Abstract
The mechanism behind the positive action of introns upon transcription and the biological significance of this positive feedback remains unclear. Functional ablation of splice sites within an HIV-derived env cDNA significantly reduced transcription that was rescued by a U1 snRNA modified to bind to the mutated splice donor (SD). Using this model we further characterized both the U1 and pre-mRNA structural requirements for transcriptional enhancement. U1 snRNA rescued as a mature Sm-type snRNP with an intact stem loop II. Position and sequence context for U1-binding is crucial because a promoter proximal intron placed upstream of the mutated SD failed to rescue transcription. Furthermore, U1-rescue was independent of promoter and exon sequence and is partially replaced by the transcription elongation activator Tat, pointing to an intron-localized block in transcriptional elongation. Thus, transcriptional coupling of U1 snRNA binding to the SD may licence the polymerase for transcription through the intron.
INTRODUCTION
The splicing of eukaryotic mRNA transcripts coincides with transcriptional elongation and the two processes are tightly coupled and coordinated by the C-terminal domain (CTD) of RNA Polymerase (Pol) II (1). The CTD is a long and unstructured domain containing, in humans, 52 repeats of the consensus heptapeptide YS2PTS5PS7 (2). Tightly regulated post-translational modification of this unit is termed the CTD code (3) and affects the binding and activity of RNA processing factors. For example, phosphorylation at Ser 5 by the CDK7 subunit of the initiation factor TFIIH (4) recruits the capping enzyme (5) and facilitates promoter clearance (6). After CDK7 action, Ser 2 of the CTD is phosphorylated by the CDK9 subunit of the positive elongation factor P-TEFb promoting the release of Pol II from promoter proximal pausing by clearance of NELF (7) and phosphorylation of DSIF (8). Truncation of the CTD reduces splicing efficiency (9) and the ability of the phosphorylated CTD to bind a suite of splicing factors (10,11) may facilitate exon definition in higher eukaryotes by tethering the exons to Pol II during transcription (12,13). Thus the CTD of Pol II represents a crucial interface between the transcription machinery and regulation of mRNA splicing.
A number of studies provide evidence for a reciprocal influence of splicing upon transcription. It was known early that introns within trans-genes of transgenic mice increase transcription from 10- to 100-fold (14). Later the splice donor (SD) was shown to be responsible for this effect (15–17) and requires the binding of U1 small nuclear RNA (snRNA) (18,19), a factor better known for its crucial role in initiation of spliceosome formation. Engineering the SD to a site more distal from the promoter reduces the positive effect on mRNA accumulation (15,16) which may be due to a reduction in 5′ cap proximal recruitment of U1 snRNA (20) or spatial interactions with re-initiating Pol II. Splicing can also activate elongation processivity as the splicing factor SC35 enhances elongation of some mammalian genes by interacting with P-TEFb (21) while spliceosomal U small nuclear ribonucleoproteins (snRNPs) can associate with the elongation factor Tat-SF1 to increase transcription from Adenovirus, HIV and β-globin templates in HeLa nuclear extract (22). Moreover, P-TEFb is dispensable for transcription of intronless genes like U2 snRNA and histone H2b (23) suggesting intron-containing genes require highly processive elongation.
Following demonstration that the promoter proximal SD1 in an HIV-derived, intron-containing transcript enhanced transcription (16), an in vitro interaction between U1 snRNA and Cyclin H was shown to enhance TFIIH-dependent reinitiation (19,24). Analogous to SD1, SD4 is essential for efficient accumulation of mRNA transcripts from a transiently transfected HIV env-derived cDNA (18). Nuclear run on analysis from an integrated env cDNA construct showed that SD4 increases transcription (17). U1snRNA enhancement of transcription is not specific to viral genes as mutation of the SD in the first intron of the bi-intronic β-globin gene triggers a similar transcriptional defect (17). But U1 snRNA binding is not a universal requirement for efficient gene expression as many cDNAs can be expressed without an intron.
We have examined the requirements for U1 snRNA enhancement of transcription using U1 snRNA-mediated rescue of Env expression from an HIV-1 env cDNA construct with mutated SD4. In addition to proper biogenesis through Sm binding, stem loop II was vital for U1-rescue. Local pre-mRNA sequence context for U1 snRNA binding was also crucial as a promoter proximal intron failed to rescue transcription of the splice mutant. The ability of U1 snRNA to facilitate efficient transcription was independent of promoter and exon sequence and could be replaced by the HIV-1 transcription elongation activator Tat. We therefore propose that U1 snRNA engagement overcomes a checkpoint for elongation present in the intron.
MATERIALS AND METHODS
Plasmid construction
The plasmid pHIV-Env has been previously described (25). pCMV-Env and pCMV-Rev were made by inserting the BssHII-BlpI fragment from pHIV-Env and pNLrev1 (25) into pHIS-HIV-B (26) respectively. pCMV(NoIntron)-Env was created by cutting pCDNA3 (Invitrogen) with NcoI+AseI, blunt ending then inserting this 248-nt fragment into pCMV-Env which had been cut with NcoI+NotI and end-filled. pCMV-Tat was derived by inserting the PstI-BamHI fragment from pCMV-Tat2x described in (25) into pCMV-Env. K51A from pTat K51A (27) was introduced into pCMV-Tat using MfeI and BamHI. pCMV-Empty was made by cutting pCMV-Env with BssHII+BlpI, end-filling and re-ligating. pCMV-GFP was created by inserting the PstI-NotI fragment from pEGFP-N1 (BD Biosciences Clonetech) into pCMV-Empty. pHIV-gp140uncGFP was created using a ‘three-way’ stitch PCR protocol to insert the GFP-coding sequence in-frame with a truncated envelope, gp140unc. The 5′ fragment used primers Odp.1124 and Odp.1854 (see Supplementary Table S1 for sequence of these and all primers named in this study) with template pHIV-Env(unc), ‘unc’ mutation described in (28). The middle fragment used Odp.1855 and Odp.1856 with template pEGFP-N1. The 3′ fragment was made using Odp.1857 and Odp.1665 with both wild-type and SA7 mutant pHIV-Env templates. The second round PCR used gel purified first round PCR fragments in equimolar amounts with primers Odp.1124 and Odp.1665. This fragment was then digested with MfeI and BlpI for ligation into either wild-type or mutSD4 containing pHIV-Env. pCMV-gp140uncGFP was made by inserting the MfeI-BlpI fragment from pHIV-gp140uncGFP into pCMV-Env. pUCB-U1 is described in (29). GFP(SD4SA7) was created using ‘three-way’ stitch PCR to insert the WT and mutant SD4-SA7 intron into pCMV-EGFP. The 5′ fragment used pCMV-GFP as template and primers Odp.241 and Odp.1548 or Odp.1745 for the GFP(SD4SA7) and GFP(mutSD4SA7) constructs, respectively. The middle fragment used Odp.1550 and Odp.1551 on pHIV-Env or Odp.1746 and Odp.1747 on pHIV-Env containing mutSD4SA7. The 3′ fragment used Odp.773 and Odp.1549 or Odp.1748 on pCMV-GFP for the GFP(SD4SA7) and GFP(mutSD4SA7) constructs respectively. The stitched fragment was digested with PstI and NotI for insertion into pCMV-GFP. All mutations introduced by PCR mutagenesis are shown in Supplementary Table S2.
