Abstract
Dihydrodiol dehydrogenases are a family of aldo-keto reductases (AKR1Cs) involved in the metabolism of steroid hormones and xenobiotics. Herein, we have cloned and characterized the proximal promoter region of the human AKR1C1 gene. The 5’ flanking proximal promoter region of the AKR1C1 gene consists of a TATA box and an inverted CCAAT binding site. Deletion analysis of the 5’-flanking, ~3.0 kb region of the human AKR1C1 gene identified the region between −128 to −88 as the minimal proximal promoter essential for basal transcription of AKR1C1 in human ovarian (2008 & 2008/C13*), lung (H23 & A549) and liver carcinoma (HepG2) cells. Site-directed mutagenesis studies indicated that the transcription factor binding sites for NF-Y/CEBP were involved in controlling the basal transcription of AKR1C1 in all the cancer cells studied. Electrophoretic mobility shift (EMSAs) and gel supershift assays demonstrated that the transcription factor NF-Y preferentially binds to the inverted CCAAT box at −109ATTGG−105 of the AKR1C1 gene. Chromatin immunoprecipitation (ChIP) analysis confirmed the in vivo association between NF-Y and human AKR1C1 gene promoter in human ovarian, lung and liver carcinoma cells. Ectopic expression of NF-Y’s increased the AKR1C1 gene transcription, whereas expression of a dominant-negative NF-YA or suppression of NF-YA decreased the AKR1C1 gene transcription. A 2-fold increase in AKR1C1 transcription was observed specifically in cisplatin-treated 2008 cells that was CCAAT box-dependent. These results indicate that the NF-Y regulates the basal transcription of AKR1C1 in human ovarian, lung and liver carcinoma cells and the cisplatin-induced transcription in human ovarian carcinoma cells.
Keywords: Hydroxysteroid dehydrogenase, dihydrodiol dehydrogenase, promoter regulation, ovarian carcinoma, lung adenocarcinoma, liver hepatoblastoma
1. INTRODUCTION
Human dihydrodiol dehydrogenases (AKR1Cs) are a family of cytosolic NADP(H)-dependent oxidoreductases involved in the de novo detoxification of xenobiotics, steroids and polycyclic aromatic hydrocarbons (PAH) (Penning and Byrns, 2009; Jin and Penning, 2007; Burczynski et al., 1999). Four different isoforms of AKR1C have been cloned and characterized in human liver, viz., i) AKR1C1/DDH1 (20α (3α)-hydroxysteroid dehydrogenase (HSD)), ii) AKR1C2/DDH2 (Type 3 3α-HSD), iii) AKR1C3/DDH3 (Type 2 3α (17β)-HSD) and iv) AKR1C4/DDH4 (Type 1 3α-HSD) (Penning et al., 2000). Of the four isoforms AKR1C1 and AKR1C2 share nearly 98% amino acid sequence homology with a difference of only seven amino acids (Dufort et al., 1996; Lou et al., 2006). AKR1C1 - AKR1C3 are also expressed in the small intestine, lung, mammary gland and prostate, while the expression of AKR1C4 is restricted to the liver (Jin and Penning, 2007; Ji et al., 2005). Each of the AKR1C isoforms has a unique substrate specificity and function. Thus, AKR1C1 is involved in inactivation of progesterone to 20α-hydroxyprogesterone, while AKR1C2 breaks down 5α-dihyrotestosterone (5α-DHT) to 3α-androstanediol (3α-diol), AKR1C3 converts 17-ketosteroid to testosterone and AKR1C4 converts 3-ketosteroids to 3α-hydroxysteroids (Penning and Byrns, 2009).
Altered expression of AKR1Cs has been observed in a variety of primary human cancers. Overexpression of AKR1C1 in non-small-cell lung cancer (NSCLC) patients has been associated with poor prognosis (Hsu et al., 2001). Overexpression of AKR1C1 has also been observed in patients with esophageal and breast cancer (Wang et al., 2004; Zhang et al., 2005). Reports by various authors have indicated increased expression of AKR1C3 in prostate and breast tumors (Stanbrough et al., 2006; Penning et al., 2006; Sasano et al., 2008). In contrast, reduced expression of AKR1C1 and AKR1C2 has been associated with the development of prostrate and ovarian cancer (Ji et al., 2003, Ji et al., 2005). Development of anticancer drug resistance has also been associated with altered expression of the AKR1C isoforms. Thus, increased expression of AKR1C2 was observed in HeLa cervical carcinoma cells resistant to doxorubicin and cisplatin (Lee et al., 2002). Studies from our laboratory have demonstrated an association between increased expression of AKR1C1 and AKR1C2 (at the mRNA and the protein level) and the development of cisplatin resistance in human ovarian carcinoma cells (Deng et al., 2002). Further, ectopic expression of full length AKR1C1 cDNA in parental, cisplatin-sensitive human ovarian (2008), lung (Calu-6, H23), germ cell (Tera-2) and cervical cancer (A431) cells resulted in development of cisplatin-resistance (Deng et al., 2004; Hung et al., 2006). Also, inherent cisplatin-resistance of the human lung adenocarcinoma (A549) cells was found to be associated with an increased expression of AKR1C1, AKR1C2 and AKR1C3 (Deng et al., 2004). Moreover, the overexpression of AKR1C1 has also been associated with development of cisplatin-resistance in ovarian cancer patients treated with cisplatin (Chen et al., 2005). However, the identity of the transcriptional machinery that controls the basal as well as induced expression of AKR1C1 in human cancer cells (and thus the development of anticancer drug resistance) remains elusive.
A review by Torigie et al., (2005) describes the potential role of various transcription factor(s) involved in the development of cisplatin resistance. Recent observations by Selga et al., (2008) suggest that the transcription factor Sp1 regulates the overexpression of AKR1C1 in HT29 human colon cancer cells resistant to methotrexate. Induction of AKR1C in the HT29 cells treated with ethacrynic acid has also been observed (Ciaccio et al., 1994). In human HepG2 cells, nuclear factor-erythroid-derived 2-related factor (NRF2) was found to regulate the transcription of AKR1C2 by binding the antioxidant response element (ARE), a cis-acting element, in response to phase II inducers (Lou et al., 2006). However, transcription factor(s) involved in regulating the transcription of AKR1C3 gene in response to ethacrynic acid (EA) were not identified (Ciaccio et al., 1996).
Considering the growing evidence of the important role of AKR1C in the development of tumor cells resistance to anticancer drugs, it is important to identify and characterize the factors that control their transcription. In this study, utilizing functional deletion constructs, we have demonstrated that the region between −128 to −88 of the AKR1C1 gene promoter was essential for its basal transcription in human ovarian, lung and liver cancer cells. Importantly, we have demonstrated that the inverted CCAAT box binding transcription factor NF-Y regulates the basal transcription of the human AKR1C1 gene in human ovarian, lung and liver carcinoma cells. We have also demonstrated that NF-Y controls the cisplatin-induced transcription of the AKR1C1 gene in a cell type-dependent manner.
2. MATERIALS AND METHODS
2.1 Cell culture
The human ovarian carcinoma cell lines 2008 and 2008/C13* were grown in RPMI-1640 supplemented with 10% fetal calf serum (HyClone, Waltham, MA) and gentamicin (Lonza, Basel, Switzerland) at a final concentration of 10µg/ml. The lung adenocarcinoma cell lines H23 and A549 were maintained in RPMI-1640/Ham’s F12K medium supplemented with 10% fetal calf serum and gentamicin at final concentration of 10µg/ml and the human liver hepatoblastoma cell line HepG2 was maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum and gentamicin at a final concentration of 10µg/ml. All cells were grown at 37°C in a 5% CO2 incubator.
2.2 Construction of plasmid vector and 5’ deletion analysis
The BAC clone RP-379P14 (168053–171109) which contains the 5’ flanking sequence of AKR1C1 gene (GeneBank Accession No: AB032150) was utilized for construction of AKR1C1 promoter - luciferase plasmids. PCR fragments generated using Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany) were cloned into the reporter vector at the KpnI, Nhe I or XhoI polycloning site by incorporating corresponding restriction endonuclease sites in the forward primer and Hind III restriction endonuclease site in the reverse primers. The −2954/+59 bp luciferase promoter which consists of the 2954bp upstream and 59bp downstream of transcription start site (Nishizawa et al., 2000) was cloned in the NheI-HindIII sites of pGL3-basic vector (Promega, Madison, WI). Similarly, the −2567/+59, −1648/+59 PCR fragments were cloned in the NheI-HindIII polycloning site in pGL3-basic vector. In case of −2301/+59, the DNA fragment was inserted in the XhoI-HindIII site of the luciferase vector. PCR amplified promoter fragments −641/+59, −349/+59, −180/+59, −152/+59, −128/+59, −88/+59 and −49/+59 were cloned in the KpnI-HindIII sites of pGL3-basic vector. The nucleotide sequence of the DNA fragments inserted into the luciferase vector was verified by sequencing.
2.3 Transient transfection and Luciferase assay
Cells were seeded in 6-well plates at a density of 8×105 cells/well for 2008, 2008/C13* and 5 × 105 cells/well for H23, A549 and HepG2 24h before transfection. Transfection was accomplished with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) using the manufacturer’s protocol. In each experiment, 2µg of the pGL3 basic reporter plasmid (without the 2954bp insert, empty vector) or the effector plasmid (the 2954bp insert or the sequentially deleted PCR fragments of human AKR1C1 promoter region) was transfected along with 0.25–0.5µg of pRL-SV40 (Renilla luciferase, Promega, Madison, WI) as an internal standard. Following incubation with the DNA complex for 4h (H23, A549, HepG2) and 24h (2008, 2008/C13*), cells were washed and incubated in fresh growth medium for an additional 44 and 24 hours, respectively. The pGL3-basic vector and pGL3-control vector (Promega, Madison, WI) were used as negative and positive controls, respectively.
In experiments designed to directly assess the role of NF-Y in regulating the transcription of AKR1C1 gene promoter, we utilized NF-YA, NF-YB, NF-YC cDNA expression vectors (Δ4YA-13, Δ4YB and Δ4YC) or the NF-YA dominant-negative expression vector (Δ4YAm29, obtained from Dr. Robert Mantovani, University of Milan, Italy, Mantovani et al., 1994). The human ovarian carcinoma cell lines (2008, and 2008/C13*) were seeded at a density of 5×105 cells/well, while human lung adenocarcinoma (H23 and A549) and liver hepatoblastoma cell line (HepG2) were seeded at a density of 3×105 cells/well 24h before transfection. Initially, 2µg of the dominant-negative NF-YA construct Δ4YAm29 or the wild type NF-YA, -YB and -YC constructs was transfected into the cells using Lipofectamine 2000 according to the manufactures instruction. After 24h, cells were co-transfected wit 2µg of pAKR1C1 −180/+59 and 125ng of pRL-SV40 as internal control.
To assess the effects of cisplatin exposure on the AKR1C1 promoter activity, cells were treated with 10µM concentrations of cisplatin (Sigma, St. Louis, MO) during the final 16–18h of incubation. Thus, 48 hours (in basal and cisplatin inducted promoter activity studies) post-transfection, cells were washed with 1x phosphate-buffered saline (PBS) and lysed in 250µl/well of 1x passive lysis buffer (Promega, Madison, WI). Luciferase activity was assayed using 20µl of the cell supernatant and the Dual-luciferase reporter plasmid system (Promega, Madison, WI) with a Turner single tube luminometer Model 20/20. Luciferase activities were normalized against Renilla luciferase (internal standard pRL-SV40) activity to determine the relative luciferase activity and the fold activation. Cells transfected with the pGL3 basic vector and treated with cisplatin were considered as negative control for cisplatin induction studies.
