Abstract
Many factors required for chromosomal DNA replication have been identified in unicellular eukaryotes. However, DNA replication in complex multicellular organisms is poorly understood. Here, we report the identification of GEMC1, a novel vertebrate protein required for chromosomal DNA replication. GEMC1 is highly conserved in vertebrates and is preferentially expressed in proliferating cells. Using Xenopus egg extract we show that Xenopus GEMC1 (xGEMC1) binds to checkpoint and replication factor TopBP1, which promotes xGEMC1 binding to chromatin during pre-replication complex (pre-RC) formation. We demonstrate that xGEMC1 directly interacts with replication factors such as Cdc45 and Cdk2-CyclinE by which it is heavily phosphorylated. Phosphorylated xGEMC1 stimulates initiation of DNA replication whereas depletion of xGEMC1 prevents DNA replication onset due to impairment of Cdc45 loading onto chromatin. Likewise, inhibition of GEMC1 expression by morpholino and siRNA oligos prevents DNA replication in embryonic and somatic vertebrate cells. These data suggest that GEMC1 promotes initiation of chromosomal DNA replication in higher eukaryotes by mediating TopBP1 and Cdk2 dependent recruitment of Cdc45 onto replication origins.
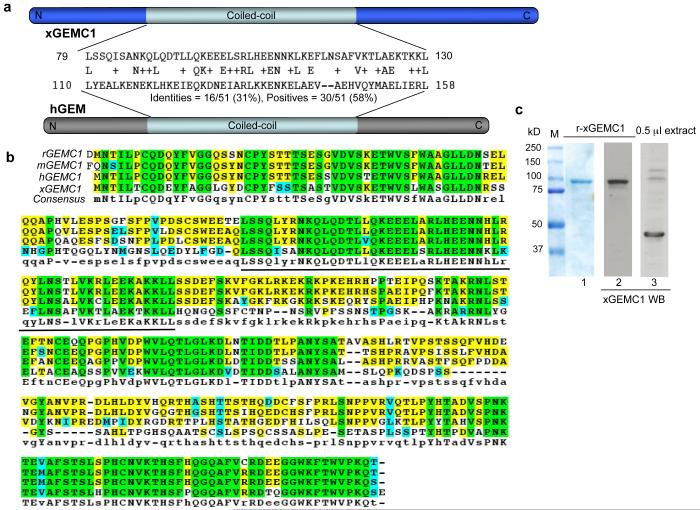
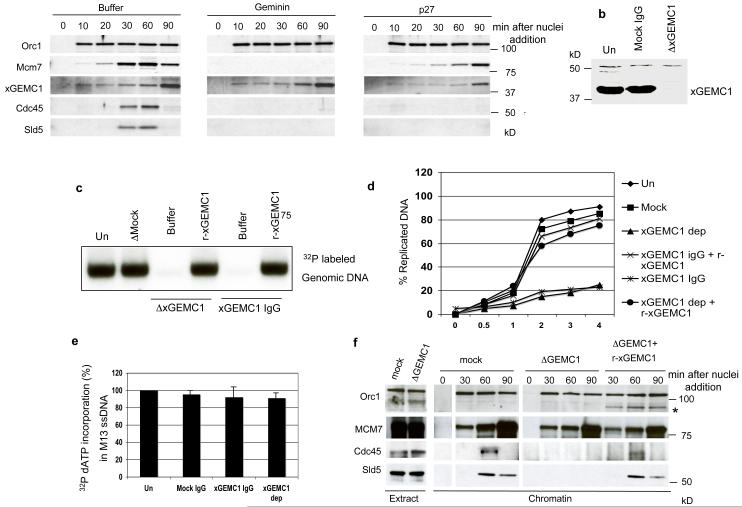
To initiate DNA replication complexes such as ORC1-6 and the MCM2-7 helicase together with Cdc6 and Cdt1 are loaded onto chromatin to assemble the pre-RC. Origin firing is triggered by Cdk2-CyclinE and Cdc7-Dbf4 kinases that promote the loading of Cdc45 and of polymerases in the presence of Mcm10, TopBP1 and the GINS complex1, 2. To identify novel proteins with a potential role in chromosomal DNA replication in complex organisms we performed database searches looking for open reading frame (ORFs) containing degenerate signature motifs present in known replication factors. We identified an ORF, which we named GEMC1 (GEMinin Coiled-coil containing protein 1), containing a region similar to the coiled-coil domain of geminin, an essential vertebrate replication protein in which the coiled-coil domain is required for its function3 (Fig. 1a and Supplementary Fig 1a). GEMC1 is highly conserved in vertebrates as close homologues can be found in H. sapiens (hGEMC1), M. musculus (mGEMC1), R. norvegicus (rGEMC1) and Xenopus laevis (xGEMC1) (Fig 1b). However, only some of the aminoacid residues critical for geminin function are conserved in GEMC14. In addition, comparison of the predicted GEMC1 structure with geminin revealed an interruption in the coiled-coil domain of GEMC1 (Supplementary Fig 1b). To investigate the role of GEMC1 in DNA replication we isolated xGEMC1 cDNA from Xenopus mRNA and used the Xenopus egg extract system5, 6. We tested whether, similar to geminin, recombinant xGEMC1 was able to inhibit DNA replication3. No significant inhibition of chromosomal DNA replication was observed when physiological amounts of xGEMC1 were added to egg extract (Supplementary Fig 1c) suggesting that xGEMC1’s role is different from geminin. To uncover xGEMC1’s function we generated polyclonal antibodies against recombinant xGEMC1 fusion proteins (Fig. 1c). xGEMC1 is expressed in most Xenopus tissues (Supplementary Fig 1d) with its expression pattern partially overlapping other replication factors such as MCM7, Cdk2 and TopBP1 (data not shown) and is enriched in proliferating cells from skin and gut, although it was also detected