasma leptin levels by 52%, suggesting that the weight-reducing effect of the drug is explained mostly by increased energy expenditure (Table 1). This is consistent with the recent finding that disruption of adipose mTORC1 signaling through raptor knockout increased energy expenditure (16). Accordingly, we found that rapamycin did not affect the amount of feces calories per gram of food ingested, indicating that intestinal lipid absorption was not affected by the drug (data not shown). Whereas the absolute mass of all tissues sampled was significantly decreased in rapamycin-treated rats (supplementary Table 3) on a relative mass basis (tissue mass/body weight), only white adipose tissue was significantly reduced compared with control rats, suggesting that rapamycin treatment preferentially impaired adipogenesis beyond its effect on body growth (supplementary Table 3 and Fig. 1A). This is supported by both the strong tendency toward smaller adipocyte diameter (P = 0.057, Fig. 1B and C) and significantly fewer adipocytes (lower DNA content per depot, Fig. 1B and C) in the rapamycin-treated group relative to control. In line with these observations, chronic rapamycin treatment increased plasma triglycerides (32%) and nonesterified fatty acids (65%) (Table 1), suggesting that adipose triglyceride deposition was impaired.
TABLE 1.
Effect of 15-day rapamycin treatment on weight, food intake, and metabolic parameters of treated rats
| Control | Rapamycin | |
|---|---|---|
| Body weight (g) | 303.6 ± 3.68 | 226.3 ± 4.34*** |
| Weight gain (g) | 91.4 ± 3.57 | 12.8 ± 3.87*** |
| Food intake (g) | 256.4 ± 6.53 | 217.7 ± 8.53** |
| Food efficiency (body weight gain/food intake) | 0.117 ± 0.004 | 0.019 ± 0.006*** |
| Glucose (mmol/l) | 8.5 ± 0.33 | 13.3 ± 2.53* |
| Insulin (pmol/l) | 351.5 ± 58.7 | 1,265.1 ± 395.1* |
| Glucagon (ng/l) | 127.25 ± 10.65 | 130.47 ± 14.33 |
| Triglycerides (mmol/l) | 0.644 ± 0.050 | 0.848 ± 0.060* |
| NEFAs (mmol/l) | 0.115 ± 0.020 | 0.190 ± 0.040* |
| Glycerol (mg/dl) | 1.025 ± 0.084 | 1.177 ± 0.107 |
| Lactate (mmol/l) | 0.904 ± 0.050 | 0.852 ± 0.039 |
| Adiponectin (μg/ml) | 11.89 ± 1.53 | 12.11 ± 0.83 |
| Leptin (ng/ml) | 9.48 ± 1.86 | 4.94 ± 1.51* |
Data are average ± SE (n = 12 control, n = 10 for rapamycin). Refed plasma data are average ± SE (n = 6–12 for control, n = 6–10 for rapamycin).
*P ≤ 0.05,
**P ≤ 0.01,
***P ≤ 0.001 vs. control. NEFAs, nonesterified fatty acids.
FIG. 1.
Chronic rapamycin treatment decreases adiposity. Sprague-Dawley rats were treated with vehicle or rapamycin (2 mg/kg/day) for 15 days. A: Relative retroperitoneal fat weight. Total DNA tissue content and adipocyte diameter (μm) (B) and representative images of retroperitoneal fat (C) from control and rapamycin-treated rats (magnification ×10) D: Representative Western blots of adipose tissue lysates are shown for phosphorylated S6 (Ser240/244), Akt (Ser473 and Thr308), GSK-3α/β (Ser21/9), and total proteins (two representative animals of six). The graphs depict densitometric analysis of normalization of phospho-Akt/Akt protein. E: Adipose tissue proteins (500 μg) were immunoprecipitated with total Akt antibody. Immunoprecipitates were analyzed for Akt activity. The graphs depict densitometric analysis of total Akt activity. n = 6 for each group. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. CTRL, control; RAP, rapamycin.
Because clinical studies have reported that rapamycin treatment causes hyperlipidemia and reduces body fat mass, it was deemed important to explore the potential underlying mechanisms by which the mTORC1 inhibitor may affect adipose tissue growth and metabolism. mTORC1 and, more recently, raptor have been shown to control adipogenesis (17) through Akt-mediated phosphorylation of tuberous sclerosis complex 2 (18). However, although rapamycin is rather selective for mTORC1, prolonged treatment with the drug can also inhibit mTORC2 and Akt (19). We thus explored whether chronic rapamycin treatment decreased adipogenesis indirectly through inhibition of mTORC2 and Akt. As expected, the phosphorylation of S6 (Ser240/244), a downstream substrate of the mTORC1/S6K1 pathway, was blunted in adipose tissue by rapamycin (Fig. 1D). Furthermore, Akt phosphorylation on Ser473 was severely inhibited in rapamycin-treated rats (Fig. 1D) indicating that mTORC2 may also have been inhibited by chronic drug exposure. In contrast, however, Akt Thr308 phosphorylation was not decreased in adipose tissue of rapamycin-treated rats (Fig. 1D) and Akt kinase activity was actually increased (Fig. 1E), suggesting that phosphorylation on Thr308 more than compensates for the reduction in Ser473 phosphorylation. This is supported by the fact that phosphorylation of the Akt substrate glycogen synthase kinase-3 (GSK-3) was not reduced in adipose tissue of rapamycin-treated rats (Fig. 1D). Thus, chronic rapamycin administration likely affected adiposity via mTORC1 but not mTORC2 inhibition.
