Abstract
Eukaryotic secretory pathway cargo fold to their native structures within the confines of the endoplasmic reticulum (ER). To ensure a high degree of folding fidelity, a multitude of covalent and noncovalent constraints are imparted upon nascent proteins. These constraints come in the form of topological restrictions or membrane tethers, covalent modifications, and interactions with a series of molecular chaperones. N-linked glycosylation provides inherent benefits to proper folding, and creates a platform for interactions with specific chaperones and Cys modifying enzymes. Recent insights into this timeline of protein maturation have revealed mechanisms for protein glycosylation, and iterative targeting of incomplete folding intermediates, which provides nurturing interactions with molecular chaperones that assist in the efficient maturation of proteins in the eukaryotic secretory pathway.
Keywords: endoplasmic reticulum, protein folding, carbohydrates, molecular chaperones, quality control
Anfinsen’s seminal studies demonstrated that proteins contain all the information in their primary sequence required to fold into their proper 3-dimensional structure [1]. Yet elevated temperatures and protein concentrations reaching 300 mg/ml provide unfavorable folding conditions in live cells. Strikingly, protein folding that is reconstituted in the test tube is generally less efficient than in the cell because cells utilize alternative mechanisms, contain factors that assist proteins in reaching their native states and destroy proteins that fail to fold properly. Approximately one third of the entire eukaryotic proteome is predicted to traffic through the secretory pathway [2, 3]. For these proteins, protein folding and maturation begin co-translationally as the protein is translocated into the endoplasmic reticulum (ER) lumen. The ER functions as a protein folding factory, harboring a large array of molecular chaperones, oxidoreductases and quality control machinery. The maturation process that includes covalent modifications, processing, folding and assembly continues post-translationally until the native structure is acquired. Cellular protein folding assistance occurs via a number of constraints imparted upon the maturing protein to streamline the folding pathway. This results in a restriction of the vast number of possible conformations and avoidance of off-pathway intermediates to promote the efficient acquisition of the native state. The fidelity of the maturation process is monitored by quality control machinery that sorts native proteins for anterograde trafficking and defective proteins for retention and subsequent destruction.
Co-translational translocation and folding in the ER
Protein folding in living cells can begin during synthesis [4, 5]. The ‘Levinthal paradox’ demonstrates that protein folding is not a random walk since there is insufficient time to sample all possible conformations available for a protein to reach its native state [6]. Protein folding in the cell is subjected to a variety of temporal, spatial and physical constraints, providing important restrictions that support a more direct route to the native state. A co-translational folding process creates a vectorial folding reaction and provides a mechanism for the cell to control and organize the environment for the vulnerable nascent chain [7, 8]. Tethering the termini of the nascent chain to the ribosome and the membrane further limits the freedom of the nascent chain or the number of available folding intermediates. In addition, the bulky ribosome separates nascent chains of a polysome, preventing nonproductive collisions or aggregation. Together, these mechanisms act to optimize the folding reaction in the apparent suboptimal folding conditions within the cell
Coupling protein folding and synthesis tapers the possible ensemble of folding intermediates available to the shorter nascent chains during the progression to the native state, thereby minimizing the available nonproductive options [9]. By allowing folding to commence co-translationally or prior to the completion of synthesis, the amount of conformational space that may be sampled by the nascent chain is greatly reduced, allowing short distance or local folding events to occur first. This early restriction in nascent chain folding streamlines the folding pathway by reducing the conformational heterogeneity that is the result of a long fully synthesized unfolded protein precursor [10]. In addition, by initiating folding prior to the completion of synthesis, aggregation and non-native interactions of nascent chains are prevented since the constraints also reduce the mobility or diffusion rate of unfolded precursors in the ER [4, 9, 11]. Thus, the vectorial nature of co-translational folding helps to fold proteins rapidly and efficiently.
Signal sequences on proteins destined for the ER are generally located at the N-terminus of proteins, and are first recognized by the cytosolic signal recognition particle (SRP)[12]. SRP delivers the ribosome/mRNA complex to the surface of the ER through interactions with the SRP receptor. Here, the ribosome associates with translocon machinery, facilitating entry of the nascent chain into the ER lumen. Protein translocation into the ER occurs through Sec61, a large multi-protein complex that interacts with the ribosome and allows nascent proteins to transit through its pore [13, 14]. The ribosome, Sec61 and their associated proteins can create a privileged environment that sterically protects the nascent chain from nonproductive interactions to favor proper protein maturation [15].
The hydrophobic nature of signal sequences supports their integration into ER membranes. This places a constraint on the maturing protein by tethering its N-terminus to the membrane and restricting the degree to which nascent chains can sample conformational space. This allows newly synthesized proteins to fold in a controlled manner that protects against premature folding events that could result in nonproductive local minima until the signal sequence is cleaved. Signal sequence cleavage is critical for proper protein maturation. In the case of the viral envelope protein, vesicular stomatitis virus glycoprotein G, the failure of the signal sequence to be cleaved results in the ER retention of the misfolded viral protein [16]. The timing and efficiency of signal sequence cleavage is substrate specific; though it generally appears to occur co-translationally, after the synthesis of ~140 amino acids [17–19]. However, the delayed post-translational cleavage of signal sequences has been implicated in the functional activity of a number of proteins including human cytomegalovirus protein US11 and human immunodeficiency virus glycoprotein 120 [20, 21]. Furthermore, the timing of co-translational folding events has been proposed to be dictated in part by signals embedded within the amino acids of the signal sequence, likely controlled by directing the timing of signal sequence cleavage and release of the membrane tethering constraint [22].
While local structures such as α-helices have been shown to form in the ribosome and translocon tunnels, global folding occurs upon entrance into the ER lumen through the Sec61 translocon [5, 23–26]. Covalent constraints such as disulfide bond formation and modification with sugar chains occur as proteins are being synthesized, and these modifications can also significantly contribute to the efficient maturation of secretory cargo [5, 27].
N-linked glycosylation
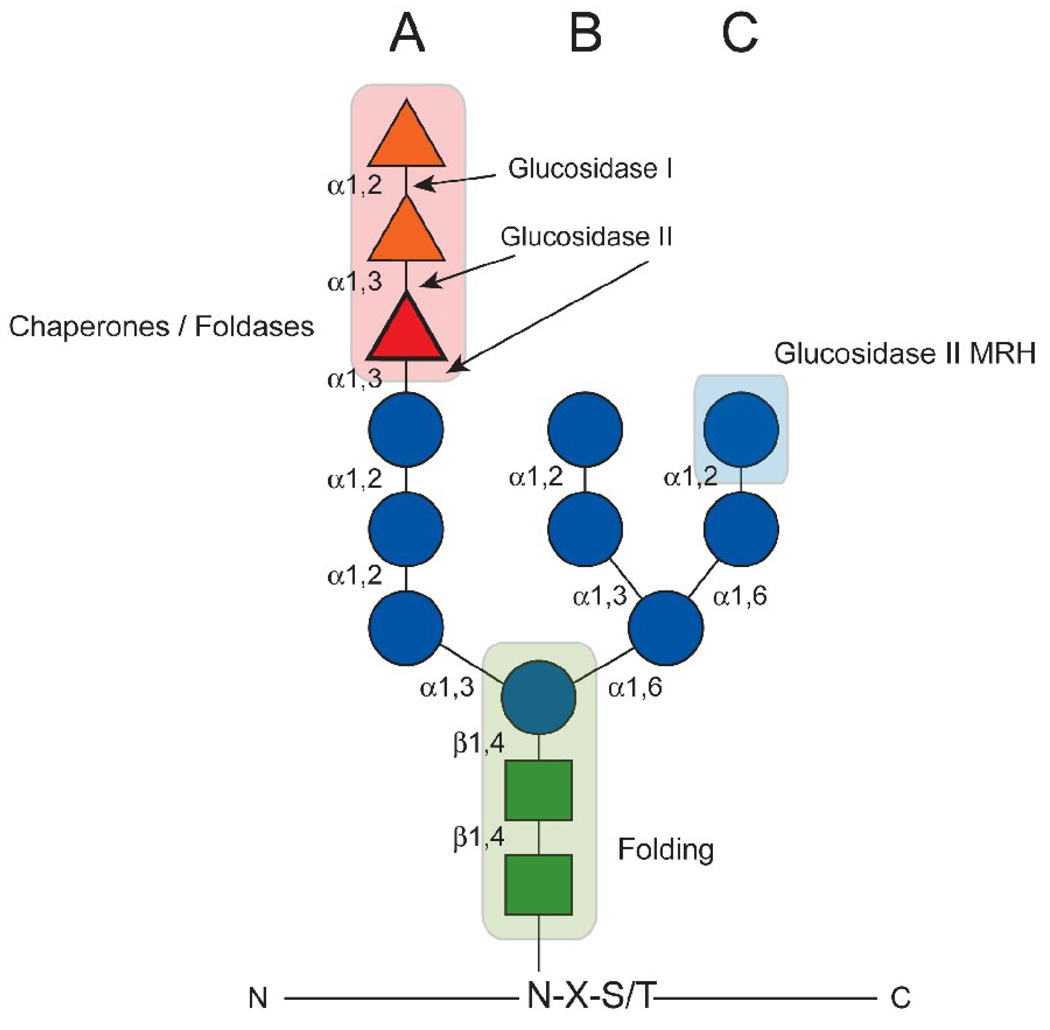
The glycosidic linkage of preformed 14-member carbohydrate chains onto Asn residues in the consensus sequence Asn-X-Ser/Thr (where X can be any amino acid except Pro) are added to the majority of proteins that traverse the secretory pathway [28–30]. Glycans can greatly impact the folding of these proteins. The N-linked glycan is comprised of 2 N-acetyl glucosamines (GlcNAc) and 9 mannoses (Man) arranged in 3 branches (A, B, and C) with 3 glucoses (Glc) attached to the A-branch mannose residue (Fig. 1). The N-glycan is transferred preassembled from dolichol-P-linked lipids by the oligosaccharyltransferase (OST) that is associated with the Sec61 translocon (Fig. 2, step 1)[28]. There are two forms of the OST that contain different isoforms of the catalytic subunit, STT3 [31]. A recent study has shown that the STT3A-containing OST can support co-translational glycosylation of nascent glycoprotein substrates, whereas the STT3B isoform can additionally facilitate the post-translational glycosylation of substrates [32]. These results suggest that the addition of glycans does not necessarily occur in linear succession. In addition, the OST subunit Ost3/6p has been found to contain an active thioredoxin-like domain possessing oxidoreductase activity [33]. Since oxidation of substrate proteins has been shown to negatively impact glycosylation efficiency [34, 35], the covalent tethering of nascent chains to the Ost3/6p subunit can impact glycosylation transfer efficiency [33].