Cells and transfection
HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% (v/v) heat inactivated Foetal Bovine Serum (Bovogen), streptomycin (100 U/ml) and penicillin (100 U/ml). Totally 2.5 × 105 cells (6-well plates) or 7.5 × 105 cells (T-25 flasks) were transfected using LipofectamineTM 2000 (Invitrogen) as per the manufacturer’s instructions. Transfections containing 2 µg pHIV-Env, also used the following; 20 ng pCMV-Tat, 100 ng pCMV-Rev and 1.5 µg pUCB-U1 if indicated in figure legends. For pCMV-Env, pCMV-Tat was omitted. Mock transfections contained 2 µg pHIS-Empty. For western blotting, pCMV-EGFP was included to measure transfection efficiency.
Western blotting
Cells harvested 48 h post-transfection using trypsin, washed in PBS and assessed for transfection efficiency by counting the percentage GFP positive cells by flow cytometry. Transfections were deemed successful if percentage GFP positive cells was >60% and did not vary more than 20% between samples. Cell viability was assessed by 7-AAD staining. Cells were lysed in 50 µl of buffer [1× TTS; 10 mM Tris, pH 8.0, 0,5% (v/v) Triton-X 100, 150 mM NaCl, 2 mM PMSF, 2 µg/µl aprotinin]. An amount of 100 µg total protein was resolved using SDS–PAGE and membranes probed with a monoclonal antibodies to HIV-1 gp41 (Chessie 8, NIH) or to β-actin (Sigma-Aldrich).
RNA isolation
Twenty-four hours post-transfection cells were rinsed with PBS, trypsinized, washed and re-suspended in 320 µl cold CF buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 40 U/ml RNasin (Promega), 1 mM DTT). After 10 min incubation on ice, cells were centrifuged at 500× g for 5 min at 4°C and RNA extracted from 300 µl with LS Trizol (Invitrogen) according to the manufacturer’s instructions. The pellet was washed and re-suspended in 320 µl of NER buffer (CF buffer plus 40 mM EDTA) and treated as per the cytoplasmic fraction. This fractionation step strips membrane-bound polysomes from the nuclear endoplasmic reticulum (NER) (30). The nuclear pellet was washed then re-suspended in 200 µl of Trizol (Invitrogen). Total RNA extractions were performed on cell pellets resuspended in Trizol (Invitrogen).
Northern blotting
RNA samples in formamide were subject to Northern blot as described previously (31). Probes were made by PCR incorporation of [α-32P]dCTP (Perkin Elmer) using the primers indicated in figure legends. Probed membranes were exposed to a phosphorimager screen and visualized using the FLA 3000, Fuji. Bands were quantified by Image Gauge Ver.4.0 (Fujifilm).
RT–PCR
cDNA was made from 1 µg of R fraction or Total RNA with random hexamer and AMV reverse transcriptase (Promega), according to manufacturer’s instructions, amplified by Phusion DNA Polymerase (Finnzymes) to 35 cycles using primers indicated in figure legends. Nucleotide sequence of indicated cDNA PCR products was determined by gel extraction, cloning and sequencing.
Real-time RT–qPCR
An amount of 1 µl cDNA was added to Stratagene Brilliant II FAST SYBR QPCR Master mix with the appropriate primers (Supplementary Table S3) and amplified on a Mx3000P QPCR machine according to manufacturer’s instructions. Each PCR was run in duplicate along with no RT enzyme and no template controls. On the same plate three reference gene amplicons; GAPDH (Acc: NM_002046), YWHAZ (Acc: NM_003406) and HPRT1 (Acc: NM_000194) were also amplified. The geometric average of the relative expression of these genes was calculated using the algorithm outlined in (32) and used to normalize expression of 4-kb env mRNA.
FACS
Thirty-six hours post-transfection, cells were trypsinized, washed and resuspended in PBS for analysis of GFP fluorescence by a BD-FACSCalibur cell sorter. Data was analysed using Weasel v2.5 (Walter & Eliza Hall Institute, Melbourne, Australia) and the relative expression measured by multiplying the percentage mock gated cells with the mean fluorescence of those gated cells.
RESULTS
The requirement for an active SD for efficient expression of intron-containing genes has been observed using viral (15–18,29), mammalian (14,17) and yeast expression systems (16). In this study, the HIV-1 env mRNA, previously shown to require the binding of U1 snRNA for efficient mRNA accumulation (18), was used as a model to study the role of U1 snRNA in transcription.
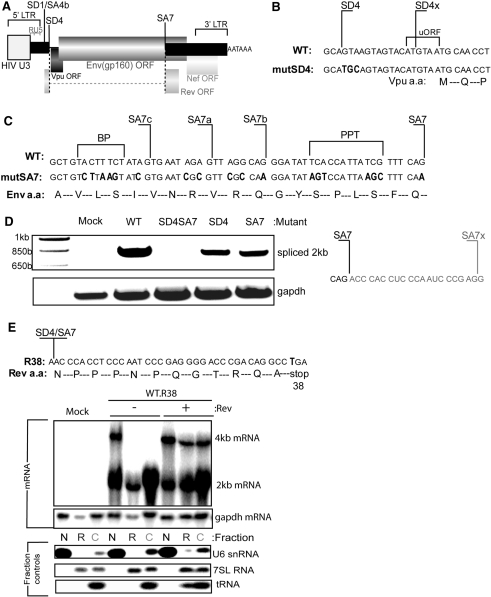
Mutations were introduced into pHIV-Env (Figure 1A) to prevent U1 snRNA binding at the SD4 site (Figure 1B). This construct expresses HIV Env, Rev and minimal amounts of Vpu protein (25). The extent of splicing of the env mRNA was used as a surrogate measure of U1 snRNA binding. Assessment of splicing by RT–PCR showed cryptic splicing induced by a single mutation at SD4 (Figure 1D, lane mutSD4). The cryptic donor site, SD4x, which was previously identified upon mutation of SD4 (33) was confirmed by sequencing of the spliced cDNA product. Inactivating SD4x was complicated by the overlapping minimal uORF and Vpu start codon that may influence Env translation (34). However, we could prevent the use of SD4x through a series of mutations at SA7 and adjacent cryptic acceptor sites, SA7c, a and b (35). The AG dinucleotide at SA7, branch point (BP), and polypyrimidine tract (PPT) which bind U2AF35, SF1 (36) and U2AF65 (37) respectively, were mutated to prevent formation of the early spliceosome complex (38). This effectively ablated splicing as shown by the absence of 2-kb spliced mRNA (Figure 1D). The cryptic site used by the SA7 mutant was also identified by sequencing and was found to be 20 nt downstream from SA7 (Figure 1D).
Figure 1.