in ovary, brain and lung tissues (Supplementary Fig 1d). Analysis in egg extract revealed that xGEMC1 is able to bind chromatin at early stage of DNA replication, although its accumulation progresses more slowly than other replication factors (Fig. 2a). xGEMC1 binding is independent of the MCM2-7 complex as it can be detected on the chromatin in the presence of recombinant geminin, which suppresses MCM2-7 loading without affecting binding of other factors such as TopBP12, 3, 7 (Fig 2a). xGEMC1 loading was minimally affected by Cdk2 inhibitor p272, 3, 7 (Fig 2a). Depletion of Cdc45 did not impair xGEMC1 binding to chromatin although it prevented loading of the GINS complex (data not shown). To investigate xGEMC1 role in DNA replication we depleted xGEMC1 from egg extract (Fig 2b) or supplemented extract with affinity-purified antibodies specific for xGEMC1 to interfere with xGEMC1 function. These treatments inhibited chromosomal DNA replication and did not affect origin independent replication of the single stranded M13 phage (Fig 2c, 2d, 2e and Supplementary Fig 1e). DNA replication inhibition was rescued by the addition of recombinant xGEMC1 to depleted egg extract or by out-competing anti xGEMC1 neutralizing antibodies with an excess of recombinant xGEMC1 (r-xGEMC1) (Fig 2c, 2d and Supplementary Fig 1e). The anti xGEMC1 antibodies did not cross-react with geminin (Supplementary Fig 1f). In addition, nuclear membrane formation, which is required for chromosomal DNA replication8, was not affected by anti xGEMC1 antibodies (Supplementary Fig 2a). xGEMC1 depletion prevented chromatin binding of Cdc45 and Sld5 of the GINS complex and this was restored by recombinant xGEMC1 (Fig 2f). Consistently, anti xGEMC1 neutralising antibodies prevented Cdc45 chromatin loading (Supplementary Fig 2b). TopBP1, Cdc7, ORC1-6 and MCM2-7 complex loading was instead unaffected by xGEMC1 depletion or anti xGEMC1 antibodies (Fig 2f and Supplementary Fig 2c). These data indicate that following its binding to chromatin xGEMC1 is required for Cdc45 and GINS loading onto replication origins. We then performed experiments to identify xGEMC1-binding partners involved in DNA replication. A pull-down with recombinant xGEMC1 fused to Maltose Binding Protein (MBP) and incubated in egg extract co-precipitated Cdc45, CyclinE and Cdk2 (Fig 3a). These interactions were specific as xGEMC1 did not interact with other factors such as Cdc7 and Sld5 (Supplementary Fig 2d). Surprisingly, xGEMC1 was not able to interact with geminin partner Cdt1 (Fig 3a). This is consistent with the fact that aminoacid residues required for Cdt1 binding4 were absent in xGEMC1. We then asked whether any of these interactions were direct using recombinant proteins. We found that recombinant xGEMC1 was able to directly interact with Cdc45 and Cdk2-CyclinE complex in vitro (Supplementary Fig. 3a and 3b). These interactions were also present endogenously as shown by co-precipitation of xGEMC1 with anti Cdc45 antibodies (Supplementary Fig. 3c) and with p13-Suc1 affinity resin for Cdks9 (Supplementary Fig. 3d). We then tested whether xGEMC1 is able to bind TopBP1 (Mus101 in Drosophila, Dpb11 in budding yeast), which is an essential component of the DNA replication machinery2, 7. We found that recombinant xGEMC1 interacts with TopBP1 in extract and in vitro (Fig. 3b). This interaction could also be detected endogenously in xGEMC1 and TopBP1 immunoprecipitates (Supplementary Fig. 3e and 3f).