Figure 1.
The composition and roles of N-linked glycans in the ER. The 14-member carbohydrate is covalently linked to Asn residues of the consensus sequence Asn-X-Ser/Thr as depicted. The glycan is comprised of 3 glucoses (triangles), 9 mannoses (circles), and 2 N-acetlyglucosamines (squares). The mannoses are arranged in three branches A, B, and C. The orientations of the glycosidic bonds are indicated. The positions at which glucosidases I and II cleave the glucoses are designated. The glucose that is transferred through GT1 activity and is involved in lectin chaperone binding is highlighted. The regions of the N-glycan that are critical for chaperone interaction, glucosidase II MRH binding, and folding kinetics are shaded.
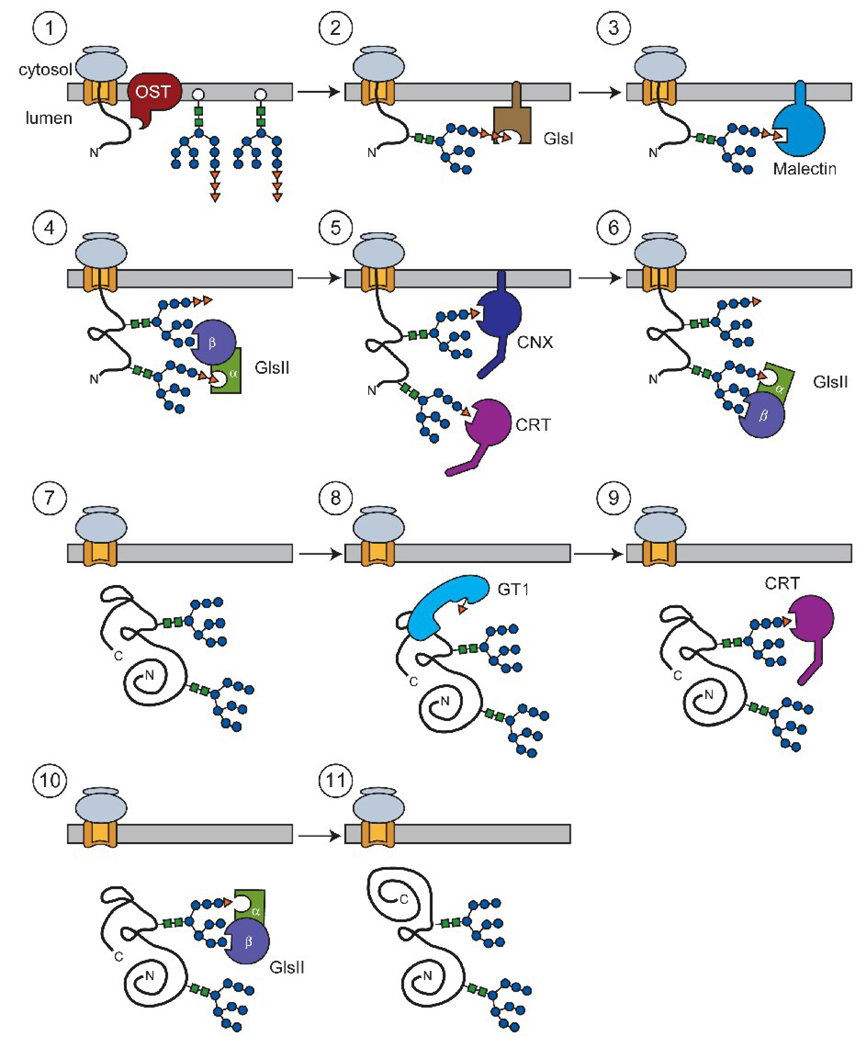
Figure 2.
Modification of N-glycans during glycoprotein maturation. Nascent secretory proteins co-translationally enter the ER lumen. At this time, the membrane integrated oligosaccharyltransferase (OST) transfers 14-member carbohydrates to nascent chains from lipid-linked precursors (step 1). Nearly immediately following glycan transfer, the membrane-associated glucosidase I (Gls I) removes the terminal glucose, generating a Glc2Man9GlcNAc2 glycoform (step 2). The exposure of the second glucose allows the type I membrane protein and lectin, malectin, to associate with nascent chains (step 3). In direct competition with malectin binding is the removal of the second glucose by glucosidase II. Glucosidase II is comprised of an MRH domain containing β-subunit and an enzymatic α-subunit. The β-subunit MRH domain docks glucosidase II to glycoproteins via interactions with auxiliary glycans. This in turn allows the α-subunit to remove the second glucose of the target glycan (step 4). Exposure of the innermost glucose permits interactions with the lectin chaperones calnexin and calreticulin (step 5). However, glucosidase II competes with the lectin chaperones for binding to the substrate glycans. In this step, glucosidase II liberates the final glucose via docking of its MRH subunit to mannoses within the target glycan (step 6). The fully deglucosylated glycan is incapable of being bound by the lectin chaperones and is free to fold to its native state (step 7). If non-native conformations persist, the folding sensor GT1 docks to misfolded regions and reglucosylates target glycans (step 8). This results in reassociation with lectin chaperones for additional rounds of folding or retention (step 9). Eventually, the glucose that was added by GT1 is removed by glucosidase II (step 10). At this point, the protein is once again free to fold to its native state. If properly folded, it will traffic to its final functional destination. It may re-enter the lectin chaperone cycle via GT1 activity if non-native conformations remain present. If terminally misfolded, it will eventually be sorted for retrotranslocation and degradation (step 11).
