Abstract
How DNA repair machineries detect and access, within the context of chromatin, lesions inducing little or no distortion of the DNA structure is a poorly understood process. Removal of oxidized bases is initiated by a DNA glycosylase that recognises and excises the damaged base, initiating the base excision repair (BER) pathway. We show that upon induction of 8-oxoguanine, a mutagenic product of guanine oxidation, the mammalian 8-oxoguanine DNA glycosylase OGG1 is recruited together with other proteins involved in BER to euchromatin regions rich in RNA and RNA polymerase II and completely excluded from heterochromatin. The underlying mechanism does not require direct interaction of the protein with the oxidized base, however, the release of the protein from the chromatin fraction requires completion of repair. Inducing chromatin compaction by sucrose results in a complete but reversible inhibition of the in vivo repair of 8-oxoguanine. We conclude that after induction of oxidative DNA damage, the DNA glycosylase is actively recruited to regions of open chromatin allowing the access of the BER machinery to the lesions, suggesting preferential repair of active chromosome regions.
INTRODUCTION
Cellular components are continually exposed to oxidative stress arising from sources both environmental, such as chemicals or radiation, and intracellular, through normal metabolism (1). In DNA, reactive oxygen species (ROS) induce a plethora of lesions, including oxidized bases, abasic (AP) sites and strand breaks. If left unrepaired, these DNA damages can compromise cell viability by blocking essential processes such as transcription or replication. Alternatively, DNA lesions can induce mutations, the accumulation of which can lead to cancer. Among base lesions, 7,8-dihydro-8-oxoguanine (8-oxoG), an oxidized form of guanine, is a major product. Although this modified base does not create strong distortion of the DNA helix, it is highly mutagenic due to its capacity to pair with adenine during replication (2). The presence of 8-oxoG can also lead to transcriptional mutagenesis (3), a phenomenon that could be particularly important in slowly growing or terminally differentiated cells (4).
Base excision repair (BER) is the main pathway for the removal of modified bases or AP sites from DNA in organisms from bacteria to humans (1). For altered bases the repair process is initiated by a DNA glycosylase that recognizes the modified base and excises it leaving an AP site. This intermediate, as well as spontaneously arisen AP sites, is further processed by an AP endonuclease in order to provide a suitable substrate for DNA synthesis and ligation steps. In mammals, the main DNA glycosylase for 8-oxoG is the OGG1 protein and the major AP endonuclease is APE1. XRCC1, a scaffolding protein without known enzymatic activity, also participates throughout BER by physically interacting with all the involved enzymes and coordinating their activities (5–10). In vitro reconstitution experiments as well as work on cell extracts have shown that the limiting step in BER is generally the one performed by the DNA glycosylase. Crystallographic studies have helped to bring into light the underlying mechanism that allows OGG1 to discriminate an 8-oxoG from its normal counterpart, evoking diffusion or scanning models that would explain DNA-glycosylase damage recognition (11,12). However, structural and biochemical analyses do not take into account the high degree of nuclear DNA condensation imposed by chromatin structure. In eukaryotes chromatin is packaged by histones in a highly organized hierarchy. Thus, chromatin can act as an impediment to the access to DNA of enzymatic machineries responsible for transcription, replication or repair. In 1991, Smerdon proposed a model of ‘access–repair–restore’ to highlight the importance of the chromatin context in the DNA repair process (13). It was later shown that reactivation by the photolyase of UV-induced damage is inhibited by the presence of nucleosomes (14). Similarly, UV lesions present on nucleosomal DNA are less efficiently repaired than those in naked DNA (15). For strand break repair the consensus is that the access of proteins to DNA is accompanied by the action of chromatin-remodelling factors capable of displacing histones from the region of the lesion (16–18). There is now clear evidence that the nucleosomal structure also inhibits, with varying efficiency, the initial steps of BER (19–22). A large number of proteins act on chromatin regulating its structure mostly through histone acetylation, phosphorylation, ubiquitination and methylation (23). These modifications can alter DNA–histone interactions within and between nucleosomes and in such way allow DNA repair proteins to overcome the nucleosome barrier. Similarly, addition of chromatin remodelling factors relieves OGG1 inhibition on chromatinized substrates (21). Beyond nucleosomes, higher-order chromatin structure constitutes a probable barrier for repair proteins access to damaged DNA. Two main types of domains compose chromosomes. In general, heterochromatin is inaccessible to DNA-processing proteins and considered essentially transcriptionally silent. Large heterochromatic domains are found encompassing chromosome structures such as centromeres and telomeres, whereas smaller heterochromatic regions are interspersed throughout the chromosome (24). Euchromatic domains, in contrast, define more accessible regions of the genome and are generally associated with active transcription (25,26). Although the link between the degree of chromatin compaction and gene transcription is well established, much less is known for DNA repair processes for which most of the evidence comes from experiments showing that silencing of yeast loci interferes with the repair of UV damage (27–29). A similar situation was recently described for the repair of double-strand breaks present in heterochromatin (30). In particular, very little is known on how the level of chromatin compaction affects BER efficiency. We had previously shown that after UVA treatment of human cells, a fraction of the nuclear OGG1 is specifically recruited to the nuclear speckles through a ROS–mediated mechanism (31). However, the number of 8-oxoG induced in those experiments was very small, impeding to establish the correlation between the relocalization of the protein and the repair of its cognate lesion.
Here, with the aim of defining the mechanisms allowing the access of the human BER machinery to lesions, in particular with respect to higher-order chromatin organization, we investigated the repair by and subnuclear redistribution of the proteins initiating BER after induction of an oxidative stress generating large amounts of 8-oxoG in chromosomal DNA.
MATERIALS AND METHODS
Plasmid construction, cell culture and treatments
Human OGG1 (wild-type and mutant versions K249Q and F319A) and APE1 fusions to fluorescent proteins were described previously (31). For the construction of XRCC1–YFP, the open reading frame of XRCC1 was amplified by PCR and subcloned into pEYFP-N1 (Clontech). Transient transfections were done with Effectene Transfection Reagent (Qiagen) according to the manufacturer's; instructions. Stable transfectants were selected in DMEM containing 800 µg/ml G418 and kept in 400 µg/ml G418.
All cell lines were cultured in DMEM (Lonza) containing 10% of foetal bovine serum at 37°C with 5% CO2. Cells at about 80% of confluence were treated with 40 mM potassium bromate (KBrO3; Sigma) diluted in DPBS (Cambrex), for 30 min at 37°C. Cells were then allowed to recover in DMEM for the indicated periods of time before fixation or extraction. When mentioned, DMEM was supplemented with 250 mM sucrose.
For transcription blockage, cells were incubated at 37°C for 2 h before KBrO3-treatment and during the recovery period with 50 µg/ml α-amanitin (Sigma) or 1 µg/ml actinomycin D (Sigma) diluted in DMEM.