Chronic rapamycin treatment coordinately downregulates genes required for lipid uptake and storage in adipose tissue.
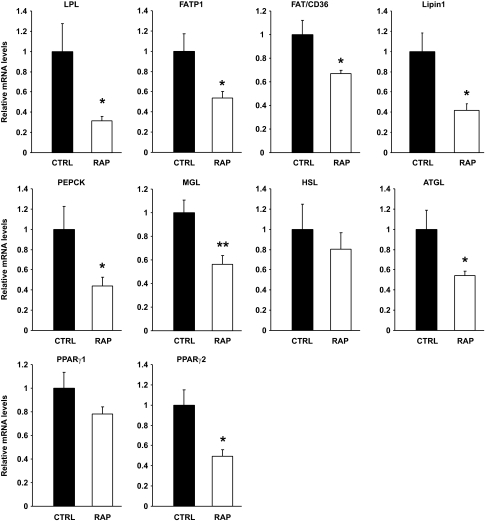
To better understand the mechanism by which chronic rapamycin treatment impairs adipogenesis and causes hyperlipidemia, we next measured the mRNA levels of several genes involved in the clearance of lipids from the circulation and their deposition in adipose tissue as triglycerides. Retroperitoneal adipose tissue expression of lipoprotein lipase (LPL), which catalyzes the hydrolysis of lipoprotein-bound triglycerides supplying fatty acids and glycerol for adipose depot uptake (20), along with the fatty acid transporters (FATP1 and FAT/CD36) were all downregulated by rapamycin treatment (Fig. 2). In addition, phosphoenolpyruvate carboxykinase (PEPCK), which generates the glycerol 3-phosphate backbone for fatty acid esterification (21), and lipin 1, an important enzyme involved in triglyceride synthesis (22), were also significantly reduced by rapamycin treatment. We also evaluated the mRNA levels of the enzymes adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoacyglycerol lipase (MGL), which catalyze the sequential hydrolysis of triglycerides to fatty acids and glycerol (23). Chronic rapamycin treatment markedly reduced the expression of ATGL and MGL without affecting those of HSL, excluding possible participation of increased lipolysis to the adipose tissue phenotype and hyperlipidemia. Interestingly, the reduced expression of genes involved in lipid uptake and storage and lipolysis was associated with a significant decrease in the mRNA levels of peroxisome proliferator-activated receptor-γ2 (PPARγ2), a master regulator of adipogenesis, lipogenesis, and lipolysis (Fig. 2) (13,20,24).
FIG. 2.
Chronic rapamycin treatment coordinately downregulates genes required for triglyceride hydrolysis, fatty acid transport, and esterification in adipose tissue. Rats were treated with rapamycin as described in the legend to Fig. 1, and adipose tissue was sampled and processed as described in the research design and methods section for determinations of LPL, FATP1, FAT/CD36, Lipin1, PEPCK, MGL, HSL, ATGL, PPARγ1, and PPARγ2 mRNA expression. The graphs depict mRNA expression in the adipose tissue of target genes corrected for the expression of 36B4 as a control gene. n = 12 for each group. *P ≤ 0.05, **P ≤ 0.01.
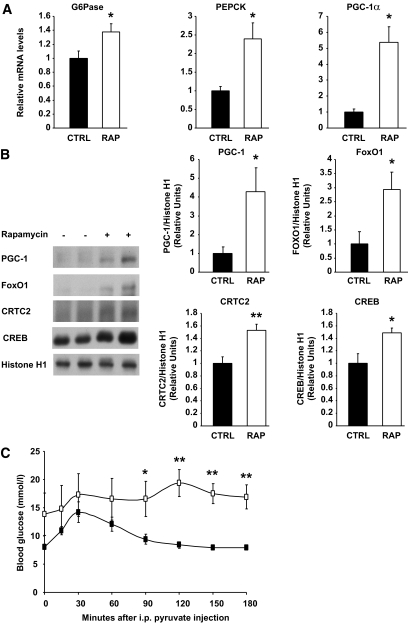
Chronic rapamycin treatment impairs glucose tolerance, β-cell mass, insulin clearance, and insulin sensitivity and results in higher expression of crucial elements of the gluconeogenic pathway in the liver.
