Abstract
In this study we investigated the regulation of FOXM1 expression by estrogen receptor α (ERα) and its role in hormonal therapy and endocrine resistance. FOXM1 protein and mRNA expression was regulated by ER-ligands, including estrogen, tamoxifen (OHT), and fulvestrant (ICI182780; ICI) in breast carcinoma cell lines. Depletion of ERα by RNA interference (RNAi) in MCF-7 cells down-regulated FOXM1 expression. Reporter gene assays demonstrated that ERα activates FOXM1 transcription through an estrogen-response element (ERE) located within the proximal promoter region. The direct binding of ERα to the FOXM1 promoter was confirmed in vitro by mobility shift and DNA pull-down assays and in vivo by chromatin immunoprecipitation (ChIP) analysis. Our data also revealed that upon OHT treatment ERα recruits histone deacetylases (HDACs) to the ERE site of the FOXM1 promoter, which is associated with a decrease in histone acetylation and transcription activity. Importantly, silencing of FOXM1 by RNAi abolished estrogen-induced MCF-7 cell proliferation and overcame acquired tamoxifen resistance. Conversely, ectopic expression of FOXM1 abrogated the cell cycle arrest mediated by the anti-estrogen OHT. OHT repressed FOXM1 expression in endocrine sensitive but not resistant breast carcinoma cell lines. Further, qRT-PCR analysis of breast cancer patient samples revealed there was a strong and significant positive correlation between ERα and FOXM1 mRNA expression. Collectively, these results demonstrate FOXM1 to be a key mediator of the mitogenic functions of ERα and estrogen in breast cancer cells, and also suggest that the deregulation of FOXM1 may contribute to anti-estrogen insensitivity.
Introduction
Breast cancer is the second most prevalent cause of cancer death in the western hemisphere and displays a complex aeitology. The forkhead box (FOX) family member FOXM1 has previously been reported to be elevated in breast, cancer as well as in carcinomas of other origins (Pilarsky et al., 2004). FOXM1 is expressed in proliferating adult tissues and in response to injury or repair (Korver et al., 1997a; Korver et al., 1997b; Leung et al., 2001; Wang et al., 2002; Ye et al., 1999). FOXM1 plays a critical role in cell cycle progression; expression of FOXM1 increases at G1 to S phase transition, and reaches maximal levels in G2 to M phase, promoting M phase entry (Laoukili et al., 2005; Leung et al., 2001). FOXM1 controls the expression of cell cycle regulatory proteins, such as cyclin B1 and cyclin D1 (Laoukili et al., 2005; Leung et al., 2001; Wang et al., 2001), in addition to genes essential for faithful chromosome segregation and mitosis, such as Nek-2, KIF20A, Cdc25B, Aurora B kinase, Survivin, centromere protein A (CENPA), and CENPB (Wang et al., 2005). Thus, knockdown of FOXM1 with siRNA results in delay in G2 and mitotic spindle defects, and induces mitotic catastrophe.
Previous work has demonstrated that FOXM1 regulates ERα expression in breast cancer cell lines (Madureira et al., 2006). Consistently, expression of the FOXM1 target cyclin D1 is associated with ERα positivity in breast cancer (Butt et al., 2005). The strong correlation between the expression patterns of ERα and FOXM1 in breast cancer cells also suggested the possibility that FOXM1 could be an ERα-regulated gene (Madureira et al., 2006). Consistent with this hypothesis, a cDNA microarray also identified FOXM1 as one of 344 estrogen-responsive genes in breast cancer cells (Cicatiello et al., 2004). Estrogen is generally considered to be mitogenic (Brunner et al., 1989), and exerts its action in the mammary epithelial cells primarily via ERα (Ali and Coombes, 2002). The expression of ERα is a good prognostic factor in breast cancer, as about two-thirds of ERα-positive patients respond to treatment with anti-estrogens such as OHT, ICI, or aromatase inhibitors (Elkak and Mokbel, 2001; Gapinski and Donegan, 1980; Osborne and McGuire, 1979). Treatment of breast cancer cells with anti-estrogens results in cell cycle arrest at the G1/S phase and in some cases, cell death (Lykkesfeldt et al., 1986). However, approximate half of the patients that initially respond to hormonal therapy develop resistance and relapse, following long-term treatment (Ali and Coombes, 2002; Goss et al., 2008; Yamashita, 2008). Our previous observation that ERα-positive breast cancer cell lines express higher levels of FOXM1 protein (Madureira et al., 2006) led us to hypothesise that FOXM1 expression is also regulated by ERα. In an effort to elucidate the mechanisms of anti-estrogen action and resistance to endocrine therapy, we investigated the regulation of FOXM1 by ERα and its role in endocrine sensitivity and resistance in breast cancer cells.
RESULTS
ERα ligands modulate the expression of FOXM1 protein and mRNA levels
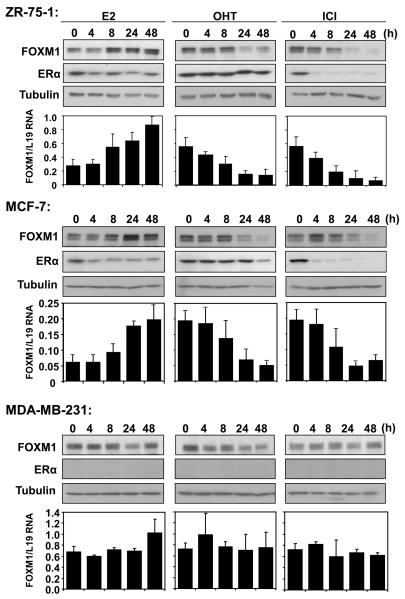
To investigate whether the FOXM1 protein and mRNA levels are regulated by ERα, the ERα-positive breast cancer cell lines ZR-75-1 and MCF-7, and ERα-negative line MDA-MB-231 were treated with the ERα ligands E2, OHT and ICI for the times indicated and harvested for Western blotting and qRT-PCR analysis. Treatment of estrogen-starved ZR-75-1 and MCF-7 cells with E2 enhanced the expression of FOXM1 protein and mRNA (Fig. 1). Within 8 h of E2 treatment, FOXM1 protein and mRNA levels peaked, and remained high for at least 48 h. By contrast, E2 treatment did not cause significant changes in either FOXM1 protein or mRNA levels in the ERα-negative control MDA-MB-231 cells. Treatment of both the ZR-75-1 and MCF-7 cells with the partial antagonist OHT resulted in a reduction in FOXM1 protein levels that was noticeable within 4 h, reaching minimal levels within 24 h post-treatment. Yet again, treatment with OHT had little effect on the expression of FOXM1 protein and mRNA in MDA-MB-231 cells. Similar to OHT, incubation with the pure antagonist ICI, which causes the degradation of ERα, resulted in a reduction in the FOXM1 expression, with both the protein and mRNA levels completely vanished within 24 h post-treatment. We also studied the ability of OHT or ICI to antagonize E2-regulated FOXM1 up-regulation, and the result showed that the effects of OHT or ICI following E2 stimulation are similar to those under normal growth conditions (Fig. S1). In summary, the FOXM1 protein and mRNA levels were up-regulated by ERα agonists and down-regulated by ERα antagonists in ERα-positive but not ERα-negative breast cancer cell lines, suggesting that FOXM1 is regulated by ERα.
Figure 1. Expression of FOXM1 and ERα in response to E2, OHT and ICI treatments in breast cancer cell lines.