Immunofluorescence and microscopy
Immunofluorescence protocols have been previously described (31). Primary antibodies used were anti-HP1α (1H5 Euromedex), anti-H3K9me2 (07-441 Upstate), anti-H3K4me2 (07-030 Upstate), anti-RNA polymerase II (H5 Eurogentec). Secondary antibodies used were coupled to Alexa 594 (Molecular probes). Nuclear DNA or RNA were counterstained with 1 µg/ml 4′,6′-diamidino-2-phenylindole (DAPI) or 1 µg/ml propidium iodide (PI). When indicated, cells were previously extracted for 5 min on ice with cytosqueleton (CSK) buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 6.8, 3 mM MgCl2, 0.5% Triton X-100 and protease inhibitors) before fixation with 4% para-formaldehyde for 20 min at room temperature (RT). For DNase and RNase treatments, cells were incubated with CSK buffer containing 0.5 U/µl DNase I or 5 µg/ml RNase I respectively for 30 min at 37°C prior to fixation. Coverslips were mounted in Dako Fluorescent Mounting Medium.
For visualization of 8-oxoG in situ, cells on coverslips were fixed in acetone:methanol (1:1) and air dried. Cells were hydrated for 15 min in phosphate-buffered saline (PBS), and DNA was denatured by incubating cells in 1.5 N HCl for 15 min at RT. Cells were washed three times in PBS and neutralized with 0.1 M Na–borate pH 8.5 for 5 min before proceeding to the immunofluorescence protocol, as previously described, using the mouse anti 8-OhdG (Japan Institute for the Control of Aging) as a primary antibody.
Image acquisition was performed with a Leica confocal microscope SPE (Wetzlar, Germany), using ACS APO 40.0 × 1.15 OIL or ACS APO 63.0 × 1.30 OIL lenses. Image treatment and analysis were done with Leica and ImageJ softwares. Plot profiles, cytofluorograms and GFP intensity levels were obtained using ImageJ software (Rasband, 1997, ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/) (32). For GFP fluorescence measurement in kinetics experiments, a mask for each cell was created, thanks to DAPI staining, and reported on the corresponding GFP image. GFP intensity was calculated for each cell as arbitrary units. For each condition, GFP intensity of about 250–500 cells was measured.
Western blots and chromatin extraction
Cell pellets (about 5×106 cells) were incubated for 10 min at 4°C in 1 ml ice-cold CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES, pH 6.8, 1 mM EDTA, 3 mM MgCl2, 1 mM DTT) containing 0.5% triton X-100 and protease inhibitors. After centrifugation at 5000 rpm for 5 min, the supernatant containing the soluble proteins was recovered (S1 fraction). Pellets were washed twice with 1 ml ice-cold CSK. The resulting pellets (P1) were resuspended in Laemmli buffer and boiled to be analysed by western blotting, or treated with 0.5 U/µl DNAse I diluted in a digestion buffer (50 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 6.8, 3 mM MgCl2, 0.5% triton X-100) for 30 min at RT. After centrifugation at 5000 rpm for 5 min, the pellets were washed twice in CSK buffer and the last pellets (P2) were resuspended in Laemmli buffer and boiled to be analysed by western blotting.
S1, P1 and P2 fractions were electrophoresed on 10% sodium dodecyl sulphate-polyacrylamide gels (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat dry milk in PBS supplemented with 0.1% Tween-20 (PBS-T) for at least 1 h and incubated with primary antibodies in PBS-T containing 1% blocking reagent (Roche Diagnostic) for 2 h at RT. After three 10-min washes with PBS-T, membranes were incubated with a horseradish-peroxidise-conjugated secondary antibody diluted in PBS-T containing 1% blocking reagent for 45 min. Proteins were revealed with ECL (Amersham Biosciences); protein amounts were quantified using a G:BOX ChemiXL (Syngene) and associated softwares GeneSnap and GeneTools.
Primary antibodies used were: anti-lamin B1 (ZL5 Abcam), anti-GFP (11814460001, Roche), anti-HP1α (1G9 Euromedex), anti-H3K4me2 (07-030 Upstate), anti-RNA polymerase II (H5 Eurogentec) and anti-Sm (MS-450-P1, NeoMarkers). For endogenous OGG1 detection, we used an anti-OGG1 (PA3) described previously (33).
Heterochromatin and euchromatin fractionation
Subnuclear fractions were obtained according to Frenster et al. (34). Briefly, cells were washed three times in CSK buffer containing 0.1% triton X-100 in order to remove soluble nucleoplasmic and cytoplasmic fraction. The resulting nuclear pellets were washed with ice-cold washing buffer I (0.25 M sucrose, 3.3 mM CaCl2) and resuspended in 0.2 ml of buffer A (0.1875 M sucrose, 20 mM glucose, 24 mM Tris–HCl (pH 7.1), 12.8 mM NaCl, 3.3 mM CaCl2). The suspension was incubated at 37°C for 30 min and centrifuged at 3100 rpm for 5 min after addition of 0.8 ml of cold buffer A. The washed nuclear pellet was extracted three times with 1 ml of buffer B (10 mM Tris–HCl, pH 7.1, 3.3 mM CaCl2). The pellet obtained by centrifugation at 3100 rpm for 5 min was resuspended in 1 ml of cation-free 0.25 M sucrose and subjected to sonication. The suspension was centrifuged at 1400 rpm for 5 min after addition of 0.5 ml of cation-free 0.25 M sucrose. The supernatant was further centrifuged at 4500 rpm for 10 min. The resulting pellet was kept as the heterochromatin (H) fraction. The supernatant fraction was centrifuged at 7700 rpm for 30 min. The pellet was kept as the intermediate (I) fraction. The resultant supernatant was again centrifuged at 44 000 rpm for 60 min and the pellet fraction was the euchromation (E) fraction. The H, I and E pellets were directly resuspended in the same volume of Laemmli buffer 1×, boiled for 5 min at 95°C and vortexed before proceeding to western blot analysis.
Quantification of oxidative purine modifications
A modified version (35) of the alkaline elution assay originally described by Kohn et al. (36) was used to quantify Fpg-sensitive oxidative purine lesions. The sum of DNA modifications sensitive to repair endonucleases (Fpg) and single-strand breaks was obtained from experiments in which the cellular DNA was incubated for 60 min at 37°C with Fpg protein (1 µg/ml) immediately after cell lysis. Under these conditions, the incision by Fpg at endonuclease-sensitive modifications has been shown to be saturated (35). The numbers of lesions incised specifically by Fpg were obtained by subtraction of the number of single-strand breaks observed in experiments without enzymatic treatment. Elution curves obtained with γ-irradiated cells were used for calibration, assuming that 6 Gy generate 1 single-strand break/106 bp (36). Induced modifications were obtained after subtraction of endogenous lesions quantified in untreated control cells.