2.4 NF-Y overexpression, NF-YA knockdown (by siRNA) and AKR1C1 expression analysis
To asses the effect of overexpression of NF-Y in regulating the AKR1C1 mRNA expression levels, 2008 (human ovarian carcinoma cells) were transiently transfected with 1µg of NF-YA, -YB and -YC cDNA expression vector or pCMV vector as negative control using lipofectamine 2000. After 24h, RNA was isolated from cells using Trizol (Invitrogen, CA) as described by the manufacturer.
Alternatively, the 2008 cells were transiently transfected with 500 picomoles of human NF-YA siRNA (siGENOME smart pool, Dharamacon, IL) or the control si-RNA (siGENOME Non-targeting siRNA pool#1, Dharamacon, IL). Transfection was performed utilizing siPORT NeoFX transfection reagent (Ambion, CA). RNA was isolated 24h after transfection using Trizol reagent (Invitrogen, CA) as instructed by the manufacturer.
One microgram of the RNA was used in the reverse transcription reaction along with 4 units of Omniscript reverse transcriptase (Qiagen, CA), 1µM oligo-dT primer (Applied Bio-systems, CA), 0.5mM dNTP (Roche), 10 units of RNase inhibitor (Applied Biosystems, CA) and 1x RT buffer (Qiagen, CA). Reverse transcription was performed at 37°C for 1h with a final incubation at 93°C for 5 min for inactivation of reverse transcriptase. Two microliters of the RT-product was used in the real-time PCR reaction. The Quantitect SYBR green PCR kit (Qiagen, CA) was utilized and PCR was performed according to manufactures instructions using the Realplex Eppendorf master cycler (Hamburg, Germany). Each PCR reaction consisted of 50% (v/v) of 2x SYBR green master mix, 4mM MgCl2, and 0.2µM gene-specific forward and reverse primers (Table 2). Quantification of glyceraldehyde-6-phosphate dehydrogenase was used to normalize the relative expression levels of AKR1C1 mRNA under different treatment conditions. Each experiment was performed in duplicates and repeated at least twice.
Table 2.
List of primers utilized for RT-PCR analysis
| Gene | Forward | Reverse | Annealing temperature |
|---|---|---|---|
| GAPDH | 5’-acccactcctccacctttg-3’ | 5’-ctcttgtgctcttgctggg-3’ | 55°C |
| AKR1C1 | 5’-gtaaagctttagaggccac-3’ | 5’-ataaggtagaggtcaacataa-3’ | 55°C |
| NFYA | 5’-gtccagaccctccaggtagt-3’ | 5’-gggaccaactgtatttgctg-3’ | 58°C |
GAPDH- Glyceraldehyde-6-phosphate dehydrogenase; AKR1C1- Aldo keto-reductase 1C1; NF-YA- Nuclear factor Y alpha. The PCR was performed with an initial denaturation at 95°C for 15 mins, followed by 45 cycles of 94°C for 15 s, annealing for 30s and extension at 72°C for 30s.
2.5 Computational analysis of AKR1C1 promoter
Potential transcription factor(s) binding sites within the AKR1C1 gene promoter were screened using the ALIBABA 2.0 program (www.gene-regulation.com). The program searches putative transcription binding sites against the TRANSFAC database.
2.6 Site-directed mutagenesis
The −180/+59 AKR1C1 promoter fragment was used to generate mutant clones of AKR1C1 promoter. The Quickchange lightning site-directed mutagenesis kit (Stratagene, La Jolla, CA) was utilized to generate mutants Mut1 pAKR1C1–180/+59, Mut2 pAKR1C1–180/+59 and Mut3 pAKR1C1–180/+59. Primers for introduction of point mutation were designed as instructed by the manufacturer. The nucleotide sequence of the mutated clones was verified by sequencing. The promoter activity of the mutated clones was assayed by transient transfection and luciferase assay as detailed in section 2.3.
2.7 Preparation of nuclear extracts
Nuclear extracts were prepared according to a method described by Suzuki et al., (1992) with some modifications. Cells were seeded at a density of 3×106 (2008 and 2008/C13* cells) or 4×106 (H23, A549 and HepG2 cells) in 100mm culture dishes 24h before extraction. Trypsinized cells were washed with ice-cold 1X PBS and centrifuged at 268 × g for 10 minutes. The cells were resuspended in 400µl of buffer containing 10mM HEPES, pH 7.8; 10mM KCl; 2mM MgCl2; 1mM DDT and Protease inhibitor cocktail (1:20 v/v) with 50µl of 1% NP-40, vortexed for 15s and then centrifuged at 11337 × g for 30s. The resulting cell pellet was washed with 400µl of buffer containing 10mM HEPES, pH 7.8; 10mM KCl; 2mM MgCl2; 1mM DDT and resuspended in 75µl of buffer containing 50mM HEPES, pH 7.8; 50mM KCl; 300mM NaCl; 0.1mM EDTA; 1mM DTT; 10% glycerol and Protease inhibitor cocktail (1:20 v/v) and rotated at room temperature (RT) for 20 min. The nuclear extract (supernatant) was recovered by a final centrifugation at 11337 × g for 2 min, and the protein concentration was determined by the Bradford’s colorimetric reagent (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as the standard.
2.8 Electrophoretic Mobility Shift Assays (EMSAs)
The wild-type and mutant probe were synthesized as double-stranded oligonucleotide (Integrated DNA Technology) from the −120 to −91 region of the AKR1C1 gene. Consensus oligonucleotide for CBF/NF-Y (CCAAT binding factor/Nuclear factor-Y), Sp1, C/EBP (CCAAT/enhancer binding protein) and NF-1 (CCAAT Transcription factor) were synthesized based on the sequence data from Santa Cruz Biotechnology (Santa Cruz, CA). The sequences of the probes utilized were; WT- 5’-CTCTCACATGCCATT GGTTAACCAGCAGACT-3’; Mutant- 5’- CTCTCACATGCCTTCAGTTAACCAGC AGACT −3’, (mutated bases shown as bold and underlined); CBF/NF-Y- 5’-AGACCGTACGTGATTGGTTAATCTCTT-3’; Sp1-5’-ATTCGATCGGGGCGGGGC GAGC-3’; C/EBP-5’-TGCAGATTGCGCAATC TGCA-3’; NF-1-5’-TTTTGGATTGA AGCCAATATGATAA-3’. All the probes were labeled with biotin using the Biotin 3’-end DNA labeling kit (Pierce Chemical, Rockford, IL) according to the manufacturer’s instruction. Eight micrograms of the nuclear extract was utilized for the binding reactions. The EMSA binding reactions were performed at room temperature for 30 min and consisted of the nuclear extract in 1x binding buffer (50% glycerol, 100mM MgCl2, 1µg/µl Poly (dI-dC), 1% NP-40, 1M KCl, 200mM EDTA and 50 pmol DNA probe). The mixture was fractionated on 8% nondenaturing polyacrylamide gels in 0.5X Tris borate-EDTA buffer at 170V. The protein-DNA complex was then transferred to Hybond-N+ nylon membrane using the Trans-Blot semi-dry method (Bio-Rad, Hercules, CA) and crosslinked using Spectrolinker XL-1000 UV crosslinker (Spectronics Corporation, Westbury, NY). Detection of biotin labeled DNA was performed using the LightShift chemiluminescent EMSA kit (Pierce Chemicals, Rockford, IL) and visualized by exposure to a charge-couple device camera (Fujifilm LAS 3000, Tokyo, Japan).
In competition EMSA, 100-fold molar excess of the cold, mutant or consensus oligonucleotides were added to the EMSA binding reaction. For the gel-supershift assay, following the incubation of the nuclear extracts with the 30bp WT AKR1C1 promoter fragment for 30 min, 2µg of NF-YA polyclonal antibody, 2µg of NF-YB polyclonal antibody and/or the three polyclonal antibodies (anti-NF-YA, -NF-YB and -NF-YC) (Santa Cruz Biotechnology, Santa Cruz, CA) were added to the binding reaction and the mixture incubated at RT for an additional 1h. The rabbit pre-immune IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was utilized as negative control in the supershift assay. The mixture was fractionated on a 5% nondenaturing polyacrylamide gel. Transfer and detection was performed as described above.
2.9 Chromatin Immunoprecipitation Assay (ChIP)
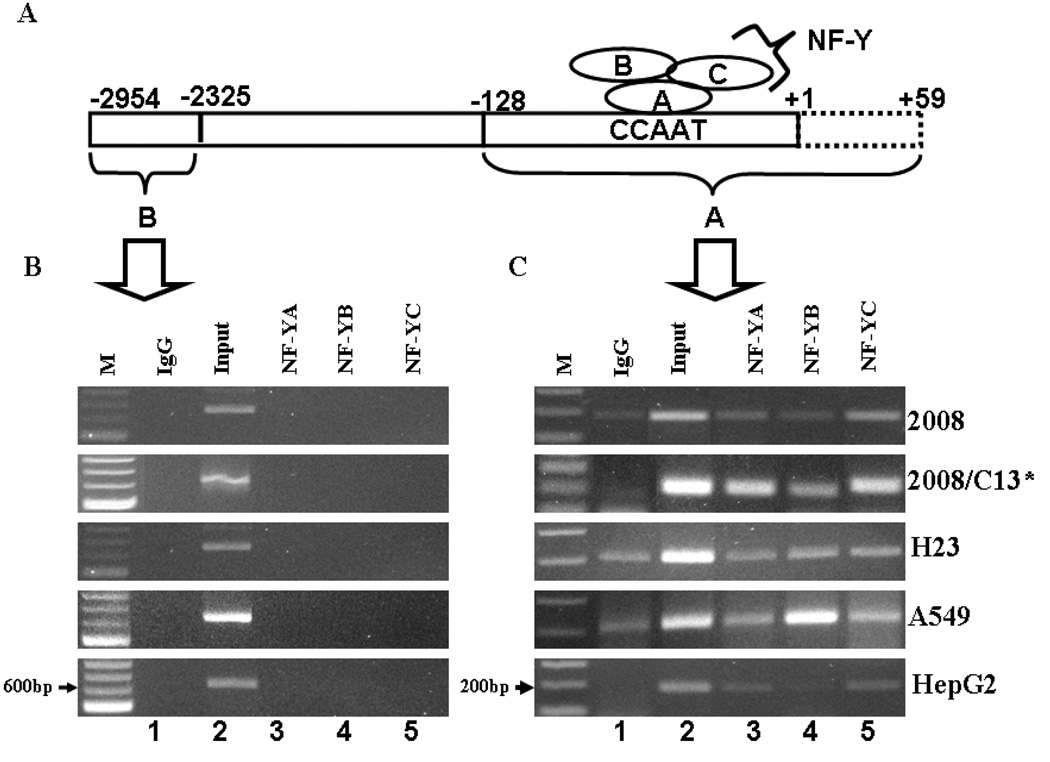
In order to analyze the in vivo association of NF-Y with the AKR1C1 gene promoter, chromatin immunoprecipitation analysis was performed. 1–2×107 cells of human ovarian, lung or liver carcinoma cells were treated with 1% formaldehyde (final concentration, v/v) for 30 mins at 37°C to cross-link proteins to DNA. After cross-linking the cells were washed twice with 1X ice cold PBS containing protease inhibitor cocktail (Sigma, MO). The cells were scrapped from tissue culture plates and centrifuged at 700 × g for 2 min, resuspended in 1mL of SDS-lysis buffer with protease inhibitor cocktail. The cells were then sonicated (Sonifier Cell disruptor 350, Branson Sonic Power Co, MA) at 30% of maximum power, 3 times for 15s each, with a 30s cooling interval. The cell lysate was centrifuged at 10,000 × g for 10 min at 4°C and the supernatant further diluted 10-folds in the ChIP dilution buffer containing 0.01% SDS, 1.1% Triton X-100, 1.2mM EDTA, 16.7mM Tris-HCl (pH 8.1), 167mM NaCl, and protease inhibitor cocktail. 25µl of the diluted DNA fraction was kept aside as input for PCR. The remaining DNA fraction was precleared using 75µl of protein G agarose beads (50% slurry) for 2h at 4°C. Immunoprecipitation was performed by adding 10µg of NF-YA, NF-YB, NF-YC or normal rabbit IgG as negative control. The immunocomplex was precipitated by incubation with 60µl of protein A sepharose beads for 2h at 4°C. The protein/DNA complex was eluted using 200µl of elution buffer (1% SDS, 0.1M NaHCO3) from the beads. Cross-linking of protein-DNA was reversed by incubation at 65°C overnight. The DNA was purified using spin columns and 2µl of the DNA was used in the PCR reaction for amplification of 187bp (−128/+59) or 629bp (−2954/-2325) of the AKR1C1 promoter region (see Fig 5A). PCR reactions were performed utilizing the sense and antisense primers (5’-CTGTCCCTCCTCTCACAT-3’; 5’-CAAGCCGACCAGTATGAT-3’) for region −128/+59 harboring the CCAAT transcription factor binding site, while the primers utilized for region −2954/-2325 were (forward- 5’-CACCACACTGACTTCCAC A-3’; reverse- 5’-CAAGCCGACCAGTATGAT-3’). Taq DNA polymerase (Roche, IN) was used to perform PCR with the following conditions: Initial denaturation for 95°C for 5 minutes followed by 30 cycles of denaturation of 95°C for 15s, annealing at 55.5°C for 30s, extension at 72°C for 30s with a final extension of 72°C for 10 min. The PCR products were fractionated on 1.5% agarose gels and visualized by ethidium bromide staining.