The addition of N-glycans to nascent proteins has an immediate effect on the folding pathway of the substrate and its eventual fate. N-glycans are highly flexible and hydrophilic structures that mask hydrophobic stretches on proteins and greatly impacting the sampling of conformational space during folding [36, 37]. Examples are available that demonstrate the intrinsic effects of glycosylation on protein stability and oligomerization, with glycans providing a chaperone-like effect on protein folding [38]. N-glycans have been shown to prevent the undesired association of hydrophobic stretches, which can lead to aggregation [39, 40]. Indeed, glycosylation has been implicated in maintaining the prion protein in a non-fibril conformation [41]. The flexibility of N-glycans is exploited to protect proteins from proteases and shield pathogens from immune detection [42–44]. Recent studies have revealed tight interactions between the base trisaccharide core (Man1GlcNAc2) and the folding polypeptide that impacts the kinetics of protein folding [45]. The majority of this contribution to protein folding thermodynamics and kinetics is actually derived from the single innermost GlcNAc residue. Analysis of the of the Human Gene Mutation Database has revealed that gain-of-glycosylation mutations occur in a much higher prevalence than previously anticipated, while mutation of glycosylation consensus sites are rarer than predicted [46]. This finding highlights the positive influence of the inherent properties of glycans on protein folding in the cell. The mutational introduction of a glycosylation site into a protein can effectively destabilize the unfolded state by restricting conformational sampling, coaxing the unfolded protein towards native state conformations [45].
In addition to the inherent influence of N-glycans on protein folding, glycans can also recruit folding factors to assist in protein maturation depending upon the composition of the N-linked glycans [29, 47]. After the transfer of the 14-member carbohydrate by the OST, the first glucose is removed by the membrane-bound glucosidase I (Fig. 1 and 2, step 2). This deglucosylation reaction occurs very rapidly, since it can be detected concomitantly with glycan transfer [48]. The Glc2Man9GlcNAc2 glycan composition prevents reassocation with the OST, acting as a commitment step to commence folding [49, 50]. Until recently, there were no known ER proteins that interacted exclusively with the diglucosylated glycoform. However, recent studies have identified malectin as a highly conserved ER-localized membrane-bound Glc2-specific lectin with unknown roles in glycoprotein maturation (Fig. 2, step 3)[51]. Malectin has been found to associate with endogenous aquaporin-2 in a large-scale proteomics study, suggestive of its involvement in glycoprotein maturation, however the scope of proteins that act as malectin substrates is unknown [52]. Malectin lacks a canonical ER retention motif but remains ER localized at steady state [51]. Thus, it is likely that malectin is retained in the ER as a component of an unidentified complex. Presumably, malectin acts co-translationally on substrate glycoproteins, since both the glycosidase action of glucosidase I that precedes malectin binding and the subsequent trimming of glucoses by glucosidase II can occur co-translationally [5, 48]. However, there is no direct evidence for the interaction of malectin with substrates as they co-translationally emerge into the ER.
The diglucosylated glycan (Glc2Man9GlcNAc2) is subsequently trimmed by glucosidase II (Fig. 1 and 2, step 4). Glucosidase II is a soluble luminal enzyme that is comprised of an 110-kDa α-subunit and a 60-kDa β-subunit [53]. Glucosidase activity is localized to the α-subunit of the heterodimer, while the β-subunit in humans contains a C-terminal HDEL ER retention signal. The glucosidase II β-subunit is present in S. cerevisiae, yet it lacks an ER retention signal and is not required for maintaining the localization of the α-subunit [54]. In contrast, the β-subunit of S. pombe has a C-terminal VDEL sequence that is important for localization of the catalytic subunit [55]. Catalytic activity is exclusive to the α-subunit, since it alone displays in vitro enzymatic properties [56]. Glucosidase II removes two glucoses from glycans; hydrolysis of the Glcα1,3Glc bond to generate Glc1Man9GlcNAc2 glycans, and hydrolysis of the Glcα1,3Man bond to produce a fully deglucosylated glycan (Fig. 1 and Fig. 2, steps 4 and 6). The initial cleavage of the Glcα1,3Glc bond occurs much more rapidly than the second cleavage of the Glcα1,3Man residues [57, 58]. The Glcα1,3Glc and Glcα1,3Man moieties are virtually indistinguishable structurally. Rather, the differences in their catalytic cleavages are likely due to the Glcα1,3Glc and Glcα1,3Man cleavage epitopes residing on opposite faces of the glycan structure [48, 59, 60]. Since there is only a single active site within glucosidase II, the slow and nonprocessive digestion of the Glc1Man9GlcNAc2 glycoform is proposed to be the result of reorientation of the active site of the α–subunit or the repositioning of the glycosylated substrate to accommodate the opposing glycosidic linkages.
Until recently the only function attributed to the glucosidase II β–subunit was in the localization of the glucosidase II heterodimer to the ER lumen via its C-terminal ER retention signal. In addition to the C-terminal ER retention signal, the glucosidase II β-subunit contains a Mannose-6 phosphate Receptor Homology (MRH) domain [48, 61]. MRH domains are typically utilized for lectin-like interactions, as observed for the ER resident proteins XTP3-B and OS-9 that are involved in glycoprotein degradation [61, 62]. Helenius and colleagues observed that multiple glycans present on substrates greatly accelerated the cleavage of the Glcα1,3Glc bond. They proposed a model whereby the MRH domain of the β-subunit binds to a mannose branch of a neighboring glycan to position the α-subunit in such a way that the Glcα1,3Glc of the substrate glycan is efficiently recognized and hydrolyzed (Fig. 2, step 4). Subsequently, the MRH domain can recognize a mannose branch within the substrate glycan to trim the final glucose of the Glcα1,3Man linkage (Fig. 2, step 6)[48]. A detailed biochemical study revealed that the lectin interaction of the MRH domain with substrates occurs via the C-branch mannoses [58]. This is in support of earlier work that suggested that glucosidase II catalytic activity decreased as mannoses were removed from substrate glycoproteins [63]. This model explains the observation that multiple glycans were required for the transition of substrate glycans from Glc2 to Glc1 glycoforms and that glucosidase II can directly compete for substrate binding with other lectins in the ER [48, 58]. It is important to note that the auxiliary glycan utilized in the transition from Glc2 to Glc1 state does not have to be localized on the substrate glycoprotein in this model. There could be trans interactions occurring between glycosylated maturation machinery or other newly synthesized glycoproteins to facilitate the cleavage of the Glcα1,3Glc bond. This model supports the findings that glucosidase II trimming occurs in a nonprocessive manner, interrupted by critical chaperone interactions.
Further studies highlight the importance of the β-subunit to glucosidase II catalytic activity. The β-subunit confers substrate specificity, is critical for the cleavage of the Glcα1,3Glc bond and is essential for the final digestion of the Glcα1,3Man bond to yield a fully deglucosylated substrate, although there is evidence that S. pombe glucosidase II α–subunit can generate fully deglucosylated substrate in the absence of the β–subunit, albeit slowly [54, 55, 64, 65]. The trimming of glycans from Glc2 to Glc1 can occur in the absence of the β–subunit; however, this cleavage is greatly accelerated in the presence of the β–subunit [55, 64]. The MRH domain of the β–subunit is essential for this kinetic enhancement of glucose trimming and the removal of the final glucose since functionally deficient mutations displayed inefficient and negligible conversion to the unglucosylated state. These findings lead to an amendment to the current model for the contribution of the β–subunit to glucosidase II activity. Previous work has shown that the α–subunit has a greater affinity for Glc2Man9GlcNAc2 glycoforms than monoglucosylated glycans [66]. The MRH domain is critical for reorientation of the active site for the nonprocessive hydrolysis of the Glcα1,3Glc and Glcα1,3Man bonds [48]. In addition, the lectin-like binding of the MRH domain to the C-branch mannoses of substrate glycoproteins greatly enhances the kinetics of glucose trimming, enhancing the affinity of the alpha subunit for the conversion of substrates from a Glc2 to a Glc1 and eventually a fully glucose trimmed state [55, 64].
The trimming of glycoproteins from Glc2Man9GlcNAc2 to Man9GlcNAc2 does not occur in a processive manner. The exposure of this innermost glucose initiates interactions with a discrete subset of chaperones including calnexin and calreticulin that aid in the folding of secretory glycoproteins (Fig. 1 and Fig. 2, step 5)[67–70]. The lectin nature of the MRH domain of glucosidase II competes with these lectin chaperones for binding to substrates, controlling the persistency of their protective interaction [58].