Figure 1. Identification of GEMC1.
a) Homology of the coiled-coil domain of xGEMC1 with human geminin (hGEM). b) Conservation of GEMC1 in vertebrates (R. Norvegicus rGEMC1 [XP_573306.2], M. Musculus mGEMC1 [NP_001013783], H. Sapiens hGEMC1 [NP_001140158.1] and X. laevis xGEMC1. Aminoacid identity in all species is in green. Line indicates coiled-coil domain. c) Expression of recombinant xGEMC1 (r-xGEMC1) fused to Maltose Binding Protein (MBP) monitored with coumassie stain (lane 1) or Western Blot (WB) (lane 2) using anti xGEMC1 antibodies. WB of endogenous xGEMC1 in egg extract is shown in lane 3. See Supplementary Fig 6 for uncropped gels shown in this figure.
Figure 2. Analysis of GEMC1 role in DNA replication.
a) Chromatin binding of Orc1, MCM7, Cdc45, xGEMC1 and Sld5 at the indicated times after addition of sperm nuclei (3000 n/μl) to interphase egg extracts supplemented with buffer (Control), 80 nM geminin (GEM) or 1 μM p27 protein. The sample collected at 0 minutes was DNA free. b) WB of egg extract depleted with mock (ΔMock) or affinity purified anti xGEMC1 antibodies (ΔxGEMC1). c) DNA replication assay showing incorporation of α32P-dCTP in genomic DNA incubated in egg extract for 2 hours. From left to right sperm nuclei (1000 n-μl) were replicated in egg extracts that were untreated (Un), depleted with mock IgGs (Mock IgG, 80 ng/μl), depleted with affinity purified anti xGEMC1 IgGs (ΔxGEMC1) and supplemented with buffer (Buffer) or with 10 ng/μl xGEMC1 (r-xGEMC1) or incubated with affinity purified anti xGEMC1 IgGs (xGEMC1 IgG, 80 ng/μl) and supplemented with buffer (Buffer) or an excess of recombinant xGEMC1 (r-xGEMC1). DNA was isolated, run on agarose gel that was dried and exposed. d) Amount of replicated DNA measured by quantifying α32P-dATP incorporation over time using TCA precipitation (see Supplementary methods) in extracts treated as indicated in (c). The graph shows a typical result. e) Amount of α32P-dCTP incorporated in single stranded M13 phage DNA relative to untreated sample. Extracts were untreated (Un), supplemented with mock IgGs (Mock IgG, 80 ng/μl), with affinity purified anti xGEMC1 immunoglobulins (xGEMC1 IgG, 80 ng/μl) or xGEMC1 depleted (xGEMC1 dep). Experiment was repeated 3 times (n=3). Average results are shown. Error bars indicate SD. f) Chromatin binding of indicated proteins at different times after addition of sperm nuclei (3000 n/μl) to egg extracts that were mock (ΔMock) or xGEMC1 depleted in the absence (ΔxGEMC1) or in the presence (ΔxGEMC1+r-xGEMC1) of 10 ng/μl r-xGEMC1. (*) indicates binding of r-xGEMC1 to chromatin, which was probed with anti MBP antibodies. See Supplementary Fig 6 for uncropped gels shown in this figure.
