Abstract
In budding yeast, we have found that sister rDNA arrays marked with fluorescent probes can be visualized as two distinguishable strands during metaphase. Upon anaphase, these arm loci are drawn into the spindle, where they adopt a cruciform-like structure and stretch 2.5-fold as they migrate to the poles. Therefore, while sister rDNA arrays appear separated in metaphase, mechanical linkages between sister arm loci persist throughout anaphase in yeast, as shown in grasshopper spermatocytes (Paliulis and Nicklas 2004). These linkages are partially dependent on the protector of cohesin, SGO1. In anaphase, the spatially regulated dissolution of these mechanical linkages serves to prevent premature sister separation and restrain the rate of spindle elongation. Thus, sister separation is temporally controlled and linkages between sister chromatids contribute to the regulation of anaphase spindle elongation.
Introduction
Metaphase chromatid arms are organized along their length into closely juxtaposed yet visibly distinct rods (Paliulis and Nicklas 2004; Nakajima et al. 2007) until anaphase, when they segregate to opposite poles. Though they are visibly distinct, sister chromatid arms are mechanically linked in metaphase. Using microneedles to physically pull sister chromatid arms apart, Paliulis and Nicklas showed that cohesion is released gradually along the length of a chromatid arm during anaphase with sister centromeres being released first and sister telomeres last. Thus, mechanical linkage between sister chromatid arms persists until after anaphase onset (Paliulis and Nicklas 2004), suggesting a spatial regulation of chromatid cohesion dissolution. This feature of the temporal control of sister segregation might aid in chromosome arm segregation or perhaps act as a governor for the rate of spindle elongation.
At the center of sister chromatid cohesion is the four-protein cohesin complex. This complex is modified by several proteins, including SGO1 which resides at the centromeres of meiotic chromosomes in fission yeast (Kitajima et al. 2004). Since its discovery, studies have localized SGO1 to the centromeres/kinetochore of both mitotic and meiotic chromosomes (Katis et al. 2004). Emerging evidence suggests that SGO1 function extends to chromatid arms. Mammalian SGO1 is present along chromosome arms from cells arrested in metaphase (Nakajima et al. 2007). Additionally, chromatids from mammalian cells depleted of SGO1 using RNA interference and subsequently arrested in metaphase display an increase in completely separated sister chromatids (Nakajima et al. 2007). These pieces of evidence suggest that SGO1 plays an essential role in cohesion maintenance along the length of sister chromatids throughout prometaphase and metaphase.
Although morphologically distinct sister chromatid arms have been observed in higher eukaryotes, they have not been observed in budding yeast. Unlike other eukaryotic systems, direct visualization of single chromosomes in live yeast cells is not possible using light microscopy. Yeast chromosome visualization has been limited to the integration of lac operator (Escherichia coli lacO) arrays that are bound by green fluorescent protein (GFP)-tagged lac repressors (lacI). In live cells, these arrays appear as diffraction-limited spots. Unless these arrays are separated by more than 0.25 μm, structural changes in sister chromatid separation may go undetected. Ribosomal DNA (rDNA) in budding yeast represents a unique region of the chromosome arm that can be exploited for visualization using fluorescent probes. Using rDNA and telomere markers, this study dissects the role chromatid organization and cohesion play in spindle mechanics during metaphase and anaphase.
Results
Sister rDNA repeat arrays appear as visibly distinct strands in metaphase
To visualize the structure of mitotic chromosomes, we utilized three probes for the 1.5 Mb repeating array of rDNA on the arm of chromosome XII. Two of the probes were endogenous proteins tagged with a fluorescent protein. One was the CDC14 phosphatase and the other was CDC14's binding partner NET1. The third probe was a lacO array integrated into each repeat of the 35S gene within the rDNA locus. The lacO arrays were visualized with lacI-GFP. Cells containing CDC14-GFP, NET1-GFP, or lacO/lacI-GFP were mated to cells containing a fluorescently tagged core kinetochore component (NUF2-GFP) or spindle pole body protein (SPC29-RFP). Surprisingly, when using CDC14-GFP or NET1-GFP to mark the rDNA array during the first mitosis after mating, rDNA arrays from both mated cells are observed (Fig. 1b; Supplementary Figure 2). This is most likely due to a mobile fraction of CDC14-GFP and NET1-GFP. When using lacO integrations to mark the rDNA locus, only one of the mated cells contains the lacO integrations, and thus, we observe only one pair of fluorescent strands (Fig. 1h). With all probes, rDNA arrays appear as distinct filaments along an axis perpendicular to the mitotic spindle in the first mitosis after mating (Fig. 1b, h, closed arrows; Supplementary Figure 2). Strands were visible before spindle elongation (Fig. 1; Supplementary Figure 2). Fluorescent strands visualized with CDC14-GFP are separated on average by 0.53±0.26 μm, strands marked by NET1-GFP were separated by 0.40±0.12, and strands marked using the lacO integrations are separated by a distance of 0.40±0.07 μm (Table 1; Supplementary Table 1). The length of the CDC14-GFP strand was 1.86± 0.52 μm, NET1-GFP strands were 2.04±0.50, and lacO strands were 1.12±0.22 μm (Table 1; Supplementary Table 1).