The lectin chaperones and their associated oxidoreductase, ERp57
Calnexin and calreticulin are glycan-dependent molecular chaperones that promote proper folding in substrate proteins by stabilizing folding events, preventing aggregation, and facilitating the catalysis of disulfide bond formation [71–74]. Monoglucosylated N-linked carbohydrates (Glc1ManxGlcNAc2) created by the sequential action of glucosidases I and II are recognized by these carbohydrate-binding chaperones (Fig. 2, step 5). Calnexin is a type I membrane protein and is comprised of a single globular carbohydrate binding domain and an extended proline-rich repeat domain, termed the P-domain [75]. Similarly, calreticulin contains both a carbohydrate binding and P-domain, but is a soluble protein with a C-terminal ER retention motif [76, 77]. These chaperones bind monoglucosylated proteins in a Ca2+-dependent fashion [75, 78, 79]. Binding of the lectin chaperones aids in glycoprotein folding by slowing folding events and allowing them to occur in a controlled manner [69, 71]. This is also evident from studies in calnexin and calreticulin deficient cell lines, where folding was found to be accelerated, however the subsequent folding efficiency was diminished [80].
Lectin chaperone association represents a critical constraint for glycoprotein folding. Studies using ribosome-arrested chains have found that there is a predetermined order of events when nascent glycoproteins first enter the ER lumen through the Sec61 translocon. The membrane-bound calnexin is found in close association with the translocon complex. Phosphorylation of the cytoplasmically exposed C-terminal tail of calnexin has been proposed to regulate its association with the ribosome/translocon complex [81]. Calnexin interacts initially with the nascent chain, while associations with the soluble calreticulin can closely follow depending upon the organization of glycans on the substrate [18, 19]. There is also a coupling between the lectin chaperone system and BiP, the ER-resident Hsp70 family member. BiP interacts with hydrophobic stretches of newly synthesized protein prior to the addition of hydrophilic N-glycans [19, 82]. The distance the glycans are located from the N-terminus can dictate whether BiP initially interacts with a nascent chain co-translationally [83]. BiP is less likely to be involved in the maturation of glycoproteins that contain glycans near the N-terminus since they are hydrophilic thus inhibitory for BiP binding and they support the early diversion to the lectin chaperone system [19, 83, 84].
The topology of the lectin chaperones and their substrate is also involved in determining which chaperones bind a particular substrate. Calnexin is situated to interact with membrane proximal glycans, either on membrane-associated proteins or with proteins still entering the ER lumen. Since calreticulin is a soluble protein, it is organized to interact with glycans that emerge deeper into the ER lumen, on either membrane or soluble substrates [85–87].
The lectin chaperones can also facilitate disulfide bond formation and isomerization on substrate proteins via their noncovalent association with the oxidoreductase ERp57 [73, 74, 88, 89]. ERp57 is an ER resident protein disulfide isomerase family member with four thioredoxin domains, two of which contain catalytic Cys pairs. It docks on the lectin chaperones through an interaction of a positively charged patch on a noncatalytic thioredoxin domain and the negatively charged tip of the extended P-domain of the lectin chaperones [76]. It is thought that there is very little free ERp57 in the ER lumen since it predominantly exists in a 1:1 association with each lectin chaperone. In addition, there is a secondary pool associated with tapasin, a protein involved in class I antigen assembly and processing, however this localization appears to be independent of ERp57 redox function [90–92].
Disulfide bond formation represents an additional constraint imparted upon nascent glycoproteins during co-translational folding. Disulfides can limit the conformational flexibility, increase structural rigidity and stabilize proteins once they are exposed to the harsh extracellular environment. Disulfides can also be used as transitory elements during folding, ensuring that a critical folding intermediate is reached, and subsequently isomerized to achieve native conformations [93, 94]. In conjunction with the lectin chaperones, ERp57 can perform oxidation, reduction, and isomerization reactions of Cys pairs in glycoprotein substrates [95]. ERp57 has been found mainly in a reduced form in cells at steady state [96, 97]. The reduced form can only perform isomerization and reduction reactions for substrate proteins. However, it has been found associated with ERO1α, the protein that generates oxidizing equivalents in the ER, and ERp57 has been shown to possess oxidase activity in vivo, suggesting that it is a fully functional oxidoreductase [95, 98]. While the deletion of ERp57 is not lethal to cells, it is essential for mouse development as knockout animals die in utero [99, 100]. There could be multiple PDI family proteins that can substitute for ERp57 in its absence, since there are currently nearly 20 ER-resident members [101]. In recent studies of ERp57, it has been shown to be intimately involved in the late-stage rearrangement of disulfides in complex glycoproteins [100]. This highlights an additional mode of glycoprotein folding that occurs after nascent proteins have fully emerged into the ER lumen.
The folding of glycoproteins generally occurs, at least for distal interactions, after their release from the lectin chaperones that is triggered by glucosidase II-mediated cleavage of the final glucose [69, 71]. Glycoproteins that have failed to achieve their native conformations following their release into the ER lumen are subjected to a quality control process [30, 102]. If non-native conformations persist, the quality control sensor GT1 (UDP-glucose: glycoprotein glucosyltransferase 1) detects these instabilities and transfers a glucose back onto deglucosylated glycoproteins, regenerating monoglucosylated glycans (Fig. 2, step 8)[103–106]. These reglucosylated substrates then re-associate with the lectin chaperones and ERp57 to reinitiate folding events in a process termed the ‘calnexin cycle’ (Fig. 2, step 9)[68–70, 104]. Persistent binding to the lectin chaperones assists protein folding, prevents aggregation through sequestration, retains immature or unassembled structures in the ER and potentially aids in the sorting of malformed substrates for degradation [72, 107–109].
GT1 reglucosylation and quality control of glycoprotein maturation
GT1 is a primary folding or quality control sensor of the ER. It is currently the only known ER enzyme that alters the glycan composition of a protein based on structural features of the modified protein [104–106, 110–112]. In vitro studies have revealed that GT1 detects malformed glycoproteins via surface-exposed hydrophobic patches, indicative of protein immaturity or misfolding [104, 106]. The selectivity of GT1 for exposed hydrophobic stretches in substrate proteins allows for discrete malformed regions to be modified by reglucosylation of proximal glycans, addressing protein misfolding at the domain level (Fig. 3B, steps 3 and 4)[110, 113]. Biophysical studies have demonstrated that GT1 targets nearly native molten globule proteins [111, 114]. GT1 has the ability to distinguish minor structural perturbations, since in vitro reglucosylation increases significantly with the introduction of aberrant structures [113, 115]. In addition, GT1 substrates may not necessarily be malformed at the level of monomers, since GT1 can target and reglucosylate orphan oligomeric subunits of soybean agglutinin that exhibit native conformations in vitro [116]. Finally, a cellular study revealed that the folding sensor capabilities of GT1 can function in the case of natively folding substrate or when protein folding is transiently or terminally disrupted (Fig. 3B, steps 6 and 7)[117].
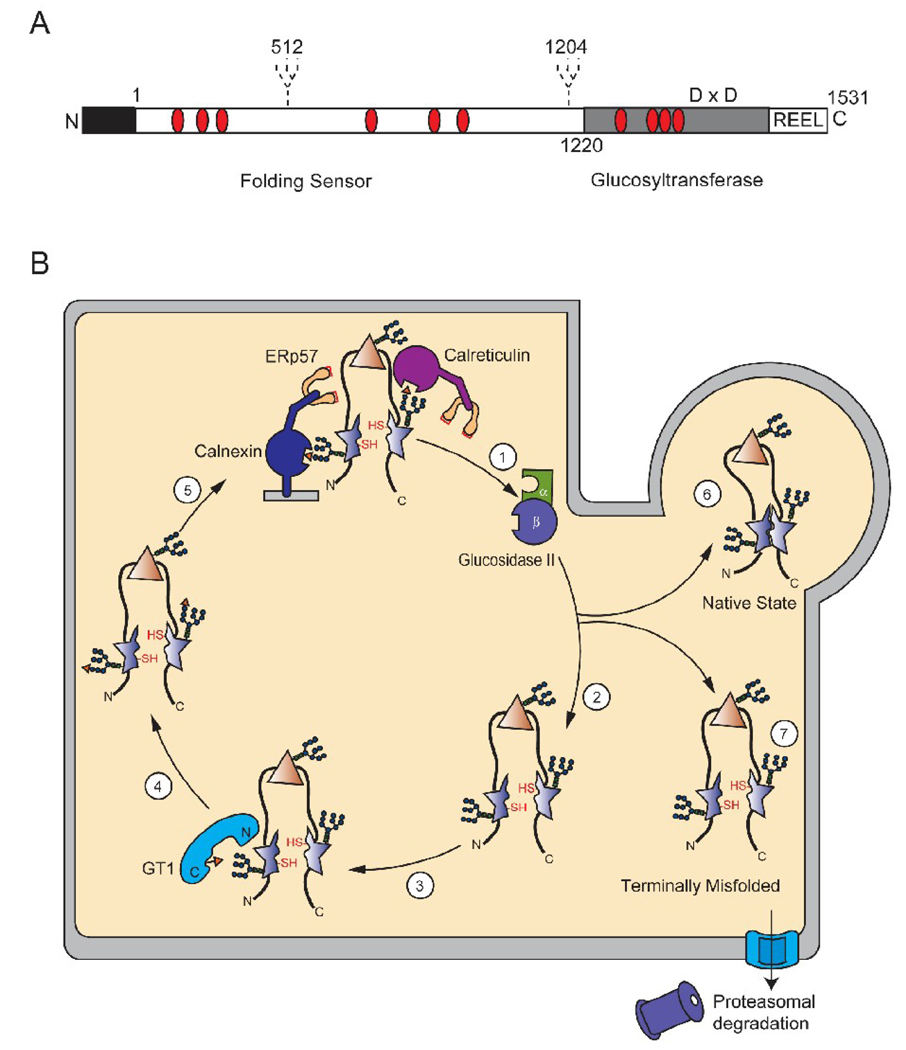
Figure 3.