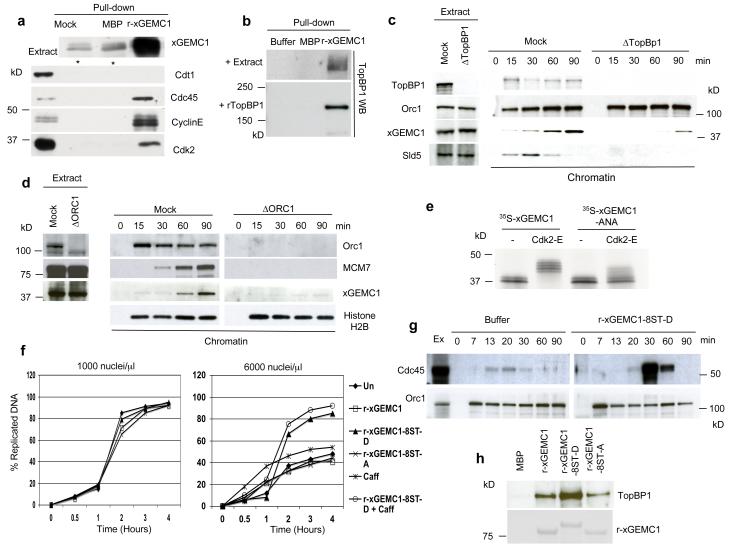
Figure 3. Identification of GEMC1 interacting proteins.
a) WB of indicated proteins on pull-downs performed with amylose resin that was untreated (Mock), pre-bound to MBP (MBP) or to xGEMC1 fused to MBP (r-xGEMC1) and subsequently incubated in egg extract. * indicates non-specific band. b) WB of TopBP1 on pull-downs performed with amylose resin that was untreated (Mock), pre-bound to MBP (MBP) or to GEMC1 fused to MBP (r-GEMC1) and incubated with extract (+ Extract) or with recombinant TopBP1. c) Chromatin binding of the indicated proteins at different times after addition of sperm nuclei (3000 n/μl) to egg extracts that were mock (Mock) or TopBP1 (ΔTopBP1) depleted. d) Chromatin binding of the indicated proteins at different times after addition of sperm nuclei (3000 n/μl) to interphase egg extracts that were mock (Mock) or Orc1 depleted (ΔORC1). e) Autoradiograph of 35S-labelled xGEMC1 (35S-GEMC1) or 35S-labelled xGEMC1 mutated in the Cyclin binding site (35S-xGEMC1-ANA) produced in reticulocyte lysates and incubated with buffer (−) or recombinant Cdk2-CyclinE complex (Cdk2-E). f) Amount of replicated DNA measured by quantifying α32P-dATP incorporation over time using TCA precipitation (see methods) at different times after the addition of 1000 or 6000 nuclei/μl to egg extracts supplemented with 300 ng/μl recombinant xGEMC1 (r-xGEMC1), 300 ng-μl xGEMC1-8ST-D (r-xGEMC1-8ST-D) carrying the serine and threonine residues phosphorylated by Cdk2 mutated to aspartate, 300 ng/μl xGEMC1-8ST-A protein (r-xGEMC1-8ST-A) with the same residues mutated to alanine, with 3 mM caffeine (Caff) or with 3 mM caffeine in the presence of 300 ng/μl xGEMC1-8ST-D protein (xGEMC1-8ST-D + Caff). The graphs show data from one representative experiment. xGEMC1 proteins used in these experiments were defective for the Cyclin-binding domain (R198NL to A198NA mutation) to avoid titration of endogenous Cdk2-Cyclin E complex. g) Cdc45 chromatin binding. Egg extracts were supplemented with buffer (Buffer) or 300 ng/μl xGEMC1-8ST-D mutant protein (r-xGEMC1-8ST-D). Orc1 was used as loading control. h) WB of TopBP1 (top panel) on pull-downs performed with amylose resin that was pre-bound to MBP (MBP), to MBP fused to wild type xGEMC1 (r-xGEMC1), to aspartate (r-xGEMC1-8ST-D) or to alanine substituted xGEMC1 (xGEMC1-8ST-A) (bottom panel, coumassie) and incubated with recombinant TopBP1. See Supplementary Fig 6 for uncropped gels shown in this figure.