ZR-75-1, MCF-7 and MDA-MB-231 cells were cultured in 5% double-charcoal striped FCS and phenol red free medium for 24h before stimulated with E2. Breast cancer cells cultured in 10% FCS and phenol red medium were also treated with OHT or ICI. At times indicated, cells were collected and analysed for FOXM1, ERα and tubulin expression by western blotting. FOXM1 mRNA levels of these cells were also analysed by qRT-PCR,, and normalized with L19 RNA expression. Total RNA (2 μg) isolated using the RNeasy Mini kit (Qiagen, Crawley, UK) was reverse transcribed using the Superscript III reverse transcriptase and random primers (Invitrogen, Paisley, UK), and the resulting first strand cDNA was used as template in the real-time PCR. All experiments were performed in triplicate. The following gene-specific primer pairs were designed using the ABI Primer Express software: FOXM1-sense: 5′-TGCAGCTAGGGATGTGAATCTTC-3′ and FOXM1-antisense: 5′-GGAGCCCAGTCCATCAGAACT-3′; ERα-sense: 5′-CAGATGGTCAGTGCCTTGTTGG-3′ and ERα-antisense: 5′-CCAAGAGCAAGTTAGGAGCAAACAG-3′; L19-sense 5′-GCGGAAGGGTACAGCCAAT-3′ and L19-antisense 5′-GCAGCCGGCGCAAA-3′. Specificity of each primer was determined using NCBI BLAST module. Real time PCR was performed with ABI PRISM 7700 Sequence Detection System using SYBR Green Mastermix (Applied Biosystems, Brackley, UK). The qRT-PCR results shown are representative of 3 independent experiments.
The FOXM1 promoter responds to ERα and ligands
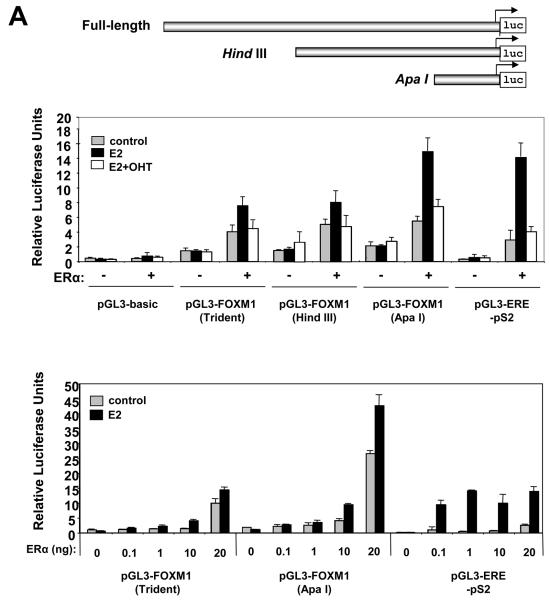
To investigate if the regulation of FOXM1 by ERα is at the promoter level, ERα-negative COS-1 cells were transiently transfected with the ERα expression construct pHEGO, with a luciferase reporter gene under the control of the proximal 2.4kb FOXM1 promoter (WT-Trident), or its truncation mutants HindIII (1.4kb), and ApaI (300bp) (Fig. 2A). We observed a 2-fold increase of the WT-Trident-length FOXM1-luc activity after E2 treatment, and this induction by E2 was repressed in the presence of OHT (Fig. 2A). The highest increase (2.7-fold) by E2 was observed for the ApaI-luc activity (Fig. 2A). The HindIII-luc activity was up-regulated to a lesser extent (1.5-fold) by E2 (Fig. 2A). The response of the FOXM1 promoter to ERα was further verified by co-transfection of COS-1 cells with titrated amounts of pHEG0 and the WT-Trident-luc or ApaI-luc reporter plasmids (Fig. 2A). Both the WT-Trident and ApaI promoters were induced in a dose-dependent manner by increasing amounts of ERα in the presence of E2 (Fig. 2A). It is notable that the control pS2 promoter showed maximum E2-stimulation with very low levels of ERα expression, supporting the notion that pS2 may be one of the most E2-sensitive genes (Masiakowski et al., 1982). Collectively, these results indicate that the ApaI truncation is the minimal proximal FOXM1 promoter responsive to ERα, consistent with a previous study identifying a similar region of the FOXM1 promoter that responds to serum stimulation (Korver et al., 1997a).
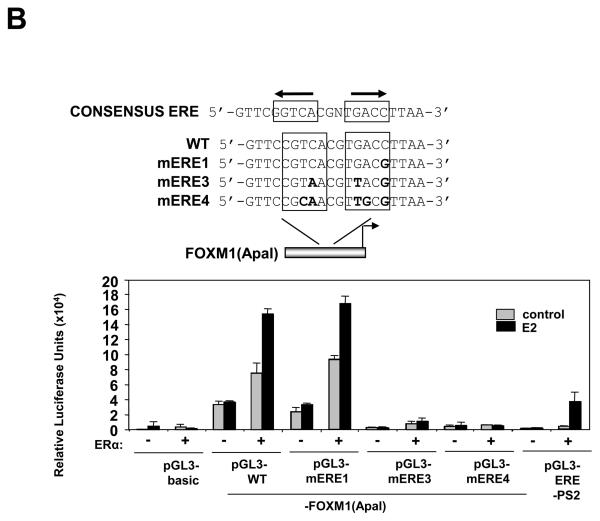
Figure 2. ERα induces the transcriptional activity of the human FOXM1 gene through a ERE consensus proximal to the transcription start site.
A) Effect of treatment with E2 and expression of ER on FOXM1 promoter activity. Schematic representation of the full-length, HindIII and ApaI FOXM1-luciferase reporter constructs. In upper panel, COS-1 cells cultured in 5% double-charcoal striped FCS and phenol red free medium were transiently transfected with 20 ng of either the empty pGL3-basic, pGL3-FOXM1(Trident), pGL3-FOXM1(ApaI), or the control pGL3-ERE-pS2 promoter/reporter and 0 ng or 10 ng of ERα expression vector (pHEGO) in the absence or presence of E2 and with OHT treatment in the presence of E2 induction (E2+OHT). Cells were harvested 24 h after transfection and assayed for luciferase activity. All relative luciferase activity values are corrected for cotransfected Renilla activity. All data shown represent the averages of data from three independent experiments, and the error bars show the standard deviations. In lower panel, COS-1 cells were transfected with pGL3-FOXM1(Trident), pGL3-FOXM1(ApaI), or pGL3-ERE promoter/reporter constructs, together with increasing amounts (0, 0.1, 1, 10, and 20 ng) of ERα expression vector (pHEGO), and processed as described above. B) Schematic representation of the ApaI FOXM1-luciferase reporter construct, showing the consensus, the wild-type, and the mutant ERE (mERE) sequences. COS-1 cells were transfected with pGL3-basic, pGL3-FOXM1(ApaI) wild-type (WT) or mutant ERE, or the control pGL3-ERE-PS2 promoter/reporter and with or without E2 treatment and 20 ng of ERα expression vector. The transfected cells were processed and assayed as described above.