Fig. 1.
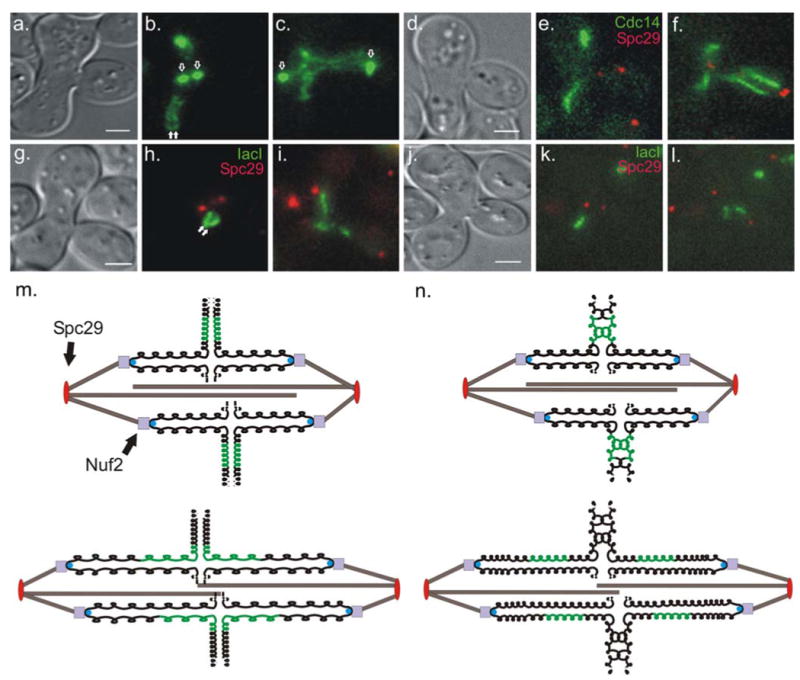
rDNA segregation trajectory in mating cells. Strands of rDNA (labeled with Cdc14-GFP) from both wild-type parental types are distinguishable and lie perpendicular to the mitotic spindle in the first metaphase after mating (b, filled arrows). In anaphase, strands elongate and adopt a cruciform-like structure (c). lac operator (lacO) arrays were integrated in the rDNA region of one parental type. In metaphase these arrays can be seen as two bars lying perpendicular to the spindle (h, filled arrows). In anaphase the bars elongate and adopt a half-cruciform-like structure (i). In mated sgo1Δ cells Cdc14-GFP appears as a single strand during the first metaphase after mating (e). In anaphase, rDNA strands destined for the daughter bud appear to stretch more than those that will stay in the zygote (f), but less than sister rDNA regions in SGO1 zygotes. Using lacO/lacI to visualize the rDNA locus in mated sgo1Δ strains show single strands of fluorescence during metaphase (k). In anaphase, the lacO/lacI-marked rDNA sisters stretch less than sisters in SGO1 zygotes. The kinetochore protein Nuf2-GFP (b and c, unfilled arrows) and the spindle pole body protein Spc29-RFP (e, f, h, i, k, and l, red foci), markers of spindle elongation, were used to distinguish metaphase from anaphase. Scale bars in (a), (d), (g), and (j) represent 2 microns. A schematic of the possible mechanism behind trajectory is shown in (m). In metaphase, the chromosomes (black) are arranged such that rDNA regions (green) from each parental type lie on opposite sides of the spindle (m, top). In anaphase, chromosomes take on a cruciform-like shape during their segregation (m, bottom). Dashed lines represent chromosome arms. (n) schematic of metaphase (top) and anaphase (bottom) of sister rDNA arrays in sgo1Δ zygotes. Sister rDNA arrays do not get organized leading to visually indistinguishable sisters in metaphase. In anaphase, cohesion between sister rDNA arrays is decreased in these zygotes and less stretching occurs
Table 1.