Fig. 5.
In vivo association between NF-Y and human AKR1C1 gene promoter. (A) Schematic representation of the nuclear factor-Y subunits A, B and C association with the CCAAT pentanucleotide sequence in the AKR1C1 gene proximal promoter region. (B) and (C) Chromatin immunoprecipitation assay for detection of the in vivo association between NF-Y and the human AKR1C1 gene promoter. (B) PCR product amplified from primers utilized from an unrelated part of the AKR1C1 gene which is void of the CCAAT pentanucleotide binding site for NF-Y transcription factor binding. (C) The immunoprecipitated DNA obtained from ChIP is amplified utilizing PCR primers specific for a region containing the NF-Y transcription factor binding site in the AKR1C1 gene promoter.
3.0 Statistical analysis
Statistical analysis was performed for calculating the significant differences in luciferase activity between constructs and cisplatin treatment by one way randomized analysis of variance (ANOVA) and Newmans-Keuls test with significance level of p<0.05.
3. RESULTS
3.1 Nucleotide sequence of AKR1C1 gene
The AKR1C1 nucleotide gene sequence (GenBank Accession No AB032150) was utilized to construct the 5’-3 kb basal promoter region. Based on the sequence, the first exon region was identified to be present between +1 to +83 nucleotides (Fig. 1A). The 5’ flanking region upstream of the +1 transcriptional start site (TSS) was tentatively considered to harbor the promoter for transcriptional regulation of AKR1C1 gene.
Fig. 1.
Sequence of the 5’ flanking region of the human AKR1C1 gene and construction of 5-deletion clones of the human AKR1C1 gene promoter. (A) The transcription start site (TSS), +1 is indicated. The sequence has been numbered with reference to the TSS. Putative, potential transcription factor(s) binding sites identified using the Ali-baba 2.0 computational software are shown in boxed regions. The region between −128 to −88 has a cluster of transcription factor(s) binding sites which are believed to be important for basal expression of human AKR1C1 gene. (B) Diagrammatic representation of the full length and sequentially deleted AKR1C1 promoter constructs used in this study. The transcription start site (TSS) is numbered as +1 and the constructs are numbered with reference to the TSS from 5’-3’ ends.
3.2 Characterization of human AKR1C1 proximal promoter
To determine the transcriptional elements in the 5’-flanking region of human AKR1C1 promoter, a series of PCR deletion clones were constructed in a pGL3-basic luciferase vector (Fig.1B) and assayed for promoter activity. Initially all the clones were assayed for promoter activity in the HepG2 cells in comparison with pGL3-basic vector (negative control). The DNA fragments −641/+59 and −1648/+59 showed the highest (11-fold) increase in relative luciferase activity (RLA) (Table 1). As shown in Fig.1B, a deletion construct containing the −128/+59 region displayed a 4-fold increase in activity compared to the pGL3 basic vector in HepG2 cells. Moreover, the pAKR1C1-88/+59 clone showed no activity above background. The activity of the construct containing the −128/+59 region of the AKR1C1 gene was similar to the full length construct pAKR1C1 −2954/+59. These results imply that the clone pAKR1C1−128/+59 contains the minimal proximal promoter of the human AKR1C1 gene. Clones pAKR1C1 −2954/+59, pAKR1C1-2567/+59, pAKR1C1-2301/+59, pAKR1C1-349/+59, pAKR1C1 −180/+59 and pAKR1C1 −152/+59 showed luciferase activity comparable to the pAKR1C1 −128/+59 clone (Table 1).
Table 1.
Identification of the human AKR1C1 basal proximal promoter.
| Fold increase in relative luciferase activity (RLA) | |||||
|---|---|---|---|---|---|
| 5’ Deletion Constructs | HepG2 | 2008 | 2008/C13* | H23 | A549 |
| pGL3 Easic | 1 | 1 | 1 | 1 | 1 |
| pAKRICI −88/+59 | 1.1 ± 0.2 | 1.4 ± 0.4 | 1.3 ± 0.8 | 1 ± 0.3 | 2.8 ± 1 |
| pAKRICI −128/+59 | 4.5 ± 2.8 | 8.2 ± 0.6 | 3.3 ± 0.6 | 7.7 ± 1.0 | 22.0 ± 9.8 |
| pAKRICI −152/+59 | 6.6 ± 2.0 | - | - | - | - |
| pAKRICI −180/+59 | 7.1 ± 2.5 | 10.5 ± 3.6 | 7.3 ± 1.0 | 10.7 ± 1.5 | 17.8 ± 1.7 |
| pAKRICI −349/+59 | 8.8 ± 3.2 | - | - | - | - |
| pAKRICI −641/+59 | 11.2 ± 6.5 | 13.7 ± 2.6 | 11 ± 3.0 | 15.8 ± 3.0 | 33 ± 3.8 |
| pAKRICI −1648/+59 | 11.0 ± 3.0 | - | - | - | - |
| pAKRICI −2301/+59 | 6.7 ± 1.6 | - | - | - | - |
| pAKRICI −2567Z+59 | 6.5 ± 1.7 | - | - | - | - |
| pAKRICI −2954/459 | 5.8 ± 1.8 | 13.5 ± 8.0 | 22.9 ± 8.5 | 36.4 ± 19.2 | 29.2 ± 1.8 |
Human liver carcinoma cells (HepG2), ovarian carcinoma cells (2008, 2008/C13*) were transfected with various AKR1C1 promoters constructs shown in Fig 1B and assayed for luciferase activity after 48h as described in Materials and Methods. Fold increase in relative luciferase activity (RLA) was compared with that observed with pGL3 basic (set as 1). Normalization in transfection efficiency was performed by co-transfection with pRL-SV40 (Renilla expression vector). The mean ± S.D are from three different experiments, each performed in triplicate.
To assess the AKR1C1 gene transcription in 2008, 2008/C13*, H23 and A549 cells, promoter constructs harboring DNA fragments −88/+59, −128/+59, −180/+59, −641/+59 and −2954/+59 of the AKR1C1 gene were utilized (Fig 1B). A 3- to 10-fold increases in relative luciferase activity (RLA) was observed in 2008 and 2008/C13* cells (Table 1) while 8- to 19-fold increase in RLA was observed in H23 and A549 cells (Table 1) containing the pAKR1C1 −128/+59 construct compared to pAKR1C1 −88/+59 whose activity was same as the negative control (pGL3 Basic). The luciferase activity of all the tested clones was similar to the pAKR1C1 −128/+59 construct. These results identify the −128 to −88 region as the minimal proximal promoter region sufficient and essential for the transcription of the human AKR1C1 gene in human ovarian, lung and liver carcinoma cells.
3.3 Identification of potential transcription factor(s) binding sites
The proximal promoter region (−128 to −88) of the AKR1C1 gene was analyzed for potential transcription factor(s) binding sites using the ALIBABA 2.0 software (www.gene-regulation.com). A tandem of transcription factor(s) binding sites for Sp1, NF-Y (Nuclear factor-Y), C/EBP (CCAAT enhancer binding protein), p40X and USF (Upstream stimulating factor) were identified within this region of −128 to −88 of the AKR1C1 gene (Fig. 1). Sequences of transcription factor(s) binding sites are provided in Fig. 2A.
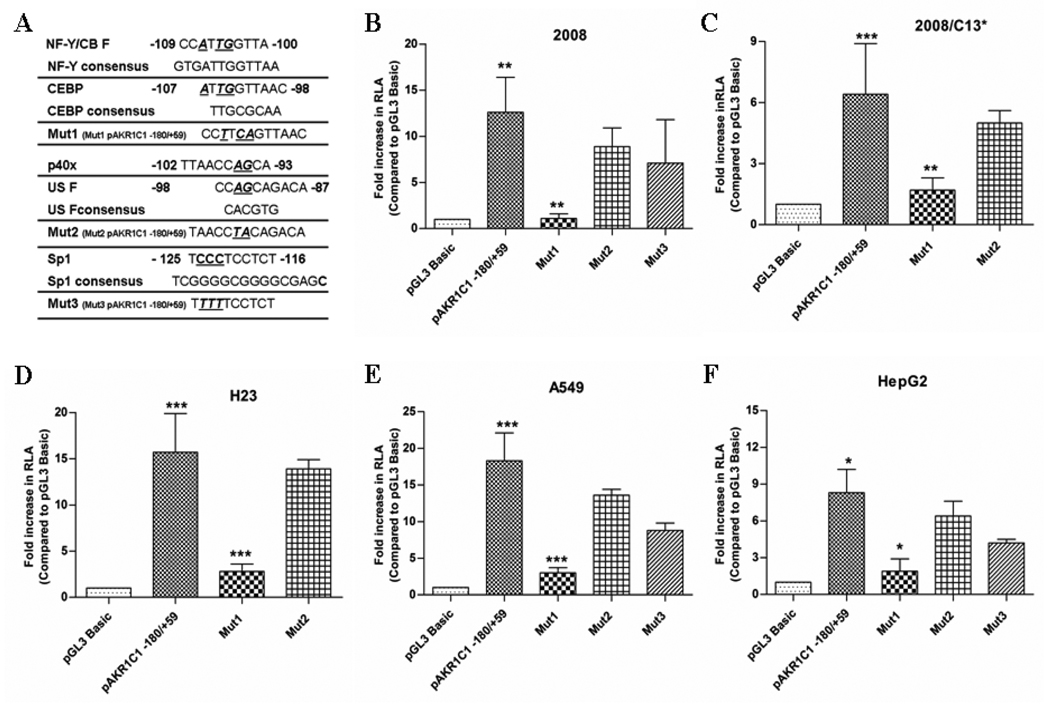
Fig. 2.