The ERE-like element at −45bp of the FOXM1 promoter confers responsiveness to ERα ligands
Analysis using the Transcription Element Search System (TESS,http://www.cbil.upenn.edu/cgi-bin/tess/tess) (Schug, 2008) revealed an ERE-like element (Bourdeau et al., 2004) located at −45bp from the transcription start site. We next focused our studies on the characterisation of the ERE-like element at −45bp in the ApaI promoter fragment and introduced point mutations using site-directed mutagenesis by base-substitution in both arms of the ERE-like palindrome of the ApaI truncation (Bourdeau et al., 2004) (Fig. 2B). For comparison, a single point mutation that does not grossly disrupt the ERE-consensus was also introduced in one arm of the ERE-like element in mERE1 (Fig. 2B). The activity of the wild-type as well as the mutated ApaI-luc constructs mERE1, mERE3 and mERE4 was examined by co-transfection assays in COS-1 cells with and without pHEG0, in the presence or absence of E2 (Fig. 2B). The results showed that the mERE1-luc construct demonstrated similar responsiveness to ERα and E2 induction as the wild-type ApaI-luc. In contrast, both the mERE3-luc and mERE4-luc mutants lost the majority of their responsiveness to E2 (Fig. 2B). Together these co-transfection results indicate that the ERE-like element located at −45bp confers the responsiveness to ERα, further confirming that FOXM1 is a target gene of ERα.
ERα binds directly to the ERE-like element of the FOXM1 promoter in vitro
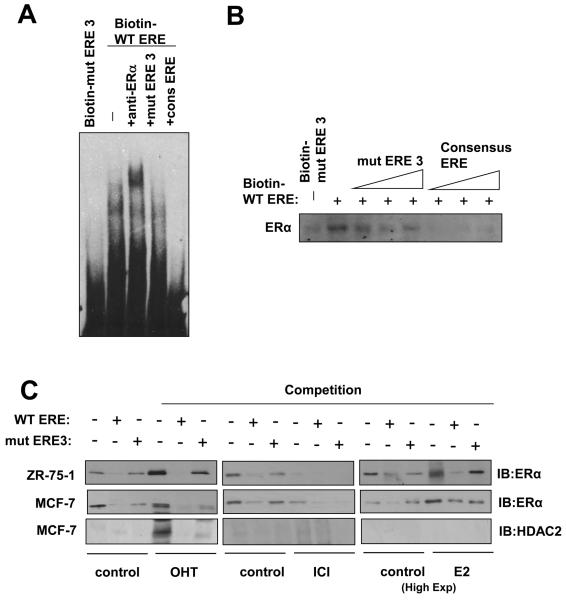
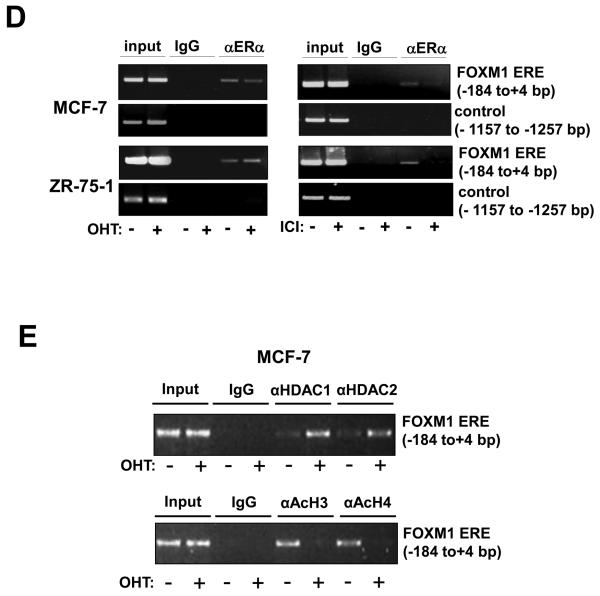
We next tested the in vitro binding of ERα to the ERE-like site by electrophoretic mobility shift assay (EMSA) with nuclear lysate from MCF-7 cells. From the EMSA, it was clear that ERα binds to the wild-type ERE-like site of ApaI, but not the mutated mERE3 site (Fig. 3A). This complex of ERα and WT ERE biotin-oligonucleotide was further supershifted with a specific anti-ERα antibody, indicating that the complex contains ERα (Fig. 3A). This ERα complex could be competed off by excess amounts of a classical consensus ERE oligonucleotide, but not by a mERE3 ApaI site oligonucleotide. Finally, unlabelled FOXM1 WT ERE oligonucleotide was successful in competing off the ERα binding on the consensus ERE oligonucleotide. To demonstrate that ERα binds to the ERE-like site of ApaI we used the biotin-labelled oligonucleotides in pull-down assays. ERα binding was analysed by immunoblotting with an anti-ERα antibody. As shown in figure 3B, ERα had much higher affinity for the wild-type ERE than the mutant mERE3. ERα-binding to the FOXM1 ERE could be competed away by molar excess of wild-type ERE, but not the mutant mERE. We next extended our pull-down assays to MCF-7 and ZR-75-1 cells in the absence or presence of OHT, ICI and E2 treatments (Fig. 3C). Western blot analysis was first performed to establish the expression patterns of ERα in cytoplasmic and nuclear fractions of MCF-7 and ZR-75-1 cells, also with or without OHT, ICI, or E2 treatment (Fig. S2). The results confirmed our previous data that both OHT and ICI inhibit ERα activity, while ICI, but not OHT, represses ERα expression. In the pull-downs, ERα binding on the biotin-WT ERE was effectively competed by 10x molar excess of unlabelled WT ERE, and not mERE3, oligonucleotides. We also probed for the recruitment of HDAC to the ERE site upon OHT, ICI or E2 treatment in MCF-7 cells, and the results revealed that HDAC2 was recruited to the ERE site upon OHT but not ICI or E2 treatment (Fig. 3C). Taken together these results showed that ERα binds specifically to the ERE-like element of the FOXM1 promoter in vitro and that HDAC is recruited to the ERE site upon OHT treatment.
Figure 3. ERα binds directly to the ERE on the FOXM1 promoter.
A) Biotinylated wild-type ERE or the mutant ERE3 oligonucleotides were incubated with MCF-7 cell lysates in the presence or absence of 10x molar excess of non-biotinylated ERE3 or consensus ERE oligonucleotides or an anti-ERα antibody. In brief, gel shift assays (20μl total volume) contained 3μg nuclear extracts and 50ng/μl poly (dI-dC) in 1x binding buffer and 20pmol unlabelled oligonucleotides. For the supershifts, 1μl of the anti-ERα (Santa Cruz; H222) antibody were included in the reaction mix. The mixtures were pre-incubated at room temperature for 15 min, followed by addition of 100fmol of biotin-labelled double stranded estrogen response element (ERE-conc. 5′-GCCGATTGGCGACGTTCGGTCACGCTGACCTTAACGCTCCGCCGGCG-3′ 5′-CGCCGGCGGAGCGTTAAGGTCACGCTGACCGAACGTCGCCAATCGGC-3′), (ERE-wt 5′-GCCGATTGGCGACGTTCCGTCACGTGACCTTAACGCTCCGCCGGCG-3′, 5′-CGCCGGCGGAGCGTTAAGGTCACGTGACGGAACGTCGCCAATCGGC-3′), or (mERE3 5′-GCCGATTGGCGACGTTCCGTAACGTTACGTTAACGCTCCGCCGGC-3′, 5′-CGCCGGCGGAGCGTTAACGTAACGTTACGGAACGTCGCCAATCGGC-3′), and incubation at room temperature for 20 min, followed by a 30 min incubation on ice. Protein-DNA complexes were separated on 5% 0.5 × TBE polyacrylamide gels. The electrophoretic transfer of the binding reactions to the nylon membrane, and the detection of the biotin labelled DNA by chemiluminescence, were performed according to the PIERCE kit's protocols. B) Nuclear extracts from MCF-7 cells were incubated with biotinylated oligonucleotides representing region of the FOXM1 promoter containing the ERE or the mutated ERE3 site in the absence or presence of molar excess of non-biotinylated ERE3 or consensus ERE oligonucleotides. Proteins binding to the biotinylated oligonucleotides were pulled-down using streptavidine agarose beads and analysed by western blot using specific antibodies as indicated. C) The nuclear extracts (Fig. S1) from MCF-7 and ZR-75-1 cells with or without OHT or ICI for 24 h were also examined by pull-down assays using biotinylated wild-type or mutant (mERE3) oligonucleotides as described above. D) Chromatin immunoprecipitation (ChIP) analysis of the human FOXM1 promoter. MCF-7 and ZR-75-1 cells untreated or treated with ICI or OHT for 24 h were used for ChIP assays using IgG, anti-ERα antibodies as indicated. After crosslink reversal, the co-immunoprecipitated DNA was amplified by PCR using primers amplifying the FOXM1 ERE containing region (−184/+4) and a control region (−1157/−1257), and resolved in 2% agarose gel. E) MCF-7 untreated or treated with OHT for 24 h were used for ChIP assays using IgG, antibodies against acetylated H3 and H4, HDAC1 and HDAC2 as described above. Representative data from three independent experiments are shown.