Engineering a U1 snRNA dependent expression system using the HIV unspliced env mRNA. (A) Diagram of the pHIV-Env model construct. An HIV U3 region promoter (grey box) drives transcription through R and U5 regions, all three of which make up the long terminal repeat (LTR). The cDNA has the SD1/SA4b intron removed and contains two exons, shown in black rectangles and one intron bounded by SD4 and SA7. The Env (gp160) open reading frame (ORF) expressed from the unspliced mRNA is shown, the Rev ORF expressed from spliced mRNA is also shown, Vpu and Nef ORFs are present but do not express significant protein in this spliced isoform. (B) Mutations made to SD4 shown in red, a second cryptic SD SD4x overlaps the minimal upstream ORF (uORF) and Vpu start. (C) Use of SD4x was prevented by mutations shown in red at the 3′ splice site, SA7. The branch point (BP), polypyrimidine tract (PPT) and cryptic acceptors were mutated in addition to SA7 itself. All mutations introduced were silent with respect to the Env-coding sequence. (D) Splice site mutations ablated splicing. RT–PCR using Odp.2 and Odp.40 was performed on RNA extracted from cells transfected with the indicated mutant. PCR products were extracted from the gel, cloned and sequenced. Sequence of SA7x is shown to the right. (E) The spliced product of the pHIV-Env transcript makes the Rev protein which controls the export of unspliced 4-kb mRNA and hence Env protein expression. The amino acid sequence is shown at the SD4/SA7 exon/exon junction. The Rev ORF was inactivated using a stop codon at Rev amino acid 38. This mutant was assessed by northern blotting of 4-kb (unspliced) and 2-kb (spliced) mRNA present in nuclear (N), nuclear-associated rough endoplasmic reticulum (R) and cytoplasmic (C) fractions. The 4- and 2-kb probe was made using Odp.1409 and Odp.1410. Probes were designed to bind to gapdh (Acc: NM_002046) for loading, U6 snRNA (Acc: X07425), 7SL (Acc:NR_002715) and mitochondrial tRNAlys (Acc:X93334, tRNA 14) to control for the efficiency of N, R and C fractions respectively.
SD4 and SA7 are used to construct the completely spliced 2-kb class of mRNAs that express the Rev protein. Rev binds to the rev responsive element (RRE) within the SD4-SA7 intron and is necessary for export of unspliced 4-kb env mRNA (39). Also, Rev has been shown to influence mRNA stability (40) and splicing (41) so is crucial for efficient Env expression. In order to examine the effects of SD4 upon Env expression whilst maintaining constant Rev protein we inactivated the Rev ORF within the env cDNA constructs by introducing a stop codon at Rev amino acid 38 (R38) (Figure 1E). We tested the phenotype of this mutation by examining the splicing and cellular distribution of mRNA. RNA from transfected cells was fractionated and analysed by Northern blotting and the integrity of the various fractions confirmed by probing for U6 snRNA [Nuclear (N)], 7SL [nuclear-associated endoplasmic reticulum (R)] and mitochondrial tRNA [cytoplasmic (C)] (Figure 1E, lower panels). The small amounts of U6 snRNA present in the cytoplasmic fractions probably derive from the contents of mitotic or nonviable cells. Notably, 4-kb mRNA was found in the R fraction, consistent with the cellular biogenesis of the Env glycoprotein which requires passage through the ER. The R38 mutation caused nuclear restriction of 4-kb env mRNA and this was rescued upon co-transfection of pCMV-Rev (Figure 1E), which showed cytoplasmic accumulation matching that seen upon transfection of pHIV-Env without the R38 mutation (Supplementary Figure S1). This allowed us to compare Env expression from R38 modified wild-type (WT.R38) and splice site modified (mutSD4SA7) env cDNAs with equal amounts of Rev supplied from a separate plasmid.
U1 snRNA enhances transcription independent from splicing
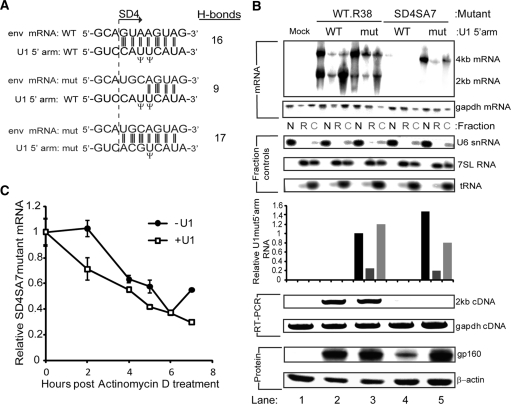
We then aimed to assess the role of U1 snRNA binding in transcription using a splice mutant env mRNA, mutSD4SA7, which had access to the same amount of Rev as the WT.R38 construct but could not bind U1 snRNA and showed reduced levels of nuclear accumulation by northern blot (Supplementary Figure S2). In the nucleus and co-incident with transcription by Pol II, U1 snRNA binds to the SD through its 5′ arm to initiate spliceosome assembly. The 5′ arm contacts the donor by making a duplex of at least 11 nt (42). We modified the 5′ arm of U1 snRNA in the plasmid pUCB-U1 so that it made 17 H-bond contacts with the SD4 mutant site instead of nine predicted for the binding of wild-type U1 snRNA to the SD4 mutant (Figure 2A). Compared to co-transfection of the modified U1 snRNA (U1mut5′arm) which cannot bind to the WT.R38 env mRNA, U1 WT 5′arm co-transfected with WT.R38 showed a relatively increased proportion of 2-kb mRNA which may result from enhanced splicing of a target mRNA with intact splice sites due to over expression of U1 snRNA from pUCB-U1 (Figure 2B, upper panels). Unlike the WT.R38 construct, U1mut5′arm increased the SD4SA7 mutant 4-kb env mRNA accumulation in the nucleus so that levels were equivalent to WT.R38. The expression and cellular distribution of U1 snRNAmut5′arm was validated by real time PCR, showing an expected nuclear and cytoplasmic distribution largely un-altered by the presence of SD4SA7 mutant env mRNA. Also, rescue of mRNA accumulation by the modified U1 snRNA did not cause an increase in splicing as detected by northern blot and the more sensitive RT–PCR (Figure 2B, lower panels). Thus the enhancement effect of U1 snRNA on nuclear mRNA accumulation occurs independently of splicing catalysis.
Figure 2.
