Abstract
Linkage studies, candidate gene and whole-genome association studies have resulted in a tremendous amount of putative risk genes for Alzheimer's disease (AD). Yet, besides the three causal genes—amyloid precursor protein and presenilin 1 and 2 genes—and one risk gene apolipoprotein E (APOE), no single functional risk variant was identified. Discussing the possible involvement of rare alleles and other types of genetic variants, this review summarizes the current knowledge on the genetic spectrum of AD and integrates different approaches and recent discoveries by genome-wide association studies.
INTRODUCTION
It was estimated that, in 2010, 35.6 million people worldwide will be living with a diagnosis of dementia. This number of diagnosed dementia patients is projected to nearly double every 20 years, leading to 65.7 million patients in 2030 (1). Also, it is known that the actual number of dementia patients is much higher since several more people live with dementia but never receive a clinical diagnosis. These numbers indicate that dementia is rapidly becoming a major threat to healthcare in our societies.
Alzheimer's disease (AD) is by far the most common form of dementia with prevalence estimates ranging from 4.4% in persons aged 65 years to 22% at ages 90 and older (2). Increased age and a positive family history of dementia are the two major risk factors of AD. AD is clinically characterized by insidious onset and progressive impairment of memory and other cognitive functions (3), ultimately resulting in complete dependency and death of the patient. As clinical AD symptoms overlap substantially with other disorders of the central nervous system {such as frontotemporal dementia [FTLD (4)]}, a definite diagnosis of AD can only be obtained after pathological brain examination. The key features of AD brains are neuronal and synapse loss, extracellular plaques composed of amyloid-β (Aβ) peptides and intraneuronal neurofibrillary tangles consisting of hyperphosphorylated tau protein (5), although other lesions such as TDP-43 immunoreactivity and ischaemia are common observations.
In general, two subgroups are recognized upon the age at which the first clinical symptoms become apparent; early-onset AD (onset age <65years) and late-onset AD (onset age >65years). Although most patients develop AD at later age, it is mainly the research performed on the rare autosomal-dominant early-onset form of AD that provided valuable insights into disease pathogenesis. Fully penetrant (causal) mutations leading to early-onset familial AD were identified within three genes; the amyloid precursor protein gene (APP) and the two presenilin genes (PSEN1 and PSEN2). While the heritability for the more common late-onset form of AD is predicted to be as high as 80% based on twin studies (6), over the last decades only the apolipoprotein E gene (APOE) has been irrefutably recognized as a major risk factor for late-onset AD (7). Nonetheless, APOE ε4 does not account for all genetic variation in AD (8,9). The complex late-onset form is most likely caused by multiple genetic and environmental susceptibility factors. High-throughput genomic association studies on extensive populations have opened up new avenues in detecting susceptibility factors for late-onset AD. Recently, three novel risk genes have been identified [CLU, CR1 and PICALM1 (10,11)].
MONOGENIC FORMS OF AD: CONSTITUTION OF Aβ CASCADE
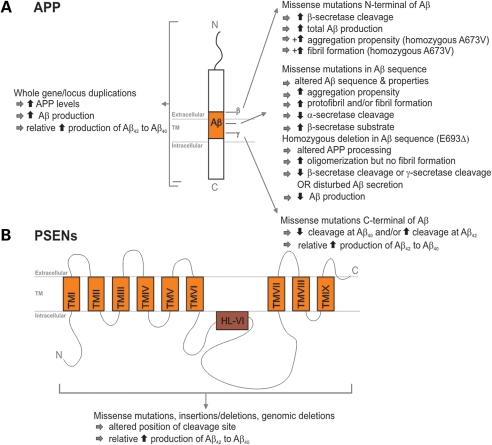
Since the identification of the first APP missense mutation in hereditary cerebral hemorrhages with amyloidosis (HCHWA-D) (12), 23 APP missense mutations have been identified in 77 AD families (for up-to-date information, see AD Mutation Database, http://www.molgen.vib-ua.be/ADMutations/). APP mutations account for <0.1% of AD patients (13). Following the amyloidogenic pathway in neurons, APP is proteolytically cleaved by β-secretase and subsequently by γ-secretase generating full-length Aβ40 or Aβ42 (14,15). Interestingly, all missense mutations influence APP processing since they are positioned in or near the Aβ coding exons (APP exons 16 and 17) (AD Mutation Database) (Fig. 1). In addition to dominant APP mutations, two recessive mutations causing disease only in the homozygous state were identified: a trinucleotide deletion E693Δ segregating in one Japanese family proportionally decreased Aβ40 and Aβ42 with no change in their ratio (16), and A673V in one other family (Fig. 1) (17). Although very rare, it does suggest that other disease-causing mutational mechanisms can occur in this well-studied gene, which might explain at least some seemingly sporadic patients with early-onset AD. Additionally, the mutation spectrum extended to APP locus duplications underscoring the importance of APP gene dosage in AD. Duplicated APP regions containing several genes (18–20) or APP only (21) were clinically linked to early-onset AD often with extensive cerebral amyloid angiopathy (22). Depending on the ethnic population under study, APP duplications accounted for <2–18% of autosomal-dominant early-onset AD families (19,21,23,24).
Figure 1.
Effect of different causal APP and PSEN1 mutations on APP processing and Aβ generation. (A) Schematic presentation of the APP protein structure. Right, the effect of APP mutations on APP processing is given according to their location relative to the Aβ peptide. The additional effects of the N-terminal recessive mutations A673V and E693Δ are indicated by the ‘+’ symbol. Left, the effect of whole APP gene or locus duplications is depicted. (B) Schematic presentation of the PSEN protein structure. Boxes represent the transmembrane regions that are separated by hydrophilic loops. The effect of different mutations scattered throughout the protein is summarized.