ERα and HDAC bind specifically on the ERE-like element of ApaI in vivo
We further tested the in vivo binding of ERα on the FOXM1 promoter in the MCF-7 and ZR-75-1 cell lines by chromatin immunoprecipitation assays (ChIP), following treatment of the cells with either OHT or ICI (Fig. 3D). The DNA immunoprecipitated by an anti-ERα antibody was amplified using the FOXM1 ERE primers to show in vivo binding of ERα on the ERE-like site of FOXM1 promoter at −45bp, in agreement with the in vitro results. Occupancy of FOXM1 promoter by ERα was enhanced in ZR-75-1 but not MCF-7 cells following treatment with OHT, and a reduction in ERα binding was observed in MCF-7 and ZR-75-1 cells following treatment with ICI. The ChIP assays also showed that upon OHT treatment there was an increase in HDAC1 and HDAC2 recruitment and a corresponding decrease in acetylated histones H3 and H4 associated with the FOXM1 promoter, indicating that OHT treatment caused the recruitment of HDAC, which confers transcriptional repression to the ERE region of the FOXM1 promoter (Fig. 3E),
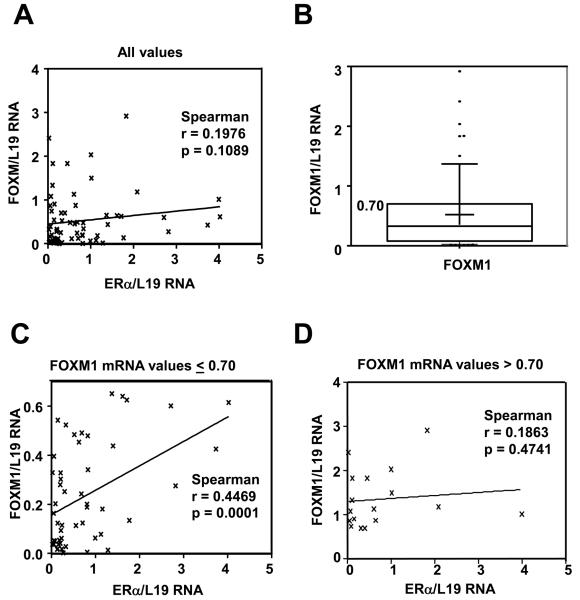
Significant positive correlation between FOXM1 and ERα mRNA levels in breast cancer patient samples
To determine the clinical relevance of the relationship between ERα and FOXM1 expression, we analysed the FOXM1 and ERα mRNA levels in 69 frozen archival clinical samples, of which 21 were from benign disease patients, 39 from patients with breast tumours and 9 from healthy individuals (Fig. S3). Preliminary studies demonstrated that despite FOXM1 mRNA expression being associated with tumour status, grade, size, and menopausal status, the differences were not significant, possibly reflecting the relatively small sample size (Fig. S4). Analysis of unadjusted data revealed no significant correlation between the expression levels of FOXM1 and ERα mRNA (Fig. 4A). This is not entirely unexpected as high levels of FOXM1 mRNA expression (upper quartile) have previously been shown be associated with breast cancer progression, poor prognosis and endocrine resistance in a number of microarray studies (Martin et al., 2008). We next re-evaluated the data after excluding patient samples with high levels of FOXM1 expression. By leaving out patient samples with FOXM1 mRNA levels above the 75th percentile value (FOXM1/L19mRNA=>0.700) (Fig. 4B), we found a significant correlation between FOXM1 and ERα mRNA expression (Fig. 4C). In cases where there were high levels of FOXM1 mRNA expression, we also found that FOXM1 expression is independent of ERα mRNA levels, suggesting FOXM1 is no longer regulated by ERα (Fig. 4D). These results validate our tissue culture findings in breast patient samples, confirming the crucial relationship between ERα and FOXM1 expression. In breast cancer samples with high FOXM1 expression, the relationship between ERα and FOXM1 mRNA expression is uncoupled, suggesting ERα no longer controls FOXM1 expression. This also supports the conjecture that uncoupling of the regulation of FOXM1 by ERα may have a role in endocrine resistance.
Figure 4. Significant correlation between ERα and FOXM1 expression in human breast samples.
Expression of ERα mRNA and FOXM1 mRNA in non-cancerous breast biopsies and malignant breast epithelial tissues. RNA was isolated from epithelial cells purified from normal breast tissues and primary tumours and subjected to qRT-PCR with FOXM1, ERα and L19 primers. A) Graph shows the FOXM1 and ERα mRNA levels of the tumour samples after normalisation with L19 RNA levels. No significant correlation is seen using Pearson correlation when considering all values (n=69; Spearman r=0.198; p=0.109). Significance is defined as p<0.05. Line is linear regression, shown for illustrative purposes. B) Box and whisker plot showing the distribution of values for FOXM1. Box edges represent 25th and 75th percentiles; middle line is the median, while plus shows the mean. The whiskers represent the 10th and 90th percentiles, while outliers are shown as dots. The 75th percentile for FOXM1 mRNA values is 0.7002. C) The right-hand graph shows the FOXM1 and ERα mRNA levels of the tumour samples with FOXM1/L19 mRNA levels below the upper quartile. Correlation analysis was performed between FOXM1 and ERα mRNA. A significant and positive association was found between FOXM1 and ERα mRNA levels in breast patient samples with FOXM1 mRNA level below the upper quartile (n=52; Spearman r=0.447; p=0.0001). P < 0.05 was considered statistically significant. D) The lower left-hand graph shows the FOXM1 and ERα mRNA levels of the tumour samples with FOXM1/L19 mRNA levels in the upper quartile (0.7) of high levels of FOXM mRNA expression. No significant correlation is found using Pearson correlation (n=17; Spearman r=0.186; p=0.474). Significance is defined as p≤0.05.