U1 snRNA enhances transcription independent of splicing (A) Hydrogen bonding (H-bonding) of U1 snRNA to SD4. The 5′arm of U1 snRNA contacts SD4 through 16 hydrogen-bonds (H-bonds), while mutSD4 makes only nine H-bonds to U1 snRNA. H-bonding was recovered by co-transfection of a modified U1 snRNA mutated (shown in large bold font) to make 17 H-bond contacts with mutSD4. (B) U1mut5′arm rescues nuclear accumulation of mutSD4SA7 sufficient to rescue protein expression in the absence of splicing. Cells were transfected with 2 µg of pHIV-Env with the indicated mutations, 20 ng of pCMV-Tat, 100 ng of pCMV-Rev and 1.5 µg of pUCB-U1 with the indicated modification to the 5′arm. These cells were either fractionated and RNA analysed by northern blotting for 4- and 2-kb mRNA and RT–PCR for 2-kb cDNA or they were lysed and protein analysed by western blotting for Env (gp160). U1mut5′arm was also detected in the various RNA fractions using RT–qPCR with primers specific to the mutated 5′ arm and normalisation to gapdh mRNA as described in the methods. Lanes are as follows, Lane 1—Mock, Lane 2—WT.R38 + WT 5′ arm, Lane 3—WT.R38 + mut 5′ arm. Lane 4—mutSD4SA7 + WT 5′ arm, Lane 5—mutSD4SA7 + WT 5′ arm. (C) U1 snRNA does not enhance mRNA stability. Transcription was inactivated with 5 µg/ml of Actinomycin D and 4-kb mRNA levels from mutSD4SA7 in the presence of U1 snRNA WT 5′arm (−U1) and U1 snRNA mut5′arm (+U1) assessed by RT–QPCR. 4-kb mRNA levels were normalized to the geometric mean of three reference genes as described in the methods. Error bars represent standard deviation of the mean from three transfections performed on different days.
Finally, the functionality of the mRNA produced from the U1-rescued SD4SA7 mutant was assessed by detection of gp160 protein by western blotting. Expression from the SD4SA7 mutant in the absence of U1 binding was significantly reduced but detectable. The absence of detectable substrate mRNA in the cytoplasm for this sample (Figure 2B, upper panel, lane 4) probably results from reduced sensitivity of cell fractionation and northern blotting. Importantly, the increase in nuclear accumulation induced by U1mut5′arm was sufficient to restore wild-type levels of Env protein (Figure 2B, lane 5, lowest panel) demonstrating that U1-rescued unspliced mRNA is efficiently exported and translated and that U1 snRNA binding alone and not the generation of an intron is responsible for diminished env mRNA and protein synthesis.
Thus U1 snRNA binding is essential for efficient nuclear mRNA accumulation. It was unclear however if U1 snRNA was acting to increase transcription and/or mRNA stability. We assessed the stability of U1-bound versus U1-unbound mutSD4SA7 env mRNA by halting transcription using Actinomycin D and measuring the decay rate of env mRNA by RT–qPCR. U1-bound env mRNA levels were found to decline more rapidly upon Actinomycin D treatment compared with U1-unbound mRNA (Figure 2C), demonstrating that mRNA stability was not increased by U1 snRNA binding and therefore not responsible for increased nuclear accumulation, confirming that U1 snRNA enhances transcription.
The Sm domain and stem loop II are important for U1 snRNA enhancement of transcription
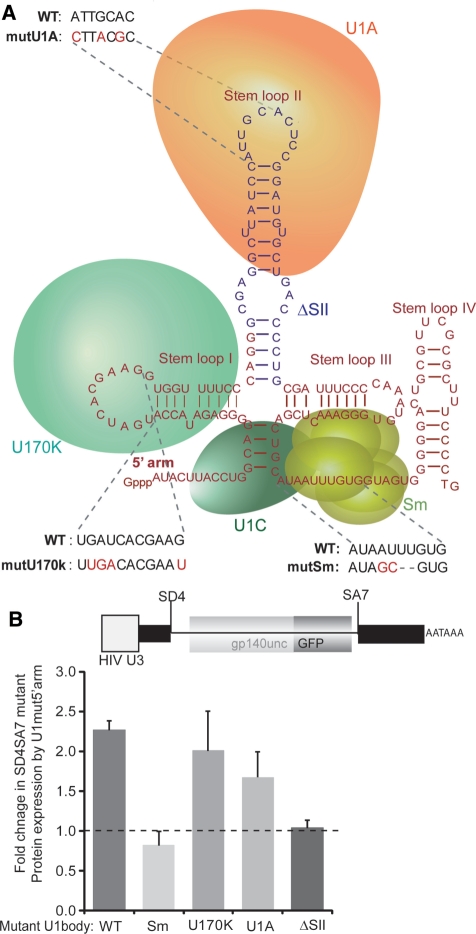
U1 snRNA exists as a highly structured RNA bound to proteins forming a small nuclear ribonucleoprotein (snRNP) (Figure 3A). We mutated the binding sites for U170k, U1A and Sm to assess the relative importance of these proteins in enhancement of transcription by U1 snRNA. We used a fluorescent Env-GFP fusion protein reporter, gp140uncGFP, where the coding sequence of GFP was placed in-frame with an un-cleaved and truncated form of Env. The Env protein was truncated immediately prior to the transmembrane domain to create a soluble Env analogue and facilitate the correct folding and fluorescence of GFP. This truncation of Env does not compromise the rev responsive element (RRE), which lies between the end of gp120 and gp140, or the splicing signals at SA7. The gp160 cleavage motif was mutated to prevent disassociation into gp120 and gp41-GFP (28). Fluorescence-activated cell sorting (FACS) of cells transfected with gp140uncGFP containing the SD4SA7 mutation was used to report the fold-increase in expression induced by U1 snRNA which could bind to mutSD4SA7 env mRNA through mut5′arm. This enabled a quantitative assessment of mutations made to the U1 snRNA body. The wild-type U1 body with mutant 5′ arm increased expression from the SD4SA7 mutant reporter by 2.4-fold (2.4 ± 0.2, n = 4) (Figure 3B), confirming the western blot data (Figure 2B, lowest panel, compare lanes 4 and 5).
Figure 3.
U1 snRNA enhancement of transcription requires the Sm domain and stem loop II (A) U1 snRNA secondary structure and location of U170k, U1A, U1C and Sm proteins in the small ribonucleoprotein (snRNP). Mutations to disrupt binding of U170k, U1A, Sm are shown in red as well as the deletion made to stem loop II in blue. (B) Sm domain and stem loop II are important for U1 snRNA rescue. Wild-type U1 snRNA and the four U1 snRNA body (nt 12–164) mutants with either a wild-type or 5′arm (nt 1–11) mutant were co-transfected with a gp140uncGFP reporter (shown) containing the SD4SA7 mutation in addition to 20 ng of pCMV-Tat and 100 ng of pCMV-Rev. Values represent the mean of the fold difference between the wild-type and mut5′arm from four transfections performed on different days. Error bars represent the standard deviation of the mean.
During the biogenesis of U1 snRNA, Sm proteins assemble on the U1 snRNA in the cytoplasm acting as an adapter for Importin-β mediated transport of the mature U1 snRNA into the nucleus (43). Sm binds to the single-stranded uridine tract in the presence of a conserved adenosine (44). When the assembly of the Sm core was blocked using the mutations shown in Figure 3A, Env expression of the SD4SA7 mutant could not be rescued (Figure 3B), indicating that U1 snRNA acts in the nucleus as a mature U1 snRNP to increase transcription. This failure to rescue expression was not due to toxicity induced by over expression of the mutant U1 snRNA as cell viability by 7-AAD staining was unaffected (Supplementary Figure S3).