Fluorescent strand dimensions in the first metaphase after mating
| Probe | Genotype | Distance between strands (μm) | % Separated in metaphase | Number | Rest length (metaphase; μm) | Compaction ratio (times B-form) | Number |
|---|---|---|---|---|---|---|---|
| Cdc14-GFP | SGO1 | 0.53±0.26 | 91 | 12 | 1.86±0.52 | 274× | 24 |
| sgo1Δ | – | 7 | 14 | 2.00±0.32 | 255× | 14 | |
| lacO-rDNA | SGO1 | 0.40±0.07 | 100 | 9 | 1.12±0.22 | 455× | 18 |
| sgo1Δ | – | 0 | 11 | 1.59±0.51 | 320× | 11 |
The appearance of rDNA strands prior to anaphase is not unique to the first metaphase after mating. Using lacO arrays integrated at every repeat within the rDNA locus, we have observed this region during vegetative mitosis by inducing expression of lacI fused to GFP. In metaphase, this rDNA probe can appear as strands of fluorescence (Fig. 2b). As the spindle elongates, the rDNA takes on a half-cruciform-like structure (Fig. 2c).
Fig. 2.
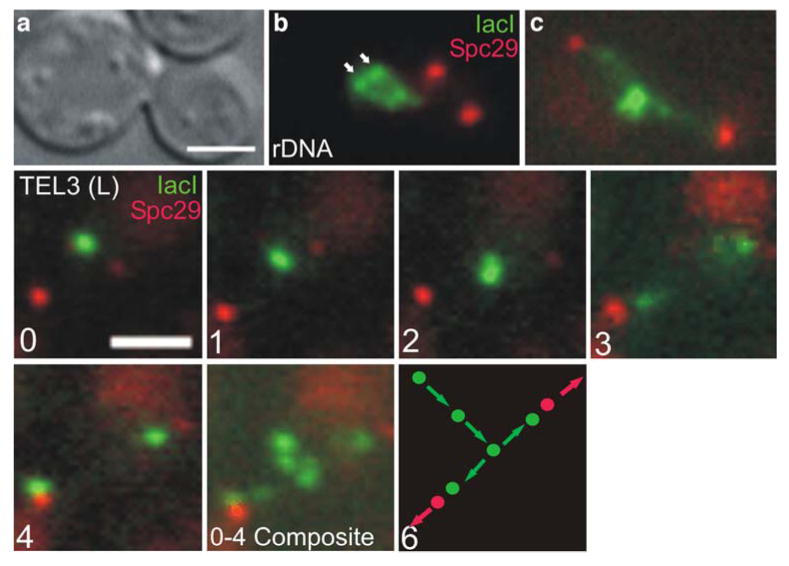
Segregation trajectory in vegetative cells. rDNA was visualized in vegetatively growing cells using lacO arrays integrated into the rDNA region. In metaphase cells, bars can be seen approximately 0.55 μm from the spindle axis (B, arrow heads). The spindle pole protein Spc29-RFP was used to mark spindle length and monitor the progression from metaphase to anaphase. In anaphase, the rDNA elongates and adopts a half-cruciform-like structure (C). Sequential time-lapse images of a lacO array integrated at the telomere of chromosome III are shown in panels 0-4 (images taken at 1 min intervals). Sister lacO arrays appear as single focus 0.8 μm from spindle axis (panel 0) and migrate towards the spindle axis in panels 1-2 as the spindle elongates. Upon reaching the spindle axis, lacO arrays separate (panel 3) each moving toward its respective spindle pole until the pole is reached (panel 4). When panels 0-4 are combined into a composite image, the entire telomere segregation trajectory can be seen (0-4 Composite). A diagram of the segregation trajectory is shown in panel 6. Scale bars in (a) and (0) represent 2 microns
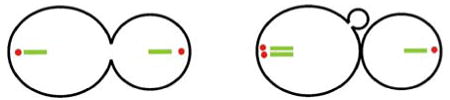
Previous studies have observed that the rDNA array adopts a “loop-like” structure in G2 and metaphase (Guacci et al. 1994; Lavoie et al. 2004; Sullivan et al. 2004; Machin et al. 2005). These studies propose that the sister rDNA arrays are not resolvable prior to anaphase onset, but instead, each sister forms a loop-like structure. If the rDNA array adopts a loop-like structure, one prediction is that the loop would be observable before and after DNA replication. To test this prediction, we have developed an assay to identify the moment of DNA replication within a single cell. Using a strain containing lacO integrations in every repeat of the RDN1 multigene locus, we measured the integrated intensity of lacI-GFP signal at two points in the cell cycle. In a large-budded cell with lacO arrays at the poles, we observed a mother/daughter (bud) ratio of 0.91 (Table 2) indicating that these fluorescent signals represented equal amounts of DNA. After cytokinesis and bud emergence, the mother/daughter ratio increased to 1.74 (Table 2). Thus, the mother cell now contained almost twice the fluorescence (in concordance with the doubling of rDNA content). The loops/strands are only observed following replication.