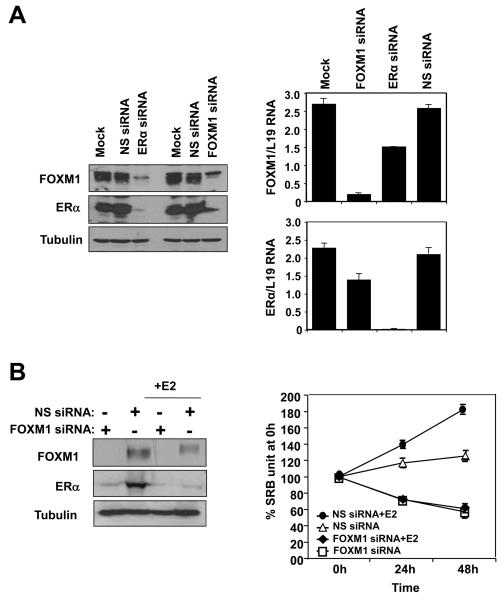
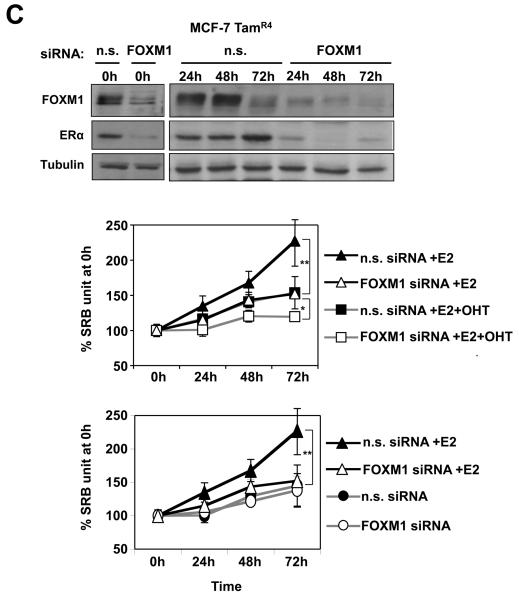
ERα silencing down-regulates FOXM1 expression while FOXM1 knock-down inhibits estrogen-induced cell proliferation
To provide further evidence for the regulation of FOXM1 by ERα, we investigated the effect of ERα silencing on FOXM1 expression in proliferating MCF-7 cells. As shown in Figure 5A, the ERα siRNA effectively knocked down the expression of ERα, which in turn also resulted in a down-regulation of FOXM1 at the protein level. Conversely, FOXM1 knockdown also resulted in a decrease in ERα expression, consistent with our previous finding (Madureira et al., 2006). qRT-PCR analysis of the knocked-down cells also revealed a substantial decrease in FOXM1 mRNA level (Fig. 5A). FOXM1 mRNA expression was not completely abolished by ERα silencing; this is likely to be due to the fact that FOXM1 transcription is regulated by other proliferative signals as well (McGovern et al., 2009). The complete down-regulation of FOXM1 at the protein level suggests the possibility of alternative mechanisms of regulation of FOXM1 expression by ERα (Luscher-Firzlaff et al., 2006; Ma et al., 2005; Major et al., 2004; Wierstra and Alves, 2006b). We next investigated the role of FOXM1 in mediating the proliferative function of ERα by studying the effects of FOXM1 knockdown on estrogen-dependent growth in MCF-7 cells. MCF-7 cells were estrogen-starved for 48 h then induced to proliferate with E2 in the presence or absence of FOXM1 siRNA. Western blot analysis showed that FOXM1 was successfully silenced by the siRNA, and that in the FOXM1 knocked-down cells there was also a significant decrease in ERα expression (Fig. 5B). This is in agreement with our previous finding that FOXM1 regulates ERα expression (Madureira et al., 2006). The results of the cell proliferation assay showed that FOXM1 silencing completely abolished the mitogenic effects of E2 in the ERα positive MCF-7 cells, indicating that FOXM1 is essential for the estrogen-dependent growth of MCF-7 cells, and mediates the proliferative function of ERα (Fig. 5B). To investigate the potential role of FOXM1 in hormonal resistance, we silenced FOXM1 expression using siRNA in the MCF-7 TAMR4 cells, an OHT-resistant MCF-7 clone (Lykkesfeldt et al., 1986; Lykkesfeldt et al., 1994), and studied its effects on cell proliferation (Fig. 5C). Interestingly, the proliferation assays showed that the MCF-7 TAMR4 was responsive to E2 re-stimulation, and knockdown of FOXM1 decreased the ability of E2 to induce the proliferation of the MCF-7 TAMR4 cells. Notably, these cells with FOXM1 knockdown cultured in the absence or presence of E2 had the same proliferation rate as steroid-depleted cells, suggesting E2 targets FOXM1 to induce proliferation in this system. In addition, our data also demonstrated that OHT could antagonise the proliferative function of E2, and that FOXM1 knockdown could be combined with OHT to cause further decreases in the rate of cell proliferation. Together these results clearly show that the knockdown of FOXM1 sensitizes the resistant cells to OHT and diminishes the responsiveness to E2.
Figure 5. Effects of ERα and FOXM1 silencing on the expression of FOXM1 and response to E2 induction in MCF-7 cells.
A) MCF-7 cells were transiently transfected with ERα, FOXM1 or control smart pool siRNA, and 72 h after transfection cells were analysed by western blot using specific antibodies as indicated and by qRT-PCR. B) MCF-7 cells were transiently transfected with smart pool siRNA against FOXM1, incubated with E2 and analysed by western blotting. SRB assays were also performed on these cells, indicating that the knockdown of FOXM1 decreases the cell proliferation rate and renders MCF-7 cells unresponsive to E2 stimulation. C) OHT-resistant TAMR4 MCF-7 cells were transiently transfected with smart pool siRNA against FOXM1 or control siRNA pool (non-specific/n.s. siRNA) and analysed by western blotting (upper panel). These transfected cells were incubated with or without E2 (middle panel), and with or without OHT in the presence of E2 (lower panel). SRB assays were performed on these cells, indicating that the knockdown of FOXM1 sensitizes the resistant TAMR4 MCF-7 cells to OHT and diminishes their responsiveness to E2 stimulation. Statistical analysis was performed on the proliferation results at 72 h. ** denotes very significant, P<0.01 and * significant, P<0.05. The results show mean+SEM of triplicate measurements.