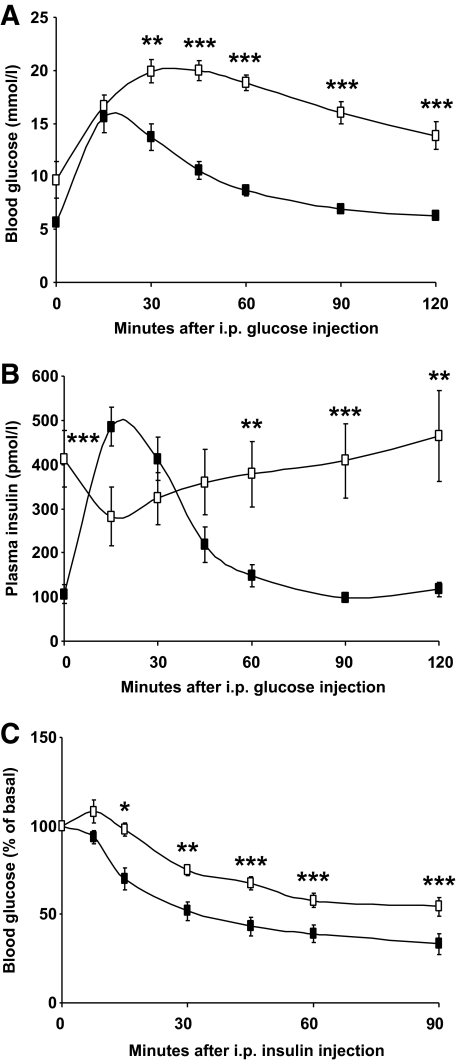
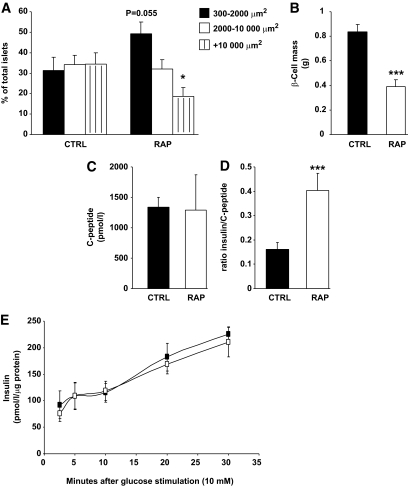
Treatment with rapamycin markedly affected glucose and insulin homeostasis as reflected by hyperglycemia and hyperinsulinemia (Table 1). Furthermore, rapamycin led to an exaggerated response to glucose administration during a glucose tolerance test, indicative of impaired glucose tolerance (Fig. 3A). The insulin levels during the glucose tolerance test were very high in the rapamycin-treated group and did not return to basal levels as they did in the control group (Fig. 3B). The glycemic response to insulin was also reduced in rapamycin-treated rats (Fig. 3C). However, plasma adiponectin, glucagon, glycerol, and lactate were not altered by the inhibitor (Table 1). The insulin response during the glucose tolerance test suggested a possible defect in islet function. Indeed, as shown in Fig. 4A, islet histology showed that rapamycin-treated rats had more small islets and fewer large islets compared with control rats as well as reduced β-cell mass (Fig. 4B). Nevertheless, plasma C-peptide levels were unchanged 3 h after refeeding, although the elevated ratio of insulin/C-peptide suggests that a marked impairment of hepatic insulin clearance underlies the hyperinsulinemic state of rapamycin-treated rats (Fig. 4C and D). To test whether rapamycin directly affected β-cell function, we also tested the effect of the drug on glucose-induced insulin secretion in MIN6 cells but found no alterations (Fig. 4E).
FIG. 3.
Chronic rapamycin treatment induces glucose and insulin intolerance in rats. Rats were treated with rapamycin as described in the legend to Fig. 1 and fasted for 6 h before intraperitoneal tolerance tests. Plasma glucose (A) and insulin levels (B) were measured during a glucose tolerance test. C: Plasma glucose levels were measured during an insulin tolerance test. n = 12 for each group. Black squares: CTRL; white squares: RAP. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
FIG. 4.
Chronic rapamycin treatment impairs β-cell mass and insulin clearance in rats. Rats were treated with rapamycin as described in the legend to Fig. 1. Pancreatic sections were stained with an antibody against insulin for the determination of islet size and β-cell mass as described in the research design and methods section. A: Size distribution of islets expressed as percentage of total islets. n = 10 for each group. B: Pancreatic β-cell mass. n = 6 for each group. C: Plasma C-peptide levels. n = 12. D: Ratio of insulin over C-peptide, a measure of insulin clearance. n = 12 for each group. E: Insulin secretion was determined in MIN6 cells treated with rapamycin (25 nmol/l; white squares) or vehicle (black squares) for 24 h followed by glucose stimulation (10 mmol/l) for various time points. n = 3 independent experiments. *P ≤ 0.05, ***P ≤ 0.001.
Given the severity of glucose intolerance, we first focused on the contribution of the liver to the diabetes-like phenotype of rapamycin-treated rats. We studied the mRNA and protein expression of transcriptional regulators of hepatic gluconeogenesis and key enzymes under their control. Notably, chronic rapamycin treatment resulted in greatly elevated liver expression of PPARγ coactivator-1α (PGC-1α) mRNA as well as higher expression of the two key gluconeogenic enzymes, PEPCK and glucose-6-phosphatase (G6Pase) (Fig. 5A). Nuclear localization of forkhead box O1 (FoxO1), PGC-1, cAMP response element-binding protein (CREB)–regulated transcription coactivator 2 (CRTC2; also known as TORC2), and CREB was also significantly enhanced after chronic rapamycin treatment (Fig. 5B). We also performed a pyruvate tolerance test to provide functional evidence that the increased gluconeogenic gene expression was associated with elevated gluconeogenesis. As shown in Fig. 5C, the blood glucose level of rapamycin-treated rats was different from control rats for the last 90 min of the test, suggesting increased hepatic gluconeogenesis. Neither liver glycogen (data not shown) nor triglyceride (Table 2) content was altered by rapamycin treatment. To confirm that rapamycin could directly alter hepatic glucose metabolism, we treated FAO hepatoma cells with rapamycin for 24 h and observed a marked augmentation of glucose production in the basal state as well as in the presence of insulin (supplementary Fig. 1A and B).