Table 2.
Mother/daughter integrated fluorescence intensity ratios of lacI-GFP bound to rDNA-imbedded lacO arrays
 | |||||
|---|---|---|---|---|---|
| Genotype | Telophase Mother/Daughter Ratio | Mother G2/Daughter G1 Ratio | Separated Upon Replication | Spindle Length (μm) | n |
| SGO1 | 0.91 +/- 0.18 | 1.74 +/- 0.14 | 63% | 0.85 +/- 0.52 | 11 |
| sgo1Δ | 0.96 +/- 0.22 | 1.75 +/- 0.58 | 31% | 0.80 +/- 0.47 | 16 |
Fluorescence was measured for the rDNA strand in the mother and in the daughter and a ratio of the two was calculated. The ratio was then calculated again following cytokinesis and bud emergence from the original mother cell. Red spots in the graphic represent the spindle poles and the green represents the rDNA locus
To address whether both sister chromatids are organized in a single loop, we have performed a fluorescent bleaching assay. Using a laser pulse, we specifically bleached a single fluorescent strand of NET1-GFP in metaphase (Supplemental Figure 3). After following the zygote through anaphase, the bleached strand was found in only the mother cell or the daughter cell, depending on which strand was bleached. These data suggest that the resolvable strands in metaphase are sister rDNA arrays.
Segregation trajectory and stretching of the rDNA repeat array during anaphase
We have followed DNA segregation through anaphase of the first zygotic division after mating in wild type cells. The most striking characteristic of sister rDNA array segregation is their adoption of a cruciform-like structure in midanaphase (Fig. 1c, i; Supplementary Figure 2c). It is in the cruciform stage that strands become elongated beyond their “rest” length in metaphase. The stretching of the mother-bound sister is slightly less than that of the daughter-bound sister. Based on CDC14-GFP, NET1-GFP, and lacO/lacI-GFP visualization, mother-bound arrays stretched 1.44-, 1.41-, and 2.15-fold their rest length in metaphase, respectively. Daughter strands stretched 2.91-, 2.13-, and 2.09-fold (Table 3; Supplementary Table 2). The segregation trajectory and extension of sister rDNA arrays during anaphase suggests that rDNA strands are mechanically linked in anaphase even though they appear as individual strands in metaphase.
Table 3.