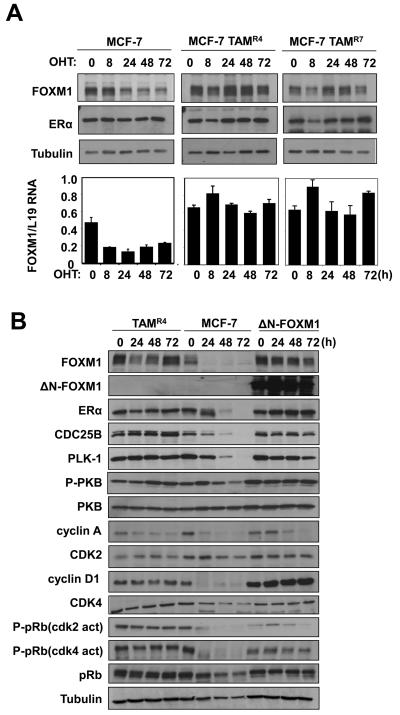
Dysregulated FOXM1 is associated with breast cancer endocrine resistance
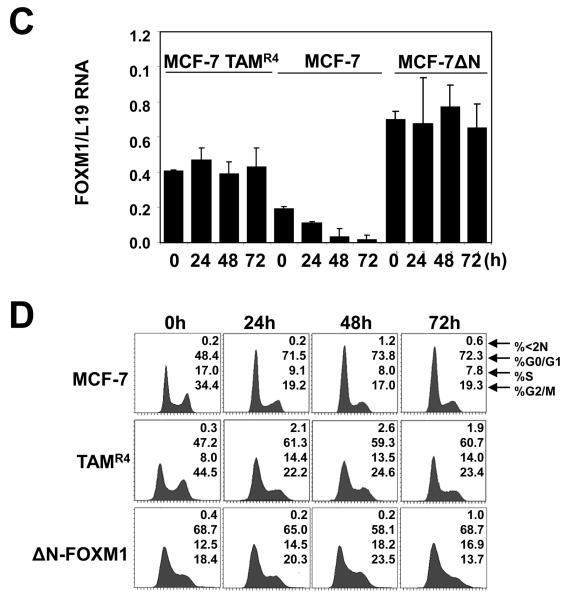
Since FOXM1 is essential for ERα's proliferative function, uncoupling FOXM1 from ERα regulation could result in resistance to endocrine therapy in breast cancer cells. To test this hypothesis, we studied the expression of FOXM1 in hormone sensitive as well as resistant MCF-7 cells, after treatment with OHT (Fig. 6A). Consistent with our earlier results, OHT treatment of the sensitive MCF-7 cells caused a drastic reduction in FOXM1 protein level over 72 h. In contrast, despite a small transient decline in FOXM1 level at 8 h, no significant decreases in FOXM1 protein level were observed in MCF-7 TAMR4 and MCF-7 TAM R7 (two independently generated) OHT-resistant clones over the time course. Consistent with the Western blot results, qRT-PCR analysis showed that OHT repressed FOXM1 mRNA expression in the parental MCF-7 cells but not in the MCF-7 TAMR4 and MCF-7 TAMR7 lines. These results suggested a role for FOXM1 in mediating the endocrine resistance in breast cancer cells. To examine this further, we next investigated if a constitutively active FOXM1 (ΔN-FOXM1) could confer resistant to OHT in MCF-7 cells (Fig. 6B). The parental MCF-7 cells and one of the resistant lines, MCF-7 TAMR4, were also included as controls. Western blot analysis revealed that the FOXM1 level declined in the parental MCF-7 cells on treatment with OHT, but not in the resistant cells or MCF-7 cells expressing the deregulated FOXM1. Consistently, the expression of FOXM1 targets, including Polo-like kinase-1 (PLK-1) and CDC25B, mirrored the expression patterns of FOXM1 in the three MCF-7 cell lines. It is notable that there were no significant changes in either the ERα or the endogenous FOXM1 in the OHT-resistant cell line and the MCF-7 harbouring the active FOXM1. This is likely to be due to the positive feedback mechanism between ERα and FOXM1, described here and previously (Madureira et al., 2006). In agreement with the Western blot results, qRT-PCR analysis showed that OHT repressed FOXM1 mRNA expression in MCF-7 cells but not in the MCF-7-TAMR4 and MCF-7-ΔN-FOXM1 cells. The Western blot results also showed that cyclin A and not its catalytic partner CDK2 was down-regulated by 48 h in MCF-7 cells, and the decline in cyclin A levels occurred in a comparatively slower kinetics in TAMR4 and ΔN-FOXM1 cells. Consistent with this, the CDK2 activity specific pRB(Thr821) antibody showed that the repression of cyclin A/E–CDK2 activity coincided with the down-regulation of cyclin A expression in these MCF-7 cells. Given that cyclin A-CDK2 phosphorylation of FOXM1 can activate its transcriptional activity (Laoukili et al., 2008a; Laoukili et al., 2008b; Park et al., 2008; Wierstra and Alves, 2006a), this finding suggests another potential mechanism by which FOXM1 activity can be downregulated by OHT. Since cyclin A and -E functions downstream of cyclin D1, (Fung and Poon, 2005; Myatt and Lam, 2007; Tashiro et al., 2007), which is a transcription target of FOXM1, it is likely that the constitutively active ΔN-FOXM1 up-regulates cyclin D1 to sustain cyclin A expression and G1 to S cell cycle phase progression upon OHT treatment in the ΔN-FOXM1 MCF-7 cells. Consistent with this, the Western blot analysis showed that the cyclin D1 level and its associated activity revealed by the CDK4/6 phospho-pRB(Ser807/811) antibody, were overexpressed in the ΔN-FOXM1- and maintained in the TAMR4-MCF-7 cells. In addition, ectopic expression of FOXM1 also increases the expression levels of cyclin D1 in both MCF-7 (Fig. S5) and MDA-MB-231 cells (Fig. S6). Given the well-documented role of cyclin D1 in endocrine resistance (Finn et al., 2009; Lundgren et al., 2008; Wang et al., 2008; Yamashita et al., 2009; Zwart et al., 2009) and G1/S transition (Fung and Poon, 2005; Myatt and Lam, 2007; Tashiro et al., 2007), our data also support a role for FOXM1 in mediating breast cancer endocrine sensitivity and resistance at least in part through modulating cyclin D1 expression. We performed cell cycle analysis of the MCF-7, MCF-7TAMR4 and MCF-7 ΔN-FOXM1 cells following treatment with OHT (Fig. 6C). The results showed that OHT caused a predominantly G1 cell cycle arrest in the parental MCF-7 cells, but had comparatively little effects on the cell cycle distribution of the MCF-7TAMR4 and MCF-7 ΔN-FOXM1 cells. We also generated MCF-7 cells stably transfected with the wild-type FOXM1 (MCF-7-FOXM1) and studied the effects of OHT on the transfected pool as well as individual clones of MCF-7-FOXM1 cells (Fig. S5). Western blot analysis showed that overexpression of the full-length FOXM1 prevented the down-regulation of FOXM1 targets such as CDC25b and PLK in response to tamoxifen treatment. The cell cycle analysis demonstrated that these cells underwent the cell cycle arrest at G1, concommitant with FOXM1 down-regulation, at a slower kinetics compared with MCF-7 cells transfected with the empty vector. This further suggests that down-regulation of FOXM1 is important for the G1/S arrest and that FOXM1 may also be regulated by tamoxifen at post-transcriptional levels. Collectively, these results indicate that FOXM1 has a role in mediating the anti-proliferative effects of OHT via ERα, and that its deregulated expression contributes towards the development of endocrine resistance.
Figure 6. Expression of FOXM1 and cell cycle regulation in wild-type, OHT-resistant, and constitutively active ΔN-FOXM1 expressing MCF-7 cells in response to OHT treatment.
A) MCF-7 and the resistant MCF-7-TAMR4 and -TAMR7 lines cultured in 10% FCS and phenol red medium were treated with OHT in a time course of 72 h. Cell lysates were prepared at the times indicated, and the expression of FOXM1, ERα and tubulin was analyzed by Western blotting. B) MCF-7, MCF-7 TAMR4 and MCF-7 ΔN-FOXM1 cells were treated with OHT in a time course of 72 h. Cell lysates were prepared at the times indicated, and the expression of FOXM1, ERα, CDC25B, P-PKB, total PKB and Tubulin was analyzed by Western blotting. C) FOXM1 mRNA levels of these cells were also analysed by qRT-PCR and normalized to L19 RNA expression. D) Cells were fixed at 0, 24, 48, and 72 h after treatment, and cell cycle phase distribution was analyzed by flow cytometry after propidium iodide staining. Percentage of cells in each phase of the cell cycle (sub-G1, G1, S, and G2/M) is indicated. Representative data from three independent experiments are shown.