FIG. 5.
Chronic rapamycin treatment induces glucose intolerance by upregulating gluconeogenesis in rats. Rats were treated with rapamycin as described in the legend to Fig. 1. A: G6Pase, PEPCK, and PGC-1α mRNA expression. The graphs depict mRNA expression in the liver of target genes corrected for the expression of 36B4 as a control gene. B: Representative Western blots of PGC-1, FoxO1, CRTC2, and CREB proteins in nuclear extracts prepared from liver samples (two representative animals of six are shown). The graphs depict densitometric analysis of normalization of total protein/Histone H1 protein. n = 6 for each group. C: Plasma glucose levels measured during a pyruvate tolerance test on rats fasted for 12 h followed by 3 h of refeeding. n = 6 for each group. Black squares: CTRL; white squares: RAP. *P ≤ 0.05, **P ≤ 0.01.
TABLE 2.
Effect of 15-day rapamycin treatment on muscle and liver triglycerides
| Control | Rapamycin | |
|---|---|---|
| Muscle triglycerides (mmol/g tissue) | 9.52 ± 0.92 | 10.40 ± 0.93 |
| Liver triglycerides (mmol/g tissue) | 18.69 ± 4.02 | 12.94 ± 0.95 |
Data are average ± SE (n = 6 for each group).
Chronic rapamycin treatment improves insulin signaling despite enhancing hepatic gluconeogenesis.
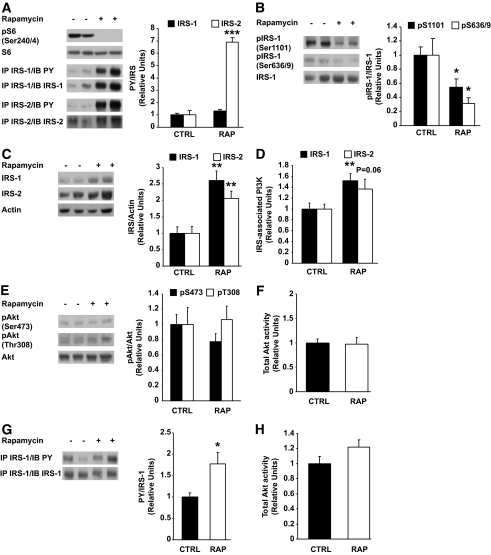
Insulin inhibits hepatic glucose production at least in part through its ability to bind and activate the insulin receptor, leading to the phosphorylation of IRS proteins and subsequent activation of PI 3-kinase and Akt. Because Akt negatively modulates hepatic gluconeogenesis through phosphorylation of FoxO1 (25), it was of major interest to test whether insulin signaling to Akt was impaired in the liver of rapamycin-treated rats. We found, however, that rapamycin-mediated inhibition of the mTORC1/S6K1 pathway, as revealed by complete loss of S6 Ser240/244 phosphorylation, improved insulin signaling in the liver, as shown by increased tyrosine phosphorylation of both IRS-1 and IRS-2 (Fig. 6A) and, conversely, diminished inhibitory phosphorylation of IRS-1 on serine residues (Ser1101 and Ser636/639) (Fig. 6B). IRS-1 and IRS-2 protein expression was also increased in the liver of rapamycin-treated rats (Fig. 6C). Further downstream in the insulin signaling cascade, we found that hepatic IRS-1 (P < 0.01)– and IRS-2 (P = 0.06)–associated PI 3-kinase activity was increased by rapamycin (Fig. 6D), whereas Akt phosphorylation on Ser473 and Thr308 as well as Akt kinase activity were not affected (Fig. 6E and F). We also found that tyrosine phosphorylation of IRS-1 was increased (Fig. 6G), whereas Akt kinase activity was not affected (Fig. 6H) in skeletal muscle of rapamycin-treated rats. Moreover, rapamycin treatment did not affect muscle triglyceride content (Table 2).
FIG. 6.