In situ hybridization with oligo(dT)
Poly(A)+ RNA was detected using a 30 mer-oligo(dT)-5’Cy3 as a probe. Briefly, non-treated (NT) or KBrO3-treated cells are allowed to recover in fresh medium for 2 h before CSK washing and fixation. Cells were subjected to successive washes on ice with 100% methanol for 10 min, 75% ethanol for 10 min and Tris 1 M pH 8.0 for 5 min. oligo(dT) probe (50 ng/µl) was diluted in hybridization buffer (1 mg/ml yeast tRNA, 0.005% BSA, 10% sulphate dextran, 25% deionized formamide, 2×SSC in DEPC water), added to coverslips and incubated for at least 1 h at 42°C. After hybridization, cells were washed once with 4×SSC for 10 min at 42°C and once with 2×SSC for 10 min at 42°C. Cells were further incubated for 1 h at RT in 2 × SSC + 0.1% triton X-100 containing the anti-GFP at a dilution 1/500. Cells were then washed three times with 2 × SSC and incubated with anti-mouse-Alexa488 for 30 min RT in 2 × SSC + 0.1% triton X-100 buffer supplemented with DAPI (1 µg/ml). Cells were finally washed twice in 2 × SSC prior to observation.
Quantification of 8-oxoG glycosylase enzymatic activity
Total cell extracts were prepared by sonication of cell pellets in 20 mM Tris–HCl, pH 7.5, 250 mM NaCl, 1 mM EDTA containing proteases inhibitors. Homogenates were centrifuged at 13 000 rpm for 15 min at 4°C and supernatants were used for 8-oxoG glycosylase activity assay. We used the 8-oxoG DNA glycosylase assay described previously (33).
RESULTS
In vivo repair of oxidative DNA lesions induced by KBrO3
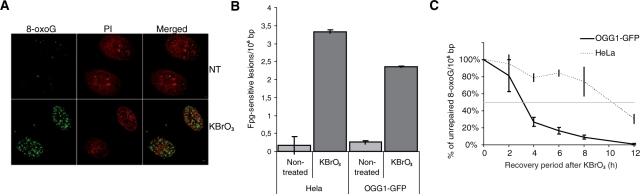
To explore the cellular DNA repair mechanisms required for the removal of 8-oxoG from chromosomal DNA we used KBrO3, a carcinogenic agent known to induce oxidative stress in eukaryotic cells. Genotoxicity requires reduction of bromate by thiols (as glutathione or reduced cysteines) and induces predominantly 8-oxoG lesions in DNA (37). As displayed in Figure 1A, immediately after a 30 min treatment with 40 mM KBrO3, HeLa cells immunostained with an antibody against 8-oxoG showed a strong fluorescent signal compared to NT cells. Fpg-sensitive lesions, mostly 8-oxoG, were quantified by alkaline elution. The steady-state level of endogenous Fpg-sensitive sites was ∼0.21 lesions/106 bp in both HeLa and HeLa cells expressing an OGG1–GFP fusion protein. KBrO3 treatment induced a 10-fold increase in Fpg-sensitive lesions (3.33 and 2.36 lesions/106 bp in HeLa and OGG1–GFP cells, respectively; Figure 1B). Because extracts from cells expressing the OGG1 fusion have about 10-fold more 8-oxoG DNA glycosylase activity, the difference in the number of induced lesions is probably due to a beginning of repair during the 30 min treatment. The DNA glycosylase activity is not affected by the exposure to KBrO3 (Supplementary Figure S1).
Figure 1.
Overexpression of OGG1–GFP accelerates repair of 8-oxoG induced in KBrO3-treated cells. (A) 8-OxoG were stained with anti-8-oxoG antibody; DNA was stained with PI after RNAse A digestion. Scale bars, 2 µm. (B) 8-OxoG in untreated and KBrO3-treated cells were quantified as Fpg-sensitive sites by alkaline elution in HeLa and OGG1–GFP cells. (C) Kinetics of repair of 8-oxoG in HeLa and OGG1–GFP cells. After KBrO3 treatments cells were allowed to recover for the indicated periods of time and analysed by alkaline elution to measure the remaining 8-oxoG lesions.
We next determined the kinetics for the repair of KBrO3-induced Fpg-sensitive sites in OGG1–GFP overexpressing and control HeLa cells using the alkaline elution assay. As expected, repair was faster in cells overexpressing OGG1 in which it reached its maximum rate between 2 and 4 h after treatment while control HeLa cells were much slower, requiring >8 h to eliminate 50% of the lesions (Figure 1C). This result shows that adding a GFP tag at the C-terminus of OGG1 protein does not interfere with the capacity of the enzyme to access chromatin and to efficiently repair 8-oxoG.
OGG1 recruitment to chromatin
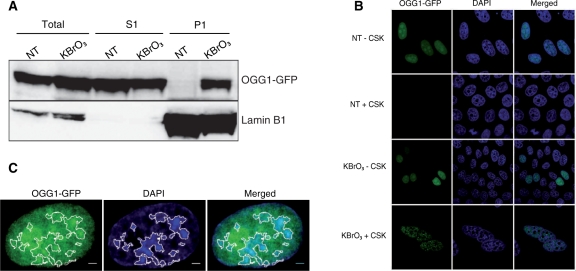
To determine OGG1 localization during the repair period, after the 30 min KBrO3 treatment cells were allowed to recover for 3 h in fresh medium before proceeding to subcellular fractionation. The soluble pool of proteins (S1), corresponding to the cytoplasm and nucleoplasm, was obtained by washing the cells with CSK buffer containing 0.5% triton. The resulting pellet (P1) corresponded to chromatin- and matrix-associated proteins. As revealed by western blot analysis of the S1 fraction, the majority of OGG1 remained soluble after KBrO3. However, whereas no retention of OGG1 could be observed in NT cells, KBrO3 induced the association of a fraction of hOGG1 with chromatin and nuclear matrix (Figure 2A).
Figure 2.
After KBrO3 OGG1 is relocalized to foci resistant to detergent-containing buffer. (A) Subcellular fractionation of NT and KBrO3-treated OGG1–GFP cells. Cells were separated into fractions S1 (soluble proteins) and P1 (chromatin fraction) and different fractions analysed by western blot using an anti-GFP antibody. Lamin B1 was used as a loading control. (B) Distribution patterns of OGG1–GFP in NT and KBrO3-treated cells. Prior to fixation, soluble proteins were removed with CSK-0.5% triton when indicated. (C) NT cells expressing OGG1–GFP were directly fixed, DAPI stained and analysed by confocal microscopy. DAPI staining was used to define heterochromatin regions (white outlines). Scale bars, 2 µm.