Model of GT1 reglucosylation of a malformed glycoprotein. (A) Schematic representation of human GT1. Human GT1 is a 174-kDa protein with the mature protein consisting of 1531 amino acids after signal sequence cleavage. The N-terminus contains the protein recognition folding sensor domain, which represents 75% of the primary sequence and the cleavable signal sequence (black). The C-terminal domain contains the catalytic glucosyltransferase activity and the ER-retention signal R-E-E-L. In addition, the positioning of the two hypothetical glycosylation sites in human GT1 are depicted as dashed lines and Cys residues are marked with red ovals. The Asp-X-Asp motif is shown in the C-terminal domain (D × D) (B) Schematic of GT1-mediated reglucosylation in the ER. Glycoproteins are extracted from the lectin chaperone cycle by glucosidase II, which removes the last glucose in the A-branch (step 1). The chaperone released protein can attempt to fold to its native state, with certain domains being properly folded (triangle) and others being non-native or malformed (star) (step 2). GT1 can reglucosylate glycans situated near these non-native domains through docking to exposed hydrophobic patches (step 3). The substrate glycoprotein is now monoglucosylated (step 4). The newly reglucosylated protein can re-enter the calnexin and calreticulin binding cycle (step 5). The affinity of the lectin chaperones will decrease as the glycan is further trimmed by mannosidases and the protein will either be able to attain its native functional form (step 6) or be designated for degradation (step 7).
GT1 is a 174-kDa glycoprotein comprised of two main functional domains [118, 119]. The N-terminal domain of GT1 contains nearly 75% of its primary sequence and functions as the sensor of substrate misfolding, while the C-terminal domain contains the glucosyltransferase catalytic activity. The C-terminal domain is highly conserved amongst species (65%) and is homologous to proteins of the glycosyltransferase family 8 (Fig. 3A). The large N-terminal domain has a much lower similarity between species that may indicate a means of substrate selection, however domain swapping experiments of the S. pombe and D. melanogaster glucosyltransferases resulted in functional reglucosylation activity on misfolded substrates [119]. Despite numerous efforts, a structure of GT1 remains elusive.
GT1 catalyzes the transfer of glucose from the donor UDP-glucose to the terminal A branch mannose of misfolded glycoproteins, regenerating the Glcα1,3Man signal (Fig. 1) [120]. This step occurs after the initial recognition of misfolding, as determined by in vitro competition assays, where saturating levels of nonglycosylated misfolded protein could prevent reglucosylation of a glycosylated substrate [113]. Regions near the C-terminus of GT1 were found to be strictly conserved amongst species and other glycosyltransferase enzymes. Mutation of the Asp-X-Asp motif, an amino acid sequence critical for glycosyltransferase activity nearly completely inhibited reglucosylation (Fig. 3A)[121, 122]. Initially, it was believed that the Asp-X-Asp motif was not involved in catalysis; rather it was utilized in binding the UDP-glucose donor. However, recent evidence from studies on other glycosyltransferases has revealed that the Asp-X-Asp motif is involved in metal coordination that is required for UDP-sugar docking and indirectly enhances catalytic activity [123]. The actual catalytic residues of GT1 are currently uncertain. In order to complete the transferase reaction, GT1 docks to the base N-acetylglucosamine and in more detailed chemical assays, has been shown to interact with the Man1GlcNAc2 base unit [105, 124].
Both mice and humans have two distinct isoforms of the glucosyltransferase termed GT1 and GT2 [125]. Human GT1 and GT2 share 55% identity in their amino acid sequences, however the majority of this high degree of identity is localized to the C-terminal catalytic domain (83%)[118, 125]. The cellular localization of GT1 and GT2 is identical when overexpressed, but differences arise in their ability to reglucosylate misfolded glycoproteins. Functional protein chimeras created from the N- and C-terminal domains of GT1 and GT2 only resulted from the combination of the GT1 N-terminal domain and either the C-terminal catalytic region of GT1 or GT2 [118]. Both GT1 and GT2 have the ability to facilitate glucose transfer but only GT1 displays an affinity for misfolded conformers of substrate proteins. Additionally, only GT1 expression is upregulated by the unfolded protein response (UPR), supporting the importance of GT1 for protein folding in the ER [118].
The localization of GT1 within the ER lumen has important implications in its function. Unstable non-native conformers can have deleterious effects on the successful maturation of newly synthesized proteins in the active folding environment. Thus, it is likely that the ‘calnexin cycle’ takes place in an environment somewhat secluded, so that active folding events can continue uninterrupted. GT1 reglucosylation occurs post-translationally, placing its activity outside of the crowded confines of the ER translocon (Fig. 2, step 8 and Fig. 3B)[117]. GT1 has been localized to late stage folding suborganellar regions in the ER by indirect immunoelectron microscopy and quantitative proteomics [126, 127]. Since not all substrates modified by GT1 are capable of achieving a native fold, it would be beneficial for degradation machinery to reside in a similar locale. Indeed, a component of the putative retrotranslocon pore, Derlin-1, is also primarily located in smooth ER membranes [127].
GT1 may not act alone in glycoprotein quality control as recently a selenocysteine-containing oxidoreductase of 15-kDa (Sep15) was found to bind GT1 with high affinity in a 1:1 complex (Kd < 20 nM). GT1 and Sep15 also appear to share a common subcellular localization and the ability to be transcriptionally upregulated by stress [127, 128]. The binding of Sep15 to GT1 does not involve any of its catalytic oxidoreductase residues, potentially leaving these available for interaction with GT1 substrates [129, 130]. A role for Sep15 in rearranging non-native disulfide bonds in an effort to reduce complications for the calnexin cycle can be envisioned. In addition, the combination of the misfolding sensor capabilities of GT1 and the reduction activity of Sep15 could be involved in preparing terminally malformed proteins for degradation.
The positioning of glycans on a substrate is important for both lectin chaperone binding and GT1-mediated reglucosylation. In vitro studies using a glycopeptide library found that GT1 reglucosylation occurred most frequently on glycopeptides that contained dual proximal hydrophobic patches located C-terminal to the glycan [112]. A more recent in vitro study used synthetic glycans and substrates to demonstrate that moving the hydro-phobic moiety closer to the glycosylation site increased GT1 activity [124]. A cellular study determined that GT1 specifically targeted glycans located on the membrane proximal slow folding domain of influenza hemagglutinin to direct lectin chaperone post-translational re-binding to the immature or aberrant region [117]. GT1 can utilize this quality control selection process to play roles in assisting native protein folding events, triggering prolonged chaperone interactions in difficult to fold regions (Fig. 3B, step 4). Additionally, it can drive persistent chaperone association to retain misfolded proteins for additional rounds of folding or delivery to degradation machinery. This adaptability and response towards many folding situations is why GT1 is so critical to secretory protein maturation in higher order organisms and likely explains why its knockout is embryonic lethal in mice [131, 132]. However, GT activity is not found in all organisms as it is missing in S. cerevisiae [133]. There is likely a restricted population of proteins that require an intact lectin chaperone binding cycle and some of these proteins might be needed for animal development and survival. The complexity and load of secretory cargo of multicellular organisms far exceeds that for unicellular organisms. Extracellular materials are frequently large complex multidomain glycosylated proteins with intricate disulfide networks. These types of proteins are strong candidates for utilizing and requiring an intact lectin chaperone binding cycle for efficiently acquiring their native states.