Mutations made to the U170k-bound stem loop I were based upon previously described modifications that prevent the binding of in vitro transcribed U1snRNA to U170k in HeLa nuclear extracts (45). The secondary structure of U1 snRNA, as predicted using M-fold RNA-folding software (46) (Supplementary Figure S4), was preserved to ensure binding of SMN for proper U1 snRNA maturation (47). Upon transfection, mutations to the binding site for U170k did not significantly affect rescue of mutSD4SA7 by U1mut5′arm (2.0 ± 0.5, n = 4) (Figure 3B), suggesting that the U170k-binding site does not play an important role in transcriptional enhancement by U1 snRNA.
Point mutations made to the U1A-binding site on stem loop II were also based on in vitro binding data (48). Purines at positions 1, 4 and 6 of the loop sequence are critical for binding (49) however mutations at these positions did not significantly affect rescue of mutSD4SA7 expression in our system (1.7 ± 0.3, n = 4) (Figure 3B). In contrast, the complete deletion of stem loop II (SII), shown previously to disrupt the binding of Cyclin H in vitro (24), resulted in a significant decrease in Env rescue implicating this stem loop in the mechanism of U1 snRNA enhancement of transcription.
Heterologous promoter and upstream intron do not alter the requirement for U1 snRNA binding at SD4
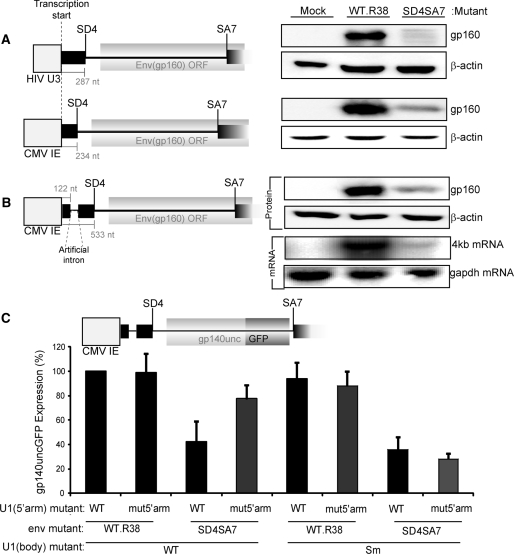
Expression of Env protein from the U1 snRNA binding WT.R38 cDNA was compared to the SD4SA7 mutant, which fails to engage U1 snRNA, using constructs with an alternative promoter and upstream heterologous intron. We first exchanged the HIV U3 promoter with the Cytomegalovirus (CMV) immediate early promoter. In the presence of a non-HIV promoter, Env expression was reduced 3-fold by the splice site mutations demonstrating that the requirement for active splice sites to recruit U1 snRNA is not specific to the HIV promoter (Figure 4A).
Figure 4.
The requirement for U1 snRNA binding is independent of promoter and an upstream artificial intron. (A) The promoter does not alter the requirement for U1 snRNA recruitment. pHIV-Env and pCMV-Env are shown with their promoter, 5′ exon and intron sequence drawn to scale. Cells were transfected with these constructs containing either the WT.R38 or mutSD4SA7 mutations, 20 ng of pCMV-Tat for the pHIV construct and 100 ng of pCMV-Rev, harvested for protein and Western blotted for Env (gp160) and β-actin. (B) An upstream artificial intron cannot rescue protein or mRNA expression from mutSD4SA7. pCMV-Intron-Env is shown drawn to scale relative to pHIV-Env and pCMV-Env. Cells were transfected with either WT.R38 or mutSD4SA7 along with 100 ng of pCMV-Rev. The northern probe was made using Odp.1126 and Odp.92 (C) U1 snRNA rescue of mutSD4SA7 is independent of the promoter and an upstream intron. The GFP reporter from Figure 3B was expressed from pCMV-Intron as shown. Values represent the GFP fluorescence from the reporter shown above, normalized to WT.R38 plus U1 5′arm WT. The Sm U1 snRNA mutant was used as a negative control. Error bars represent the standard deviation of the mean for three transfections performed on different days.
We investigated if expression from the SD4SA7 mutant construct could be rescued by inclusion of a promoter proximal intron consisting of the SD from CMV intron A with a strong U1 snRNA-binding site and the SA from murine IgG heavy chain. Constitutive splicing of the hybrid intron was validated using RT–PCR amplification of the spliced product (Supplementary Figure S5). Surprisingly, Env expression levels were not improved by inclusion of this upstream intron and mRNA accumulation remained compromised (Figure 4B). Thus U1 snRNA binding to the heterologous SD could not substitute for U1 snRNA binding at SD4 demonstrating that the local mRNA context of SD4 is important for U1 snRNA enhancement of transcription. To confirm this, we used the Env GFP reporter, gp140uncGFP, to assess the ability of U1mut5′arm to rescue expression from the SD4SA7 mutant. Protein expression from the SD4SA7 mutant was reduced to 42% ± 17 of wild-type (Figure 4C), indicating that the heterologous hybrid intron could not substitute for U1 snRNA binding to the authentic SD4. Upon co-transfection of the modified U1 snRNA, protein expression was rescued to ∼77% ± 11 of wild-type, while the Sm mutant could not rescue Env expression from the SD4SA7 mutant.
Transfer of the SD4-SA7 intron into a heterologous context maintains U1 snRNA responsiveness
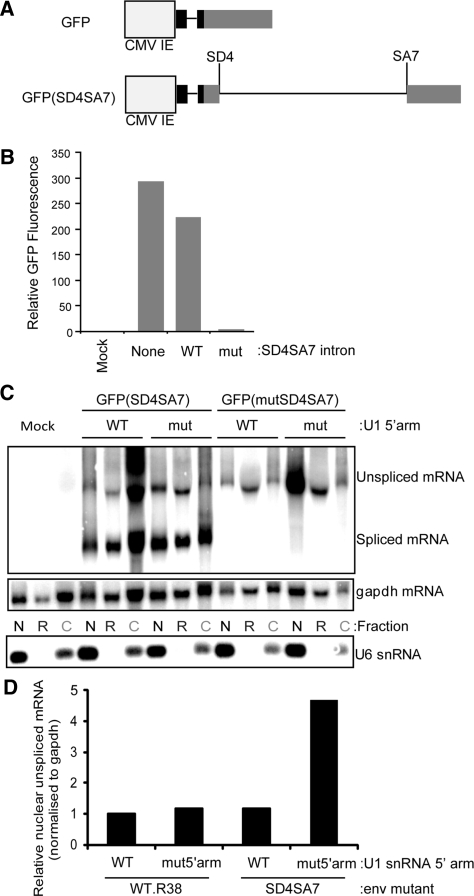
Binding sites for the SR-proteins SF2/ASF and SRp40 exist upstream of SD4 (50) and may confer dependence upon U1 snRNA binding on account of their exon definition role and interaction with the CTD of Pol II (10,12,13). To test if the SR-binding, exonic sequence confers the transcriptional requirement for U1 snRNA binding, we replaced HIV exon sequence with GFP-coding sequence to make GFP(SD4SA7) (Figure 5A). Insertion of the SD4-SA7 intron only reduced GFP fluorescence by 20%, indicating that the intron is spliced accurately and efficiently to produce GFP (Figure 5B, cf. GFP to GFP(SD4SA7). The small decrease in fluorescence may be due to retention of the SD4-SA7 intron from inefficient splicing and the presence of Rev. The incorporation of SD4 and SA7 mutations (analogous to those in Figure 1B and C) reduced GFP fluorescence to near background levels confirming the abolition of splicing [Figure 5B, GFP(mutSD4SA7)].