The NF-Y/C-EBP binding site controls the transcription of the human AKR1C1 gene. (A) Three different mutants where obtained as explained in Materials and Methods. Transcription factor binding sites mutated in the AKR1C1 promoter region are indicated in bold, italics and are underlined. Mut1 pAKR1C1 −180/+59 (Mut1) targeted the binding sites for transcription factor(s) NF-Y and CEBP, Mut2 pAKR1C21 −180/+59 (Mut2) targeted the binding sites for transcription factor(s) p40X and USF, while Mut3 pAKR1C1 −180/+59 (Mut3) was targeted towards the Sp1 binding site. (B) 2008, (C) 2008/C13*, (D) H23, (E) A549 and (F) HepG2 cells were transfected with Mut1, Mut2, Mut3, or the wild-type promoter (pAKR1C1-180/+59) and assayed for luciferase activity after 48h as described in Table 1. Transfection efficiency was normalized by co-transfection with pRL-SV40 (Renilla expression vector). The mean ± S.D are from three different experiments, each experiment performed in triplicate (***p<0.001, **p<0.01, *p<0.05 with pAKR1C1 −180/+59 compared to the control pGL3 Basic vector and Mut1 compared to pAKR1C1 −180/+59 wild-type construct).
3.4 Screening of specific transcription factor(s) by site-directed mutagenesis
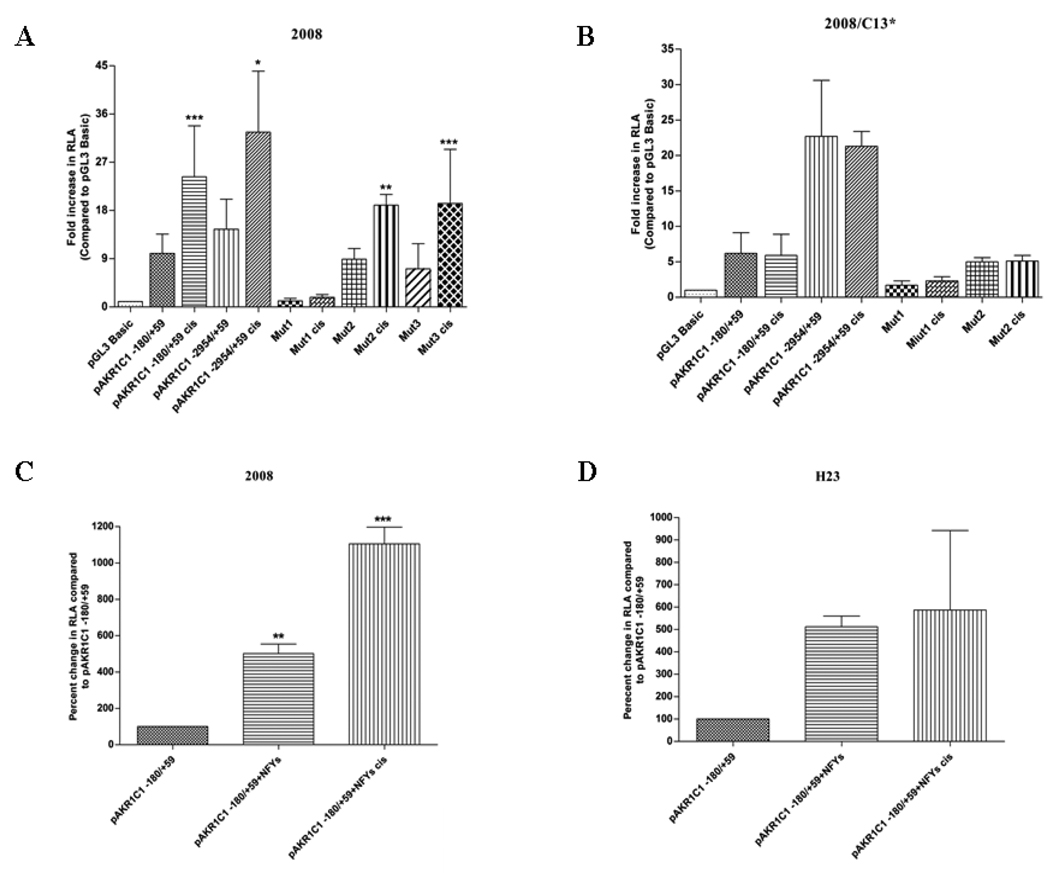
Results from the 5’ deletion analysis and computational screening identified potential binding sites for Sp1, NF-Y, C/EBP, p40X and USF transcription factor in the region between −128 to −88 of the AKR1C1 gene (the basal proximal promoter region). Utilizing PCR based site-directed mutagenesis, point mutations were introduced within the transcription factor(s) binding site(s) (Fig. 2A). Thus, the tandem transcription factor binding site for NF-Y and C/EBP in pAKR1C1–180/+59 clone was mutated to generate the construct Mut1 pAKR1C1 −180/+59 using the forward and reverse primers 5’-GTCCCTCCTCTCACATGCCTTCAGTTAACCAGCAGACAGTGT-3’; 5’- ACACTG TCTGCTGGTTAACTGAAGGCATGTGAGAGGAGGGAC. Similarly, the tandem transcription factor binding site for p40X and USF was mutated using the forward and reverser primers 5’- ACATGCCATTGGTTAACCTACAGACAGTGTGCTCAGGG-3’; 5’- CCCTGAGCACACTGTCTGTAGGTTAACCAATGGCATGT-3’ to generate the Mut2 pAKR1C1 −180/+59 construct, while the binding site for Sp1 in pAKR1C1 −180/+59 were mutated using 5’- TCACAGCTTGTGTAAGACTGCCTCTGTTTTTCCT CTCACATGCC-3’ forward and 5’- GGCATGTGAGAGGAAAAACAGAGGCAGTCT TACACAAGCTGTGA-3’ reverse primers to generate the construct, Mut3 pAKR1C1 −180/+59, respectively. Transfection of the Mut1 pAKR1C1 −180/+59 construct displayed a 12-fold reduced activity in the 2008 cells and a 4-fold reduction in activity in the 2008/C13* cells compared to the wild-type pAKR1C1 −180/+59 construct (Fig. 2B and C). In contrast, the mutation in the p40X and USF binding site (Mut2 pAKR1C1 −180/+50 construct) did not affect the basal luciferase activity in the 2008 or the 2008/C13* cells (ovarian carcinoma) compared to the wild-type construct pAKR1C1 −180/+59 (Fig. 2B and C) Similarly, mutation in the Sp1 binding site (Mut3 pAKR1C1 −180/+59) did not affect the transcription of the AKR1C1 gene promoter in the 2008 cells (Fig. 2B). Moreover, the activity of the Mut1 pAKR1C1 −180/+59 construct was also reduced by 6-fold in the H23 and A549 cells (lung adenocarcinoma) and 4-fold in the HepG2 cells (liver hepatoblastoma), compared to the activity of the wild-type pAKR1C1 −180/+59 construct. (Fig. 2D, E and F) respectively. In contrast, the activity of the Mut2 pAKR1C1 −180/+59 construct and the Mut3 pAKR1C1 −180/+59 construct in these cells was similar to the activity observed with the wild-type pAKR1C1 −180/+59 construct. (Fig. 2D, E and F). These results suggest that the tandem transcription factor(s) binding site for NF-Y/CEBP but not p40X, USF or Sp1 is critical for the basal, constitutive transcription of AKR1C1 gene in human ovarian, lung and liver cancer cells.
3.5 Binding of transcription factor(s) to AKR1C1 minimal proximal promoter
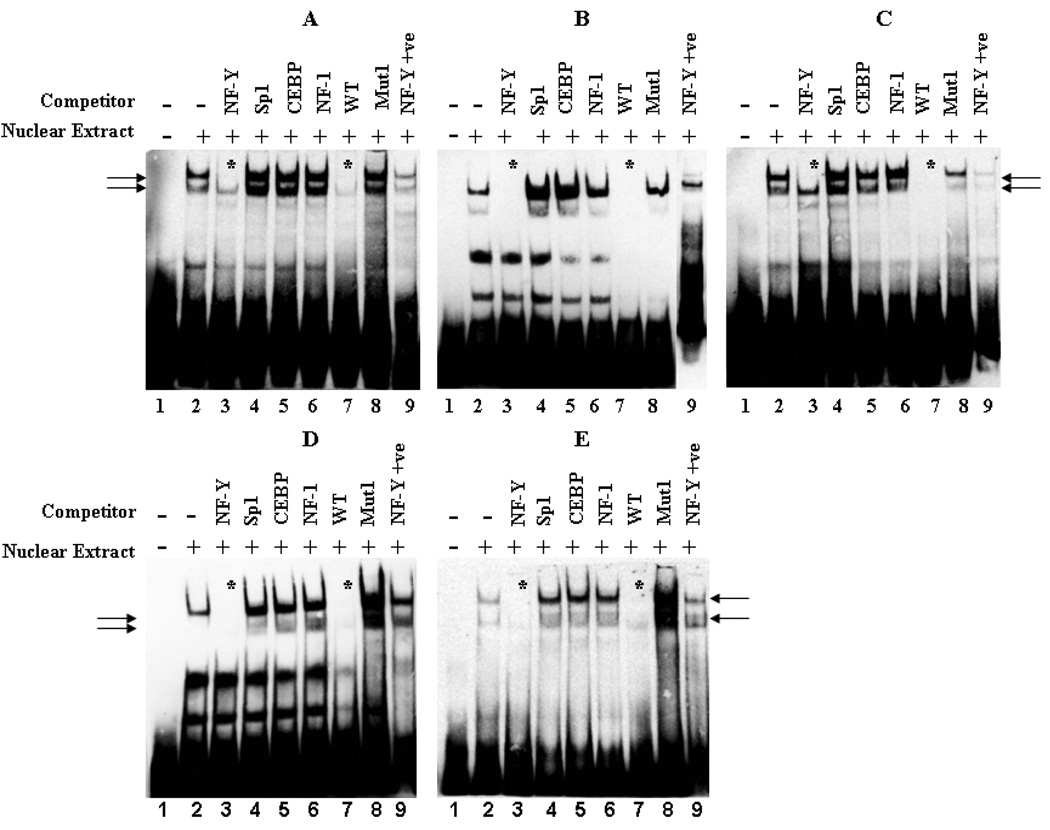
Based on the observations from the site-directed mutagenesis study, a wild-type (WT) probe (containing the binding sites for NF-Y and C/EBP as well as p40X, USF and Sp1) from −131 to −81 of the AKR1C1 gene promoter region was synthesized. Consensus oligonucleotide probes for Sp1, NF-Y, C/EBP and NF-1 were also synthesized. Preliminary EMSA study utilizing nuclear extracts from HepG2 cells displayed the presence of 3 protein-DNA complexes (unpublished observations). Competition with 100-fold molar excess of CBF/NF-Y consensus oligonucleotide inhibited the formation of the 2 slower migrating protein-DNA complexes observed with the wild-type (WT) probe. In contrast, 100-fold molar excess of the Sp1 consensus oligonucleotide inhibited the formation of the fastest moving protein-DNA complexes (unpublished observation). Since mutation of the Sp1 binding site (Mut3 pAKR1C1 −180/+59) did not affect the basal activity in any of the cell lines studied, we synthesized a 30bp WT probe (−120 to −91) from the AKR1C1 gene promoter region devoid of the Sp1 binding site and containing the NF-Y/CEBP binding sites (Fig. 3). EMSA analysis with this probe in the presence of nuclear extracts from human ovarian, lung and liver cancer cells led to the formation of 2 protein-DNA complexes (Fig. 3A – E, lane 2). Competition with 100-fold molar excess of the unlabelled WT probe completely abolished the formation of the observed protein-DNA complex (Fig. 3A – E, lane 7). Similarly, competition with 100-fold molar excess of NF-Y consensus oligonucleotide resulted in a complete inhibition of the protein-DNA complex formation (Fig. 3A – E, lane 3), whereas competition with Sp1, C/EBP or NF-1 consensus oligonucleotides did not abolish the protein-DNA complex formed by the WT probe (Fig. 3A – E, respectively, lanes 4 – 6). To further reconcile the EMSA results with the observations of the luciferase study, competition experiments with 100-fold molar excess of the NF-Y/CEBP mutant AKR1C1 promoter fragment probe were also performed. The mutant probe had changes in nucleotide sequence corresponding to those of the Mut1 pAKR1C1 −180/+59 construct; i.e. an altered NF-Y/CEBP binding site. As expected, the mutant probe did not compete with the DNA binding activity of the AKR1C1 WT (−120 to −91) probe (Fig. 3A – E, lane 8). In addition, the pattern of protein-DNA complexes observed with the labeled NF-Y consensus probe were similar to those observed with the WT probe of the AKR1C1 gene (−120 to −91 region of the AKR1C1 gene) (Fig. 3A – E, compare lane 2 with lane 9). These results indicate that the transcription factor NF-Y, but not CEBP, binds to the −120 to −91 region of the AKR1C1 gene and regulates its transcription in human ovarian, lung and liver cancer cells.