Depletion of TopBP1 inhibited xGEMC1 loading onto chromatin during early stages of pre-RC formation indicating that TopBP1 is directly required for xGEMC1 binding to chromatin (Fig 3c). Taken together our data indicate that TopBP1 dependent loading of xGEMC1 during pre-RC assembly promotes subsequent Cdc45 binding to chromatin. Residual TopBP1 independent binding of xGEMC1 to chromatin taking place after DNA replication initiation was also observed suggesting that xGEMC1 has additional binding partners on DNA not directly involved in replication initiation (Fig 3c). Importantly, xGEMC1 binding to chromatin was dependent upon the ORC1-6 complex as ORC1 depletion prevented xGEMC1 chromatin binding (Fig 3d). The interaction of xGEMC1 with Cdk2-CyclinE prompted us to verify whether xGEMC1 is also a substrate for Cdk2. We found that xGEMC1 is heavily phosphorylated in vitro by Cdk2-CyclinE (Fig 3e) and by Cdk2-CyclinA (Supplementary Fig 3g). Multiple phosphorylated forms of xGEMC1 sensitive to lambda phosphatase treatment and roscovitine10 (data not shown) were observed when 35S-labelled xGEMC1 was incubated in egg extract (Supplementary Fig 3h). These results suggest that xGEMC1, similar to other replication proteins1, 11, is a Cdk substrate in egg extract. We then tested whether the Cyclin-binding site R198NL (Arginine-X-Leucine)12 present at the C-terminus of xGEMC1 was required for the phosphorylation. Upon mutation of R198NL to A198NA (Alanine-X-Alanine) xGEMC1-ANA phosphorylation by Cdk2 was strongly reduced (Fig. 3e and Supplementary Fig 3g and 3h). The Cyclin-binding domain was also required for the interaction between xGEMC1 and Cdk2-CyclinE complex as this was abolished by the A198NA mutation without affecting the interaction with Cdc45 (Supplementary Fig 3i). Phosphorylation assays using xGEMC1 peptide arrays revealed a major Cdk2 phosphorylation site on Threonine 153 (Supplementary Fig 4a). Antibodies against phosphorylated Threonine 153 (Supplementary Fig 4b) recognized endogenous xGEMC1 phosphorylation on Threonine 153 (Supplementary Fig 4c). However, xGEMC1 lacking Threonine 153 could still be phosphorylated by Cdk2 in vitro (Supplementary Fig 4d). Therefore, we mutated 7 of the 8 putative Cdk sites that could be phosphorylated by Cdk2-CyclinE and Cdk2-CyclinA in the absence of Threonine 153 (Supplementary Fig 4d). As expected for a Cdk target bearing a strong Cyclin-binding domain, only combined mutations of the additional Cdk sites completely suppressed xGEMC1 phosphorylation by Cdk2 (Supplementary Fig 4e). As we detected multiple phosphorylated forms of xGEMC1 protein in egg extract (Supplementary Fig 3h) it is likely that these sites are phosphorylated by Cdk2 also in vivo. We then assessed the relevance of xGEMC1 phosphorylation by Cdk2 for DNA replication. Extracts have a limited ability to support full replication of a high amount of sperm nuclei (Fig 3f and reference 13, 14). This is due to limiting Cdk activity14 and to the activation of the ATM-ATR checkpoint that represses Cdk2 dependent origin firing15.
Since xGEMC1 is a Cdk2 target required for DNA replication we tested whether xGEMC1 is a limiting factor for replication of a high amount of sperm nuclei. To this end we supplemented egg extract with an excess of recombinant xGEMC1 bearing putative Cdk2 serine-threonine sites mutated to aspartic (xGEMC1-8ST-D) or alanine (xGEMC1-8ST-A) residues to mimic constitutively phosphorylated and un-phosphorylatable proteins, respectively. We then monitored the replication of the absolute amount of DNA template added to egg extract. In these conditions xGEMC1-8ST-D, but not xGEMC1-8ST-A, was able to stimulate efficient replication of a high concentration of sperm nuclei (Fig 3f). xGEMC1-8ST-D dependent stimulation was also observed in the presence of the ATM-ATR inhibitor caffeine15, indicating that it was not due to down-regulation of the ATM-ATR checkpoint (Fig 3f). The increase in DNA replication was accompanied by an enhanced loading of Cdc45 (Fig 3g). DNA replication kinetics (Fig 3f) and replication fork rate measured by DNA fiber spreading analysis (Supplementary Fig 5a) were not significantly affected by xGEMC1-8ST-D. In addition, analysis of DNA replication products on CsCl gradients excluded the presence of re-replication in the presence of xGEMC1-8ST-D (Supplementary Fig 5b). Collectively, these results suggest that xGEMC1-8ST-D-mediated increase of DNA replication is due to enhanced origin firing.
Intriguingly, xGEMC1-8ST-D showed higher affinity for TopBP1 whereas xGEMC1-8ST-A binding to TopBP1 was decreased, although it was not abolished (Fig 3h). To confirm the relevance of xGEMC1 phosphorylation we monitored DNA replication in extracts depleted of xGEMC1 and supplemented with physiological amounts of xGEMC1-8ST-A or xGEMC1-8ST-D mutant proteins. xGEMC1-8ST-A could not restore DNA replication, whereas xGEMC1-8ST-D induced a significant DNA replication increase (Supplementary Fig 5c and 5d). xGEMC1-8ST-D was unable to completely bypass the requirement for Cdk2 in DNA replication as it did not restore DNA replication in extract in which Cdk2 activity was inhibited by roscovitine10 (data not shown). This suggests that xGEMC1 is not the only Cdk2 target required for initiation of DNA replication.