Fluorescent strand dimension in the first anaphase after mating
| Probe | Genotype | Maximum extension (anaphase) | Fold extension | Compaction ratio | Number (of each) | |||
|---|---|---|---|---|---|---|---|---|
| Mother (μm) | Daughter (μm) | Mother (max. extension/rest length) | Daughter (max. extension/rest length) | Mother (times B-form) | Daughter (times B-form) | |||
| Cdc14-GFP | SGO1 | 2.56±0.72 | 5.03±1.06 | 1.44 | 2.91 | 197× | 100× | 12 |
| sgo1Δ | 1.89±0.34 | 3.24±1.02 | 0.96 | 1.58 | 270× | 157× | 14 | |
| lacO-rDNA | SGO1 | 2.48±0.95 | 2.48±1.07 | 2.15 | 2.09 | 205× | 205× | 8 |
| sgo1Δ | 1.55±0.64 | 1.89±0.62 | 0.97 | 1.23 | 329× | 270× | 10 | |
Differences in fold extension between SGO1 and sgo1Δ have p values of 0.01 (CDC14-GFP) and 0.002 (lacO/lacI-GFP)
Sister telomere–proximal lacO arrays follow a segregation trajectory and rapidly migrate to their respective poles in anaphase
To characterize the separation of sister telomeres, we measured the separation kinetics of a telomere–proximal lacO array using Spc29-RFP to track spindle elongation. Prior to spindle elongation, the sister lacO arrays appear as a single diffraction-limited spot displaced from the spindle axis (defined by Spc29-RFP foci) by 0.8 μm. After anaphase onset, as the spindle elongates, the average distance between the spot and the spindle axis decreases but sister lacO arrays cannot be visibly distinguished. Sister lacO arrays first appear as two spots when they are within 0.5 μm of the spindle axis, approximately 100 s after the initiation of spindle elongation. An example of this segregation trajectory is shown in Fig. 2. Images in Fig. 2 0–4 were taken at 1 min intervals. After anaphase onset and seconds prior to their appearance as two spots, sister lacO arrays are not equidistant from the spindle poles. Sister lacO arrays are closer to one of the spindle poles, but there is no bias as to mother or daughter (5/11 fast acquisition movies of anaphase show sisters closer to the mother pole, 6/11 show sisters closer to the daughter pole). These results suggest that sister telomeres are mechanically linked after anaphase onset and is consistent with the observation of TEL-linked lacO arrays initiating pole-ward migration after CEN-linked lacO arrays (Straight et al. 1997). This also indicates that sister telomeres are not separated upon anaphase onset or that they are separated but closer than the limit of resolution in the light microscope. Once juxtaposed on the spindle axis, sister lacO arrays rapidly separate at a rate of 2.96± 1.08 μm/min (Table 4). The rate of telomere separation is approximately five times faster than spindle pole separation (0.58±0.19 μm/min) over the same period. Rapid separation of telomeres suggests that pole-ward migration of the telomeres is not mediated solely by spindle elongation (Table 4; Fig. 3). This also suggests that chromatid arms are under tension during anaphase.
Table 4.
Rate of sister telomere-proximal lacO array separation
| Genotype | Rate of sister TEL separation (μm/min) | Rate of spindle pole separation (μm/min) | Length of spindle at onset (μm) | Number |
|---|---|---|---|---|
| SGO1 | 2.96±1.08 | 0.58±0.19 | 4.07±0.83 | 10 |
| sgo1Δ | 1.91±0.96 | 0.92±0.23 | 3.95±0.50 | 11 |
Statistical analysis reveals significant difference between SGO1 and sgo1Δ in spindle elongation (p=0.001) and sister telomere separation rate (p=0.028)
Fig. 3.
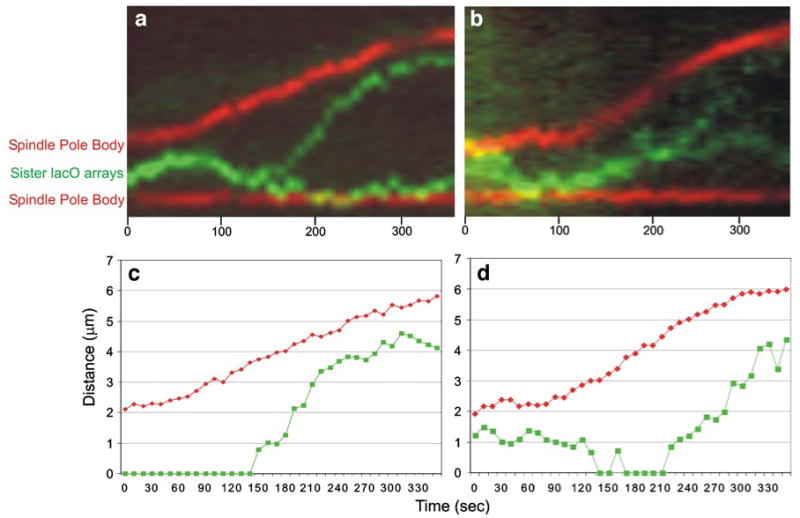
Sister telomere-proximal lacO array separation kinetics. Kymographs showing sister telomere separation (green) and spindle pole body separation (red) are shown in (a) and (b). (a) depicts an SGO1 cell, (b) depicts a sgo1Δ cell. Anaphase is evident from the increasing distance between the spindle poles. In SGO1 cells, telomere III lies between the poles and separates 100 sec after anaphase B initiation. The steeper slope of the green line in (a) relative to the red line depicts the rapid separation of sister telomeres. sgo1Δ mutants show sister telomere separation prior to anaphase onset as shown by the thickness of the green line in (b). Shortly before pole-ward migration, sister telomeres are indistinguishable in this particular sgo1Δ cell. Sister separation in sgo1Δ mutants occurs at a rate 1.5× slower than SGO1 cells. The rate of spindle elongation in sgo1Δ mutants is 1.6× that in SGO1 cells as shown by the steeper slope of the red line in (b). Graphical representations of (a) and (b) are seen in (c) and (d), respectively. The distances between spindle pole bodies (red) and distances between sister lacO arrays (green) were measured over time