Fig. 3.
Binding of NF-Y to the proximal promoter region of the AKR1C1 gene. Electrophoretic mobility shift assay (EMSA) was performed with nuclear extracts (8µg) from (A) 2008, (B) 2008/C13*, (C) H23, (D) A549, and (E) HepG2 cells that were incubated with the wild-type (WT) probe (−120 to −91) from human AKR1C1 gene as described in Material and Methods. In competition experiments, a 100- fold molar excess of unlabelled NF-Y, Sp1, CEBP, NF-1 consensus sequences, wild-type and the Mut1 probe (the nucleotide bases were mutated similar to that of Mut1 pAKR1C1-180/+59 construct) were utilized to demonstrate the specificity of each binding reaction. Arrows indicate the formation of specific protein-DNA complexes. Efficient loss of protein-DNA complex formation by competition is denoted by an asterisk. Each of the experiments was repeated at least three times with similar results.
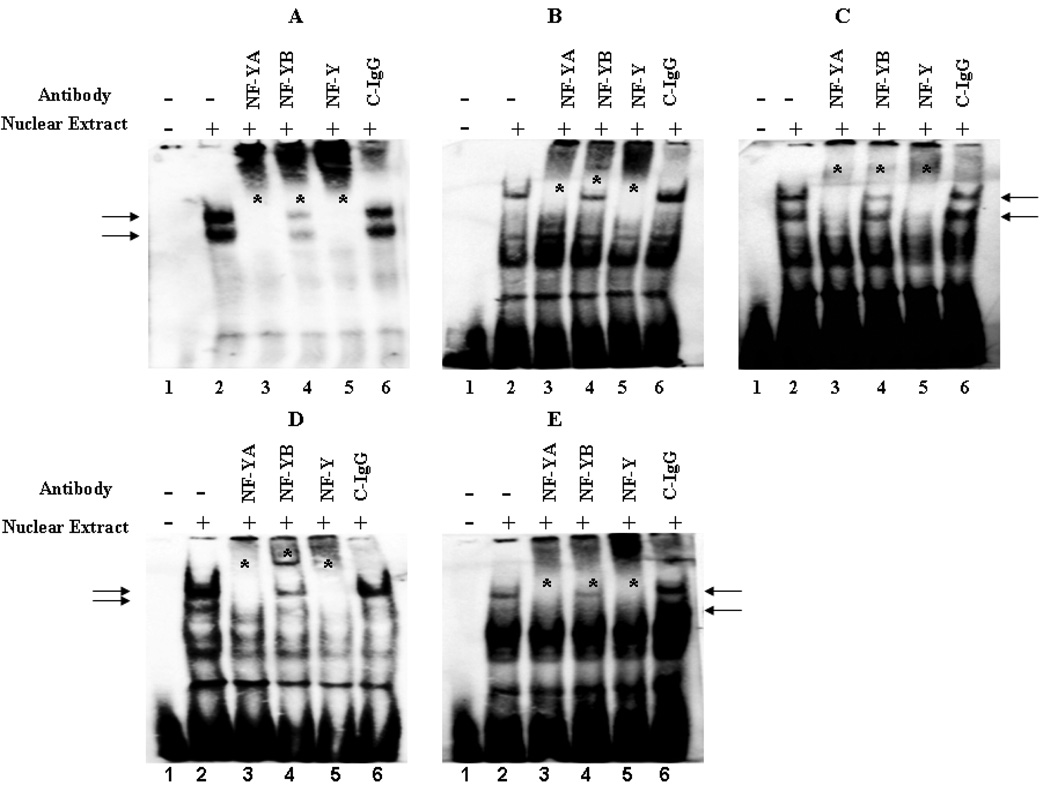
To further corroborate the EMSA observations, gel-supershift analysis was carried out utilizing a polyclonal antibody against the A, B and C subunit of NF-Y (Fig. 4). As a negative control, rabbit pre-immune IgG was also utilized in the assay (Fig. 4A – E, lane 6). Addition of the NF-YA, NF-YB or NF-YA+NF-YB+NF-YC, polyclonal antibody to the binding reaction decreased the migration of the protein-DNA complexes, with a concomitant disappearance of the protein-DNA complex I and II observed in the absence of the antibody (Fig. 4A – E, compare lanes 3 – 5 with lane 2). In contrast, the rabbit pre-immune IgG did not have any effect on the migration and/or formation of the protein-DNA complexes (Fig. 4A – E, lane 6). These results confirm that CBF/NF-Y binds to the inverted CCAAT sequence (−105 to −109) in AKR1C1 promoter region and regulates the basal, constitutive transcription of the AKR1C1 gene in human ovarian adenocarcinoma, lung adenocarcinoma and liver hepatoblastoma cells.
Fig. 4.
Confirmation of NF-Y binding to the proximal promoter region of the AKR1C1 gene. NF-Y is composed of three subunits NF-YA, NF-YB and NF-YC that are required for interaction with the CCAAT pentanucleotide sequence in the AKR1C1 gene promoter. Nuclear extracts (8µg) from (A) 2008, (B) 2008/C13*, (C) H23, (D) A549, and (E) HepG2 were incubated with WT probe (−120 to −91) from AKR1C1 gene along with 2µg of polyclonal antibody directed towards NF-YA, NF-YB or NF-Y(NF-YA+NF-YB+NF-YC). The negative control consisted of rabbit pre-immune control IgG (C-IgG). The formation of DNA-protein complexs is designated by solid arrows and the loss of protein-DNA complex’s and/or gel super-shift in the presence of the antibody is denoted by an asterisk. Each of the experiments was repeated at least three times with similar results.
3.6 In vivo association of NF-Y with AKR1C1 gene promoter through the CCAAT box binding site
To assess the in vivo association of NF-Y to the AKR1C1 gene promoter, ChIP analysis was performed. The cross-linked protein-DNA was immunoprecipitated with antibody against each of the individual NF-Y subunits (NF-YA, -YB or -YC; Fig 5A) and immunoprecipitation with the rabbit pre-immune IgG was utilized as negative control. The amplification of 187bp fragment harboring the CCAAT box in the AKR1C1 gene promoter was clearly seen in immunoprecipitates obtained using antibodies directed towards the NF-YA, -YB or -YC subunits in human ovarian (2008, 2008/C13*), lung (H23 and A549) and liver carcinoma cells (HepG2) (Fig. 5 C lane 3–5). To confirm the specificity of ChIP, a set of primers that amplify a 5’ flanking region not containing any NF-Y binding sites in the AKR1C1 gene, 2954bp downstream of the ATG start site, was utilized. The 629bp fragment was visible in the input sample obtained from all the cell lines (Fig. 5B, lane 2) while amplification was not seen in DNA obtained with antibody precipitation in any of the cell lines (Fig. 5B, lanes 1, 3–5). These results clearly demonstrate that the nuclear factor-Y subunits A, B and C associate with AKR1C1 gene promoter, in vivo, in human ovarian, lung and liver carcinoma cells.
3.7 NF-Y directly regulates the basal transcription of AKR1C1 gene promoter in human ovarian, lung and liver carcinoma cells
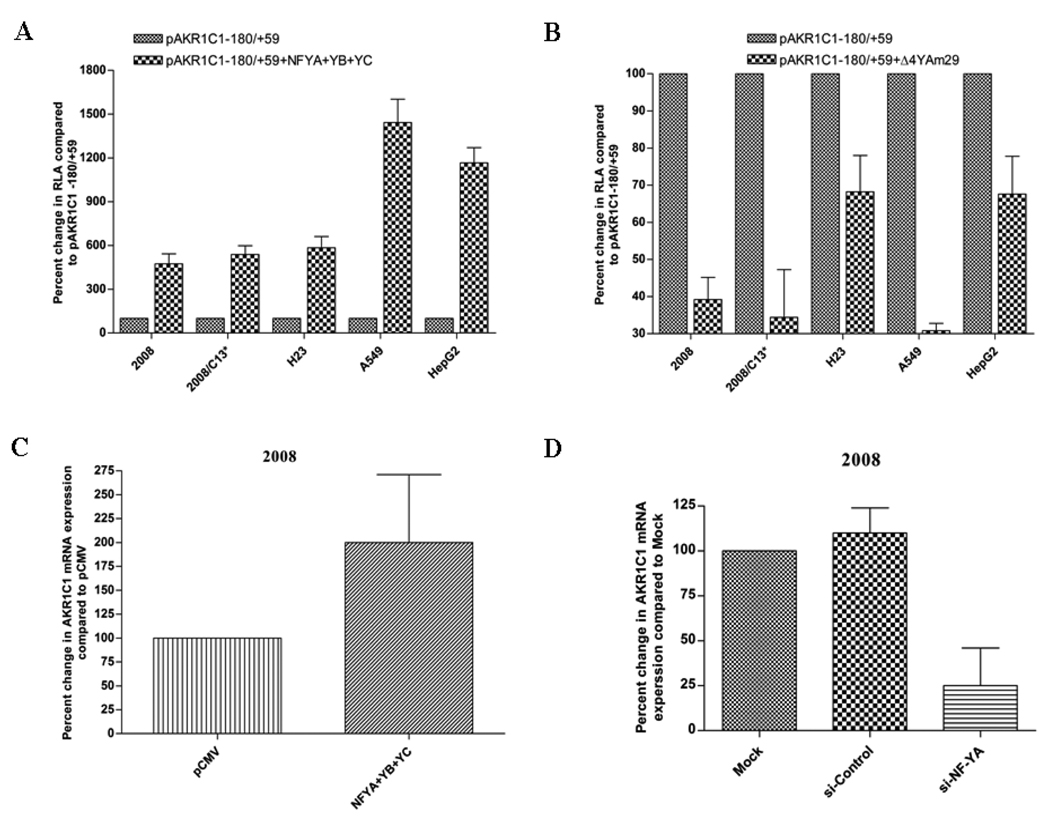
Results from site-directed mutagenesis, EMSA, gel-super shift and ChIP demonstrated the in vitro and in vivo association of NF-Y transcription factor to AKR1C1 gene promoter through the CCAAT pentanucleotide sequence. In order to analyze directly the role of NF-Y in regulating the transcription of AKR1C1 gene, human ovarian, lung and liver carcinoma cells were transiently transfected with NF-YA, B and C cDNA expression vectors along with the AKR1C1 proximal promoter pAKR1C1 −180/+59. Simultaneous expression of NF-YA, -YB and -YC, led to a 5- to 14- fold increase in basal AKR1C1 promoter activity in human ovarian, lung and liver carcinoma cells (Fig. 6A) compared to cells transfected with empty vector. In contrast, the expression of dominant-negative NF-YA resulted in suppression (1.3–3.3 fold) of AKR1C1 promoter activity in 2008, 2008/C13*, H23, A549 and HepG2 cells (Fig. 6B).