Figure 5.
U1 snRNA transcriptional enhancement depends on in the intron sequence following the U1-bound SD. (A) Plasmid map of the GFP and GFP(SD4SA7) constructs. (B) The SD4-SA7 intron is efficiently and accurately spliced from GFP(SD4SA7) to produce GFP protein. Fluorescence from GFP, GFP(SD4SA7) and GFP(mutSD4SA7) constructs was measured using FACS. GFP(mutSD4SA7) contains the mutations outlined in Figure 1B and C and these transfections included 100 ng of pCMV-Rev. (C) U1 snRNA increases the nuclear accumulation of unspliced mRNA in the absence of HIV exonic sequence. Cells were transfected with the indicated constructs, including 100 ng of pCMV-Rev, and fractionated into nuclear (N), nuclear-associated rough ER (R) and cytoplasmic (C) fractions. RNA was northern blotted using a probe to GFP made from Odp. 445 and Odp.1250. The purity of fractions was assessed by probing for U6 snRNA. (D) Nuclear unspliced mRNA band intensities were quantified from the blot shown above and normalized to the band intensities of gapdh. Values represent the fold difference in mRNA levels expressed compared to GFPi plus U1WT5′arm.
The ability of U1 snRNA to increase nuclear accumulation of unspliced mRNA in the context of GFP exons was examined by northern blotting (Figure 5C). Since Rev was co-transfected with this construct, unspliced mRNA could be detected in nuclear-associated ER and cytoplasmic fractions indicating that Rev, even in the presence of non-native exonic sequences, can facilitate the export of unspliced mRNA containing the SD4-SA7 intron. The downward size shift of unspliced mRNA in the nuclear-associated ER fraction may result from deadenylation, induced by the nonsense-mediated decay pathway (51) as the intron contains numerous stop codons. Nuclear accumulation of unspliced mRNA was not reduced by the SD4SA7 mutation after being normalized to gapdh mRNA levels (Figure 5D). However, co-transfection of U1mut5′arm with GFP(mutSD4SA7) resulted in an almost 5-fold increase in nuclear mRNA accumulation (Figure 5D), indicating that exon sequence identity is not crucial for U1 snRNA enhancement.
The elongation activator, HIV Tat, can partially rescue the SD4SA7 mutant expression
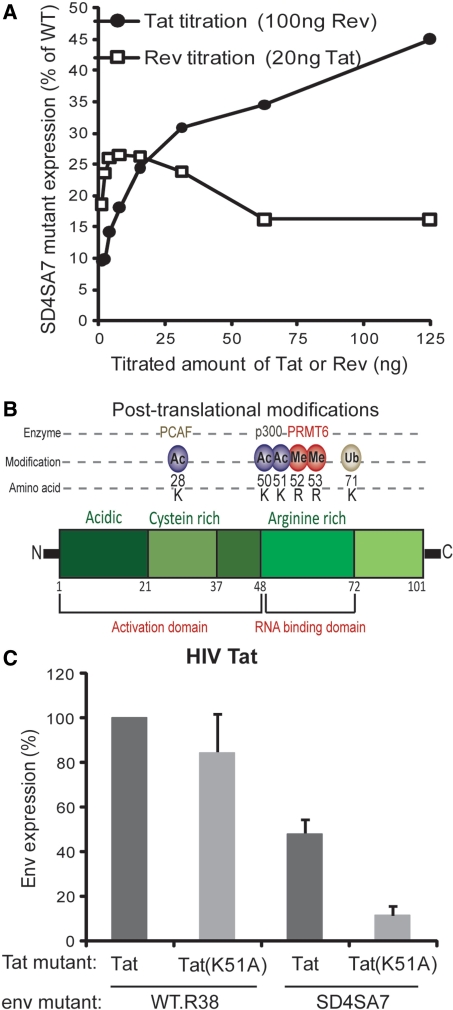
HIV is one of the most well characterized viral expression systems and studies using this virus have lead to key insights into higher eukaryotic molecular biology (52). We used the HIV proteins Tat and Rev to probe into the mechanism of U1 snRNA enhancement of expression. Tat activates elongation from the HIV LTR promoter by interacting with the trans-activation response (TAR) element in the first 50 nt of the HIV transcript. This interaction enhances the binding of Cyclin T1 of the P-TEFb complex to TAR which allows phosphorylation of the CTD of Pol II at Ser 2. In contrast to Tat, Rev acts post-transcription to allow the export of unspliced env mRNA to the cytoplasm for translation. We titrated these two regulatory proteins to assess their role in the expression of Env in the absence of U1 snRNA recruitment by splicing signals. Increasing concentrations of Tat increased expression from the SD4SA7 mutant construct relative to WT.R38 from 9.5% at 1 ng, up to 45% at 125 ng (Figure 6A). In contrast, Rev did not show such a dose response with relative expression compared to the WT.R38 construct remaining below 26.5%. Furthermore, Tat was able to trans-activate the SD4SA7 mutant more than 200-fold compared to only 18-fold for WT.R38 (Supplementary Figure S6). Thus the elongation activator Tat is able to partially replace the activity of U1 snRNA binding, suggesting that U1 snRNA may influence transcriptional elongation.
Figure 6.
Tat acetylation is important for transcription in the absence of U1 snRNA recruitment. (A) Tat and not Rev partially replaced the need to bind U1. gp140uncGFP with indicated mutations reported a dose-response from pCMV-Tat and pCMV-Rev. Values represent the percentage expression level of mutSD4SA7 relative to WT.R38. (B) Post-translational modifications of Tat and locations of acetylation sites. Lysine 51 (K51) in the RNA-binding domain is acetylated by p300. (C) Lysine 51 is important only for expression in the absence of U1 snRNA recruitment by splicing signals. Tat mutant K51A was tested for trans-activation activity by co-transfection with WT.R38 or mutSD4SA7. Protein expression was assessed by FACS analysis of gp140uncGFP. Error bars represent the standard deviation of the mean of five independent transfections.
We investigated this further by using a Tat mutant, K51A, which removes an acetylation site at lysine (K) 51 but still allows efficient trans-activation of a simple LTR-GFP reporter (data not shown). Acetylation at K50 and K51 in the RNA-binding domain of Tat is performed by p300 and permits dissociation of the Tat:TAR:Cyclin T1 complex (53) (Figure 6B). When Tat K51A was used to trans-activate transcription of the WT.R38 construct, there was no significant change in reporter expression (Figure 6C). In contrast, when Tat K51A was co-transfected with the SD4SA7 mutant, expression was reduced by 87% ± 4.3. Thus, in the absence of U1 snRNA binding, acetylation at lysine 51 is critical for transcription through the distal intron sequence.