To further characterize the relocalization of OGG1, its distribution was followed by confocal microscopy. As previously reported (31), the OGG1–GFP protein was exclusively nuclear and remained soluble in NT cells, as all the fluorescence signal was removed after CSK buffer wash (Figure 2B). As expected for an enzyme possessing DNA-binding affinity, in NT cells OGG1 was concentrated on condensed DNA patches corresponding to heterochromatin (Figure 2C). In the case of KBrO3-treated cells, a fraction of the OGG1–GFP protein resisted the detergent washes (Figure 2B), confirming an association with chromatin. To rule out an effect of the fused GFP protein, we used OGG1 tagged with a FLAG sequence in which case the protein was detected by immunofluorescence. The same results were obtained (data not shown). Because we used non-synchronous cell populations, the observation of the relocalization of OGG1 to the chromatin fraction in all cells suggests that this phenomenon does not depend on the cell-cycle status of the cells. This was further confirmed by the analysis of OGG1 recruitment to the insoluble fraction in individual cells for which the cell-cycle phase was determined with specific markers. Indeed, the recruitment of OGG1 was confirmed to happen in cells going through the different cell-cycle phases (data not shown).
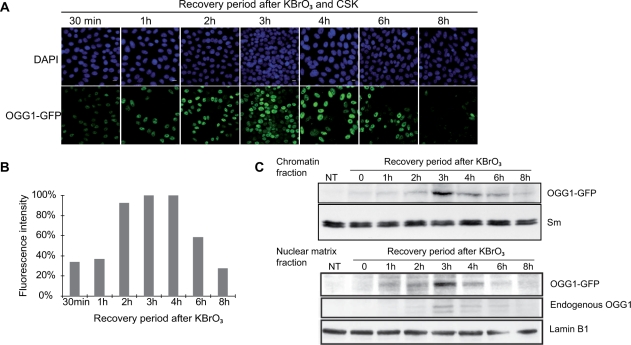
In order to determine the kinetics of OGG1 recruitment to chromatin after KBrO3 treatment, cells expressing OGG1–GFP were washed with CSK after different recovery times prior to analysis by confocal microscopy. As soon as 30 min after treatment, 70–90% of the cells showed a detergent-resistant OGG1 fraction that increased with time to peak between 3 and 4 h after the end of the treatment (Figure 3A and B). The fraction of OGG1 associated with chromatin subsequently decreased to reach basal levels after 8 h of recovery. Subcellular fractionation and immunoblotting analysis confirmed the recruitment kinetics (Figure 3C, upper panel) and allowed to establish that the endogenous OGG1 was also recruited to the insoluble fraction with similar kinetics (Figure 3C, lower panel). Interestingly, the maximum accumulation of OGG1 in the chromatin fraction coincided with the maximum rate of repair of 8-oxoG (Figure 1C), suggesting a link between BER initiated by the DNA glycosylase and its recruitment to chromatin.
Figure 3.
OGG1 is dynamically recruited to chromatin after KBrO3 treatment. (A) Confocal analysis of OGG1–GFP recruitment to chromatin fraction in KBrO3-treated cells. Cells were allowed to recover in fresh medium for the indicated times before CSK washing for the removal of the soluble fraction. Scale bars, 10 µm. (B) Fluorescence intensities of cells showed in (A). Values represent an average of fluorescence intensity of cells (n > 50). (C) Upper panel, western blot with an anti-GFP antibody shows OGG1–GFP transient accumulation in the chromatin fraction (P1) after KBrO3 treatment. Sm was used as a loading control. The lower panel shows the accumulation of overexpressed and endogenous OGG1 proteins in the nuclear matrix fraction obtained after DNAse digestion. OGG1–GFP and endogenous OGG1 were visualized with anti-GFP and anti-OGG1 (PA3) primary antibodies, respectively. Lamin B1 served as a loading control.
Other BER proteins are recruited to chromatin together with OGG1
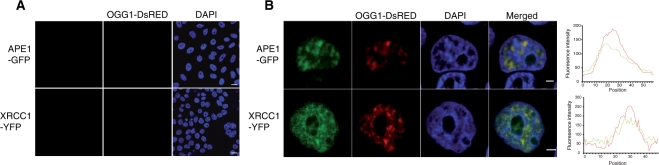
After recognition and excision of the modified base by the DNA glycosylase, other proteins need to be recruited to the sites of repair to assure complete restoration of the DNA strand. We asked whether proteins participating in post-excision steps of BER, such as XRCC1 and APE1, were also recruited to the chromatin fraction in KBrO3-treated cells. To answer such question, co-localization analyses were performed after co-transfection of plasmids expressing OGG1–DsRED with constructs expressing either APE1–GFP or XRCC1–YFP. While in NT cells APE1 and XRCC1 are both soluble in the nucleoplasm, and therefore removed by CSK washes (Figure 4A), treatment with KBrO3 induces their relocalization to the chromatin, within the same areas where is found OGG1 (Figure 4B) suggesting that complexes of BER proteins are formed in response to oxidative stress to protect the genome from mutagenic lesions.
Figure 4.
APE1 and XRCC1, together with OGG1 are relocalized to euchromatin in KBrO3-treated cells. Fusion proteins (OGG1–DsRed and APE1–GFP or XRCC1–YFP) were transiently expressed in HeLa cells. Soluble proteins were extracted by CSK washes prior to fixation and DAPI staining. (A) NT cells, Scale bar=10 µm. (B) Three hours after KBrO3 treatment. Plot profiles along the lines in the merged image reflect the co-localization of OGG1 with APE1 and XRCC1, respectively. Scale bar 2 µm. Fluorescence intensities of each channel along a line in the merged image are represented in the right panels.