Fig. 6.
Functional analysis of NF-Y in regulating the transcription of human AKR1C1 in human ovarian, lung and liver carcinoma cells. (A) Cells were transiently transfected with 2µg of NF-YA, -YB and -YC cDNA expression vector along with the proximal promoter pAKR1C1 −180/+59. (B) Cells were transiently transfected with 2µg of the NF-YA dominant-negative expression vector along with the proximal promoter pAKR1C1 −180/+59. Luciferase assay was performed as described in Materials and Methods. Each experiment was performed in duplicate and repeated at least three times. To directly assess the effect of NF-Y on the expression of AKR1C1 mRNA, human ovarian carcinoma cells (2008) were transfected with (C) 1µg of NF-YA, -YB and -YC cDNA expression vector or pCMV control vector and (D) Five hundred picomoles of NF-YA siRNA or siRNA-scramble as the negative control. Twenty four hours after transfection RNA was isolated and the expression of AKR1C1 mRNA was assayed using real-time RT-PCR. Each experiment was performed in duplicates and repeated at least two times.
Ectopic expression of NF-YA, -B and -YC also led to a 2-fold increase in AKR1C1 mRNA levels in 2008 human ovarian carcinoma cells (Fig. 6C) compared to the pCMV control vector transfected cells. Furthermore, when 2008 cells were transfected with si-RNA directed against NF-YA, a 4-fold decrease in AKR1C1 mRNA expression levels was observed compared to control si-RNA (Fig. 6D). Effectiveness of si-RNA was confirmed by a 7-fold downregulation of NF-YA in 2008 cells (unpublished observation). Taken together these results confirm a direct role of NF-Y in regulating the basal transcription of AKR1C1 gene in human ovarian, lung and liver carcinoma cells.
3.8 Effect of cisplatin on the activity of the AKR1C1 proximal promoter
We have previously demonstrated (Deng et al., 2002) that the levels of AKR1C1 and AKR1C2 are constitutively increased in a cisplatin-resistant human ovarian carcinoma cell line (2008/C13*) compared to the parental 2008 cells. Moreover, cisplatin treatment of the 2008 cells led to a rapid increase in the expression of AKR1C1 and AKR1C2 mRNA. In contrast, cisplatin treatment of the drug-resistant 2008/C13* cells did not significantly change the expression of AKR1C1 and AKR1C2 mRNA. This differential control of the AKR1C1 gene transcription by cisplatin was further analyzed in this study utilizing the pAKR1C1 −2954/+59 and the pAKR1C1 −180/+59 promoter constructs. The 2008 and 2008/C13* cells were treated with 10µM cisplatin during the final 16–18 hours post- transfection with the promoter constructs. Treatment of 2008 cells with cisplatin induced a 2 to 2.5-fold increase in the luciferase activity with both the pAKR1C1 −2954/+59 and the pAKR1C1 −180/+59 promoter constructs compared to the basal activity of these constructs (Fig. 7A). Overexpression of NF-YA, -YB and -YC led to a further 11-fold increase in pAKR1C1 −180/+59 transcription in 2008 cells treated with cisplatin compared to the non-transfected 2008 cells treated with cisplatin (Fig. 7C). In addition, cisplatin treatment, while inducing the activity of the Mut2 pAKR1C1 −180/+59 constructs and Mut3 pAKR1C1 −180/+59 (2-fold), failed to induce the activity of the Mut1 pAKR1C1 −180/+59 construct in the 2008 cells (Fig. 7A). In contrast, treatment of 2008/C13* cells with cisplatin did not cause any significant alteration in luciferase activity of either pAKR1C1 promoter construct (Fig. 7B). Similarly, treatment with cisplatin did not induce any significant alteration in the AKR1C1 transcription in the human lung and liver cancer cells (unpublished observation). Moreover, there was no increase in AKR1C1 proximal promoter activity in cisplatin-treated H23 lung adenocarcinoma cells upon overexpression of NF-YA, -YB and -YC (Fig. 7D). These results suggest that the transcriptional element present in the proximal promoter region (viz., NF-Y binding sites) is sufficient to drive the constitutive as well as inducible transcription of AKR1C1 gene in cisplatin-sensitive human ovarian carcinoma cells (2008), while controlling the basal (but not the cisplatin-inducible) transcription of AKR1C1 gene in the cisplatin-resistant 2008/C13* cells and the human lung and liver cancer cells (H23, A549 and HepG2).
Fig. 7.
Effect of cisplatin in inducing the transcription of human AKR1C1 gene. Human ovarian carcinoma cells (A) 2008 (cisplatin-sensitive) and (B) 2008/C13* (cisplatin-resistant) were transfected with the AKR1C1 proximal promoter pAKR1C1 −180/+59, pAKR1C1 −3030/+59 (full length promoter), Mut1 pAKR1C1 −180/+59 (Mut1), Mut2 pAKR1C1 −180/+59 (Mut2) and Mut3 pAKR1C1 −180/+59 constructs. Cells were treated with cisplatin (10µM) for the final 16–18h, followed by the luciferase assay, which was performed as described in Table 1. Relative luciferase activity of pGL3 Basic (untreated & treated) was considered as 1. The mean ± S.D are from three different experiments, each performed in triplicate. (***p<0.001, **p<0.01, *p<0.05 compared with untreated). (C) 2008 cells and (D) H23 cells were transfected with 2µg of NF-YA, -YB and -YC cDNA expression vector along with the indicated AKR1C1 proximal promoter construct. Cells were treated with cisplatin (10µM) for the final 16–18h, followed by the luciferase assay, which was performed as described in Table 1. Relative luciferase activity of pGL3 Basic (untreated & treated) was considered as 1. The mean ± S.D are from two different experiments, each performed in duplicates (***p<0.001, **p<0.01, *p<0.05 compared with pAKR1C1 −180/+59).
4. DISCUSSION
AKR1C1 (also referred to as 20α-hydroxysteroid dehydrogenase (20 α-HSD), dihydrodiol dehydrogenase 1 (DDH1)) is a member of aldoketo reductase (AKR) family of enzymes involved in progesterone metabolism (Higaki et al., 2002). The AKR1C family of aldo-keto reductases consist of four isoforms, AKR1C1-AKR1C4, while the other members of AKR are aldehyde reductase (AKR1A1), aldose reductase (AKR1B1 and AKR1B10), steroid 5β-reductase (AKR1D1), and aflatoxin aldehyde reductase (AKR7A1 and AKR7A2) (Hyndman et al., 2003). AKR1C3 (type II AKR1C) promoter analysis has indicated the presence of positive and negative cis-acting regulatory elements such as NF-IL6, HNF-5, AP-1, AP-2 and NFκB that were thought to be involved in constitutive and inducible transcription of human AKR1C3 gene promoter (Ciaccio et al., 1996). Further studies have identified the essential role of the NF-1 transcription factor in regulating the basal as well as induced transcription of rat AKR1C9 gene; a human AKR1C1 gene homolog (Hung and Penning, 1999). Functional analysis of rat 3α-HSD/DD gene (homologous to the human AKR1C2 gene) demonstrated the importance the Oct-1 transcription factor in the control of its transcription in HepG2 cells (Lin and Penning, 1995). Furthermore, in HepG2 cells, a co-operative regulation of the AKR1C4 gene transcription by the hepatocyte nuclear factor (HNF)-4α/β and HNF-1α has been demonstrated (Ozeki et al., 2001). However, the transcriptional elements that control the transcription of the human AKR1C1 gene remain uncharacterized. Thus, in the present study, we have functionally characterized the human AKR1C1 gene basal proximal promoter and provide evidence of the role of the CCAAT binding protein, NF-Y, in the basal regulation of the AKR1C1 gene in human ovarian, lung and liver carcinoma cells
Nucleotide sequencing of the BAC clone RP-379P14 (GenBank Accession No. AB032150), which carries the 5’ flanking region of the human AKR1C1 gene promoter, identified the presence of a TATA box and several putative transcription factor(s) binding sites (Fig. 1A) upstream of the putative transcription start site of the human AKR1C1 gene. The promoters of the AKR family of genes differ in the presence/absence of a TATA box; AKR1C2, AKR1C4, AKR1B1 (aldose reductase), and AKR7A1 (aflatoxin aldehyde reductase) have a TATA box upstream of the transcription start site (Lou et al., 2006; Wang et al., 1993) whereas, the type II 3α-HSD (AKR1C3), AKR1B10 and AKR1A1 (aldehyde reductase) have TATA-less promoters (Ciaccio et al., 1996; Barski et al., 1999; Lie et al., 2009). Sequential deletion and functional analysis of an approximately 3000 bp 5’ flanking region of AKR1C1 gene promoter led us to identify the DNA construct pAKR1C1 −128/+59 as being essential for the basal transcription of AKR1C1 gene in human liver, ovarian and lung carcinoma cells (Fig. 1B and Table 1). The AKR1C1 gene shares 98% sequence homology with AKR1C2 gene and a previous study has demonstrated that the region between −117 to −33 of the 5’ flanking region of AKR1C2 proximal promoter was essential for its transcription in HepG2 cells (Lou et al., 2006). Transcription factor binding sites for C/EBPβ and AP-1 were identified in this region of the AKR1C2 proximal promoter (Lou et al., 2006). In the case of the human AKR1C1 gene, computational analysis indicated the presence of tandem binding sites for Sp1, NF-Y, C/EBP, p40x and USF transcription factors in the minimal proximal promoter region (−128/+59). Utilizing site-directed mutagenesis, point mutations were introduced in the pAKR1C1 −180/+59 construct at the consensus binding sites for the transcription factor(s) NF-Y/CEBP (Mut1 pAKR1C1 −180/+59), p40X/USF (Mut2 pAKR1C1 −180/+59) and Sp1 (Mut3 pAKR1C1 −180/+59). Mutation of the NF-Y/CEBP transcription factor binding site resulted in a 4- to 12-fold decrease in AKR1C1 basal transcription in the human ovarian, lung and liver carcinoma cells, while mutation in the p40X/USF and the Sp1 binding sites did not affect the AKR1C1 transcription in any of the cell lines (Fig. 2B–F). These results clearly demonstrate that the tandem binding site for NF-Y and C/EBP is essential for regulating the basal transcription of AKR1C1 gene in human ovarian, lung and liver carcinoma cells. However, in a methotrexate-resistant colon carcinoma cell line, Selga et al., (2008) have demonstrated that the Sp1 binding domain in the human AKR1C1 gene promoter region between −88 to −80 is important for its basal transcription. Thus, it is possible that, based on cell type, different regions of the AKR1C1 gene differentially regulate its basal transcription.