DISCUSSION
The unusual dependence upon U1 snRNA binding for efficient expression from HIV-derived cDNA constructs has been a long-standing issue and its examination here provides several new insights. First, the trans-rescue by pUCB-U1 allowed us to determine elements within U1 snRNA which permitted an increase in transcription. Fortunately, the secondary structure and binding sites for U170k, U1A and Sm have been extensively mapped in vitro (44,45,48,49). A deletion of the entire stem loop II ablated rescue of mutSD4SA7 by the U1mut5′arm. The crystal structure of U1 snRNP shows stem loop II protruding from the globular Sm core (54). Additionally, stem loop II is non-essential for E complex formation and in vitro splicing (55), but essential for interaction with Cyclin H of the TFIIH complex which increased the kinase activity of CDK7 against the CTD (24). As mutations to the binding site for U1A on loop II did not significantly inhibit U1-rescue, stem loop II (when unbound to U1A) may be free to interact with Cyclin H, Cyclin T1 [perhaps through the TAR recognition motif (56)] or indeed other components of the transcription machinery. A precedent for regulation of CTD kinase activity by non-coding RNA already exists in 7SK snRNP sequestration of the inactive P-TEFb (57).
Gene expression is predominantly regulated by transcription factor recruitment to sequence-specific DNA elements in the promoter which stimulate initiation (58). The promoter can also affect the pattern of alternative splicing (59), thus presenting a likely element connecting U1 snRNA and transcription. However, swapping the HIV promoter with a CMV promoter did not alter the influence of U1 snRNA on transcription so determinants of U1-dependence must exist within the transcript. The inclusion of a heterologous upstream intron however also did not change the requirement for U1 snRNA recruitment to SD4. Thus U1 engagement alone is insufficient to mediate enhancement of transcription and the spatial or temporal proximity to the SD4-SA7 intron may be crucial. Evidence for intron specific regulation of transcription by splicing was shown by the SC35 depletion-induced Pol II pausing in an intron of mouse Ptbp1 but not the closely related Ptb2 gene (21). Also, a heterologous intron capable of directing efficient splicing does not increase P-TEFb-dependent elongation of the intron-less U2 snRNA (23). These data indicate that gene specific elements mediate the connection between transcription and splicing and points to the existence of an intrinsic block to transcription elongation in the SD4-SA7 intron which is counteracted by U1 snRNA binding to SD4. This is supported by the maintenance of U1 snRNA responsiveness upon moving the SD4-SA7 intron to a heterologous context.
The significance of post-initiation regulation in eukaryotic gene expression was highlighted by global ChIP analysis where 75% of human genes contained initiated Pol II with no detectable transcripts (60). Furthermore, P-TEFb-dependent pausing was shown also to occur at late points along the β-actin gene (61). Transcriptional pausing can be induced by nucleosomes (62,63), RNA structure (64–66), or the negative elongation factors such as NELF and DSIF (67). Recent SHAPE analysis of HIV RNA (68), presents many possible structured or unstructured regions which could regulate elongation processivity (65). One described function for transcriptional pausing is to ensure proper capping of the transcript (69). A similar mechanism may exist at the 5′ border of some introns to afford time for proper assembly of U1 snRNP complexes.
The transcriptional elongation activator Tat, can replicate the activity of U1 snRNA binding. An association of U1 snRNA with the elongation factor Tat-SF1 may assist in recruitment of P-TEFb (22) akin to Tat recruitment of P-TEFb through TAR. Recruitment local to the SD4 site may provide the boost to elongation required to transcribe the intron. Certainly, mutation of SD1 in a CMV-driven HIV proviral clone inducing Tat-dependence (70) lends further evidence for U1 snRNA enhancement of elongation. Mutation of the acetylation site K51 resulted in a significant reduction in expression specific to the mutSD4SA7 mutant. Lysines 50 and 51 lie within the TAR-binding domain and when acetylated, disrupt formation of the Tat:TAR:Cyclin T1 ternary complex leaving Tat free to associate with the elongating Pol II (71) and/or recruit the histone acetyltransferase (HAT) PCAF (72). Transcription from DNA plasmids is enhanced by histone deacetylase inhibitors (73) and intron sequence 3′ to SD4 is occupied by nucleosomes in both an integrated HIV-1 genome and a nuclear episome suggesting primary DNA sequence determines nucleosome placement in this region (74). It is possible that both acetylated Tat and U1 snRNA increase transcription by facilitating passage of Pol II through a DNA template associated with nucleosomes. The newly identified U1-TAF15 snRNP which associates with chromatin in an RNA-dependent manner provides preliminary evidence that U1 snRNA may influence nucleosomal packaging of DNA (75).
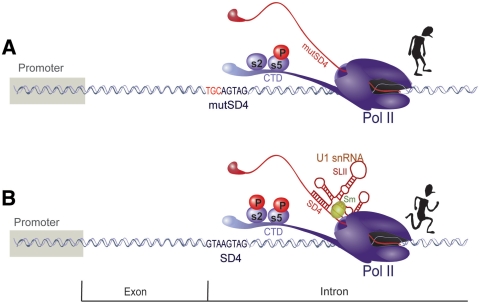
The molecular determinants allowing U1 snRNA enhancement of transcription are unclear, but the data presented could result from a transcriptional pause somewhere within the intron, which is overcome by the recruitment of an Sm-type U1 snRNA with intact stem loop II to the intron-bordering SD as modelled in Figure 7. A non-mutually exclusive possibility is that formation of the RNA:RNA helix between U1mut5′arm and mutSD4 neutralizes a transcriptional defect induced by mutSD4. Given the central role of phosphorylation of the CTD of Pol II in transcriptional control and the observed partial rescue of the splice mutant by HIV Tat, our data also raises the possibility that U1 snRNA–binding induces an increase in the phosphorylation at Ser 2, suggesting that U1 snRNA may provide positive feedback to the elongating Pol II during transcription through this intron-containing mRNA.
Figure 7.
Proposed model for U1 snRNA enhancement of transcription. (A) Transcription of pre-mRNA containing the SD4-SA7 intron is inhibited in the absence of U1 snRNA recruitment by SD4. (B) When U1 snRNA binds at the SD, transcription through the SD4-SA7 intron is efficient, possibly due to an increase in the processivity of elongation via P-TEFb phosphorylation of the Pol II CTD at Serine 2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Health and Medical Research Council. Funding for open access charge: Australian Health and Medical Research Council (Grants 400302 and 510488).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Lou Hammarskjold for donating the pUCB-U1 construct. They also thank Peter Revill, Shahan Campbell, Jane Howard, Adam Johnson, Sean Chung, Helen Christensen, Rob Ramsay, David Harrich and Jens Bohne for reagents and helpful discussions. Real-time RT–qPCR advice from Adriana Gaeguta and Liyen Loh was greatly appreciated.