OGG1 is relocalized to open chromatin regions
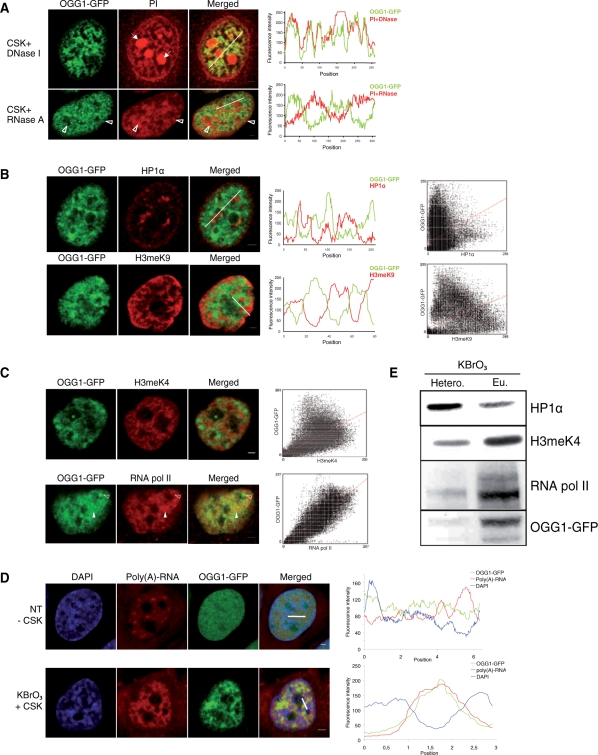
We next explored the possibility that OGG1 would be recruited to specific chromatin domains. When KBrO3-treated cells were submitted to DNAse digestion prior to fixation, the OGG1–GFP signal was still observed, suggesting that while processing DNA OGG1 is associated with an insoluble nuclear fraction. Similar observations have been reported for proteins associated with transcription and replication factories. Surprisingly, OGG1 perfectly co-localized with the PI signal, suggesting an association with RNA-rich regions of the nucleus (Figure 5A, upper panel). However, RNA digestion prior to CSK buffer wash and fixation also failed to remove the OGG1–GFP signal in KBrO3-treated cells (Figure 5A, lower panel). Interestingly, the OGG1 signal was excluded from the large patches of heterochromatin revealed by PI staining after RNAse digestion. Indeed, line scans of OGG1 (green) and PI (red) staining on Figure 5A clearly showed a complete exclusion of the DNA glycosylase from DNA-dense regions (lower panel). Taken together, these results show that KBrO3 treatment induces an active relocalization of OGG1 from a soluble pool to less-condensed DNA regions enriched in RNA.
Figure 5.
OGG1 is excluded from heterochromatin and colocalizes with euchromatin-associated proteins. Following a 3-h recovery after KBrO3 treatment, soluble proteins were removed with CSK buffer prior to fixation and analysis by confocal microscopy. (A) DNA (upper panel) or RNA (lower panel) were digested before fixation and PI staining. Solid arrows indicate ribosomal RNA in nucleoli. Open arrows show patches of heterochromatin. Positions of the line scans used for the plot profile are indicated in the merged images. (B) Heterochromatin was immunostained with HP1α (upper panel) and H3meK9 (lower panel), in red. Cytofluorogram of both merged images shows a great dispersion of points, reflecting an absence of correlation of both intensity signals. (C) RNA polymerase II (upper panel) and H3meK4 (lower panel) partially colocalize with OGG1–GFP (filled arrows), although some OGG1 foci are excluded from RNA polymerase II staining (unfilled arrows). Correlations between green and red signals are presented in the cytofluorograms. (D) In situ hybridization of mRNA with oligo(dT)5’Cy3 in NT and KBrO3-treated cells. Line scans used for the plot profiles are indicated in the merged images. Both plot profiles and cytofluorogram show a colocalization between polyadenylated RNA and OGG1 after KBrO3 and an exclusion of both signals in NT cells. (E) Heterochromatin/euchromatin fractionation of KBrO3-treated cells. Heterochromatin (HP1α) and euchromatin (H3meK4 and RNA polymerase II) markers are used as controls. Scale bars, 2 µm.
The patterns described above are consistent with a preferential recruitment of OGG1 to euchromatin regions. Dual-label experiments further confirmed this hypothesis. Co-labelling of OGG1 with either HP1α or histone 3 dimethylated on lysine 9 (H3meK9), two proteins generally associated with heterochromatin, showed total exclusion of OGG1 signal from heterochromatin patches after KBrO3 treatment (Figure 5B). The scatter plots to the right, representing the correlation degree of each pixel for fluorescence intensities, reveal an important point dispersion implying that the fluorescent variables are not correlated. We then asked if OGG1 was associated with open chromatin markers. Co-labelling experiments with proteins normally found in euchromatic regions such as H3meK4 (Figure 5C, upper panel), acetylated-histone H4 (data not shown) and hyperphosphorylated RNA polymerase II (Figure 5C, lower panel) displayed a good degree of co-localization of OGG1 with those proteins linked to open chromatin regions. Biochemical fractionation of chromatin from KBrO3-treated cells confirmed these results. Indeed, as shown on Figure 5E, OGG1 was concentrated in those fractions enriched in H3meK4 and RNA polymerase II, mainly euchromatin, while the HP1α-rich fraction (heterochromatin) presented very little DNA glycosylase accumulation.
The partial co-localization of OGG1 with RNA polymerase II (Figure 5C) and their presence in the same chromatin fraction (Figure 5E) suggest the possibility of the recruitment of OGG1 to actively transcribed chromatin regions. Further support for an association of OGG1 with regions of open chromatin arises from the high degree of co-localization found between the protein and poly(A) RNAs revealed by hybridization with fluorescent poly(dT) oligos (Figure 5D). Nevertheless, blocking transcription using specific inhibitors of RNA polymerase elongation as α-amanitin or actinomycin D did not impede OGG1 relocalization after KBrO3, suggesting that active transcription is not required for translocation of the glycosylase to the chromatin (Supplementary Figure S2).
Recognition of 8-oxoG by OGG1 is not required for the recruitment of the protein to chromatin
A possible explanation for the relocalization of OGG1 onto open chromatin domains is that DNA lesions are preferentially formed in more accessible regions. We therefore tested whether the number of lesions induced by KBrO3 was modulated by the degree of chromatin compaction. Hypertonic shock triggered by sucrose is known to induce a rapid structural change in chromatin conformation leading to condensation (38). As shown in Figure 6A, a 3-h incubation with sucrose lead to the appearance of brighter DAPI-stained patches. Quantification of DNA damage right before the oxidative treatment showed that incubation in medium supplemented with sucrose did not induce Fpg-sensitive lesions in chromosomal DNA. When KBrO3 was applied to cells in which the chromatin was highly compacted, the number of lesions induced was 1.34 Fpg-sensitive sites/106 bp, a number indistinguishable from that of the lesions induced in cells that have not been pre-incubated with sucrose (1.17 Fpg-sensitive sites/106 bp). This result suggests that KBrO3 treatment can induce lesions in DNA likely to be in highly condensed domains, challenging the assumption that 8-oxoG recognition by OGG1 triggers the DNA glycosylase relocalization observed to be mainly directed to euchromatin regions. To directly test the relevance of damage recognition by OGG1, we used an OGG1 mutant (F319A), which was shown to be correctly structured but unable to recognize its 8-oxoG substrate and therefore completely inactive as a DNA glycosylase (39). We reasoned that if affinity of OGG1 for the lesion was the driving force in the relocalization of the DNA glycosylase, the OGG1–F319A mutant should not be found associated to euchromatin after the oxidative treatment. However, after KBrO3 exposure, cells expressing the fusion protein OGG1–F319A–GFP showed the same pattern of fluorescence than those expressing the wild type OGG1 fusion as well as the recruitment of OGG1 to the chromatin fraction (P1) (Supplementary Figure S3), ruling out the recognition of the lesion by the DNA glycosylase as the signal to recruit OGG1 to the chromatin fraction.