At present, 178 different AD-related mutations in 393 families have been identified in PSEN1, while only 14 mutations in 23 families were detected in PSEN2 (http://www.molgen.vib-ua.be/ADMutations/). The majority of PSEN mutations are single-nucleotide substitutions, but small deletions and insertions have been described as well (AD Mutation Database). Mutations are scattered over the protein with some clustering within the transmembrane domains and the hydrophilic loops surrounding these domains (25). PSENs are functionally involved in the γ-secretase-mediated proteolytic cleavage of APP (26). Mutations in PSENs impair this cleavage, resulting in an increased Aβ42/Aβ40 ratio, by either an increase in Aβ42 as shown in plasma and fibroblast media of PSEN mutation carriers (27,28) or by a decrease in Aβ40, suggesting a loss-of-function mechanism rather than a gain-of-function [(29,30) Fig. 1].
Summarized, all three causal AD genes lend support to a common pathogenic AD pathway, stating a pivotal role for Aβ. According to this amyloid hypothesis, neurodegenerative processes are the consequence of an imbalance between Aβ production and Aβ clearance (31–33), suggesting that other genes involved in these pathways might be risk factors as well.
COMPLEX FORMS OF AD: CONTINUOUS SEARCH FOR NEW RISK GENES
The genetic complexity of late-onset AD is underscored by the detection of only one consistent susceptibility factor; APOE ε4 (34). Yet, genetic linkage and association studies over the last 20 years led to a plethora of putative risk genes (for up-to-date information; see AlzGene; http://www.alzgene.org/) (35). Approaches differed from hypothesis-driven (gene or pathway-based) to hypothesis-free (genome-scale) genetic studies. In a hypothesis-free setup, linkage scans on late-onset AD families and affected sib-pairs (ASP) identified candidate regions on several chromosomes. Particularly chromosomes 6, 9, 10, 12 and 21 were repeatedly found (36) and predicted to harbor susceptibility genes for late-onset AD. To date, no gene has been reported to explain these linkage peaks with high confidence.
The design of hypothesis-driven association studies has gradually shifted from studies in which only a few SNPs per gene were investigated to studies employing a more extensive linkage disequilibrium (LD)-based approach covering the complete genetic variation in a gene or a gene region, including, for example, regulatory regions. Besides the amyloid hypothesis, numerous candidate genes fitting one or another AD-related hypothesis on neurodegenerative pathways were analyzed, such as APP cleavage and trafficking, cholesterol metabolism, calcium dysregulation and so on (Fig. 2). As such, an endless record of candidate-gene-based studies was publicized. Nevertheless, none of the associated candidate genes attained an effect size similar to APOE ε4 and along with positive associations, negative replications for the same gene were described (their number will probably be higher since publication in the field is biased toward positive finding; http://www.alzgene.org/). Reasons for lack of reproducibility may include: insufficient study power to detect variants with minor contributions, biological, genetic and allelic heterogeneity, differences in study design and the presence of population substructure.
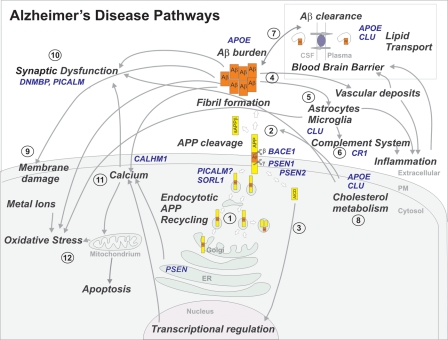
Figure 2.
Overview of several disease pathways involved in AD pathogenesis. Causal AD genes and AD risk factors are marked in blue. APP is synthesized by the endoplasmatic reticulum (ER) and the Golgi apparatus (1). Following the amyloidogenic pathway in neurons, APP is cleaved by β-secretase (BACE1) and γ-secretase (PSEN) to generate Aβ peptides and the amyloid intracellular domain [AICD] (2), which influences the transcription of several genes (3). In the APP retromer recycling pathway (1), APP is redirected to endosomes by SORL1. PICALM has a presumed role in APP endocytotic recycling (1). Aβ monomers aggregate into Aβ fibrils, causing amyloid plaques in brain parenchyma and vasculature (4). Aβ activates microglia and astrocytes, inducing the complement system, local inflammatory responses and oxidative stress (5). CR1 is the receptor of the complement C3b protein and participates in the clearance of Aβ from circulation (6). Besides causing increased Aβ endocytosis into glial cells, CLU is involved in Aβ clearance at the blood–brain barrier (7). APOE enhances amyloid plaque formation by conformational changes of Aβ. Clusterin (APOJ) and APOE are the main escorting proteins of Aβ in brain (7). Both are also important in cholesterol metabolism at the neuronal membrane (8) and high intracellular cholesterol may enhance APP amyloidogenic processing (2), which in turn can lead to membrane damage (9). Moreover, impaired cholesterol metabolism may influence synaptic dysfunction (10). Both PICALM and DNMBP are related at the synapse (10). Interaction of Aβ oligomers at the membrane is further connected to the calcium hypothesis in AD (11). Polymorphisms in the Ca2+ channel CALHM1 impair Ca2+ permeability at the plasma membrane (11). In addition, PSENs function as ER Ca2+-leak channels and several early-onset mutations impair Ca2+-leak-channel function, resulting in an excessive Ca2+ accumulation in the cytosol. An excessive Ca2+ is taken up by mitochondria, further leading to oxidative stress and apoptosis (12).
The comprehensive approach of genome-wide association (GWA) studies seemed very promising for complex disorders, as it permits the simultaneous assessment of thousands of genetic variants without prior assumption on biological pathways. Until now, 10 GWA studies have been published on AD, with some differences in terms of design (patient-control or family based), SNP selection method (LD-based or based upon biological function), ethnicity of population, number of AD patients and controls, number of SNPs and SNP genotyping platform. Apart from the fact that all GWA studies except one [which was conducted in two extended pedigrees (37)] substantiated APOE ε4 as the most significant finding, other genes were proposed as risk factors for late-onset AD [for an overview (38)]. Although none met standard criteria for genome-wide significance, several loci were replicated in independent study populations (39,40), though it should be noted that few replication studies have been published so far.