REFERENCES
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 4.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 5.Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 8.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 9.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 10.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell. Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Zeng C, Berget SM. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 2000;20:8290–8301. doi: 10.1128/mcb.20.21.8290-8301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc. Natl Acad. Sci. USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett NL, Li X, Carmichael GG. The sequence and context of the 5′ splice site govern the nuclear stability of polyoma virus late RNAs. Nucleic Acids Res. 1995;23:4812–4817. doi: 10.1093/nar/23.23.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furger A, O'S;ullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 18.Kammler S, Leurs C, Freund M, Krummheuer J, Seidel K, Tange TO, Lund MK, Kjems J, Scheid A, Schaal H. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001;7:421–434. doi: 10.1017/s1355838201001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwek KY, Murphy S, Furger A, Thomas B, O'G;orman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 20.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 21.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 23.Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'G;orman W, Thomas B, Kwek KY, Furger A, Akoulitchev A. Analysis of U1 small nuclear RNA interaction with cyclin H. J. Biol. Chem. 2005;280:36920–36925. doi: 10.1074/jbc.M505791200. [DOI] [PubMed] [Google Scholar]
- 25.Anderson JL, Johnson AT, Howard JL, Purcell DF. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J. Virol. 2007;81:4664–4676. doi: 10.1128/JVI.01028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dale CJ, De Rose R, Stratov I, Chea S, Montefiori DC, Thomson S, Ramshaw IA, Coupar BE, Boyle DB, Law M, et al. Efficacy of DNA and fowlpox virus priming/boosting vaccines for simian/human immunodeficiency virus. J. Virol. 2004;78:13819–13828. doi: 10.1128/JVI.78.24.13819-13828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Center RJ, Wheatley AK, Campbell SM, Gaeguta AJ, Peut V, Alcantara S, Siebentritt C, Kent SJ, Purcell DF. Induction of HIV-1 subtype B and AE-specific neutralizing antibodies in mice and macaques with DNA prime and recombinant gp140 protein boost regimens. Vaccine. 2009;27:6605–6612. doi: 10.1016/j.vaccine.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Lu XB, Heimer J, Rekosh D, Hammarskjold ML. U1 small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remains unspliced. Proc. Natl Acad. Sci. USA. 1990;87:7598–7602. doi: 10.1073/pnas.87.19.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pryme IF. The nuclear-associated endoplasmic reticulum. Int. J. Biochem. 1989;21:119–125. doi: 10.1016/0020-711x(89)90099-2. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. 3rd edn. Cold Spring Harbour Laboratory; 2001. [Google Scholar]
- 32.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasioulas G, Zolotukhin AS, Tabernero C, Solomin L, Cunningham CP, Pavlakis GN, Felber BK. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krummheuer J, Johnson AT, Hauber I, Kammler S, Anderson JL, Hauber J, Purcell DF, Schaal H. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology. 2007;363:261–271. doi: 10.1016/j.virol.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 35.Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guth S, Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 37.Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 38.Forch P, Merendino L, Martinez C, Valcarcel J. U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor of 65 kDa, U2AF65, can promote U1 snRNP recruitment to 5′ splice sites. Biochem. J. 2003;372:235–240. doi: 10.1042/BJ20021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 40.Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl Acad. Sci. USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kjems J, Sharp PA. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6.U5 small nuclear ribonucleoprotein in spliceosome assembly. J. Virol. 1993;67:4769–4776. doi: 10.1128/jvi.67.8.4769-4776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, Preuss T, et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–6975. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacios I, Hetzer M, Adam SA, Mattaj IW. Nuclear import of U snRNPs requires importin beta. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raker VA, Hartmuth K, Kastner B, Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surowy CS, van Santen VL, Scheib-Wixted SM, Spritz RA. Direct, sequence-specific binding of the human U1-70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell. Biol. 1989;9:4179–4186. doi: 10.1128/mcb.9.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall KB. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- 49.Benitex Y, Baranger AM. Recognition of essential purines by the U1A protein. BMC Biochem. 2007;8:22. doi: 10.1186/1471-2091-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caputi M, Freund M, Kammler S, Asang C, Schaal H. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 2004;78:6517–6526. doi: 10.1128/JVI.78.12.6517-6526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cullen BR. Viral RNAs: lessons from the enemy. Cell. 2009;136:592–597. doi: 10.1016/j.cell.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 1999;9:1489–1492. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 54.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Will CL, Rumpler S, Klein Gunnewiek J, van Venrooij WJ, Luhrmann R. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24:4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anand K, Schulte A, Fujinaga K, Scheffzek K, Geyer M. Cyclin box structure of the P-TEFb subunit cyclin T1 derived from a fusion complex with EIAV tat. J. Mol. Biol. 2007;370:826–836. doi: 10.1016/j.jmb.2007.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 58.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 59.Kadener S, Fededa JP, Rosbash M, Kornblihtt AR. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl Acad. Sci. USA. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egloff S, Al-Rawaf H, O'R;eilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol. Cell. Biol. 2009;29:4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dallinger G, Oberkofler H, Seelos C, Patsch W. Transcriptional elongation of the rat apolipoprotein A-I gene: identification and mapping of two arrest sites and their signals. J. Lipid Res. 1999;40:1229–1239. [PubMed] [Google Scholar]
- 65.Hugo H, Cures A, Suraweera N, Drabsch Y, Purcell D, Mantamadiotis T, Phillips W, Dobrovic A, Zupi G, Gonda TJ, et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer. 2006;45:1143–1154. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 66.Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 67.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess J.W., Jr, Swanstrom R, Burch CL, Weeks, K M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bohne J, Krausslich HG. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 2004;563:113–118. doi: 10.1016/S0014-5793(04)00277-7. [DOI] [PubMed] [Google Scholar]
- 71.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol. Cell. 2003;12:167–176. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 72.Ott M, Dorr A, Hetzer-Egger C, Kaehlcke K, Schnolzer M, Henklein P, Cole P, Zhou MM, Verdin E. Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation. Novartis Foundation Symposium. 2004;259:182–193; discussion 193–186, 223–185. [PubMed] [Google Scholar]
- 73.Nan X, Hyndman L, Agbi N, Porteous DJ, Boyd AC. Potent stimulation of gene expression by histone deacetylase inhibitors on transiently transfected DNA. Biochem. Biophys. Res. Commun. 2004;324:348–354. doi: 10.1016/j.bbrc.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 74.Stanfield-Oakley SA, Griffith JD. Nucleosomal arrangement of HIV-1 DNA: maps generated from an integrated genome and an EBV-based episomal model. J. Mol. Biol. 1996;256:503–516. doi: 10.1006/jmbi.1996.0104. [DOI] [PubMed] [Google Scholar]
- 75.Jobert L, Pinzon N, Van Herreweghe E, Jady BE, Guialis A, Kiss T, Tora L. Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep. 2009;10:494–500. doi: 10.1038/embor.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.