Overall, these data indicate that xGEMC1 is involved in promoting initiation of DNA replication by mediating TopBP1 and Cdk2 dependent loading of Cdc45 onto replication origins.
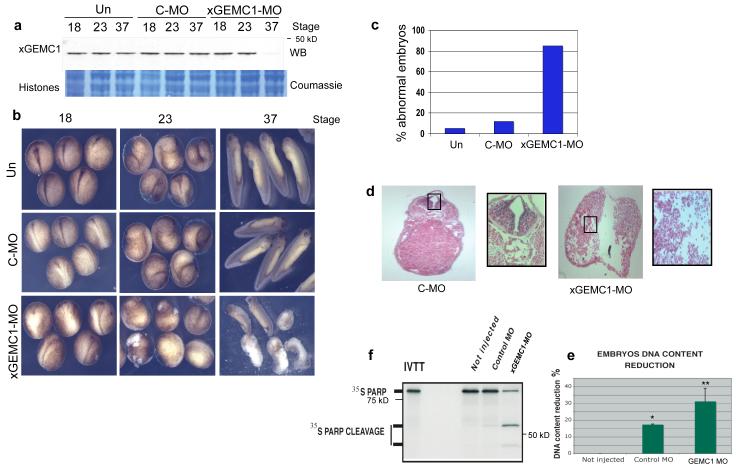
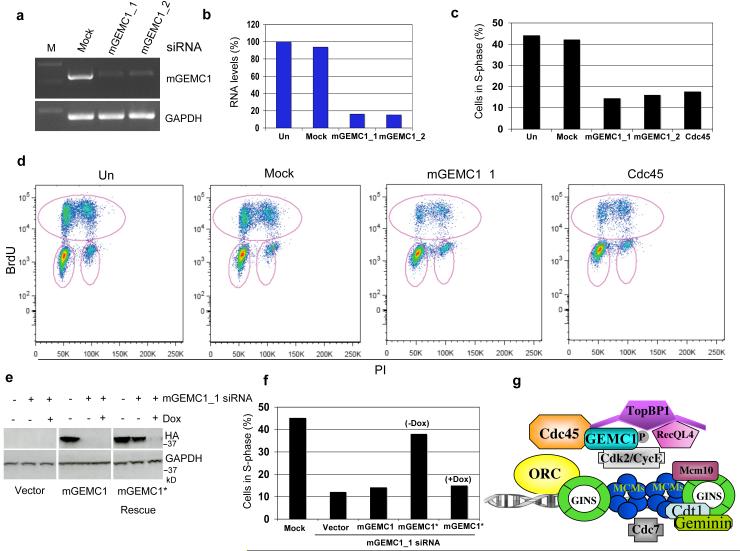
To validate these results we depleted xGEMC1 protein from Xenopus embryos by selectively inactivating xGEMC1 expression with morpholino antisense oligos (MO)16. We selected MO oligos that were able to inhibit xGEMC1 translation in the reticulocyte system (xGEMC1-MO) (Supplementary Fig 5e). When injected into fertilized eggs, xGEMC1-MO oligos inhibited xGEMC1 expression (Fig 4a), whereas control oligos (C-MO) did not affect xGEMC1 (Fig 4a). Depletion of xGEMC1 induced severe delay in embryo development and gross anatomical defects with poor yolk resorbption and no distinct structures such as the gut, the optic vesicle or the tail. These defects were detected in most of the embryos injected with xGEMC1-MO oligos (Fig 4b and 4c). The most significant effects were detected after complete down-regulation of xGEMC1 protein levels around neurula stage (Fig 4b). The delayed effects of morpholino oligos were likely due to persistence of high levels of xGEMC1 protein up to late developmental stages. Histological analysis revealed a dramatic decrease in cell density and DNA content when xGEMC1 was inhibited (Fig 4d and 4e). This was accompanied by apoptosis induction (Fig 4f). To confirm that xGEMC1 has a role in DNA replication in mammalian organisms we isolated the mouse homologue of GEMC1 (mGEMC1) whose mRNA levels were assed by RT-PCR (Fig 5a). A decrease in mGEMC1 mRNA obtained with siRNA oligos (Fig 5a and 5b) led to severe inhibition of cell proliferation (Fig 5c and 5d). The extent of inhibition was similar to the one obtained with inhibition of Cdc45 expression (Fig 5c, 5d and Supplementary Fig 5f). Ectopic expression of RNAi resistant mGEMC1 protein, obtained by introducing silent mutations in mGEMC1 encoding cDNA, which was expressed under the control of doxycycline regulated promoter, restored proliferation in mGEMC1 silenced cells (Fig 5e and 5f).