The two most recent GWA studies differ largely from all previous ones (10,11). First and foremost, they independently provided strong evidence for an association with the clusterin gene (CLU aka apoliprotein J gene, APOJ), making CLU the first consistent risk gene in AD history since the identification of APOE ε4. Secondly, major sample sizes were acquired by international collaboration (>13 000 individuals in the study of Lambert et al. and >16 000 individuals in the study of Harold et al.), surpassing the power hurdle to detect variants with a minor effect. Two other risk genes were detected, i.e. the receptor gene for the complement C3b protein, CR1, and PICALM, encoding the phosphatidylinositol-binding clathrin assembly protein. However, together the three risk genes explained only part of the genetic variance which at their discovery was even likely an overestimate of the true effect (41). Ongoing meta-analyses using raw data of GWA studies will undoubtedly reveal novel genes, given their increased power to detect alleles with a minor effect.
The three novel risk genes support the existing AD hypotheses. Several properties of CLU, an abundantly expressed apolipoprotein in brain, are directly connected to Aβ. Present in amyloid deposits (42), CLU has an increased expression in several AD brain regions (43). It acts as an Aβ chaperone blocking the aggregation of Aβ42 peptides (44,45). However, depending on the balance between Aβ and CLU, CLU can either enhance or prevent amyloid fibril formation and cytotoxicity (46), though it remains unknown whether the same holds true in AD patients. CLU mediates Aβ clearance at the blood–brain barrier (47) and by increased endocytosis into glial cells (48–50). CR1 is linked to AD through fibrillar Aβ-induced activation of the C3 complement cascade (51). Circulating Aβ42 is cleared by C3b-mediated adherence to CR1 at the erythrocyte surface, a process which is decreased in AD patients when compared with control individuals (52). Complement inhibition of C3 in transgenic mice leads to an increased Aβ deposition and neurodegeneration (53), suggesting a protective role of the complement system in AD mice. The precise role of PICALM in AD pathophysiology is unclear, but it could include a role in APP processing through the endocytotic pathway, synaptic fusion and memory formation during trafficking of vesicle-associated membrane protein 2 [VAMP2 (54,55), Fig. 2]. Previous associations were detected within the dynamin-binding protein gene (DNMBP) (55,56), underscoring that genetic variability in genes involved in synaptic functioning might contribute to AD risk. The primary genetic variants within the three novel genes CLU, CR1 and PICALM underlying the associations with risk for AD remain to be elucidated.
LESSONS FROM GENETIC EVIDENCE
The fact that late-onset AD has a complex genetic heterogeneity is reflected by the dissimilar results obtained in the hypothesis-free genome scans and GWA studies. This casts some doubt on the appropriate detection methods for AD susceptibility factors. Late-onset AD might be caused by multiple common or rare alleles, as well as by a wide spectrum of alleles of different frequencies and phenotypic effect sizes. In the common disease-common variant hypothesis, complex diseases are caused by a limited number of common variants with small predisposing effects (57–59). GWA studies restricting the use of tagSNPs to common SNPs (minor allele frequency, MAF ≥ 5%) have the potential to detect common variants since LD mapping depends on allele frequency of disease and marker locus (60–62). Even common variants with small effects (odds ratios in the range of 1.1–1.2) (63) will be detected when using very large sample sizes acquired by combining samples from many different settings.
On the contrary, rare variants will likely go undetected by GWA studies, no matter how large the sample size and the number of common SNPs being genotyped (64). Yet, rare variants might be implicated in complex pathways as previous studies suggested their contribution to common disease (65) and their importance in several disorders (66–68). Evidence suggested that rare variants in regulatory regions of causal genes contribute to AD susceptibility. For example, mutations detected within the APP 5′ regulatory region in early- and late-onset AD patients might increase APP transcriptional activity (69,70), although this was not corroborated by a subsequent study (71). Whether 3′-UTR variants exert a functional effect on APP translation by miRNAs still remains to be assessed, but a recent re-sequencing effort detected rare mutations at highly conserved predicted miRNA target sites in late-onset AD patients (72). In addition, regulatory PSEN1 promoter variants were associated with AD in multiple early-onset populations (73–75) and for some a decreasing effect on transcriptional activity was shown (75,76). A deletion polymorphism inside the PSEN2 promoter region was shown to increase PSEN2 transcription by deprivation of transcription factor repression in a Russian early-onset AD population (77), although this could not be demonstrated in early- and late-onset AD populations of other ethnicities (78–80).
Besides the known AD genes, common or rare variants in genes associated with other neurodegenerative dementias might also confer risk to AD. For instance, missense mutations in progranulin (PGRN), in which null mutations result in FTLD, are possible susceptibility factors for AD by influencing PGRN levels (81). Association findings with interesting susceptibility genes warrant further investigation of rare variants, since they might be functionally involved. Sequencing of the β-site APP-cleaving enzyme 1 (BACE1) 3′-UTR region identified rare patient-specific variants at predicted miRNA-binding sites (72). Besides the involvement of multiple genes in AD, several variants within one gene might confer risk to AD. For example, allelic heterogeneity was supported for the sortilin-related receptor (SORL1), where the association with AD was found in two distinct gene regions (82,83).
Another type of genetic variation that has been underrepresented in genetic studies of AD but gained attention over the last years given its contribution to phenotypic diversity and complex diseases (84) is copy-number variations (CNVs). CNVs are implicated in a number of neurodegenerative disorders [for review, see (85)], including AD where APP duplications result in early-onset autosomal-dominant AD (18–21). In the future, genome-wide microarray data might shed light on the contribution of CNVs to complex AD.
SUMMARY AND FUTURE PROSPECTS: WHERE TO GO?