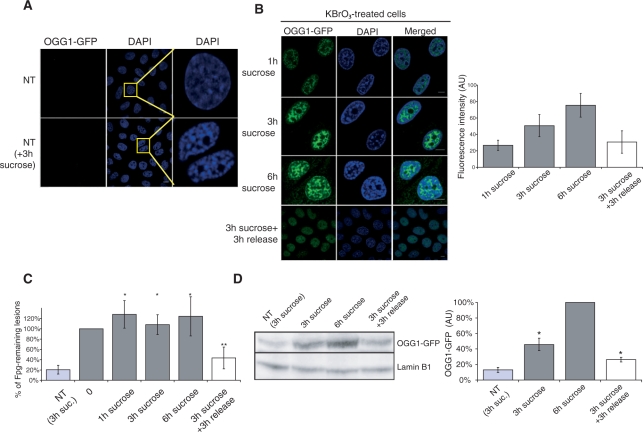
Figure 6.
Chromatin condensation by hypertonic shock impedes 8-oxoG repair. (A) NT cells pre-incubated or not with 250 mM sucrose for 3 h and washed with CSK–0,5% triton buffer prior to fixation. DNA is stained with DAPI. (B) OGG1–GFP cells are treated with 40 mM KBrO3 for 30 min and allowed to recover in DMEM supplemented with 250 mM sucrose. In the last row of images, sucrose was removed after 3 h and replaced by fresh medium for 3 h. Cells were washed with CSK buffer prior to fixation. Graph represents GFP intensity for each condition (number of nucleus > 10). Scale bar = 10 µm. (C) 8-OxoG quantification by alkaline elution of cells treated with 40 mM KBrO3 and recovered in 250 mM sucrose supplemented medium. Bilateral Student test (*P > 0.1 compared to the point 0; **P< 0.04 and P< 0.02 compared to the points 0 and 3 h sucrose respectively). (D) western blot of cells treated with 40 mM KBrO3 and recovered in 250 mM sucrose supplemented medium. Antibodies against GFP are used and lamin B1 is used as a loading control. Quantification of signals using Scan GBox is represented on the graph on the right. Bilateral Student’s test (*P < 0.05).
Open chromatin is required for efficient 8-oxoG repair
The results on the re-localization of OGG1 suggest that removal of 8-oxoG from chromosomal DNA is associated with the decondensed areas of chromatin. To analyse the impact of chromatin organization on the repair of 8-oxoG we induced chromatin condensation immediately after KBrO3 treatment in cells expressing the OGG1–GFP fusion. Incubation with sucrose to trigger condensation did not inhibit the enzymatic activity of OGG1 as determined both in extracts from cells incubated in the presence of sucrose or by the addition of 250 mM sucrose to extracts from NT cells (Supplementary Figure S1). However, analysis of the repair kinetics showed that chromatin compaction delayed repair, suggesting that in such conditions the lesions were present within compacted chromatin. Indeed, in the presence of sucrose, 6 h after their appearance Fpg-sensitive modifications induced by the oxidative treatment persisted at their highest levels (Figure 6C). These results suggest that condensed chromatin represents a barrier for OGG1 access to the lesions.
Surprisingly, chromatin condensation by sucrose not only did not prevent OGG1 recruitment to the CSK-insoluble fraction, but it stimulated its accumulation in that fraction with time (Figure 6B and D). However, the relocalization of OGG1 after oxidative stress on sucrose-treated cells was observed to still direct the protein mainly to regions excluded from highly condensed DNA (Figure 6B). These results, together with the inhibition of repair by the sucrose treatment (Figure 6C) indicate that whereas heterochromatin represents a barrier for OGG1 accession to the lesions it does not impede the recruitment of OGG1 to the BER patches, further supporting the conclusion that recognition of the lesion is not required for the recruitment of OGG1 to open chromatin regions.
Taking advantage of the fact that the hypertonic shock effects on chromatin are rapidly reversible, as can be confirmed by the recovery of a normal DAPI staining pattern 30 min after sucrose removal (Figure 6B), we asked whether decondensation of chromatin could reverse the repair blockage. Indeed, sucrose removal 3 h after KBrO3 treatment allowed repair to resume with normal kinetics as shown by the fact that >50% of the lesions has been removed 3 h later (Figure 6C). Furthermore, in agreement with the need of repair completion to release OGG1 from chromatin, 3 h after sucrose removal chromatin-associated OGG1 levels fell to 50% of those in cells kept in the presence of sucrose (Figure 6B and D).
Release of OGG1 requires completion of repair
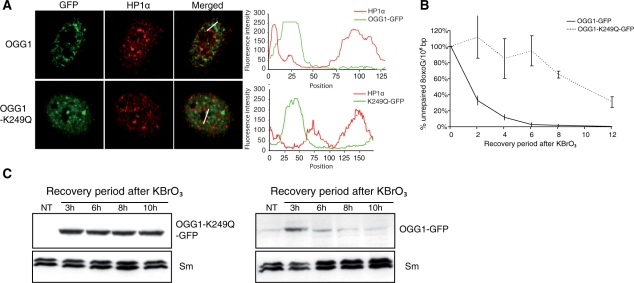
The correlation between the presence of OGG1 in euchromatin regions and the repair kinetics of 8-oxoG suggests that once repair is accomplished, the DNA glycosylase is released back to the soluble pool. We therefore hypothesized that an OGG1 mutant (K249Q) that recognizes the lesion but is unable to proceed to the excision of the modified base (40) would be retained at the chromatin level. We first confirmed by confocal microscopy that 3 h after KBrO3 treatment OGG1–K249Q was recruited to euchromatin regions in the same way as the wild-type protein (Figure 7A). We then analysed the repair and recruitment kinetics for both forms of the protein. As expected, cells expressing the inactive OGG1–K249Q–GFP were much slower to repair Fpg-sensitive lesions when compared to those expressing the wild-type fusion protein (Figure 7B). Consistent with the notion that repair of the lesion is required to release OGG1 from euchromatin, and in agreement with the presence of un-repaired lesions, western blot analysis of the P1 fractions (Figure 7C) showed that OGG1–K249Q remained tightly associated with the chromatin fraction even 10 h after the end of the treatment, thus supporting the idea that the removal of the oxidized base is required to release the DNA glycosylase from the insoluble fraction associated with euchromatin.
Figure 7.