Figure 4. Effects resulting from inhibition of GEMC1 expression in Xenopus embryos.
a) WB of xGEMC1 extracted from Xenopus embryos that were untreated (Un) or injected with control (C-MO) and morpholino oligos complementary to xGEMC1 DNA sequence (xGEMC1-MO) taken at the indicated stages. Lower panel shows coumassie staining of histone proteins from total embryo lysates. b) Morphology of Xenopus embryos that were untreated (Un), injected with control (C-MO) or anti xGEMC1 morpholino oligos (xGEMC1-MO) taken at the indicated stages. c) Quantification of experiment shown in (b). 100 embryos were counted for each treatment. Graph shows a typical result. d) Sections of fixed Xenopus embryos injected with control (C-MO) or with anti xGEMC1 morpholino oligos (xGEMC1-MO) and stained with ematossilin-eosin. Area in the rectangle shows 5x magnification. e) DNA content reduction in embryos that were uninjected (Not injected), injected with control morpholino oligos (Control MO) or with anti xGEMC1 morpholino oligos (xGEMC1-MO). Data are mean ± SD of 3 independent experiments; * and ** P <0.001 compared with not injected control, t-test) f) Cleavage of 35S-labeled PARP induced by lysates of embryos that were uninjected (Not injected), injected with control morpholino oligos (Control MO) or anti xGEMC1 morpholino oligos (xGEMC1-MO). See Supplementary Fig 6 for un-cropped gels shown in this figure.
Figure 5. Effects resulting from inhibition of GEMC1 expression in mammalian cells.
a) RT-PCR of mouse GEMC1 (mGEMC1) and GAPDH (GAPDH) mRNA derived from mouse NIH 3T3 fibroblasts that were treated with control (Mock) or mouse GEMC1 siRNA oligos (mGEMC1_1 and mGEMC1_2). b) Quantification of RNA levels shown in (a). The graph shows data from one representative experiment. c) Histograms relative to FACS analysis following incorporation of BrdU in mouse NIH3T3 cells that were untreated (Un), mock treated (Mock), transfected with two independent mouse GEMC1 siRNA (mGEMC1_1 or mGEMC1_2) or with Cdc45 siRNA oligos. The graph shows data from one representative experiment. d) Typical FACS profiles relative to experiments shown in (c). Circles in pink indicate BrdU incorporating cells. e) WB of MEF-3T3 cells stably transfected with empty TET-OFF vector (Vector), with TET-OFF vector expressing hemagglutinin epitope (HA) tagged mGEMC1 (mGEMC1) or with the same vector expressing siRNA resistant HA tagged mGEMC1 (mGEMC1* Rescue). Cells were subjected to silencing with mGEMC1_1 siRNA in the absence or presence of 1 μg/ml doxycycline (Dox), which represses the expression driven by the exogenous promoter present on the vector. After 96 hours cells were collected and processed for WB using anti HA and anti GAPDH antibodies as indicated. f) The histograms represent the percentage in S-phase of MEF-3T3 cells that were treated with mock (Mock) or mGEMC1 siRNA oligos (mGEMC1_1 siRNA). mGEMC1_1 siRNA treated cells were stably transfected with empty TET-OFF vector (Vector), with TET-OFF vector expressing HA tagged mGEMC1 (mGEMC1) or with the same vector expressing siRNA resistant HA tagged mGEMC1 (mGEMC1*) in the absence (−Dox) or in the presence (+Dox) of 1 μg/ml doxycycline. Cells were BrdUTP labelled and processed for FACS analysis. The graph shows data from one representative experiment. g) Model for GEMC1 function at replication origins (see text for explanation). See Supplementary Fig 6 for uncropped gels shown in this figure.