DISCUSSION
Expression of ERα expression in breast cancer cell lines is regulated by FOXM1 (Madureira et al., 2006). In this study, we investigated the reciprocal regulation of FOXM1 expression by ERα. Using breast carcinoma cell lines, we showed that FOXM1 protein and mRNA expression are regulated by ER-ligands, including E2, OHT, and ICI. We also found that depletion of ERα by RNA interference in MCF-7 cells leads to down-regulation of FOXM1 expression. Using reporter gene assays, we demonstrated that ERα activates FOXM1 transcription through an ERE located at −45bp upstream of the transcriptional start site (Bourdeau et al., 2004). The direct binding of ERα to the FOXM1 promoter was confirmed in vitro by mobility shift and DNA pull-down assays, and in vivo by ChIP analysis. Importantly, silencing of FOXM1 by RNA interference abolishes estrogen-mediated MCF-7 cell proliferation and overcomes tamoxifen resistance in ‘acquired resistant’ MCF-7 cells. Conversely, ectopic expression of a constitutively active FOXM1 can abrogate the cell cycle arrest mediated by OHT. Collectively, the results presented clearly confirm FOXM1 as a key mediator of the mitogenic functions of ERα and estrogen in breast cancer cells.
To address the physiological significance of our finding, we studied the FOXM1 mRNA expression and its relationship to ERα mRNA level in the breast cancer biopsy samples. After the exclusion of the data with high levels of FOXM1 expression (upper 25th percentile), a statistically significant correlation between ERα and FOXM1 mRNA expression was detected. This is in agreement with a recent breast cancer patient microarray dataset analysis indicating that high levels of FOXM1 mRNA expression (upper 25th percentile) are associated with poor prognosis in breast cancer (Martin et al., 2008). The discordance between ERα and FOXM1 mRNA expression in patient samples with high FOXM1 expression probably indicates that in these subjects FOXM1 expression is deregulated, with their control of FOXM1 transcription by ERα overridden by other mitogenic signals or genetic changes. Our findings that OHT represses FOXM1 expression in endocrine sensitive but not resistant breast carcinoma cell lines, and that ectopic expression of FOXM1 can abrogate the anti-proliferative effects of OHT, further confirm that deregulation of FOXM1 may contribute to anti-estrogen insensitivity. The observation that there were no significant changes in the ERα or the endogenous FOXM1 levels in the MCF-7 cells expressing the active FOXM1 highlights an important positive feedback mechanism between ERα and FOXM1, described here and previously (Madureira et al., 2006). This makes FOXM1 a particularly critical ERα target gene in breast cancer development and endocrine resistance, as the feedback loop will amplify the mitogenic action of estrogens. It is likely this positive feedback transcriptional mechanism is uncoupled in ERα-negative breast cancer, as ERα overexpression does not induce FOXM1 expression and vice versa in the ERα-negative endocrine resistant MDA-MB-231 breast carcinoma cells (Fig. S6). Previous studies have shown that changes in epigenetic regulation, such as DNA methylation and histone acetylation, are responsible silencing of the ERα gene in ER-negative human breast cancer cells (Yan et al., 2003). We also speculate that recruitment of ERα to target genes in vivo might require ‘pioneering factors, such as FOXA1, which are deregulated in ERα-negative cells (Carroll and Brown, 2006; Carroll et al., 2005). This positive feedback transcriptional mechanism may also account for the significant association between ERα and FOXM1 mRNA observed in breast tissue samples. Consistently, FOXM1 is one of eight overexpressed genes identified in solid tumours by comparison of different microarray datasets (Pilarsky et al., 2004). In conclusion, our results indicate that FOXM1 is a downstream target of ERα. Our findings also provide potential insights into the mechanism of anti-estrogen action and endocrine resistance, and show that ER regulation of FOXM1 may also contribute to anti-estrogen responsiveness.
MATERIALS AND METHODS
Cell culture
The human breast carcinoma MCF-7, ZR-75-1, and MDA-MB-231 cell lines, and COS-1 cells, were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, and 100 units/ml penicillin/streptomycin at 37°C. The OHT-resistant MCF-7-TAMR4 and TAMR7 cells and their growth conditions have previously been described (Lykkesfeldt et al., 1994; Madsen et al., 1997). The cells were exposed to ERα ligands 10−8 M estradiol (E2), 10−6 M 4-OHT (OHT) or 10−7 M ICI prepared in ethanol, or only ethanol (vehicle control) for the indicated times prior to harvesting. For steroid starvation, cells were cultured in phenol-free DMEM/F-12 containing 5% double charcoal-stripped FCS.
Plasmids and transfections
The human FOXM1 promoter constructs have previously been described (Korver et al., 1997b). Cells were transfected with the human FOXM1 promoter and Renilla (pRL-TK; Promega, Southampton, UK) as internal transfection control using Fugene-6 (Qiagen, Crawley, UK) as described (Kwok et al., 2008).
Western blotting, ChIP analysis and antibodies
Western blotting was performed on whole cell extracts by lysing cells in buffer as described (McGovern et al., 2009). Antibodies cyclin B (H-433), cyclin D1 (DCS-6), PLK (F8), tubulin (H235), ERα (F-10 and HC20), pS2 (FL-84), FOXM1 (C-20), Cyclin A (C-19), pRB (C-15), HDAC-1 (N-19) CDK2 (M2), CDK4 (C-22), and pRB (C-15) were from Santa Cruz Biotechnology (Wiltshire, UK). The anti-phospho-pRB (Ser807/811) and HDAC2 (2540) antibodies were purchased from New England Biolabs (Hitchin, UK), the anti-phospho-pRB (Thr821) antibody from Biosource (London, UK), and Cdc25b (DCS-162) from Abcam (Cambridge, UK). ChIP assays were performed as described (Essafi et al., 2009). Antibodies for acetylated H3 (06-599) and H4 (06-866) were from Upstate (Milton Keynes, UK).
Quantitative real-time PCR (qRT-PCR)
Frozen samples from patients who had undergone surgery at Charing Cross Hospital (London, UK) were used for RNA extraction. The procedures for purification of normal human breast epithelial cells have previously been described (Diaz-Arias et al., 1993; Fellowes et al., 2004; Kothari et al., 2003). FOXM1 and L19 transcript levels were quantified using the standard curve method. L19, a non-regulated ribosomal housekeeping gene, was used as an internal control to normalise input cDNA.
Gene Silencing with Small Interfering RNAs (siRNAs)
For gene silencing, cells were transiently transfected with 50 nM of the siRNA SMARTpool reagents purchased from Dharmacon (Lafayette, CO) using OligofectAMINE (Invitrogen) according to manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v5.0.
Site-directed Mutagenesis
Mutants of the FOXM1 promoter generated for this study were made using the QuickChange| Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer's protocol.
Gel shift and pull-down assays using biotin-labelled oligonucleotides
Gel shift assays were performed following a modified version of the manufacturer's protocol from the LightShift Chemiluminescent EMSA Kit (PIERCE) (Essafi et al., 2009). Nuclear and cytoplasmic extracts were prepared and pull-down assays performed as described previously (Essafi et al., 2005; Essafi et al., 2009).