Fewer sister rDNA arrays appear as separate strands prior to anaphase in sgo1Δ cells
To investigate the role SGO1 plays in the structure and/or mechanical linking of sister rDNA arrays, we imaged the rDNA locus in zygotes whose haploid parents were null for the SGO1 gene. During metaphase, zygotes in which both parent haploid strains are sgo1Δ display a much different rDNA array structure compared to SGO1 zygotes when probed with CDC14-GFP or rDNA-imbedded lacO arrays marked by lacI-GFP. In metaphase, sister rDNA arrays in the sgo1Δ background appeared as a single strand of fluorescence in 93% of zygotes. The length of these strands is not statistically different from strands seen in SGO1 zygotes (Table 1).
The appearance of one strand could be due to a failure to replicate the rDNA array. To determine whether or not replication occurred, integrated intensity ratios of mother to daughter were used as previously described. As in wild-type cells, an increase in fluorescence intensity is observed in mother cells after bud emergence (Table 2). However, after replication and before metaphase (as tracked using spindle pole separation), only 31% of sister rDNA arrays appeared as separated strands in sgo1Δ cells as compared to 63% of SGO1 cells (Table 2). Therefore, in the absence of SGO1, the ability to visualize distinct strands is reduced. There are two possible explanations for this phenotype: (1) either sister strands are more tightly linked along their length or (2) sister strands are separated, but not organized into two longitudinal rods, i.e., they may be separated but entangled.
Sister rDNA arrays display decreased stretching in sgo1Δ zygotes
To determine whether the observation of single strands of rDNA in sgo1Δ cells is due to more tightly linked sister loci or entangled rDNA arrays, we followed sister rDNA arrays through anaphase in the absence of SGO1. If sister rDNA arrays were more tightly linked in the absence of SGO1, we would predict that the amount of stretch of sister rDNA arrays would increase compared to wild-type zygotes due to an increase in the amount of tension on the rDNA array. However, if the sister rDNA arrays were separated but entangled, we would predict a decrease in stretching compared to wild-type zygotes. Visualization of the rDNA arrays was accomplished using both CDC14-GFP- and rDNA-embedded lacO arrays bound by lacI-GFP. During anaphase in sgo1Δ zygotes, sister rDNA arrays adopt a cruciform-like structure. Compared to wild-type sister rDNA array separation, there is a decrease in the extent which sisters are stretched. Mother-bound strands in sgo1Δ cells were 0.96 and 0.97 (visualized by CDC14-GFP and lacO/lacI, respectively) times their length in metaphase while daughter-bound rDNA arrays extended 1.58- and 1.23-fold their metaphase lengths (Table 3). Based on these data, we conclude that there are SGO1-dependent linkages between sister rDNA arrays. This is consistent with previous data that have shown that sister chromatid linkage is lost in SGO1-depleted cells (Nakajima et al. 2007). Since sister rDNA arrays are predominantly unlinked, we conclude that the sister arrays appear as a single strand in metaphase in the absence of SGO1 because they are entangled. The decrease in organization and mechanical linkage in sgo1Δ cells could lead to the increase in nondisjunction also associated with SGO1 deletion (Kitajima et al. 2004).
Sister telomere–proximal lacO arrays exhibit a reduced rate of separation in sgo1Δ cells
To investigate whether SGO1 plays a role along the entire length of a chromosome arm, we deleted SGO1 in cells containing a telomere–proximal lacO array and Spc29-RFP to mark the spindle poles. When spindle poles were approximately 1.5–2 μm apart (metaphase), sister lacO arrays appeared as two spots, evidence that sister telomere cohesion had been perturbed. In 36% (four of 11) of sgo1Δ cells, sister telomeres did appear as one spot for more than one frame of the time lapse. This appeared to be a stochastic event, with no correlation to spindle length or the timing of anaphase onset. During anaphase, sister telomere separation is approximately 1.5 times slower in sgo1Δ cells compared to SGO1 cells (Table 4). We conclude that SGO1 plays a role in mechanically linking sister telomeres during metaphase and anaphase. This result also suggests that although we cannot visibly distinguish sister rDNA strands in a sgo1Δ background during metaphase, individual sister loci within the rDNA are likely to be separated. The failure to visualize the separation between sister rDNA arrays may be due to geometry, that is, sister loci of the rDNA array are not organized longitudinally into rods.