Outlook
The proper folding of secretory proteins within the confines of the ER is governed by a number of covalent and noncovalent constraints that guide the protein towards native state conformations. The trimming of glucoses from the N-glycans of glycoproteins triggers a cascade of assistive interactions with chaperones and their associated oxidoreductase. One recently discovered lectin, malectin, provides an interesting example of temporal interactions with glycosylated substrates. Malectin interacts with Glc2 glycoforms, a glycan moiety that was thought previously to either be inert during protein maturation or provide a delay before binding to calnexin and calreticulin. It will be important to determine if malectin interacts with substrates co-translationally, and how its role in preceding calnexin and calreticulin binding assists glycoprotein folding.
The cleavage of mannoses also occurs in the ER. In functional contrast to the relation of glucose trimming to folding, loss of mannoses is thought to act as a signal for targeting for transport or degradation [29, 47, 134, 135]. A model whereby mannose trimming signals for degradation is attractive, since mannose-trimmed glycoforms display a decreased affinity for the machinery associated with proper folding, such as glucosidase II, the lectin chaperones, and GT1 [55, 64, 105, 136, 137]. However, recent studies have revealed that the mannose trimming may also occur on the quality control machinery, providing an indirect signal for localizing an aberrant substrate that is bound to an ERAD receptor to be recruited by the membrane retrotranslocation machinery [138–140].
Recent studies have elucidated a further level of complexity in the involvement of MRH domains in glycoprotein maturation. The β̃subunit of glucosidase II contains an MRH domain that binds fully mannosylated glycoforms [64]. However, the quality control degradation receptors OS-9 and XTP3-B utilize MRH domains to bind mannose-trimmed species en route to proteasomal degradation [138, 141–143]. How these MRH domains are functionally distinct remains unclear, since mutations of conserved residues are detrimental to the binding capacity of both classes of MRH domain-containing proteins.
The effect of glycans on protein folding is location dependent. The discrete positioning of glycans along the polypeptide chain to aid in vectorial folding is an overlooked determinant for proper folding. Specific glycans amongst an array of glycans localized on a substrate can elicit isolated lectin chaperone association or trigger the degradation process [86, 144, 145]. In addition, glycans can be evolutionarily selected to reside near critical Cys residues to prevent nonnative covalent association and recruit the glycoprotein-specific oxidoreductase ERp57 to the site of oxidation [18]. The situation of glycosylation consensus sites near Cys residues can also control the efficiency of glycan transfer by the OST, affecting further downstream folding events [33].
The ER is the nexus of secretory protein folding in the cell. The constraints imparted on protein folding pathways within this organelle ensure a high stringency for selection of properly formed functional folds to maintain essential cellular processes and viability. Understanding the organization of the cellular machinery in the ER responsible for eliciting these constraints using improved high-resolution microscopy techniques will provide valuable insight into how the spatial organization of the ER helps control the temporal associations with nascent chains along the maturation assembly line. Advances in our understanding of the regulation and administration of covalent and noncovalent constrictions during protein maturation in the ER will further our knowledge of these processes and provide insight into how they can be manipulated to provide potential therapies for protein misfolding diseases.
ACKNOWLEDGEMENTS
We would like to acknowledge members of the Hebert lab for fruitful discussions and critical reading of this review including Dr. Taku Tamura, Kristina Moody Giorda and Johan Sunryd, and support by US Public Health grant CA79864 and GM086874 (to D.N.H.). B. R. P. was partially supported by an NIH Chemistry-Biology Interface training grant (T32GM00815) and a predoctoral University fellowship from the University of Massachusetts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y, Gu X, Luo J. SPD--a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frydman J, Erdjument-Bromage H, Tempst P, Hartl FU. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Helenius J, Braakman I, Helenius A. Cotranslational folding and calnexin binding of influenza hemagglutinin in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levinthal C. Are there pathways for protein folding? J. Chim. Phys. 1968;65:44–45. [Google Scholar]
- 7.Schnell DJ, Hebert DN. Protein modulators: multi-functional mediators of protein translocation across membranes. Cell. 2003;112:491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 8.Clark PL. Protein folding in the cell: reshaping the folding funnel. Trends Biochem. Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Fedorov AN, Baldwin TO. Cotranslational protein folding. J Biol Chem. 1997;272:32715–32718. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- 10.Alexandrov N. Structural argument for N-terminal initiation of protein folding. Protein Sci. 1993;2:1989–1991. doi: 10.1002/pro.5560021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frydman J, Nimmesgern E, Ohtsuka K, Hartl F-U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 12.Walter P, Gilmore R, Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- 13.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 14.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Helenius A. Role of ribosome and translocon complex during folding of influenza hemagglutinin in the endoplasmic reticulum of living cells. Mol. Biol. Cell. 2000;11:765–772. doi: 10.1091/mbc.11.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw AS, Rottier PJM, Rose JK. Evidence for the loop model of signal-sequence insertion inot the endoplasmic reticulum. Proc. Natl. Acad. Acad. USA. 1988;85:7592–7596. doi: 10.1073/pnas.85.20.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, VanValkenburgh C, Liang H, Fang H, Green N. Signal peptidase and oligosaccharyltransferase interact in a sequential and dependent manner within the endoplasmic reticulum. J Biol Chem. 2001;276:2411–2416. doi: 10.1074/jbc.M007723200. [DOI] [PubMed] [Google Scholar]
- 18.Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 19.Wang N, Daniels R, Hebert DN. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: the heat shock protein 70 system hands off to the lectin-based chaperone system. Mol Biol Cell. 2005;16:3740–3752. doi: 10.1091/mbc.E05-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm A, Stern P, Ploegh HL, Tortorella D. Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. Embo J. 2001;20:1573–1582. doi: 10.1093/emboj/20.7.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Luo L, Thomas DY, Kang CY. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology. 2000;272:417–428. doi: 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 22.Rutkowski DT, Ott CM, Polansky JR, Lingappa VR. Signal sequences initiate the pathway of maturation in the endoplasmic reticulum lumen. J. Biol. Chem. 2003;278:30365–30372. doi: 10.1074/jbc.M302117200. [DOI] [PubMed] [Google Scholar]
- 23.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 24.Kowarik M, Kung S, Martoglio B, Helenius A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Molecular Cell. 2002;10:769–778. doi: 10.1016/s1097-2765(02)00685-8. [DOI] [PubMed] [Google Scholar]
- 25.Kosolapov A, Tu L, Wang J, Deutsch C. Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Mingarro I, Nilsson I, Whitley P, von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitley P, Nilsson I, von Heijne G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 1996;271:6241–6244. doi: 10.1074/jbc.271.11.6241. [DOI] [PubMed] [Google Scholar]
- 28.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 29.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 30.Hebert DN, Molinari M. In and Out of the ER: Protein Folding, Quality Control, Degradation, and Related Human Diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell. 2003;12:101–111. doi: 10.1016/s1097-2765(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulz BL, Stirnimann CU, Grimshaw JP, Brozzo MS, Fritsch F, Mohorko E, Capitani G, Glockshuber R, Grutter MG, Aebi M. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci U S A. 2009;106:11061–11066. doi: 10.1073/pnas.0812515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen S, Naim HY, Bulleid NJ. Intracellular folding of tissue-type plasminogen activator. Effects of disulfide bond formation on N-linked glycosylation and secretion. J Biol Chem. 1995;270:4797–4804. doi: 10.1074/jbc.270.9.4797. [DOI] [PubMed] [Google Scholar]
- 35.Holst B, Bruun AW, Kielland-Brandt MC, Winther JR. Competition between folding and glycosylation in the endoplasmic reticulum. EMBO J. 1996;15:3538–3546. [PMC free article] [PubMed] [Google Scholar]