A marked evolution in study design has been seen in the field of AD genetics. Whereas classical, family-based linkage studies identified three causal AD genes and the APOE susceptibility factor, gene- and genome-based association studies have produced an intractable amount of association signals for late-onset AD, though the latter studies have been dominated by inconsistent findings (AlzGene database). Identification of CLU, CR1 and PICALM as novel risk genes for late-onset AD was a major breakthrough and boost for the genetic field of late-onset AD. Besides these three novel risk genes, interesting genetic signals attaining sub-threshold significance should be examined in more detail. Conducting meta-analyses on GWA data will likely reveal new findings given the increased power to detect risk genes with smaller effect sizes. Conversely, this might bring about problems inherent to combining samples of different sources to obtain larger and powerful sample sizes, i.e. population-specific signals are diluted using mixed samples, especially taken the extensive genetic and allelic heterogeneity between populations. Furthermore, association outcomes might be different depending on the LD pattern between populations. Alternatively, studying isolated populations with a few founders might reduce the genetic heterogeneity and augment the chance to detect these otherwise ‘undetected’ signals. Another approach could be the use of intermediate phenotypes that resemble more closely the underlying disease pathogenesis and more directly interact with the genetic risk variance.
Since the failure to detect rare variants is another limitation of GWA studies, other detection methods are warranted to interrogate the complete allelic spectrum of AD. With the reducing costs of novel technologies, extensive re-sequencing efforts capturing all rare variants will eventually become possible in large AD populations. Whole exome-sequencing of probands of early-onset AD families has already started to take off, identifying a number of potential rare AD mutations [presented at ASHG2009, (86)]. Furthermore, whole-genome sequencing of AD patients will detect variations in non-coding regulatory sequences of RNA transcripts (such as miRNAs), and at an even higher level, variations in sequences mediating the expression of these regulatory sequences. Using the data of the 1000 Genomes Project (http://www.1000genomes.org), future GWA studies might include rare variants to ensure that a high proportion of rare SNPs are captured (61). The role of structural variants, such as CNVs in relation to AD, should be interrogated across the complete genome. This is facilitated by the development of new generation SNP arrays and technology tools that combine SNP level and CNV dosage associations. In follow-up studies, all statistically significant associations should be functionally assessed, although this remains a hazardous task since many genetic variants play an unknown role and the functional effect may well be very subtle (87).
As no single method will fully elucidate the genetic spectrum of AD, more comprehensive approaches are recommended. The main challenge over the next years will be to devise methods allowing the integration and joint analysis of several types of variants and data, i.e. genetic data from GWA studies and re-sequencing experiments as well as transcriptomic, proteomic, epigenomic and metabolomic data. This genomic convergence approach has already been successful in prioritization of risk genes for AD on chromosome 10 (88). Once functionally established, epistasis should be evaluated by interaction studies between different susceptibility genes. One further challenge will be the study of the contribution of environmental risk factors in susceptibility to AD, since so far not much is known about the interplay of genetic and environmental factors.
The genetic architecture of AD is far from being completely unraveled (Fig. 3). Identifying additional genetic factors will remain challenging as genetic searches are complicated at several levels. However, promising strategies and tools are being developed to optimize comprehensive approaches. Detecting new susceptibility factors with a functional impact on AD will bring about major insights into the disease pathways, and initiate new lines of research toward improved medical treatment. Though, it is still a long way to go, genetic risk profiles will eventually be translated into improved medical healthcare, including better and earlier diagnoses of AD as well as individualized care and treatments of AD patients.
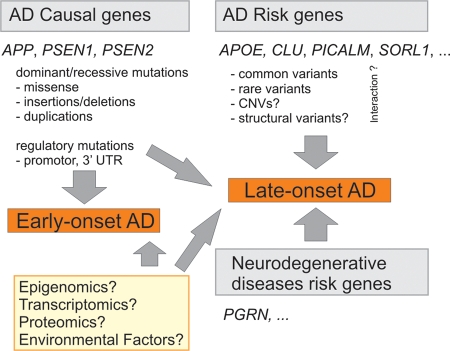
Figure 3.
Schematic summary of the current knowledge of AD molecular genetics. Mutations in causal AD genes APP, PSEN1 and PSEN2 are associated with early-onset AD. Other mutations such as regulatory variants in promoter regions or 3′ untranslated regions (3′-UTR) of causal AD genes may confer susceptibility to late-onset AD. Multiple common and/or rare variants in AD susceptibility genes confer risk to late-onset AD. The top four risk genes (Alzforum, status on 2 April 2010) are listed. The role of CNVs and other structural variants in late-onset AD remains to be clarified, likewise for the interaction between the different risk factors (indicated by question marks). Genes associated with other neurodegenerative dementias like PGRN are involved in AD genetic etiology as well. Several different approaches will likely reveal new genes involved in early and late-onset AD (yellow box).
Conflict of Interest statement. K.S. is receiving a postdoctoral fellowship and K.B. a PhD fellowship of the FWO-V.
FUNDING
The research in the author's group is in part supported by the Special Research Fund of the University of Antwerp, the Fund for Scientific Research-Flanders (FWO-V); the Foundation for Alzheimer Research (SAO/FRMA), the Interuniversity Attraction Poles (IAP) program P6/43 of the Belgian Federal Science Policy Office and a Methusalem Excellence Grant of the Flemish Government, Belgium. Funding to pay the Open Access charge was provided through the University of Antwerp.