Here we have identified GEMC1, a novel vertebrate protein required for initiation of DNA replication. GEMC1 directly interacts with TopBP1, Cdc45 and Cdk2-CyclinE. Cdk2, Cdc7, MCM10, the GINS complex and TopBP1 have been shown to promote Cdc45 recruitment onto DNA17-20. xGEMC1 is required for binding to chromatin of Cdc45, which promotes origin unwinding, recruitment of RPA and DNA polymerase α loading21, 22. Continuous accumulation of xGEMC1 on chromatin observed after Cdc45 loading suggests that xGEMC1 has also other functions besides DNA replication.
Although xGEMC1 has a coiled-coil domain weakly similar to geminin it does not behave like geminin. In S. cerevisiae a complex made by Sld2, Sld3 and Dpb11 is essential to trigger DNA replication initiation23, 24. Emerging evidence shows that these two proteins represent the minimal set of S-phase Cdk substrates required for DNA replication23, 24. Sld3 interacts with Cdc45 and is required for its chromatin binding25, 26. Sld3 phosphorylation by S-phase Cdks promote its binding to Dpb11-TopBP123, 24, 27. To date, an Sld2 homologue in vertebrates has been identified and corresponds to RecQL428, 29. Sld3 is instead poorly conserved even within fungi, suggesting rapid evolution of this protein. Although S. pombe Sld3 has coiled-coil domains we could not detect significant aminoacid similarity between either S. Pombe or S. cerevisiae Sld3 and GEMC1. However, similar to S. cerevisiae Sld3 xGEMC1 is phosphorylated by S-phase Cdks, interacts with Cdc45 and TopBP1 and is required for Cdc45 loading onto chromatin (Fig 5g). Importantly, xGEMC1 constitutively phosphorylated on Cdk sites has higher affinity for TopBP1 and is able to stimulate Cdc45 chromatin loading and initiation of DNA replication. This indicates that, similar to Sld323, 24, phosphorylation of xGEMC1 by Cdk2 is important for origin firing. It is tempting to speculate that GEMC1 represents a functional vertebrate homolog of yeast Sld3. The absence of protein sequence conservation with Sld3 might reflect the fact that during evolution the function played by Sld3 and xGEMC1 has dynamically evolved to adapt to the individual needs of origin firing control in each species. Alternatively, GEMC1 might not be the only protein with these properties as other factors together or in addition to GEMC1 might be required to mediate the interaction between TopBP1, Cdc45 and Cdk2 at replication origins. In any case the study of GEMC1 will lead to a better understanding of normal and uncontrolled cell cycle progression in vertebrate organisms.
Supplementary Material
Acknowledgements
We thank Tim Hunt, members of Clare Hall Laboratories and of the Genome Stability Unit for their comments. We thank H Mahbubani and J Kirk for technical support with Xenopus laevis, J. Gannon for technical assistance in antibody production and Mary Wu for embryos injection. This work was funded by Cancer Research UK. V. Costanzo is also supported by the Lister Institute of Preventive Medicine, the European Research Council (ERC) start up grant (N. 206281) and the EMBO Young Investigator Program (YIP).
Footnotes
Accession number:
xGEMC1 cDNA sequence is available at NCBI (Accession number:GU594151).
Methods
The Methods and their associated references appear only online.
Note: Supplementary Information is available on the Nature Cell Biology website.
Reference
- 1.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Takisawa H, Mimura S, Kubota Y. Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr Opin Cell Biol. 2000;12:690–696. doi: 10.1016/s0955-0674(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 3.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 4.Saxena S, et al. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol Cell. 2004;15:245–258. doi: 10.1016/j.molcel.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 5.Costanzo V, Gautier J. Xenopus cell-free extracts to study DNA damage checkpoints. Methods Mol Biol. 2004;241:255–267. doi: 10.1385/1-59259-646-0:255. [DOI] [PubMed] [Google Scholar]
- 6.Trenz K, Errico A, Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. Embo J. 2008;27:876–885. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. Embo J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 9.Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 10.Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- 12.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 14.Krasinska L, et al. Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. Embo J. 2008;27:758–769. doi: 10.1038/emboj.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 16.Khokha MK, et al. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- 17.Kubota Y, et al. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Hatten RA, et al. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 20.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 21.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. Embo J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. Embo J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 25.Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. Embo J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima R, Masukata H. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol Biol Cell. 2002;13:1462–1472. doi: 10.1091/mbc.02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka S, Tak YS, Araki H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2007;2:16. doi: 10.1186/1747-1028-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangrithi MN, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubota Y, Takisawa H. Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell free extracts. J Cell Biol. 1993;123:1321–1331. doi: 10.1083/jcb.123.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.