Chronic rapamycin treatment improves insulin signaling in liver and muscle. Rats were treated with rapamycin as described in the legend to Fig. 1. A: Representative Western blots of liver lysates are shown for phosphorylated S6 (Ser240/244) and phospho-tyrosine on IRS-1 or IRS-2 immunoprecipitates (1 mg) (two representative animals of six). The graphs depict densitometric analysis of normalization of phospho-tyrosine/IRS protein. B: Representative Western blots of phosphorylated IRS-1 (Ser1101 and Ser636/639) and total proteins (two representative animals of six are shown) in liver lysates. The graphs depict densitometric analysis of normalization of phospho-IRS-1/IRS-1 protein. C: Representative Western blots of total IRS-1 and IRS-2 protein in liver lysates. The graphs depict densitometric analysis of normalization of total IRS/Actin. D: Equal amounts of liver protein (1 mg) were immunoprecipitated with IRS-1 and IRS-2 antibodies. Immunoprecipitates were analyzed for PI 3-kinase activity. The graphs depict densitometric analysis of IRS-1– and IRS-2–associated PI 3-kinase activity. E: Representative Western blots of liver lysates are shown for phosphorylated Akt (Ser473 and Thr308) and total proteins (two representative animals of six). The graphs depict densitometric analysis of normalization of phospho-Akt/Akt protein. F: Liver proteins (500 μg) were immunoprecipitated with total Akt antibody. Immunoprecipitates were analyzed for Akt activity. The graphs depict densitometric analysis of total Akt activity. G: Representative Western blots of muscle lysates are shown for phospho-tyrosine on IRS-1 immunoprecipitates (1 mg). The graphs depict densitometric analysis of IRS-1 phospho-tyrosine corrected for IRS-1 protein content. n = 6 animals. H: Muscle proteins (500 μg) were immunoprecipitated with total Akt antibody. Immunoprecipitates were analyzed for Akt activity and the graph depicts the densitometric analysis of several independent determinations. n = 6 for each group. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
DISCUSSION
The mTORC1/S6K1 pathway has recently emerged as a critical signaling component in the development of obesity-linked insulin resistance by operating a negative feedback loop toward PI 3-kinase/Akt through increasing inhibitory serine phosphorylation of IRS-1 (4–6,26). Accordingly, we and others have demonstrated that acute treatment with rapamycin in vitro prevents nutrient-induced insulin resistance by interfering with this negative feedback loop and limiting phosphorylation of IRS-1 on multiple serine residues (3,5,7,27). Moreover, we recently reported that S6K1 plays a key role in hepatic insulin action through phosphorylation of both Ser1101 and Ser636/639 in hepatic cells and liver of obese insulin-resistant high-fat–fed mice (6). Other studies have shown that long-term activation of the mTORC1/S6K1 pathway also promotes insulin resistance through the proteasomal degradation of IRS-1 (28) and IRS-2 (29). In the present study, we found that chronic treatment with rapamycin also improves insulin signaling to IRS/PI 3-kinase and maintains activation of Akt in the liver, in agreement with our previous findings in hepatocytes acutely treated with the drug (4). Accordingly, we found that chronic inhibition of the mTORC1/S6K1 pathway in rats increases IRS-1 and IRS-2 protein expression, enhances their tyrosine phosphorylation while reducing IRS-1 inhibitory phosphorylation on Ser1101 and Ser636/639. This led to upregulation of IRS-dependent PI 3-kinase activation and normal Akt activity in liver, despite the development of severe whole-body glucose intolerance and insulin resistance. This apparent uncoupling between hepatic insulin signaling and glucose homeostasis is in large part explained by augmented gluconeogenesis, unraveling a novel mechanism of regulation of hepatic glucose production by the mTORC1/S6K1 pathway.
Liver gluconeogenesis is driven by the availability of gluconeogenic substrates and the activity of two important gluconeogenic enzymes, PEPCK and G6Pase (30). In the refed state, insulin suppresses gluconeogenesis by transcriptional downregulation of PEPCK and G6Pase (31). Our data indicate that rapamycin upregulates gluconeogenesis in the refed state by increasing the expression of PEPCK and G6Pase. This was observed despite hyperinsulinemia and without any change in the concentrations of circulating glucagon or major gluconeogenic substrates. We also found that rapamycin markedly increases PGC-1α mRNA expression, which likely contributes to the upregulation of the gluconeogenic enzymes, as observed in type 2 diabetes (32). Because rapamycin blocks a major nutrient-sensing pathway, it has been suggested that it mimics a starvation-like signal even in the presence of nutrients (33) and as such could explain the increase in PGC-1α expression. This is the first report of PGC-1α regulation by mTORC1/S6K1 in liver but recent studies have linked this nutrient-sensing pathway to PGC-1α and the regulation of skeletal muscle metabolism (34–36).
PGC-1α increases gluconeogenic gene expression at least in part by coactivating transcription by FoxO1 (37), possibly through its ability to target O-GlcNAc transferase for GlcNAcylation of FoxO1 (38). Conversely, insulin suppresses gluconeogenesis through Akt-dependent phosphorylation of FoxO1, leading to its nuclear exclusion and degradation (39). We found that chronic rapamycin treatment increases FoxO1 nuclear content where it activates the transcription of PEPCK and G6Pase (40). Thus, FoxO1 nuclear exclusion was not observed, despite the presence of hyperinsulinemia and the maintenance of normal hepatic insulin signaling to Akt in rapamycin-treated rats. Further studies will be needed to test whether rapamycin increases gluconeogenic gene transcription via a PGC1α-dependent GlcNAcylation of FoxO1, thus bypassing insulin-induced Akt activation and phosphorylation of FoxO1.