REFERENCES
- 1.Alzheimer's Disease International Consortium. AD International, World Alzheimer Report. 2009. Alzheimer's disease International, London. Available at http://www.alz.co.uk/ [Google Scholar]
- 2.Lobo A., Launer L.J., Fratiglioni L., Andersen K., Di Carlo A., Breteler M.M., Copeland J.R., Dartigues J.F., Jagger C., Martinez-Lage J., et al. Prevalence of dementia and major subtypes in Europe: a collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology. 2000;54:S4–S9. [PubMed] [Google Scholar]
- 3.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 4.van der Zee J., Sleegers K., Van Broeckhoven C. Invited article: the AD-frontotemporal lobar degeneration spectrum. Neurology. 2008;71:1191–1197. doi: 10.1212/01.wnl.0000327523.52537.86. doi:10.1212/01.wnl.0000327523.52537.86. [DOI] [PubMed] [Google Scholar]
- 5.Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. doi:10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 6.Gatz M., Reynolds C.A., Fratiglioni L., Johansson B., Mortimer J.A., Berg S., Fiske A., Pedersen N.L. Role of genes and environments for explaining AD. Arch. Gen. Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. doi:10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 7.Coon K.D., Myers A.J., Craig D.W., Webster J.A., Pearson J.V., Lince D.H., Zismann V.L., Beach T.G., Leung D., Bryden L., et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J. Clin. Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. doi:10.4088/JCP.v68n0419. [DOI] [PubMed] [Google Scholar]
- 8.Slooter A.J., Cruts M., Kalmijn S., Hofman A., Breteler M.M., Van Broeckhoven C., van Duijn C.M. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch. Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. doi:10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 9.Daw E.W., Payami H., Nemens E.J., Nochlin D., Bird T.D., Schellenberg G.D., Wijsman E.M. The number of trait loci in late-onset AD. Am. J. Hum. Genet. 2000;66:196–204. doi: 10.1086/302710. doi:10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. doi:10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. doi:10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 12.Levy E., Carman M.D., Fernandez-Madrid I.J., Power M.D., Lieberburg I., van Duinen S.G., Bots G.T., Luyendijk W., Frangione B. Mutation of the Alzheimer's disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. doi:10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 13.Tanzi R.E., Kovacs D.M., Kim T.W., Moir R.D., Guenette S.Y., Wasco W. The gene defects responsible for familial Alzheimer's disease. Neurobiol. Dis. 1996;3:159–168. doi: 10.1006/nbdi.1996.0016. doi:10.1006/nbdi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 14.Klafki H., Abramowski D., Swoboda R., Paganetti P.A., Staufenbiel M. The carboxyl termini of beta-amyloid peptides 1–40 and 1–42 are generated by distinct gamma-secretase activities. J. Biol. Chem. 1996;271:28655–28659. doi: 10.1074/jbc.271.45.28655. [DOI] [PubMed] [Google Scholar]
- 15.Citron M., Diehl T.S., Gordon G., Biere A.L., Seubert P., Selkoe D.J. Evidence that the 42- and 40-amino acid forms of amyloid beta protein are generated from the beta-amyloid precursor protein by different protease activities. Proc. Natl Acad. Sci. USA. 1996;93:13170–13175. doi: 10.1073/pnas.93.23.13170. doi:10.1073/pnas.93.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomiyama T., Nagata T., Shimada H., Teraoka R., Fukushima A., Kanemitsu H., Takuma H., Kuwano R., Imagawa M., Ataka S., et al. A new amyloid beta variant favoring oligomerization in Alzheimer's-type dementia. Ann. Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. doi:10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 17.Di Fede G., Catania M., Morbin M., Rossi G., Suardi S., Mazzoleni G., Merlin M., Giovagnoli A.R., Prioni S., Erbetta A., et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. doi:10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rovelet-Lecrux A., Hannequin D., Raux G., Le M.N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. APP locus duplication causes autosomal dominant early-onset AD with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. doi:10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 19.Kasuga K., Shimohata T., Nishimura A., Shiga A., Mizuguchi T., Tokunaga J., Ohno T., Miyashita A., Kuwano R., Matsumoto N., et al. Identification of independent APP locus duplication in Japanese patients with early-onset AD. J. Neurol. Neurosurg. Psychiatry. 2009;80:1050–1052. doi: 10.1136/jnnp.2008.161703. doi:10.1136/jnnp.2008.161703. [DOI] [PubMed] [Google Scholar]
- 20.Rovelet-Lecrux A., Frebourg T., Tuominen H., Majamaa K., Campion D., Remes A.M. APP locus duplication in a Finnish family with dementia and intracerebral haemorrhage. J. Neurol. Neurosurg. Psychiatry. 2007;78:1158–1159. doi: 10.1136/jnnp.2006.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sleegers K., Brouwers N., Gijselinck I., Theuns J., Goossens D., Wauters J., Del Favero J., Cruts M., van Duijn C.M., Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. doi:10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 22.Cabrejo L., Guyant-Marechal L., Laquerriere A., Vercelletto M., De la Fourniere F., Thomas-Anterion C., Verny C., Letournel F., Pasquier F., Vital A., et al. Phenotype associated with APP duplication in five families. Brain. 2006;129:2966–2976. doi: 10.1093/brain/awl237. doi:10.1093/brain/awl237. [DOI] [PubMed] [Google Scholar]
- 23.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. doi:10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Blom E.S., Viswanathan J., Kilander L., Helisalmi S., Soininen H., Lannfelt L., Ingelsson M., Glaser A., Hiltunen M. Low prevalence of APP duplications in Swedish and Finnish patients with early-onset Alzheimer's disease. Eur. J. Hum. Genet. 2008;16:171–175. doi: 10.1038/sj.ejhg.5201966. doi:10.1038/sj.ejhg.5201966. [DOI] [PubMed] [Google Scholar]