Catalytic activity of OGG1–GFP is not required for the recruitment to the chromatin fraction after KBrO3. HeLa cell lines stably expressing OGG1–GFP or the mutant version OGG1(K249Q)–GFP were treated with KBrO3, allowed to recover for 3 h and CSK pre-extracted prior to fixation (A) Heterochromatin is stained with an anti-HP1α (red). Line scans used for plot profiles are indicated in merged images. Scale bar, 2 µm. (B) Repair kinetics of 8-oxoG lesions in OGG1(K249Q)–GFP and OGG1–GFP cells lines by alkaline elution after a 20 mM KBrO3 treatment. (C) Western blot using an anti-GFP antibody showing the recruitment kinetics of OGG1(K249Q)–GFP (left panel) and OGG1–GFP protein (right panel) to the chromatin fraction.
DISCUSSION
We have analysed here the first steps of the BER in the context of the nuclear architecture. Our results show that chromatin compaction induces strong inhibition of the initial step carried out by the DNA glycosylase, the enzyme responsible for the recognition and excision of the modified base. The simplest explanation for these results is that higher order chromatin compaction represents a barrier impeding the access of the protein to the lesion. Consistent with this hypothesis, we found that after oxidative stress the DNA glycosylase OGG1 is specifically recruited to euchromatin regions.
Initiation of repair within the chromatin structure
Most of our present knowledge on the recognition of DNA lesions and the recruitment of repair proteins comes from the nucleotide excision repair (NER) and double-strand break repair (DSBR) systems, required to remove essentially toxic lesions. In both cases the paradigm states that one or several soluble factors present in the nucleoplasm and capable of detecting the lesion will rapidly diffuse to the site of damage probably directed by a perturbation of the chromatin around the lesion. This first recognition step will then trigger a cascade of events allowing the recruitment of not only the proteins directly carrying out the enzymatic removal of the lesion and subsequent reconstitution of a normal DNA strand, but also of factors such as histone modifiers and chromatin remodellers to allow access of the enzymes to DNA. This leads to the formation of foci or patches of DNA repair enzymes and factors at the site of the lesion (41,42).
Our results suggest that a different situation might occur in the case of BER. This repair mechanism has for substrate small lesions that, in most cases and in contrast with strand breaks or UV-induced damages, have little impact on the DNA helical structure. Moreover, many of the lesions recognized by the DNA glycosylases, and in particular 8-oxoG, do not compromise the normal replication or transcription of DNA, having as their main consequence the induction of mutations. These characteristics suggest that for a DNA glycosylase to recognize the lesion, the latter must be accessible. Consistently, all the in vitro evidences show that OGG1 (21), as well as other BER enzymes (19,22,43), are inhibited by the first level of chromatin organization provided by the presence of nucleosomes. Although the compacted chromatin obtained by sucrose treatment cannot necessarily be compared to normal heterochromatin, our experiments show that higher order chromatin organization constitutes a first barrier for the access of the BER machinery to the lesions in vivo. Interestingly, it has been recently shown that DSBR is slower in heterochromatic lesions and that ATM signalling is specifically required for their efficient repair, possibly by triggering a restructuring of the chromatin and therefore allowing the access of repair proteins to the lesion (30). More specifically related to BER, Bhakat et al. (44) showed that treatment of cells with the trichostatin A resulted in an accelerated repair of cellular 8-oxoG by OGG1. Besides its contribution to the modification of OGG1 itself and its activity—as shown by the use of non-acetylable OGG1 mutants (44)—this deacetylase inhibitor is known to induce chromatin decondensation, suggesting the possibility that part of the enhancement it induces in the rate of 8-oxoG removal could be due to the improved accessibility of the DNA glycosylase to DNA.
BER repair centres?
The reorganization described here for BER proteins into patches in response to an oxidative stress raises the possibility of the existence of BER ‘repair factories’. Such an organization could enhance the efficiency of the repair process by favouring the assemblage of BER complexes required for the repair, in particular considering the need for several DNA glycosylases to scan DNA in search of their cognate lesions (7,8). Compartmentalization of NER or DSBR has been well established. Indeed, DNA repair proteins are recruited to the site of UV damage or double-strand breaks to form repair foci (42,45). Interestingly, single-strand break repair (SSBR) also involves the rapid and transient formation of foci detected by poly(ADP-ribose) synthesis sites and the presence of XRCC1 protein (46,47). Consistently, XRCC1 was recently shown to be recruited to the nuclear matrix as rapidly as 10 min after H2O2 treatment in a phosphorylation-dependent process (48). Although SSBR and BER share some of their enzymatic steps, the patches we observed here for OGG1 are clearly distinct from the foci formed at single-strand break in their kinetics of formation and disassembly (46,47).
The resistance to detergent of the BER patches suggests their association with relatively stable nuclear structures such as lamina or a putative actin-based nucleoskeleton (49,50), but possibly chromatin itself could serve as attachment site for complexes involved in DNA transactions (51). The exclusion of the BER patches from heterochromatin and their co-localization with H3meK4 or acetylated histone H4 place those repair centres within euchromatin regions. Furthermore, the association of BER patches with RNA polymerase II and mRNAs suggests that they are present in transcriptionally active regions. The formation of foci by RNA polymerase II and other transcription factors has lead to definition of ‘transcription factories’ (52,53). The association of active genes with such structures would facilitate their efficient transcription (54). It is tempting to propose that the recruitment of BER to those structures, or their environment, would facilitate repair by providing an easier access to DNA. In response to an oxidative stress BER proteins would be relocated preferentially to or around transcription factories where ATP-dependent chromatin remodelling and histone modification factors are already present. This would be in contrast with UV damage or DSBR, where those factors are recruited to the lesion (18,55). The association of the BER machinery to euchromatin, a region shown to be enriched in genes (56), could provide cells with a mechanism for preferentially repairing mutagenic lesions in sequences more likely to be expressed. Similarly, the interaction of thymine DNA glycosylase with CBP/p300 found by Tini et al. (57) prompted the authors to suggest that this interaction would result in the specific recruitment of the BER machine to promoter regions. As in our case, they propose a model ensuring that transcriptionally active genes are repaired prior to transcription. This strategy differs from transcription-coupled repair of UV damage that relies on the blockage of the transcription machinery by the lesion to initiate the preferential repair of the transcribed strand (58). The consequences of the mechanism we propose for BER will be rather similar to those arising from the transcription domain-associated repair (DAR) of UV damage described originally for highly differentiated cells (59). Indeed, through this subset of global NER both DNA strands of active genes continue to be proficiently repaired while there is a general attenuation of the NER in other regions.