- 36.Imperiali B, Rickert KW. Conformational implications of asparagine-linked glycosylation. Proc Natl Acad Sci U S A. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3:643–649. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 38.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Jitsuhara Y, Toyoda T, Itai T, Yamaguchi H. Chaperone-like functions of high-mannose type and complex-type N-glycans and their molecular basis. J Biochem. 2002;132:803–811. doi: 10.1093/oxfordjournals.jbchem.a003290. [DOI] [PubMed] [Google Scholar]
- 40.Nagaya H, Tamura T, Higa-Nishiyama A, Ohashi K, Takeuchi M, Hashimoto H, Hatsuzawa K, Kinjo M, Okada T, Wada I. Regulated motion of glycoproteins revealed by direct visualization of a single cargo in the endoplasmic reticulum. J Cell Biol. 2008;180:129–143. doi: 10.1083/jcb.200704078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosques CJ, Imperiali B. The interplay of glycosylation and disulfide formation influences fibrillization in a prion protein fragment. Proc Natl Acad Sci U S A. 2003;100:7593–7598. doi: 10.1073/pnas.1232504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundra R, Kornfeld S. Asparagine-linked oligosaccharides protect Lamp-1 and Lamp-2 from intracellular proteolysis. J Biol Chem. 1999;274:31039–31046. doi: 10.1074/jbc.274.43.31039. [DOI] [PubMed] [Google Scholar]
- 43.Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 45.Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A. 2009;106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogt G, Chapgier A, Yang K, Chuzhanova N, Feinberg J, Fieschi C, Boisson-Dupuis S, Alcais A, Filipe-Santos O, Bustamante J, de Beaucoudrey L, Al-Mohsen I, Al-Hajjar S, Al-Ghonaium A, Adimi P, Mirsaeidi M, Khalilzadeh S, Rosenzweig S, de la Calle Martin O, Bauer TR, Puck JM, Ochs HD, Furthner D, Engelhorn C, Belohradsky B, Mansouri D, Holland SM, Schreiber RD, Abel L, Cooper DN, Soudais C, Casanova JL. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 47.Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Deprez P, Gautschi M, Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell. 2005;19:183–195. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 49.Hubbard SC, Robbins PW. Synthesis and processing of protein-linked oligosaccharides in vivo. J. Biol. Chem. 1979;254:4568–4576. [PubMed] [Google Scholar]
- 50.Lehrman MA. Oligosaccharide-based information in the endoplasmic reticulum quality control and other biological systems. J. Biol. Chem. 2001;276:8623–8626. doi: 10.1074/jbc.R100002200. [DOI] [PubMed] [Google Scholar]
- 51.Schallus T, Jaeckh C, Feher K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, Pieler T, Muhle-Goll C. Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell. 2008;19:3404–3414. doi: 10.1091/mbc.E08-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barile M, Pisitkun T, Yu MJ, Chou CL, Verbalis MJ, Shen RF, Knepper MA. Large scale protein identification in intracellular aquaporin-2 vesicles from renal inner medullary collecting duct. Mol Cell Proteomics. 2005;4:1095–1106. doi: 10.1074/mcp.M500049-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trombetta ES, Simons JF, Helenius A. Endoplasmic Reticulum Glucosidase II is Composed of a Catalytic Subunit, Conserved from Yeast to Mammals, and a Tightly Bound Non-catalytic HDEL-containing Subunit. J. Biol. Chem. 1996;271:27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson BM, Purswani J, Stirling CJ. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J Biol Chem. 2006;281:6325–6333. doi: 10.1074/jbc.M510455200. [DOI] [PubMed] [Google Scholar]
- 55.Stigliano ID, Caramelo JJ, Labriola CA, Parodi AJ, D'Alessio C. Glucosidase II Beta Subunit Modulates N-Glycan Trimming in Fission Yeasts and Mammals. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trombetta ES, Fleming KG, Helenius A. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry. 2001;40:10717–10722. doi: 10.1021/bi010629u. [DOI] [PubMed] [Google Scholar]
- 57.Kaushal GP, Pastuszak I, Hatanaka K, Elbein AD. Purification to homogeneity and properties of glucosidase II from mung bean seedlings and suspension-cultured soybean cells. J Biol Chem. 1990;265:16271–16279. [PubMed] [Google Scholar]
- 58.Totani K, Ihara Y, Matsuo I, Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J Biol Chem. 2006;281:31502–31508. doi: 10.1074/jbc.M605457200. [DOI] [PubMed] [Google Scholar]
- 59.Petrescu AJ, Butters TD, Reinkensmeier G, Petrescu S, Platt FM, Dwek RA, Wormald MR. The solution NMR structure of glucosylated N-glycans involved in the early stages of glycoprotein biosynthesis and folding. Embo J. 1997;16:4302–4310. doi: 10.1093/emboj/16.14.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackeen MM, Almond A, Deschamps M, Cumpstey I, Fairbanks AJ, Tsang C, Rudd PM, Butters TD, Dwek RA, Wormald MR. The conformational properties of the Glc3Man unit suggest conformational biasing within the chaperone-assisted glycoprotein folding pathway. J Mol Biol. 2009;387:335–347. doi: 10.1016/j.jmb.2009.01.043. [DOI] [PubMed] [Google Scholar]
- 61.Munro S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Cur. Biol. 2001;11:R499–R501. doi: 10.1016/s0960-9822(01)00302-5. [DOI] [PubMed] [Google Scholar]
- 62.Cruciat CM, Hassler C, Niehrs C. The MRH protein Erlectin is a member of the endoplasmic reticulum synexpression group and functions in N-glycan recognition. J Biol Chem. 2006;281:12986–12993. doi: 10.1074/jbc.M511872200. [DOI] [PubMed] [Google Scholar]
- 63.Grinna LS, Robbins PW. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J. Biol. Chem. 1980;255:2255–2258. [PubMed] [Google Scholar]
- 64.Quinn RP, Mahoney SJ, Wilkinson BM, Thornton DJ, Stirling CJ. A novel role for Gtb1p in glucose trimming of N-linked glycans. Glycobiology. 2009 doi: 10.1093/glycob/cwp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe T, Totani K, Matsuo I, Maruyama J, Kitamoto K, Ito Y. Genetic analysis of glucosidase II beta-subunit in trimming of high-mannose-type glycans. Glycobiology. 2009;19:834–840. doi: 10.1093/glycob/cwp061. [DOI] [PubMed] [Google Scholar]
- 66.Alonso JM, Santa-Cecilia A, Calvo P. Glucosidase II from rat liver microsomes. Kinetic model for binding and hydrolysis. Biochem J. 1991;278(Pt 3):721–727. doi: 10.1042/bj2780721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ou WJ, Cameron PH, Thomas DY, Bergeron JJM. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 68.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharides, glucose trimming and calnexin during glycoprotein folding in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 70.Peterson JR, Ora A, Nguyen Van P, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol. Biol. Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 72.Vassilakos A, Cohen-Doyle MF, Peterson PA, Jackson MR, Williams DB. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 1996;15:1495–1506. [PMC free article] [PubMed] [Google Scholar]
- 73.Oliver JD, van, der, Wal, Fj, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 74.Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J. Biol. Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 75.Schrag JD, Bergeron JJM, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Molecular Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 76.Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc. Natl. Acad. Sci. U S A. 2002;99:1954–1959. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kapoor M, Ellgaard L, Gopalakrishnapai J, Schirra C, Gemma E, Oscarson S, Helenius A, Surolia A. Mutational analysis provides molecular insight into the carbohydrate-binding region of calreticulin: pivotal roles of tyrosine-109 and aspartate-135 in carbohydrate recognition. Biochemistry. 2004;43:97–106. doi: 10.1021/bi0355286. [DOI] [PubMed] [Google Scholar]
- 78.Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266:21458–21465. [PubMed] [Google Scholar]
- 79.Tjoelker LW, Seyfried C, Eddy RL, Byers MG. Human, mouse, and rat calnexin cDNA cloning: idetnification of potential calcium bindinh motifs and gene localization to human chromosome 5. Biochemistry. 1994;33:3229–3236. doi: 10.1021/bi00177a013. [DOI] [PubMed] [Google Scholar]
- 80.Molinari M, Eriksson KK, Calanca V, Galli C, Cresswell P, Michalak M, Helenius A. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. doi: 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 81.Chevet E, Wong HN, Gerber D, Cochet C, Fazel A, Cameron PH, Gushue JN, Thomas DY, Bergeron JJ. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. Embo J. 1999;18:3655–3666. doi: 10.1093/emboj/18.13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething M-JH. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 83.Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 84.Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 85.Wada I, Imai S-i, Kai M, Sakane F, Kanoh H. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J. Biol. Chem. 1995;270:20298–20304. doi: 10.1074/jbc.270.35.20298. [DOI] [PubMed] [Google Scholar]
- 86.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Danilczyk UG, Cohen-Doyle MF, Williams DB. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J Biol Chem. 2000;275:13089–13097. doi: 10.1074/jbc.275.17.13089. [DOI] [PubMed] [Google Scholar]