Interestingly, rapamycin treatment increases nuclear recruitment of both CREB and CRTC2. In response to glucagon and other cAMP-elevating hormones, CRTC2 binds to CREB and stimulates the gluconeogenic program by triggering the dephosphorylation and nuclear translocation of CRTC2, whereas insulin blunts this process through activation of salt-inducible kinase 2, which phosphorylates and induces the cytoplasmic translocation and proteasomal degradation of CRTC2 (41,42). CREB may also promote gluconeogenesis through increased PGC-1α expression (43). The observation that both nuclear CRTC2 and FoxO1 remained elevated 3 h after refeeding in the liver of rapamycin-treated rats likely explains the robust activation of gluconeogenesis in these animals. A recent study by Liu et al. (44) has demonstrated the existence of a temporal switch from CRTC2 to FoxO1 as a critical inducer of gluconeogenesis during a prolonged fast. This switch is operated by a complex series of acetylation and deacetylation events that affect stability and activity of both CRTC2 and FoxO1. The fact that both of these redundant transcriptional factors are activated even after refeeding of rapamycin-treated rats, and in the presence of a fully operative hepatic insulin signaling pathway, suggests that mTORC1/S6K1 exerts a previously unsuspected powerful control over the gluconeogenic program.
We also found that rats treated chronically with rapamycin have a decreased β-cell mass. This is consistent with recent reports that chronic activation of mTOR leads to β-cell mass expansion (15,45). Despite the reduced β-cell mass, insulin secretion as revealed by C-peptide levels after refeeding was not altered in rapamycin-treated rats, suggesting that their hyperinsulinemic state is explained rather by impaired hepatic insulin clearance. Although the reduced β-cell mass could explain the lack of further insulin release during the glucose tolerance test, two observations suggest that the liver defects precede the development of β-cell dysfunction in this model. First, we found that liver gluconeogenesis was affected already after only 2 days of rapamycin treatment (supplementary Fig. 2) even before hyperglycemia develops. Second, insulin secretion by MIN6 cells is not altered by rapamycin treatment for up to 48 h. Nevertheless, the reduction in β-cell mass suggests that chronic exposure to rapamycin may eventually cause major perturbations in β-cell function and further contribute to the diabetes-like syndrome induced by the mTOR inhibitor.
Reduced accretion of body fat and hyperlipidemia constitute another important phenotypic feature of rapamycin-treated rats. The inhibitor appears to affect adiposity mainly through a reduction in cell number, with a small contribution from reduced adipocyte size. This is consistent with studies in cell lines in which chronic rapamycin treatment or small interfering RNA–mediated downregulation of either mTOR, raptor, or S6K1 can have direct effects on adipogenesis and lipid content (17,46,47). The gene expression data suggest a robust reduction in lipid flux in and out of adipose tissue. Hence, the coordinated downregulation of the expression of enzymes required for intravascular hydrolysis (LPL), cellular uptake (FAT/CD36 and FATP1), and storage of lipid (PEPCK and lipin), all known to impact adiposity (48), appeared to be counterbalanced by the downregulation of lipolytic (ATGL, MGL) enzyme expression, consistent with the near maintenance of cell size in rapamycin-treated rats. Reduced lipid uptake and fat cell number impairs the capacity of adipose tissue for plasma lipid clearance, which likely contributes to hyperlipidemia.
Several lines of evidence suggest that the rapamycin-induced decrease in the expression of lipid metabolism genes in adipose tissue might be the result of a reduced transcriptional activity of PPARγ. Indeed, all such genes are known to be positively modulated by PPARγ (13,20,24). Furthermore, rapamycin inhibits PPARγ transcriptional activity in adipocytes (46), and adipose-specific disruption of raptor in mice reduces adiposity (16). Conversely, activation of mTORC1 increases PPARγ expression in adipocytes (18). In the present study, rapamycin reduced the expression of both PPARγ2 and lipin 1, a key lipogenic enzyme and PPARγ coactivator (22). Collectively, these congruent findings strongly suggest a molecular link between mTORC1 and PPARγ transcriptional activity.
We also considered the possibility that the effects of rapamycin on PPARγ expression and adipogenesis might be linked to secondary inhibition of mTORC2, leading to a reduction of Akt activation. However, the reduced Akt Ser473 phosphorylation was possibly compensated by normal phosphorylation of Thr308 because Akt kinase activity and GSK-3 phosphorylation were not reduced in adipose tissue of rapamycin-treated rats. GSK-3 phosphorylation was recently found to be preserved despite complete loss of Akt Ser473 phosphorylation in mouse embryo fibroblasts from rictor knockout animals (49). Our conclusion that mTORC2 inhibition cannot account for the reduction of adiposity is also supported by recent studies in mice with adipose tissue–specific disruption of rictor and mTORC2 signaling, which failed to reduce adipogenesis or fat mass in chow-fed animals (50).