Cell Cycle and proliferation assays
Cell cycle analysis was performed using propidium iodide (PI) staining alone, as previously described (Collado et al., 2000). Cell proliferation was determined using the sulforhodamine B colorimetric assay (Skehan et al., 1990).
Supplementary Material
Acknowledgments
Grant support: Breast Cancer Campaign ( J. Millour and E.W-F. Lam), Cancer Research UK (S. S. Myatt, K.-K. Ho, R. C. Coombes, E.W-F. Lam), Biotechnology and Biological Sciences Research Council (M. S. C. Wilson and E.W-F. Lam).
REFERENCES
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, et al. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–27. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- Brunner N, Zugmaier G, Bano M, Ennis BW, Clarke R, Cullen KJ, et al. Endocrine therapy of human breast cancer cells: the role of secreted polypeptide growth factors. Cancer Cells. 1989;1:81–6. [PubMed] [Google Scholar]
- Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12(Suppl 1):S47–59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–14. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Scafoglio C, Altucci L, Cancemi M, Natoli G, Facchiano A, et al. A genomic view of estrogen actions in human breast cancer cells by expression profiling of the hormone-responsive transcriptome. J Mol Endocrinol. 2004;32:719–75. doi: 10.1677/jme.0.0320719. [DOI] [PubMed] [Google Scholar]
- Collado M, Medema RH, Garcia-Cao I, Dubuisson ML, Barradas M, Glassford J, et al. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J Biol Chem. 2000;275:21960–8. doi: 10.1074/jbc.M000759200. [DOI] [PubMed] [Google Scholar]
- Diaz-Arias AA, Loy TS, Bickel JT, Chapman RK. Utility of BER-EP4 in the diagnosis of adenocarcinoma in effusions: an immunocytochemical study of 232 cases. Diagn Cytopathol. 1993;9:516–21. doi: 10.1002/dc.2840090509. [DOI] [PubMed] [Google Scholar]
- Elkak AE, Mokbel K. Pure antiestrogens and breast cancer. Curr Med Res Opin. 2001;17:282–9. [PubMed] [Google Scholar]
- Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–29. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- Essafi A, Gomes AR, Pomeranz KM, Zwolinska AK, Varshochi R, McGovern UB, et al. Studying the subcellular localization and DNA-binding activity of FoxO transcription factors, downstream effectors of PI3K/Akt. Methods Mol Biol. 2009;462:201–11. doi: 10.1007/978-1-60327-115-8_13. [DOI] [PubMed] [Google Scholar]
- Fellowes VS, Husebekk A, Gress RE, Vance BA. Minimal residual disease detection in breast cancer: improved sensitivity using cytokeratin 19 and epidermal growth factor receptor RT-PCR. Int J Oncol. 2004;24:861–7. [PubMed] [Google Scholar]
- Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TK, Poon RY. A roller coaster ride with the mitotic cyclins. Semin Cell Dev Biol. 2005;16:335–42. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Gapinski PV, Donegan WL. Estrogen receptors and breast cancer: prognostic and therapeutic implications. Surgery. 1980;88:386–93. [PubMed] [Google Scholar]
- Goss PE, Muss HB, Ingle JN, Whelan TJ, Wu M. Extended adjuvant endocrine therapy in breast cancer: current status and future directions. Clin Breast Cancer. 2008;8:411–7. doi: 10.3816/CBC.2008.n.049. [DOI] [PubMed] [Google Scholar]
- Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997a;25:1715–9. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997b;46:435–42. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- Kothari MS, Ali S, Buluwela L, Livni N, Shousha S, Sinnett HD, et al. Purified malignant mammary epithelial cells maintain hormone responsiveness in culture. Br J Cancer. 2003;88:1071–6. doi: 10.1038/sj.bjc.6600866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of FOXM1 expression. Mol Cancer Ther. 2008;7 doi: 10.1158/1535-7163.MCT-08-0188. In Press. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, et al. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008a;28:3076–87. doi: 10.1128/MCB.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008b;7:2720–6. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Leung TW, Lin SS, Tsang AC, Tong CS, Ching JC, Leung WY, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:R81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher-Firzlaff JM, Lilischkis R, Luscher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580:1716–22. doi: 10.1016/j.febslet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Lykkesfeldt AE, Larsen JK, Christensen IJ. Cell cycle analysis of estrogen stimulation and antiestrogen inhibition of growth of the human breast cancer cell line MCF-7. Breast Cancer Res Treat. 1986;7(Suppl):S83–90. [PubMed] [Google Scholar]
- Lykkesfeldt AE, Madsen MW, Briand P. Altered expression of estrogen-regulated genes in a tamoxifen-resistant and ICI 164,384 and ICI 182,780 sensitive human breast cancer cell line, MCF-7/TAMR-1. Cancer Res. 1994;54:1587–95. [PubMed] [Google Scholar]
- Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- Madsen MW, Reiter BE, Larsen SS, Briand P, Lykkesfeldt AE. Estrogen receptor messenger RNA splice variants are not involved in antiestrogen resistance in sublines of MCF-7 human breast cancer cells. Cancer Res. 1997;57:585–9. [PubMed] [Google Scholar]
- Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–76. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiakowski P, Breathnach R, Bloch J, Gannon F, Krust A, Chambon P. Cloning of cDNA sequences of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982;10:7895–903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. 2009;8:582–91. doi: 10.1158/1535-7163.MCT-08-0805. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. Promiscuous and lineage-specific roles of cell cycle regulators in haematopoiesis. Cell Div. 2007;2:6. doi: 10.1186/1747-1028-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK, McGuire WL. The use of steroid hormone receptors in the treatment of human breast cancer: a review. Bull Cancer. 1979;66:203–9. [PubMed] [Google Scholar]
- Park HJ, Wang Z, Costa RH, Tyner A, Lau LF, Raychaudhuri P. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene. 2008;27:1696–704. doi: 10.1038/sj.onc.1210814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–50. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008:6. doi: 10.1002/0471250953.bi0206s21. Chapter 2: Unit 2. [DOI] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Tashiro E, Tsuchiya A, Imoto M. Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci. 2007;98:629–35. doi: 10.1111/j.1349-7006.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33:1404–14. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–6. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res. 2008;68:5628–38. doi: 10.1158/0008-5472.CAN-07-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J. FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3alpha. Biochem Biophys Res Commun. 2006a;348:99–108. doi: 10.1016/j.bbrc.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol Chem. 2006b;387:949–62. doi: 10.1515/BC.2006.119. [DOI] [PubMed] [Google Scholar]
- Yamashita H. Current research topics in endocrine therapy for breast cancer. Int J Clin Oncol. 2008;13:380–3. doi: 10.1007/s10147-008-0818-7. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Takahashi S, Ito Y, Yamashita T, Ando Y, Toyama T, et al. Predictors of response to exemestane as primary endocrine therapy in estrogen receptor-positive breast cancer. Cancer Sci. 2009;100:2028–33. doi: 10.1111/j.1349-7006.2009.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-alpha (ER) in ER-negative human breast cancer cell lines. Cancer Biol Ther. 2003;2:552–6. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–80. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Rondaij M, Jalink K, Sharp ZD, Mancini MA, Neefjes J, et al. Resistance to antiestrogen arzoxifene is mediated by overexpression of cyclin D1. Mol Endocrinol. 2009;23:1335–45. doi: 10.1210/me.2008-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.