- 25.Cruts M., Van Broeckhoven C. Presenilin mutations in Alzheimer's disease. Hum. Mutat. 1998;11:183–190. doi: 10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. doi:10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., Annaert W., Von Figura K., Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. doi:10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 27.Scheuner D., Eckman C., Jensen M., Song X., Citron M., Suzuki N., Bird T.D., Hardy J., Hutton M., Kukull W., et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat. Med. 1996;2:864–870. doi: 10.1038/nm0896-864. doi:10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 28.Martins R.N., Turner B.A., Carroll R.T., Sweeney D., Kim K.S., Wisniewski H.M., Blass J.P., Gibson G.E., Gandy S. High levels of amyloid-beta protein from S182 (Glu246) familial Alzheimer's cells. Neuroreport. 1995;7:217–220. [PubMed] [Google Scholar]
- 29.Kumar-Singh S., Theuns J., Van Broeck B., Pirici D., Vennekens K., Corsmit E., Cruts M., Dermaut B., Wang R., Van Broeckhoven C. Mean age-of-onset of familial AD caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum. Mutat. 2006;27:686–695. doi: 10.1002/humu.20336. doi:10.1002/humu.20336. [DOI] [PubMed] [Google Scholar]
- 30.Bentahir M., Nyabi O., Verhamme J., Tolia A., Horre K., Wiltfang J., Esselmann H., De S.B. Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J. Neurochem. 2006;96:732–742. doi: 10.1111/j.1471-4159.2005.03578.x. doi:10.1111/j.1471-4159.2005.03578.x. [DOI] [PubMed] [Google Scholar]
- 31.Selkoe D.J. The molecular pathology of Alzheimer's disease. Neuron. 1991;6:487–498. doi: 10.1016/0896-6273(91)90052-2. doi:10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- 32.Hardy J., Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol. Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. doi:10.1016/0165-6147(91)90609-V. [DOI] [PubMed] [Google Scholar]
- 33.Hardy J.A., Higgins G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. doi:10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 34.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial AD. Proc. Natl Acad. Sci. USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. doi:10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of AD genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. doi:10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 36.Bertram L., Tanzi R.E. Alzheimer's disease: one disorder, too many genes? Hum. Mol. Genet. 2004;13(Spec no. 1):R135–R141. doi: 10.1093/hmg/ddh077. [DOI] [PubMed] [Google Scholar]
- 37.Poduslo S.E., Huang R., Huang J., Smith S. Genome screen of late-onset Alzheimer's extended pedigrees identifies TRPC4AP by haplotype analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:50–55. doi: 10.1002/ajmg.b.30767. [DOI] [PubMed] [Google Scholar]
- 38.Bertram L., Tanzi R.E. Genome-wide association studies in Alzheimer's disease. Hum. Mol. Genet. 2009;18:R137–R145. doi: 10.1093/hmg/ddp406. doi:10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bettens K., Brouwers N., Van Miegroet H., Gil A., Engelborghs S., De Deyn P.P., Vandenberghe R., Van Broeckhoven C., Sleegers K. Follow-up study of susceptibility loci for Alzheimer's disease and onset age identified by genome-wide association. J. Alzheimers Dis. 2010;19:1169–1175. doi: 10.3233/JAD-2010-1310. [DOI] [PubMed] [Google Scholar]
- 40.Sleegers K., Bettens K., Brouwers N., Engelborghs S., van Miegroet H., De Deyn P.P., Van Broeckhoven C. Common variation in GRB-associated Binding Protein 2 (GAB2) and increased risk for Alzheimer dementia. Hum. Mutat. 2009;30:E338–E344. doi: 10.1002/humu.20909. doi:10.1002/humu.20909. [DOI] [PubMed] [Google Scholar]
- 41.Goring H.H., Terwilliger J.D., Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am. J. Hum. Genet. 2001;69:1357–1369. doi: 10.1086/324471. doi:10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi-Miura N.H., Ihara Y., Fukuchi K., Takeda M., Nakano Y., Tobe T., Tomita M. SP-40,40 is a constituent of Alzheimer's amyloid. Acta Neuropathol. 1992;83:260–264. doi: 10.1007/BF00296787. doi:10.1007/BF00296787. [DOI] [PubMed] [Google Scholar]
- 43.May P.C., Lampert-Etchells M., Johnson S.A., Poirier J., Masters J.N., Finch C.E. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer's disease and in response to experimental lesions in rat. Neuron. 1990;5:831–839. doi: 10.1016/0896-6273(90)90342-d. doi:10.1016/0896-6273(90)90342-D. [DOI] [PubMed] [Google Scholar]
- 44.Oda T., Pasinetti G.M., Osterburg H.H., Anderson C., Johnson S.A., Finch C.E. Purification and characterization of brain clusterin. Biochem. Biophys. Res. Commun. 1994;204:1131–1136. doi: 10.1006/bbrc.1994.2580. doi:10.1006/bbrc.1994.2580. [DOI] [PubMed] [Google Scholar]
- 45.Oda T., Wals P., Osterburg H.H., Johnson S.A., Pasinetti G.M., Morgan T.E., Rozovsky I., Stine W.B., Snyder S.W., Holzman T.F. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1-42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp. Neurol. 1995;136:22–31. doi: 10.1006/exnr.1995.1080. doi:10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 46.Yerbury J.J., Poon S., Meehan S., Thompson B., Kumita J.R., Dobson C.M., Wilson M.R. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. doi:10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- 47.Bell R.D., Sagare A.P., Friedman A.E., Bedi G.S., Holtzman D.M., Deane R., Zlokovic B.V. Transport pathways for clearance of human Alzheimer's amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammad S.M., Ranganathan S., Loukinova E., Twal W.O., Argraves W.S. Interaction of apolipoprotein J–amyloid beta-peptide complex with low density lipoprotein receptor-related protein-2/megalin. A mechanism to prevent pathological accumulation of amyloid beta-peptide. J. Biol. Chem. 1997;272:18644–18649. doi: 10.1074/jbc.272.30.18644. doi:10.1074/jbc.272.30.18644. [DOI] [PubMed] [Google Scholar]