- 88.Elliott JG, Oliver JD, High S. The thiol-dependent reductase ERp57 interacts specifically with N-glycosylated integral membrane proteins. J Biol. Chem. 1997;272:13849–13855. doi: 10.1074/jbc.272.21.13849. [DOI] [PubMed] [Google Scholar]
- 89.Van der Wal FJ, Oliver JD, High S. The transient association of ERp57 with N-glycosylated proteins is regulated by glucose trimming. Eur J Biochem. 1998;256:51–59. doi: 10.1046/j.1432-1327.1998.2560051.x. [DOI] [PubMed] [Google Scholar]
- 90.Peaper DR, Cresswell P. The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc Natl Acad Sci U S A. 2008;105:10477–10482. doi: 10.1073/pnas.0805044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peaper DR, Wearsch PA, Cresswell P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. Embo J. 2005;24:3613–3623. doi: 10.1038/sj.emboj.7600814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. doi: 10.1146/annurev.cellbio.24.110707.175347. [DOI] [PubMed] [Google Scholar]
- 93.Sevier CS, Kaiser CA. Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal. 2006;8:797–811. doi: 10.1089/ars.2006.8.797. [DOI] [PubMed] [Google Scholar]
- 94.Jansens A, van Duijn E, Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 95.Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, Glockshuber R, Ellgaard L. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J. Biol. Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- 96.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. Embo J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jessop CE, Bulleid NJ. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 98.Jessop CE, Chakravarthi S, Garbi N, Hammerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. Embo J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garbi N, Tanaka S, Momburg F, Hammerling GJ. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol. 2006;7:93–102. doi: 10.1038/ni1288. [DOI] [PubMed] [Google Scholar]
- 100.Solda T, Garbi N, Hammerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- 101.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 102.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Labriola C, Cazzulo JJ, Parodi AJ. Retention of glucose units added by the UDP-GLC: glycoprotein glucosyltransferase delays exit of glycoproteins from the endoplasmic reticulum. J Cell Biol. 1995;130:771–779. doi: 10.1083/jcb.130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sousa M, Parodi AJ. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc: glycoprotein glucosyltransferase. EMBO J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- 106.Trombetta ES, Helenius A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol. 2000;148:1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rajagopalan S, Xu Y, Brenner MB. Retention of unassembled components of integral membrane proteins by calnexin. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- 108.Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. Embo J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 109.Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 110.Ritter C, Helenius A. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat. Struct. Biol. 2000;7:278–280. doi: 10.1038/74035. [DOI] [PubMed] [Google Scholar]
- 111.Caramelo JJ, Castro OA, Alonso LG, de Prat-Gay G, Parodi AJ. UDP-Glc: glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc. Natl. Acad. Aca. Sci. USA. 2003;100:86–91. doi: 10.1073/pnas.262661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taylor SC, Thibault P, Tessier DC, Bergeron JJ, Thomas DY. Glycopeptide specificity of the secretory protein folding sensor UDP-glucose glycoprotein:glucosyltransferase. EMBO Rep. 2003;4:405–411. doi: 10.1038/sj.embor.embor797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24:1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J. Biol. Chem. 2004;279:46280–46285. doi: 10.1074/jbc.M408404200. [DOI] [PubMed] [Google Scholar]
- 115.Taylor SC, Ferguson AD, Bergeron JJ, Thomas DY. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 2004;11:128–134. doi: 10.1038/nsmb715. [DOI] [PubMed] [Google Scholar]
- 116.Keith N, Parodi AJ, Caramelo JJ. Glycoprotein tertiary and quaternary structures are monitored by the same quality control mechanism. J Biol Chem. 2005;280:18138–18141. doi: 10.1074/jbc.M501710200. [DOI] [PubMed] [Google Scholar]
- 117.Pearse BR, Gabriel L, Wang N, Hebert DN. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–320. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arnold SM, Kaufman RJ. The noncatalytic portion of human UDP-glucose: glycoprotein glucosyltransferase I confers UDP-glucose binding and transferase function to the catalytic domain. J Biol Chem. 2003;278:43320–43328. doi: 10.1074/jbc.M305800200. [DOI] [PubMed] [Google Scholar]
- 119.Guerin M, Parodi AJ. The UDP-glucose:Glycoprotein glucosyltransferase is organized in at least two tightly bound domains from yeast to mammals. J. Biol. Chem. 2003;278:20540–20546. doi: 10.1074/jbc.M300891200. [DOI] [PubMed] [Google Scholar]
- 120.Trombetta S, Bosch M, Parodi AJ. Glucosylation of glycoproteins by mammlian, plant, fungal and trypanosomatid protozoa microsomal membranes. Biochemistry. 1989;28:8108–8116. doi: 10.1021/bi00446a022. [DOI] [PubMed] [Google Scholar]
- 121.Wiggins CA, Munro S. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci U S A. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tessier DC, Dignard D, Zapun A, Radominska-Pandya A, Parodi AJ, Bergeron JJ, Thomas DY. Cloning and characterization of mammalian UDP-glucose glycoprotein: glucosyltransferase and the development of a specific substrate for this enzyme. Glycobiology. 2000;10:403–412. doi: 10.1093/glycob/10.4.403. [DOI] [PubMed] [Google Scholar]
- 123.Pedersen LC, Dong J, Taniguchi F, Kitagawa H, Krahn JM, Pedersen LG, Sugahara K, Negishi M. Crystal structure of an alpha 1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J Biol Chem. 2003;278:14420–14428. doi: 10.1074/jbc.M210532200. [DOI] [PubMed] [Google Scholar]
- 124.Totani K, Ihara Y, Matsuo I, Tsujimoto T, Ito Y. The Recognition Motif of the Glycoprotein-Folding Sensor Enzyme, UDP-Glc: Glycoprotein Glucosyltransferase. Biochemistry. 2009 doi: 10.1021/bi8020586. [DOI] [PubMed] [Google Scholar]
- 125.Arnold SM, Fessler LI, Fessler JH, Kaufman RJ. Two homologues encoding human UDP-glucose:glycoprotein glucosyltransferase differ in mRNA expression and enzymatic activity. Biochemistry. 2000;39:2149–2163. doi: 10.1021/bi9916473. [DOI] [PubMed] [Google Scholar]
- 126.Zuber C, Fan JY, Guhl B, Parodi A, Fessler JH, Parker C, Roth J. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc Natl Acad Sci U S A. 2001;98:10710–10715. doi: 10.1073/pnas.191359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 128.Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 129.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 130.Labunskyy VM, Ferguson AD, Fomenko DE, Chelliah Y, Hatfield DL, Gladyshev VN. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 2005;280:37839–37845. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- 131.Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ, Tate P, Dill K, Pangilinan E, Wakenight P, Tessier-Lavigne M, Skarnes WC. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 132.Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol Cell. 2005;20:503–512. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 133.Fernandez FS, Trombetta SE, Hellman U, Parodi AJ. Purification to homogeneity of UDP-glucose:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisae. J. Biol. Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- 134.Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Tren. Biochem. Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- 135.Lederkremer GZ, Glickman MH. A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 136.Spiro RG, Zhu Q, Bhoyroo V, Soling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J. Biol. Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- 137.Vassilakos A, Michalak M, Lehrman MA, Williams DB. Oligosaccharide binding characteristics of the molecular chaperone calnexin and calreticulin. Biochemistry. 1998;37:3480–3490. doi: 10.1021/bi972465g. [DOI] [PubMed] [Google Scholar]
- 138.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1?SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tamura T, Cormier JH, Hebert DN. Sweet bays of ERAD. Trends Biochem Sci. 2008;33:298–300. doi: 10.1016/j.tibs.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 140.Cormier JH, Tamura T, Sunryd JC, Hebert DN. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Molecular Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 142.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 143.Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Spear ED, Ng DT. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vanoni O, Paganetti P, Molinari M. Consequences of Individual N-glycan Deletions and of Proteasomal Inhibition on Secretion of Active BACE. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-05-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]