In conclusion, chronic inhibition of the mTORC1/S6K1 pathway by rapamycin causes glucose intolerance mainly through inducing transcriptional activation of gluconeogenic genes via the coordinated activation of PGC-1α, CRTC2, CREB, and FoxO1. The robust activation of the gluconeogenic program was observed even under hyperinsulinemia in the refed state, and despite the maintenance of normal activation of hepatic insulin signaling to Akt owing to the blockade of the mTORC1/S6K1 inhibitory feedback loop by rapamycin. The mTORC1 inhibitor has also marked effects on lipid deposition in adipose tissues that likely contribute to elevated concentrations of plasma triglycerides and nonesterified fatty acids. Our findings therefore further emphasize the critical importance of the mTORC1/S6K1 pathway in the physiological regulation of glucose and lipid metabolism and unravel a new role for this nutrient-sensing pathway in the control of hepatic gluconeogenesis through the modulation of several key transcription factors of the gluconeogenic program. This unrestrained activation of hepatic gluconeogenesis likely underlies the occurrence of a diabetes-like syndrome in patients treated with rapamycin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant (144336) from the Canadian Institutes of Health Research (CIHR). A.M. holds a CIHR/Pfizer Research Chair in the pathogenesis of insulin resistance and cardiovascular diseases. V.P.H. was supported by a PhD studentship from the Fonds de la Recherche en Santé du Québec (FRSQ). S.B. and W.T.F. were supported by a postdoctoral fellowship from the CIHR-funded Obesity Training Program of the Quebec Heart and Lung Institute Research Centre. P.-G.B. was the recipient of a PhD studentship from CIHR.
No potential conflicts of interest relevant to this article were reported.
We thank Philip White and Drs. Geneviève Pilon and Mathieu Laplante for critical reading of the article. We also thank Simon Pelletier, Sébastien Morasse, Marc Majaron, and Véronique Turcotte for their help with animal care, pyruvate tolerance test, quantitative PCR, and statistical analysis.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Wullschleger S, Loewith R, Hall MN: TOR signaling in growth and metabolism. Cell 2006; 124: 471–484 [DOI] [PubMed] [Google Scholar]
- 2.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM: DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009; 137: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremblay F, Marette A: Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway: a negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem 2001; 276: 38052–38060 [DOI] [PubMed] [Google Scholar]
- 4.Khamzina L, Veilleux A, Bergeron S, Marette A: Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology 2005; 146: 1473–1481 [DOI] [PubMed] [Google Scholar]
- 5.Tremblay F, Gagnon A, Veilleux A, Sorisky A, Marette A: Activation of the mammalian target of rapamycin pathway acutely inhibits insulin signaling to Akt and glucose transport in 3T3–L1 and human adipocytes. Endocrinology 2005; 146: 1328–1337 [DOI] [PubMed] [Google Scholar]
- 6.Tremblay F, Brûlé S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, Marette A: Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A 2007; 104: 14056–14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzatsos A, Kandror KV: Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol 2006; 26: 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruzado JM: Nonimmunosuppressive effects of mammalian target of rapamycin inhibitors. Transplant Rev (Orlando) 2008; 22: 73–81 [DOI] [PubMed] [Google Scholar]
- 9.Stallone G, Infante B, Grandaliano G, Gesualdo L: Management of side effects of sirolimus therapy. Transplantation 2009; 87: S23–S26 [DOI] [PubMed] [Google Scholar]
- 10.Morrisett JD, Abdel-Fattah G, Hoogeveen R, Mitchell E, Ballantyne CM, Pownall HJ, Opekun AR, Jaffe JS, Oppermann S, Kahan BD: Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res 2002; 43: 1170–1180 [PubMed] [Google Scholar]
- 11.Teutonico A, Schena PF, Di Paolo S: Glucose metabolism in renal transplant recipients: effect of calcineurin inhibitor withdrawal and conversion to sirolimus. J Am Soc Nephrol 2005; 16: 3128–3135 [DOI] [PubMed] [Google Scholar]
- 12.Xu E, Dubois MJ, Leung N, Charbonneau A, Turbide C, Avramoglu RK, DeMarte L, Elchebly M, Streichert T, Lévy E, Beauchemin N, Marette A: Targeted disruption of carcinoembryonic antigen-related cell adhesion molecule 1 promotes diet-induced hepatic steatosis and insulin resistance. Endocrinology 2009; 150: 3503–3512 [DOI] [PubMed] [Google Scholar]
- 13.Festuccia WT, Blanchard PG, Turcotte V, Laplante M, Sariahmetoglu M, Brindley DN, Deshaies Y: Depot-specific effects of the PPARgamma agonist rosiglitazone on adipose tissue glucose uptake and metabolism. J Lipid Res 2009; 50: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berthiaume M, Laplante M, Festuccia W, Gélinas Y, Poulin S, Lalonde J, Joanisse DR, Thieringer R, Deshaies Y: Depot-specific modulation of rat intraabdominal adipose tissue lipid metabolism by pharmacological inhibition of 11beta-hydroxysteroid dehydrogenase type 1. Endocrinology 2007; 148: 2391–2397 [DOI] [PubMed] [Google Scholar]
- 15.Hamada S, Hara K, Hamada T, Yasuda H, Moriyama H, Nakayama R, Nagata M, Yokono K: Upregulation of the mTOR complex 1 pathway by Rheb in pancreatic β cells leads to increased β cell mass and prevention of hyperglycemia. Diabetes Published online before print March 3, 2009, doi: 10.2337/db08-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN: Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 2008; 8: 399–410 [DOI] [PubMed] [Google Scholar]
- 17.El-Chaâr D, Gagnon A, Sorisky A: Inhibition of insulin signaling and adipogenesis by rapamycin: effect on phosphorylation of p70 S6 kinase vs eIF4E-BP1. Int J Obes Relat Metab Disord 2004; 28: 191–198 [DOI] [PubMed] [Google Scholar]