- 49.Cole G.M., Ard M.D. Influence of lipoproteins on microglial degradation of Alzheimer's amyloid beta-protein. Microsc. Res. Tech. 2000;50:316–324. doi: 10.1002/1097-0029(20000815)50:4<316::AID-JEMT11>3.0.CO;2-E. doi:10.1002/1097-0029(20000815)50:4<316::AID-JEMT11>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 50.Bartl M.M., Luckenbach T., Bergner O., Ullrich O., Koch-Brandt C. Multiple receptors mediate apoJ-dependent clearance of cellular debris into nonprofessional phagocytes. Exp. Cell Res. 2001;271:130–141. doi: 10.1006/excr.2001.5358. doi:10.1006/excr.2001.5358. [DOI] [PubMed] [Google Scholar]
- 51.Webster S., Bradt B., Rogers J., Cooper N. Aggregation state-dependent activation of the classical complement pathway by the amyloid beta peptide. J. Neurochem. 1997;69:388–398. doi: 10.1046/j.1471-4159.1997.69010388.x. [DOI] [PubMed] [Google Scholar]
- 52.Rogers J., Li R., Mastroeni D., Grover A., Leonard B., Ahern G., Cao P., Kolody H., Vedders L., Kolb W.P., Sabbagh M. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol. Aging. 2006;27:1733–1739. doi: 10.1016/j.neurobiolaging.2005.09.043. doi:10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 53.Wyss-Coray T., Yan F., Lin A.H., Lambris J.D., Alexander J.J., Quigg R.J., Masliah E. Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer's mice. Proc. Natl Acad. Sci. USA. 2002;99:10837–10842. doi: 10.1073/pnas.162350199. doi:10.1073/pnas.162350199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harel A., Wu F., Mattson M.P., Morris C.M., Yao P.J. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9:417–429. doi: 10.1111/j.1600-0854.2007.00694.x. doi:10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- 55.Bettens K., Brouwers N., Engelborghs S., De Pooter T., De Deyn P.P., Sleegers K., Van Broeckhoven C. DNMBP is genetically associated with Alzheimer dementia in the Belgian population. Neurobiol. Aging. 2009;30:2000–2009. doi: 10.1016/j.neurobiolaging.2008.02.003. doi:10.1016/j.neurobiolaging.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Kuwano R., Miyashita A., Arai H., Asada T., Imagawa M., Shoji M., Higuchi S., Urakami K., Kakita A., Takahashi H., et al. Dynamin-binding protein gene on chromosome 10q is associated with late-onset Alzheimer's disease. Hum. Mol. Genet. 2006;15:2170–2182. doi: 10.1093/hmg/ddl142. doi:10.1093/hmg/ddl142. [DOI] [PubMed] [Google Scholar]
- 57.Reich D.E., Lander E.S. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. doi:10.1016/S0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 58.Chakravarti A. Population genetics—making sense out of sequence. Nat. Genet. 1999;21:56–60. doi: 10.1038/4482. doi:10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 59.Yang Q., Khoury M.J., Friedman J., Little J., Flanders W.D. How many genes underlie the occurrence of common complex diseases in the population? Int. J. Epidemiol. 2005;34:1129–1137. doi: 10.1093/ije/dyi130. doi:10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- 60.Hedrick P.W. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeggini E., Rayner W., Morris A.P., Hattersley A.T., Walker M., Hitman G.A., Deloukas P., Cardon L.R., McCarthy M.I. An evaluation of HapMap sample size and tagging SNP performance in large-scale empirical and simulated data sets. Nat. Genet. 2005;37:1320–1322. doi: 10.1038/ng1670. doi:10.1038/ng1670. [DOI] [PubMed] [Google Scholar]
- 62.VanLiere J.M., Rosenberg N.A. Mathematical properties of the r2 measure of linkage disequilibrium. Theor. Popul. Biol. 2008;74:130–137. doi: 10.1016/j.tpb.2008.05.006. doi:10.1016/j.tpb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W.Y., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. doi:10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 64.Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. doi:10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorlov I.P., Gorlova O.Y., Sunyaev S.R., Spitz M.R., Amos C.I. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. doi:10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romeo S., Pennacchio L.A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H.H., Cohen J.C. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 2007;39:513–516. doi: 10.1038/ng1984. doi:10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. doi:10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 68.Cohen J.C., Pertsemlidis A., Fahmi S., Esmail S., Vega G.L., Grundy S.M., Hobbs H.H. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc. Natl Acad. Sci. USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. doi:10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brouwers N., Sleegers K., Engelborghs S., Bogaerts V., Serneels S., Kamali K., Corsmit E., De Leenheir E., Martin J.J., De Deyn P.P., et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer's disease. Brain. 2006;129:2984–2991. doi: 10.1093/brain/awl212. doi:10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- 70.Theuns J., Brouwers N., Engelborghs S., Sleegers K., Bogaerts V., Corsmit E., De Pooter T., van Duijn C.M., De Deyn P.P., Van Broeckhoven C. Promoter mutations that increase amyloid precursor–protein expression are associated with AD. Am. J. Hum. Genet. 2006;78:936–946. doi: 10.1086/504044. doi:10.1086/504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guyant-Marechal L., Rovelet-Lecrux A., Goumidi L., Cousin E., Hannequin D., Raux G., Penet C., Ricard S., Mace S., Amouyel P., et al. Variations in the APP gene promoter region and risk of AD. Neurology. 2007;68:684–687. doi: 10.1212/01.wnl.0000255938.33739.46. doi:10.1212/01.wnl.0000255938.33739.46. [DOI] [PubMed] [Google Scholar]