Chromatin recruitment and release of BER proteins
The finding that a mutant of OGG1 that does not recognize its substrate is still able to form BER patches after an oxidative stress indicates that the direct interaction of the enzyme with its substrate is not a requirement for its recruitment to BER factories. This observation raises the question of what could be the signal for triggering the re-localization of the BER proteins. Taking into account the results obtained after UVA (31), it would seem reasonable to propose that whatever mechanism underlies the OGG1 relocalization, ROS are likely involved. A possibility is that in response to an oxidative stress OGG1 is subjected to a post-translational modification that allows its interaction with some component of the transcription machinery or some structural protein in the nucleus. Several post-translational modifications, such as acetylation (44), phosphorylation (60) or oxidation (61) have been reported for OGG1. Initial analysis by 2D gels of the P1 and S1 fractions failed to detect post-translational modifications specifically associated to the recruited fraction (R.A. and J.P.R., unpublished results). Alternatively, the modification in response to an oxidative stress of proteins involved in the organization of chromatin (62) could lead to an increase in their affinity for the DNA glycosylase. The recruitment of the downstream proteins APE1 and XRCC1 is likely to rely on the assembly of repair complexes on the initially recruited DNA glycosylase (7,8).
Our kinetics experiments using the inactive mutants of OGG1 and those showing the retention of the DNA glycosylase in cells whose chromatin compaction status blocks repair indicate that disassembly of the BER patches depends on the completion of the repair process. As for NER factors (45,63), OGG1 remains associated with the repair centres for the period of time required for the removal of 8-oxoG and, once it has completed its task, it diffuses away.
CONCLUSION
The data presented in this study show that an oxidative stress induces specific re-localization of the first enzyme in the BER pathway to structures linked to euchromatin. The tight correlation between the repair of 8-oxoG and this re-localization of OGG1 and other BER proteins to transcription factories or their environment suggests a model in which oxidative stress induces the recruitment of the BER machinery to regions of open chromatin, assuring preferential repair of active regions of the genome. Recently, the observation of an increased divergence at the single nucleotide level in silenced DNA regions of budding yeast closely related species prompted the authors to propose an interference of the silencing machinery with DNA repair (64). It is tempting to speculate that the inhibition of BER on highly condensed chromatin regions contributes to such localized genetic hypervariability.
FUNDING
Commissariat à l’Energie Atomique (CEA), the Centre National pour la Recherche Scientifique and grants from the Association pour la Recherche contre le Cancer (n°1118 to J.P.R.), Electricité de France (to J.P.R.) and the DEUTSCHE Forschungsgeimeinschaft (EP11/8-1 to B.E.). R.A. was supported by fellowships from the CEA and the Fondation pour la Recherche Médicale. Exchanges between the two laboratories were supported by the PROCOPE collaborative programme between France and Germany. Funding for open access charges: CEA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of their laboratories for discussions and Ina Schulz for her help with the alkaline elution assays.
REFERENCES
- 1.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 2.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation- damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 3.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol. Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick- sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campalans A, Marsin S, Nakabeppu Y, O'C;onnor TR, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 9.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-Ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 12.David SS, O'S;hea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smerdon MJ. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 14.Wellinger RE, Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. Embo J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suter B, Livingstone-Zatchej M, Thoma F. Chromatin structure modulates DNA repair by photolyase in vivo. Embo. J. 1997;16:2150–2160. doi: 10.1093/emboj/16.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr. Opin. Genet. Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Osley MA, Shen X. Altering nucleosomes during DNA double-strand break repair in yeast. Trends Genet. 2006;22:671–677. doi: 10.1016/j.tig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 18.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 19.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl Acad. Sci. USA. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 21.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol. Cell. Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO. J. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 25.Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb. Symp. Quant. Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- 26.Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri S, Wyrick JJ, Smerdon MJ. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:1690–1700. doi: 10.1093/nar/gkp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingstone-Zatchej M, Marcionelli R, Moller K, de Pril R, Thoma F. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J Biol. Chem. 2003;278:37471–37479. doi: 10.1074/jbc.M306335200. [DOI] [PubMed] [Google Scholar]
- 29.Terleth C, van Sluis CA, van de Putte P. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci. 2007;120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- 32.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 33.Bravard A, Vacher M, Moritz E, Vaslin L, Hall J, Epe B, Radicella JP. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009;69:3642–3649. doi: 10.1158/0008-5472.CAN-08-3943. [DOI] [PubMed] [Google Scholar]
- 34.Frenster JH, Allfrey VG, Mirsky AE. Repressed and active chromatin isolated from interphase lymphocytes. Proc. Natl Acad. Sci. USA. 1963;50:1026–1032. doi: 10.1073/pnas.50.6.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pflaum M, Will O, Epe B. Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis. 1997;18:2225–2231. doi: 10.1093/carcin/18.11.2225. [DOI] [PubMed] [Google Scholar]
- 36.Kohn KW, Erickson LC, Ewig RA, Friedman CA. Fractionation of DNA from mammalian cells by alkaline elution. Biochemistry. 1976;15:4629–4637. doi: 10.1021/bi00666a013. [DOI] [PubMed] [Google Scholar]
- 37.Ballmaier D, Epe B. DNA damage by bromate: mechanism and consequences. Toxicology. 2006;221:166–171. doi: 10.1016/j.tox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Richter K, Nessling M, Lichter P. Experimental evidence for the influence of molecular crowding on nuclear architecture. J. Cell Sci. 2007;120:1673–1680. doi: 10.1242/jcs.03440. [DOI] [PubMed] [Google Scholar]
- 39.van der Kemp PA, Charbonnier JB, Audebert M, Boiteux S. Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Res. 2004;32:570–578. doi: 10.1093/nar/gkh224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nash HM, Lu R, Lane WS, Verdine GL. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem. Biol. 1997;4:693–702. doi: 10.1016/s1074-5521(97)90225-8. [DOI] [PubMed] [Google Scholar]
- 41.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 43.Ishiwata K, Oikawa A. Chromatin structure interferes with excision of abnormal bases from DNA. Biochim. Biophys. Acta. 1982;698:15–21. doi: 10.1016/0167-4781(82)90178-6. [DOI] [PubMed] [Google Scholar]
- 44.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- 46.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubota Y, Takanami T, Higashitani A, Horiuchi S. Localization of X-ray cross complementing gene 1 protein in the nuclear matrix is controlled by casein kinase II-dependent phosphorylation in response to oxidative damage. DNA Repair (Amst). 2009;8:953–960. doi: 10.1016/j.dnarep.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem. Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 50.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J. Cell Mol. Med. 2009 doi: 10.1111/j.1582-4934.2008.00676.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 52.Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: shaping the landscape of the genome. Nat. Rev. Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 53.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 54.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 55.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 58.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 59.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dantzer F, Luna L, Bjoras M, Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bravard A, Vacher M, Gouget B, Coutant A, de Boisferon FH, Marsin S, Chevillard S, Radicella JP. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol. Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl Acad. Sci. USA. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Politi A, Mone MJ, Houtsmuller AB, Hoogstraten D, Vermeulen W, Heinrich R, van Driel R. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies. Mol. Cell. 2005;19:679–690. doi: 10.1016/j.molcel.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 64.Teytelman L, Eisen MB, Rine J. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 2008;4:e1000247. doi: 10.1371/journal.pgen.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.