Further corroboration of the importance of NF-Y in controlling the basal transcription of the AKR1C1 gene was obtained using EMSA studies. Our data from gel shift analysis indicate that the nuclear factor for Y box (NF-Y) consensus sequence (containing the CCAAT box) specifically competed for nuclear protein binding with the WT AKR1C1 promoter fragment, −120 to −91 (nuclear extracts were obtained from 2008, 2008/C13*, H23, A549 and HepG2 cells) (Fig. 3A – E). However, competition with consensus oligonucleotides for Sp1, C/EBP and NF-1 binding sites did not abolish this protein-DNA complex formation. Specificity of the binding was demonstrated by utilizing a CCAAT binding sequence mutant oligonucleotide (−120 to −91) that did not decrease the formation of protein-DNA nuclear complex observed with the WT probe. The CCAAT-binding proteins include NF-Y, C/EBP and NF-1, binding sites which are present in most of the eukaryotic promoters (Bucher, 1990). Thirty percent of eukaryotic promoters carry the binding site for CCAAT box either in forward or reverse orientation within −60 to −100 nucleotides from the transcription start site (Su et al., 2006). In the present study, the CCAAT box was present in the reverse orientation in the region between −109 to −105 of the human AKR1C1 gene promoter. Supershift analysis with anti-NF-YA, anti-NF-YB and anti-NF-YC antibody confirmed the presence of the NF-Y transcription factor in the protein-DNA nuclear complex (Fig. 4A – E). ChIP analysis further confirmed the in vivo association between NF-Y and the AKR1C1 gene promoter in human ovarian (2008, 2008/C13*), lung (H23, A549) and liver (HepG2) cells was observed (Fig. 5). These results further strengthen the notion that the NF-Y transcription factor regulates the basal transcription of human AKR1C1 gene in human ovarian, lung and liver carcinoma cells.
NF-Y is a ubiquitous transcription factor involved in both constitutive and inducible transcription of various promoters harboring the CCAAT box (Sun et al., 2009; Kabe et al., 2005). The consensus sequence recognized by NF-Y is the 5’-CTGATTGGTTRR-3’ or 5’-YYRRCCAATCAG-3’ (Matuoka and Chen, 1999; Mantovani, 1998, Bi et al., 1997). Biochemical analysis reveals that NF-Y is a heterotrimeric protein consisting of subunits A, B and C. The binding of NF-YA to its consensus region requires the prior formation of a NF-YB:NF-YC protein complex sometimes referred to as the “handshake motif” (Caretti et al., 1999).The role of NF-Y in promoter regulation was first studied in MHC class II conserved Y box in Ea gene (Dorn et al., 1987). Subsequently, several other proximal promoters regulated by NF-Y binding were identified including the yeast USAS gene, α2(I) collagen gene, α1(I) collagen gene, human multidrug resistant gene (mdr1), rat liver S14 gene, phospholipid hydroperoxide glutathione peroxidase gene, human α1(XI) collage gene (COL11A1) and fibroblast growth factor receptor 2(FGFR2) (Sun et al., 2009; Goldsmith et al., 1993; Jump et al., 1997; Huang et al., 1999; Matsuo et al., 2003). Overall, our data from site-directed mutagenesis, EMSA, gel-supershift and ChIP analysis suggests that the core CCAAT pentanucleotide motif in the AKR1C1 gene promoter is sufficient for NF-Y binding leading to the activation of AKR1C1 gene transcription.
We also assessed the direct functional response of NF-Y in regulating the basal transcription of AKR1C1 gene promoter. Overexpression of NF-Y’s significantly increased the basal AKR1C1 gene transcription in all cell lines studied (Fig. 6A). Similarly, overexpression of NF-Y cDNA was found to increase the promoter activity of mouse gonadotropin releasing hormone receptor (GnRHR) and mouse proline-rich nuclear receptor coactivator 2 (PNRC2) gene (Kam et al., 2005; Zhou et al., 2005). In contrast, overexpression of NF-Y cDNA was found to repress the promoter activity of human PNRC2 gene (Zhang et al., 2008). In our study, expression of a dominant-negative NF-YA mutant (Mantovani et al., 1994) repressed the human AKR1C1 gene promoter activity (Fig. 6B). Various other authors have also observed a decrease in promoter activity of the target gene upon co-expression with the dominant-negative NF-YA mutant (Lamb et al., 1997; Liu et al., 1997; Sugiura and Takishima, 2000; Kam et al., 2005; Wageningen et al., 2008).
We also assessed the importance of the basal proximal promoter region of the AKR1C1 gene in controlling its transcription upon treatment with the anticancer drug cisplatin. Exposure of the human ovarian carcinoma cells (2008) to cisplatin, led to a two fold increase in basal and induced AKR1C1 transcription compared to the untreated cells (Fig. 7A and 7C). Further, mutation of the NF-Y/CEBP binding site in the AKR1C1 promoter region (Mut1 pAKR1C1 −180/+59) negated the cisplatin induced increase in the AKR1C1 gene transcription in the 2008 cells. These results suggest that the transcriptional elements (specifically the NF-Y/CEBP binding motif) in the region between −180/+59 of the AKR1C1 gene promoter, are also involved in controlling the drug-induced transcription of the AKR1C1 gene specifically in the 2008 cells. Surprisingly, cisplatin did not induce the transcription of the AKR1C1 gene in the cisplatin-resistant human ovarian carcinoma cells (2008/C13*- Fig. 7B), human lung adenocarcinoma cells H23 (Fig. 7D) and A549 or the human hepatoblastoma cells (unpublished data). However, these results compare well with our previous observations, wherein the expression of the AKR1C1 and AKR1C2 mRNA was found to be increased in a cisplatin-dependent manner only in the 2008 cells but not in the 2008/C13* cells (Deng et al., 2002).
Based on our observations of the specificity of NF-Y in regulating the transcription AKR1C1 gene in the cisplatin-treated 2008 cell, we further selected this human ovarian carcinoma cell line to analyze the differential expression of cellular AKR1C1 mRNA in response to overexpression or knockdown of NF-Y. Ectopic expression of NF-Y's in 2008 cells significantly increased the expression of AKR1C1 mRNA (Fig. 6C). In contrast, si-RNA mediated knockdown of NF-YA decreased the expression of AKR1C1 mRNA in the 2008 cells (Fig. 6D).
Thus, functional analysis, site-directed mutagenesis studies, EMSA, gel-shift assays, ChIP, ectopic expression analysis and siRNA studies lead us to conclude that the transcription factor NF-Y is sufficient and necessary for regulating the constitutive transcription of AKR1C1 gene in human ovarian, lung and liver carcinoma cells while the involvement of NF-Y in cisplatin-induced AKR1C1 transcription is cell-type dependant.
ACKNOWLEDGEMENTS
The study was supported by grant from Pennsylvania Department of Health (PADOH) grant # 420339-05440-02 to H.K.P and NIH grant # CA98804 to H.S. This study was presented in part as a poster [Pallai R, Simpkins H, and Parekh, HP. Characterization of human dihydrodiol dehydrogenase gene 1 promoter] at the American Association for Cancer Research annual meeting; 12–16 April 2008; San Diego, CA.
We thank Dr. Koneti Rao, Director Sol Sherry Thrombosis Center, Temple University School of Medicine for granting access to the luminometer instrumentation for luciferase assay. We also thank Dr. Satya P. Kunapuli, Department of Physiology, Temple University School of Medicine for granting access to the charge-couple device camera. We personally thank Dr. Barry Ashby, Department of Pharmacology, Temple University School of Medicine for helping in reviewing the manuscript. We are thankful to Dr. Robert Mantovani at University of Milan, Italy for the expression plasmid of NF-YA, -YB, -YC and the dominant-negative NF-YA mutant expression vector. We would also like to thank Dr. Ronald Hines at Medical College of Wisconsin, IL for the pCMV control vector.