- 72.Bettens K., Brouwers N., Engelborghs S., Van M.H., De Deyn P.P., Theuns J., Sleegers K., Van Broeckhoven C. APP and BACE1 miRNA genetic variability has no major role in risk for AD. Hum. Mutat. 2009;30:1207–1213. doi: 10.1002/humu.21027. doi:10.1002/humu.21027. [DOI] [PubMed] [Google Scholar]
- 73.van Duijn C.M., Cruts M., Theuns J., Van Gassen G., Backhovens H., van den Broeck M., Wehnert A., Serneels S., Hofman A., Van Broeckhoven C. Genetic association of the presenilin-1 regulatory region with early-onset Alzheimer's disease in a population-based sample. Eur. J. Hum. Genet. 1999;7:801–806. doi: 10.1038/sj.ejhg.5200373. [DOI] [PubMed] [Google Scholar]
- 74.Theuns J., Del-Favero J., Dermaut B., van Duijn C.M., Backhovens H., Van den Broeck M.V., Serneels S., Corsmit E., Van Broeckhoven C., Cruts M. Genetic variability in the regulatory region of presenilin 1 associated with risk for Alzheimer's disease and variable expression. Hum. Mol. Genet. 2000;9:325–331. doi: 10.1093/hmg/9.3.325. doi:10.1093/hmg/9.3.325. [DOI] [PubMed] [Google Scholar]
- 75.Lambert J.C., Mann D.M., Harris J.M., Chartier-Harlin M.C., Cumming A., Coates J., Lemmon H., StClair D., Iwatsubo T., Lendon C. The -48 C/T polymorphism in the presenilin 1 promoter is associated with an increased risk of developing Alzheimer's disease and an increased Abeta load in brain. J. Med. Genet. 2001;38:353–355. doi: 10.1136/jmg.38.6.353. doi:10.1136/jmg.38.6.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Theuns J., Remacle J., Killick R., Corsmit E., Vennekens K., Huylebroeck D., Cruts M., Van Broeckhoven C. Alzheimer-associated C allele of the promoter polymorphism -22C > T causes a critical neuron-specific decrease of presenilin 1 expression. Hum. Mol. Genet. 2003;12:869–877. doi: 10.1093/hmg/ddg098. doi:10.1093/hmg/ddg098. [DOI] [PubMed] [Google Scholar]
- 77.Riazanskaia N., Lukiw W.J., Grigorenko A., Korovaitseva G., Dvoryanchikov G., Moliaka Y., Nicolaou M., Farrer L., Bazan N.G., Rogaev E. Regulatory region variability in the human presenilin-2 (PSEN2) gene: potential contribution to the gene activity and risk for AD. Mol. Psychiatry. 2002;7:891–898. doi: 10.1038/sj.mp.4001101. doi:10.1038/sj.mp.4001101. [DOI] [PubMed] [Google Scholar]
- 78.Gacia M., Safranow K., Gabryelewicz T., Styczynska M., Peplonska B., Dziedziejko V., Jakubowska K., Chlubek D., Zekanowski C., Barcikowska M. Two polymorphisms of presenilin-2 gene (PSEN2) 5' regulatory region are not associated with Alzheimer's disease (AD) in the Polish population. J. Neural. Transm. 2008;115:85–90. doi: 10.1007/s00702-007-0846-x. doi:10.1007/s00702-007-0846-x. [DOI] [PubMed] [Google Scholar]
- 79.Quan W., Yasuda M., Hashimoto M., Yamamoto Y., Ishii K., Kazui H., Mori E., Kakigi T., Maeda K. Polymorphism of the regulatory region of the presenilin-2 gene in sporadic Alzheimer's disease: a case-control study. J. Neurol. Sci. 2006;240:71–75. doi: 10.1016/j.jns.2005.09.004. doi:10.1016/j.jns.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 80.Di Natale M., Perri M., Kawarai T., Maletta R., Tomaino C., Sato C., Nacmias B., Shibata N., Sorbi S., St George-Hyslop P.H., et al. Absence of association between AD and the regulatory region polymorphism of the PS2 gene in an Italian population. Neurosci. Lett. 2003;343:210–212. doi: 10.1016/s0304-3940(03)00335-5. [DOI] [PubMed] [Google Scholar]
- 81.Brouwers N., Sleegers K., Engelborghs S., Maurer-Stroh S., Gijselinck I., van der Zee J., Pickut B.A., Van den B.M., Mattheijssens M., Peeters K., et al. Genetic variability in progranulin contributes to risk for clinically diagnosed AD. Neurology. 2008;71:656–664. doi: 10.1212/01.wnl.0000319688.89790.7a. doi:10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 82.Rogaeva E., Meng Y., Lee J.H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C.T., Cheng R., Hasegawa H., et al. The neuronal sortilin-related receptor SORL1 is genetically associated with AD. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. doi:10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bettens K., Brouwers N., Engelborghs S., De Deyn P.P., Van Broeckhoven C., Sleegers K. SORL1 is geneticaly associated with increased risk for late-onset Alzheimer's disease in the Belgian population. Hum. Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. doi:10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 84.Yang Y., Chung E.K., Wu Y.L., Savelli S.L., Nagaraja H.N., Zhou B., Hebert M., Jones K.N., Shu Y., Kitzmiller K., et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. doi:10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J.A., Lupski J.R. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. doi:10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 86.Bertram L., Kerick M., Schjeide B.-M. M., Thomson B., Werber M., Schweiger M.R., Sudbrak R., Herrmann B., Tanzi R.E., Burgess D., Lehrach H., Timmermann B. High Throughput Sequencing of Coding Regions in Alzheimer's Disease using Next Generation Technologies. 2009 Presented at The American Society of Human Genetics, October 22, 2009. [Google Scholar]
- 87.Katsanis N. From association to causality: the new frontier for complex traits. Genome Med. 2009;1:23. doi: 10.1186/gm23. doi:10.1186/gm23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang X., Slifer M., Martin E.R., Schnetz-Boutaud N., Bartlett J., Anderson B., Zuchner S., Gwirtsman H., Gilbert J.R., Pericak-Vance M.A., Haines J.L. Genomic convergence to identify candidate genes for AD on chromosome 10. Hum. Mutat. 2009;30:463–471. doi: 10.1002/humu.20953. doi:10.1002/humu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]