- 18.Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD: Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One 2009; 4: e6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM: Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006; 22: 159–168 [DOI] [PubMed] [Google Scholar]
- 20.Laplante M, Festuccia WT, Soucy G, Gélinas Y, Lalonde J, Berger JP, Deshaies Y: Mechanisms of the depot specificity of peroxisome proliferator-activated receptor gamma action on adipose tissue metabolism. Diabetes 2006; 55: 2771–2778 [DOI] [PubMed] [Google Scholar]
- 21.Beale EG, Hammer RE, Antoine B, Forest C: Glyceroneogenesis comes of age. Faseb J 2002; 16: 1695–1696 [DOI] [PubMed] [Google Scholar]
- 22.Reue K, Brindley DN: Thematic review series: glycerolipids: multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J Lipid Res 2008; 49: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A: Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res 2009; 50: 3–21 [DOI] [PubMed] [Google Scholar]
- 24.Festuccia WT, Laplante M, Berthiaume M, Gélinas Y, Deshaies Y: PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia 2006; 49: 2427–2436 [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG: FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 2006; 281: 10105–10117 [DOI] [PubMed] [Google Scholar]
- 26.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G: Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004; 431: 200–205 [DOI] [PubMed] [Google Scholar]
- 27.Giraud J, Leshan R, Lee YH, White MF: Nutrient-dependent and insulin-stimulated phosphorylation of insulin receptor substrate-1 on serine 302 correlates with increased insulin signaling. J Biol Chem 2004; 279: 3447–3454 [DOI] [PubMed] [Google Scholar]
- 28.Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, Kobayashi M: Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol Cell Biol 2001; 21: 5050–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briaud I, Dickson LM, Lingohr MK, McCuaig JF, Lawrence JC, Rhodes CJ: Insulin receptor substrate-2 proteasomal degradation mediated by a mammalian target of rapamycin (mTOR)-induced negative feedback down-regulates protein kinase B-mediated signaling pathway in beta-cells. J Biol Chem 2005; 280: 2282–2293 [DOI] [PubMed] [Google Scholar]
- 30.Barthel A, Schmoll D: Novel concepts in insulin regulation of hepatic gluconeogenesis. Am J Physiol Endocrinol Metab 2003; 285: E685–E692 [DOI] [PubMed] [Google Scholar]
- 31.Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM: Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A 2003; 100: 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM: Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001; 413: 131–138 [DOI] [PubMed] [Google Scholar]
- 33.Peng T, Golub TR, Sabatini DM: The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol 2002; 22: 5575–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar V, Alliouachene S, Sotiropoulos A, Sobering A, Athea Y, Djouadi F, Miraux S, Thiaudière E, Foretz M, Viollet B, Diolez P, Bastin J, Benit P, Rustin P, Carling D, Sandri M, Ventura-Clapier R, Pende M: S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metab 2007; 5: 476–487 [DOI] [PubMed] [Google Scholar]
- 35.Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Rüegg MA: Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab 2008; 8: 411–424 [DOI] [PubMed] [Google Scholar]
- 36.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P: mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007; 450: 736–740 [DOI] [PubMed] [Google Scholar]
- 37.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM: Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 2003; 423: 550–555 [DOI] [PubMed] [Google Scholar]
- 38.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW: A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem 2009; 284: 5148–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A: Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 2003; 52: 642–649 [DOI] [PubMed] [Google Scholar]
- 40.Puigserver P: Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int J Obes (Lond) 2005; 1( Suppl.): S5–S9 [DOI] [PubMed] [Google Scholar]
- 41.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M: TORCs: transducers of regulated CREB activity. Mol Cell 2003; 12: 413–423 [DOI] [PubMed] [Google Scholar]
- 42.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M: Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 2007; 449: 366–369 [DOI] [PubMed] [Google Scholar]
- 43.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M: CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001; 413: 179–183 [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M: A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 2008; 456: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rachdi L, Balcazar N, Osorio-Duque F, Elghazi L, Weiss A, Gould A, Chang-Chen KJ, Gambello MJ, Bernal-Mizrachi E: Disruption of Tsc2 in pancreatic beta cells induces beta cell mass expansion and improved glucose tolerance in a TORC1-dependent manner. Proc Natl Acad Sci U S A 2008; 105: 9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JE, Chen J: regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes 2004; 53: 2748–2756 [DOI] [PubMed] [Google Scholar]
- 47.Veilleux A, Houde VP, Bellmann K, Marette A: Chronic inhibition of the mTORC1/S6K1 pathway increases insulin-induced PI 3-kinase activity but inhibits Akt2 and glucose transport stimulation in 3T3–L1 adipocytes. Mol Endo In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinstock PH, Levak-Frank S, Hudgins LC, Radner H, Friedman JM, Zechner R, Breslow JL: Lipoprotein lipase controls fatty acid entry into adipose tissue, but fat mass is preserved by endogenous synthesis in mice deficient in adipose tissue lipoprotein lipase. Proc Natl Acad Sci U S A 1997; 94: 10261–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM: Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11: 859–871 [DOI] [PubMed] [Google Scholar]
- 50.Cybulski N, Polak P, Auwerx J, Rüegg MA, Hall MN: mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A 2009; 106: 9902–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.