Abbreviations used
- AKR
aldoketo-reductase
- DDH
dihydrodiol dehydrogenase 1
- PAH
polycyclic aromatic hydrocarbons
- HSD
hydroxysteriod dehydrogenase
- NRF2
nuclear factor erythroid-derived 2-related factor
- ARE
human antioxidant response element
- EA
ethacrynic acid
- NF-Y
nuclear factor Y
- EMSA
electrophoretic mobility shift assay
- CBF
CCAAT binding factor
- C/EBP
CCAAT enhancer binding protein
- USF
Upstream stimulatory factor
- NF-1
CCAAT transcription factor
- NF-YA
Nuclear factor-Y alpha
- NF-YB
Nuclear factor Y beta
- NF-YC
Nuclear factor Y gamma
- IgG
Immunoglobulin G
- ChIP
Chromatin immunoprecipitation
- RLA
Relative luciferase activity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barski OA, Gabbay KH, Bohren KM. Characterization of the human aldehyde reductase gene and promoter. Genomics. 1999;60:188–198. doi: 10.1006/geno.1999.5915. [DOI] [PubMed] [Google Scholar]
- Bi W, Wu L, Coustry R, Crombrugghe Bde, Maity SN. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- Bucher P. Weight matrix description of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, Lin HK, Penning TM. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs). Electrophiles, and oxidative stress: Implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res. 1999;59:607–614. [PubMed] [Google Scholar]
- Caretti G, Motta MC, Mantovani R. NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms. Mol. Cell Biol. 1999;19:8591–8603. doi: 10.1128/mcb.19.12.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Yuan CC, Chow KC, Wang PH, Lai CR, Yen MS, Wang LS. Overexpression of dihydrodiol dehydrogenase is associated with cisplatin-based chemotherapy resistance in ovarian cancer patients. Gynecol. Oncol. 2005;97:110–117. doi: 10.1016/j.ygyno.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Ciaccio PJ, Jaiswal AK, Tew KD. Regulation of human dihydrodiol dehydrogenase by Michael acceptor xenobiotics. J. Biol. Chem. 1994;22:15558–15562. [PubMed] [Google Scholar]
- Ciaccio PJ, Walsh ES, Tew KD. Promoter analysis of a human dihydrodiol dehydrogenase. Biochem. Biophy. Res. Commun. 1996;228:524–529. doi: 10.1006/bbrc.1996.1693. [DOI] [PubMed] [Google Scholar]
- Deng HB, Adikari M, Parekh HP, Simpkins H. Ubiquitous induction of resistance to platinum drugs in human ovarian, cervical, germ-cell and lung carcinoma tumor cells overexpressing isoforms 1 and 2 of dihydrodiol dehydrogenase. Cancer. Chemother. Pharmacol. 2004;54:301–307. doi: 10.1007/s00280-004-0815-0. [DOI] [PubMed] [Google Scholar]
- Deng HB, Parekh HP, Chow KC, Simpkins H. Increased expression of dihydrodiol dehydrogenase induces resistance to cisplatin in human ovarian carcinoma cells. J. Biol. Chem. 2002;17:15035–15043. doi: 10.1074/jbc.M112028200. [DOI] [PubMed] [Google Scholar]
- Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Dufort I, Soucy P, Labrie F, Luu-The V. Molecular cloning of human type 3 3α-hydroxysteroid dehydrogenase that differs from 20α-hydroxysteroid dehydrogenase by seven amino acids. Biochem. Biophys. Res. Commun. 1996;228:474–479. doi: 10.1006/bbrc.1996.1684. [DOI] [PubMed] [Google Scholar]
- Goldsmith ME, Madden MJ, Morrow CS, Cowan KH. A Y-box consensus sequences is required for basal expression of the human multidrug resistance (mdr1) gene. J. Biol. Chem. 1993;268:5856–5860. [PubMed] [Google Scholar]
- Higaki Y, Kamiya T, Usami N, Shintani S, Shiraishi H, Ishikura S, Yamamoto I, Hara A. Molecular characterization of two monkey dihydrodiol dehydrogenases. Drug. Metabol. Pharmacokin. 2002;17:348–356. doi: 10.2133/dmpk.17.348. [DOI] [PubMed] [Google Scholar]
- Hsu NY, Ho HC, Chow KC, Lin TY, Shih CS, Wang LS, Tsai CM. Overexpression of dihydrodiol dehydrogenae as a prognostic marker of non-small cell lung cancer. Cancer Res. 2001;61:2727–2731. [PubMed] [Google Scholar]
- Huang H-S, Jiunn C, Chang W-C. The CCAAT box binding factor NF-Y is required for the expression of phospholipid hydroperoxide glutathione peroxidase in human epidermoid carcinoma A431 cells. FEBS. 1999;455:111–116. doi: 10.1016/s0014-5793(99)00866-2. [DOI] [PubMed] [Google Scholar]
- Hung CF, Penning TM. Members of the nuclear factor 1 transcription factor family regulate rat 3α-hydroxysteroid/dihydrodiol dehydrogenase (3α-HSD/DD AKR1C9) gene expression: A member of the aldo-keto reductase superfamily. Mol. Endocrinol. 1999;13:1704–1717. doi: 10.1210/mend.13.10.0363. [DOI] [PubMed] [Google Scholar]
- Hung JJ, Chow KC, Wang HW, Wang LS. Expression of dihydrodiol dehydrogenase and resistance to chemotherapy and radiotherapy in adenocarcinoma cells of lung. Anticancer Res. 2006;26:2949–2956. [PubMed] [Google Scholar]
- Hyndman DR, Dauman DR, Heredia VV, Penning TM. The aldo-keto reductase superfamily homepage. Chem-Biol. Inter. 2003;143:621–631. doi: 10.1016/s0009-2797(02)00193-x. [DOI] [PubMed] [Google Scholar]
- Ji Q, Aoyama C, Chen PK, Stolz A, Liu P. Localization and altered expression of AKR1C family members in human ovarian tissues, Mol. Cell Probes. 2005;19 doi: 10.1016/j.mcp.2005.03.003. 231-226. [DOI] [PubMed] [Google Scholar]
- Ji Q, Chang L, VanDenBerg D, Stanczyk FZ, Stolz A. Selective reduction of AKR1C2 in prostrate cancer and its role in DHT metabolism. Prostate. 2003;54:275–289. doi: 10.1002/pros.10192. [DOI] [PubMed] [Google Scholar]
- Jin Y, Penning TM. Aldo-keto reductases and bioactivation/detoxification. Annu. Rev. Pharmacol. Toxicol. 2007;47:263–292. doi: 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- Jump DB, Badins MV, Thelen A. The CCAAT box binding factor, NF-Y, is required for thyroid hormone regulation of rat liver S14 gene transcription. J. Biol. Chem. 1997;272:27778–27786. doi: 10.1074/jbc.272.44.27778. [DOI] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam K-Y, Jeong K-H, Norwitz ER, Jorgensen EM, Kaiser UB. Oct-1 and nuclear factor Y bind to the SURG-1 element to direct basal and gonadotropin releasing hormone (GnRH)- stimulated mouse GnRH receptor gene transcription. Mol. Endocrinol. 2005;19:148–162. doi: 10.1210/me.2004-0025. [DOI] [PubMed] [Google Scholar]
- Lamb KA, Johnson LR, Rizzino A. NF-Y binds to the CCAAT box motif of the FGF-4 gene and promotes FGF-4 expression in embryonal carcinoma cells. Mol. Rep. Dev. 1997;48:301–309. doi: 10.1002/(SICI)1098-2795(199711)48:3<301::AID-MRD1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Lee EK, Regenold WT, Shapiro P. Inhibition of aldose reductase enhances HeLa cell sensitivity to chemotherapeutic drugs and involves activation of extracellular signal-regulated kinases. Anticancer Drugs. 2002;13:859–868. doi: 10.1097/00001813-200209000-00012. [DOI] [PubMed] [Google Scholar]
- Lin HK, Penning TM. cloning, sequencing and functional analysis of the 5’-flanking region of the rat 3α-hydroxysteroid/dihydrodiol dehydrogenase gene. Cancer Res. 1995;55:4105–4113. [PubMed] [Google Scholar]
- Liu Q, Yan H, Dawes NJ, Lu Y, Zhu H. Transcriptional activation of the p34cdc2 gene by cdc2 promoter binding factor/Nuclear factor-Y in fetal rat ventricular myocytes. Circ. Res. 1998;82:251–260. doi: 10.1161/01.res.82.2.251. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhong L, Krishack PA, Robbins S, Cao JX, Ahzo UY, Chung S, Cao D. Structure and promoter characterization of aldo-keto reductase family 1 B10 gene. Gene. 2009;437:39–44. doi: 10.1016/j.gene.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Du S, Ji Q, Stolz A. Induction of AKR1C2 by phase II inducers: identification of a distal consensus antioxidant response element regulated by Nrf2. Mol. Pharmacol. 2006;69:1662–1672. doi: 10.1124/mol.105.019794. [DOI] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acid Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R, Li X-L, Pessara U, van Huisjduijnen RH, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J. Biol. Chem. 1994;32:20340–20346. [PubMed] [Google Scholar]
- Matsuo N, Hua WY, Sumiyoshi H, Takatani KS, Nagato H, Sakai K, Sakurai M, Yoshioka H. The transcription factor CCAAT-bindng factor CBF/NF-Y regulates the proximal promoter activity in the human α1 (XI) collagen gene (COL11A1) J. Biol. Chem. 2003;278:32763–32770. doi: 10.1074/jbc.M305599200. [DOI] [PubMed] [Google Scholar]
- Matuoka K, Chen KY. Nuclear factor Y (NF-Y) and cellular senescence. Exp Cell Res. 1999;253:365–371. doi: 10.1006/excr.1999.4605. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Nakajima T, Yasuda K, Kanzaki H, Sasaguri Y, Watanabe K, Ito S. Close kinship of human 20α-hydroxysteroid dehydrogenase gene with three aldo-keto reductase genes. Genes Cells. 2000;5:111–125. doi: 10.1046/j.1365-2443.2000.00310.x. [DOI] [PubMed] [Google Scholar]
- Ozeki T, Takahashi Y, Kume T, Nakayama K, Yokoi T, Numoya K-I, Hara A, Kamataki T. Co-operative regulation of the transcription of human dihydrodiol dehydrogenase (DD)4/aldo-keto reductase (AKR)1C4 gene by hepatocyte nuclear factor (HNF)-4α/γ and HNF-1α. Biochem. J. 2001;355:537–544. doi: 10.1042/0264-6021:3550537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning TM, Byrns MC. Steroid hormone transforming aldo-keto reductases and cancer. Ann. N. Y. Acad. Sci. 2009;1155:33–42. doi: 10.1111/j.1749-6632.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning TM, Burczynski ME, Jez JM, Hung CF, Lin HK. Human 3α-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation of male and female sex hormones. Biochem. J. 2000;351:67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning TM, Steckelbroeck DR, Bauman DR. Aldo-keto reductase (AKR) 1C3: Role in prostate disease and the development of specific inhibitors. Mol. Cell Endocrinol. 2006;248:182–191. doi: 10.1016/j.mce.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Sasano H, Suzuki T, Miki Y. Intracrinology of estrogens and androgens in breast carcinoma. J. Steroid. Biochem. Mol. Biol. 2008;108:181–185. doi: 10.1016/j.jsbmb.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Selga E, Noé V, Ciudad CJ. Transcriptional regulation of aldo-keto reductase 1C1 in HT29 human colon cancer cells resistant to methotrexate: Role in the cell cycle and apoptosis. Biochem. Pharmacol. 2008;75:414–426. doi: 10.1016/j.bcp.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- Su M, Lee D, Ganss B, Sodek J. Stereochemical analysis of the functional significance of the conserved inverted CCAAT and TAT elements in the rat bone sialoprotein gene promoter. J. Biol. Chem. 2006;281:9882–9890. doi: 10.1074/jbc.M508364200. [DOI] [PubMed] [Google Scholar]
- Sugiura N, Takishima K. Regulation of the gene promoter for extracellular signal-regulated protein kinase 2 by transcription factors NF-Y and Sp3. Biochem. J. 2000;347:155–161. [PMC free article] [PubMed] [Google Scholar]
- Sun F, Xie Q, Ma J, Yang S, Chen Q, Hong A. NF-Y is required for basal activation and chromatin accessibility of FGFR2 promoter in osteoblast-like cells. J. Biol. Chem. 2009;284:136–3147. doi: 10.1074/jbc.M808992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki YJ, Agarwal BB, Packer L. α-lipoic acid is a potent inhibitor of NF-κB activation in human T cells. Biochem. Biophys. Res. Commun. 1992;189:1709–1715. doi: 10.1016/0006-291x(92)90275-p. [DOI] [PubMed] [Google Scholar]
- Torigoe T, Izumi H, Ishiguchi H, Yoshida Y, Tanabe M, Yoshida T, Igarashi T, Niina I, Wakasugi T, Imaizumi T, Momii Y, Kuwano M, Kohno K. Cisplatin resistance and transcription factors. Curr. Med. Chem. 2005;5:15–27. doi: 10.2174/1568011053352587. [DOI] [PubMed] [Google Scholar]
- Wageningen S, van Nikoloski G, Vierwinden G, Knops R, van der Reijden BA, Jansen JH. The transcription factor nuclear factor Y regulates the proliferation of myeloid progenitor cells. Haematologica. 2008;93:1580–1582. doi: 10.3324/haematol.12425. [DOI] [PubMed] [Google Scholar]
- Wang K, Bohren KM, Gabbay KH. Characterization of the human aldose reductase gene promoter. J. Biol. Chem. 1993;268:16052–16058. [PubMed] [Google Scholar]
- Wang LS, Chow KC, Wu YC, Lin TY, Li WY. Inverse expression of dihydrodiol dehydrogenase and glutathione-s-transferase in patients with esophageal squamous cell carcinoma. Int. J. Cancer. 2004;111:246–251. doi: 10.1002/ijc.11650. [DOI] [PubMed] [Google Scholar]
- Zhang D, Tai LK, Wong Li, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol. Cell Proteomics. 2005;4:1686–1696. doi: 10.1074/mcp.M400221-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen B, Li Y, Chen J, Lou G, Chen LM, Zhou D. Transcriptional regulation of the human PNRC promoter by NFY in HepG2 cells. J. Biochem. 2008;143:675–683. doi: 10.1093/jb/mvn019. [DOI] [PubMed] [Google Scholar]
- Zhou D, Masri S, Ye JJ, Chen S. Transcriptional regulation of the mouse PNRC2 promoter by the nuclear factor Y (NFY) and E2F1. Gene. 2005;361:89–100. doi: 10.1016/j.gene.2005.07.012. [DOI] [PubMed] [Google Scholar]