If the rapid separation of sister telomeres is dependent on mechanical links between sister telomeres, then abolishing all sister–sister links should abolish the rapid movement of the telomeres. To test this, we observed the dynamics of an unreplicated telomere during spindle elongation. This was achieved by placing CDC6 under the control of the GAL promoter. CDC6 is an essential member of the prereplicative complex responsible for initiating replication. Cells depleted of CDC6 manage to elongate their spindles, randomly segregating their unreplicated chromosomes (Piatti et al. 1995). To directly compare the dynamics of replicated telomeres to unreplicated telomeres, lacO spot to pole movement was measured over time. This measurement was called the recoil rate. In wild-type cells, the average recoil rate was 3.52 μm/min. Telomeres in cells depleted of CDC6 show no quantitable recoil (Fig. 4e; Table 5). A lacO spot representing an unreplicated telomere appeared to associate with the mother or daughter spindle pole, not moving more that 1.5 μm from it (Fig. 4e). Based on these dynamics, we conclude that the mechanical links between sister telomeres are required for rapid telomere-to-pole movement.
Fig. 4.
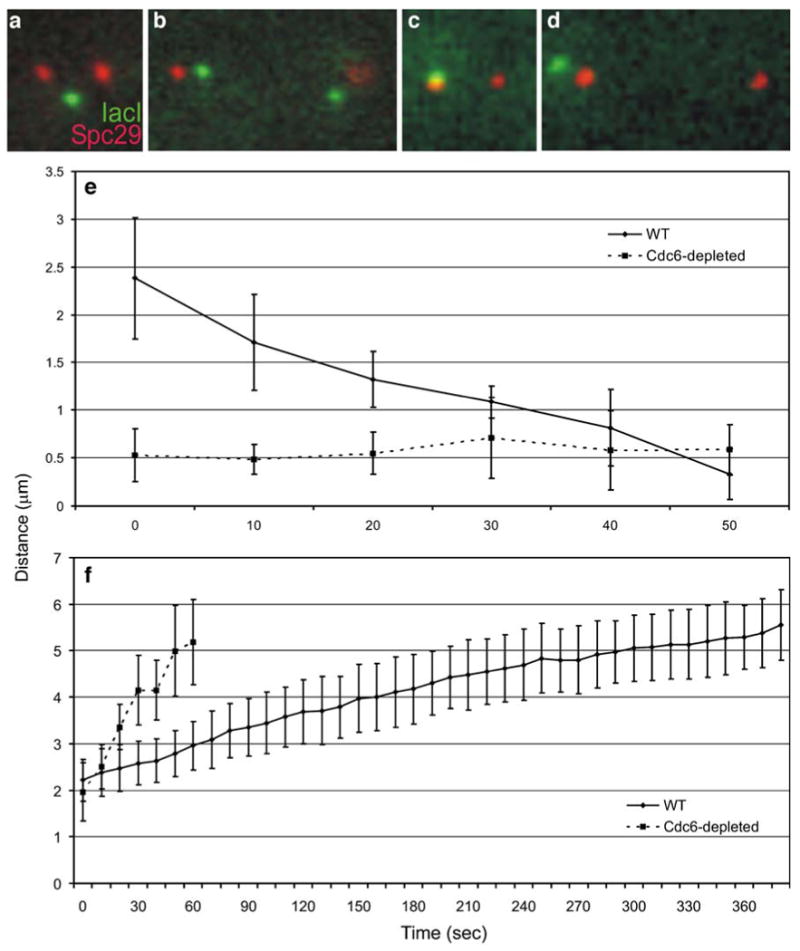
Spindle and telomere-proximal lacO array kinetics in cells with replicated and unreplicated chromosomes. (a) and (b) are images of a telomere-proximal lacO (green) in metaphase and anaphase, respectively (Spc29 shown in red). (c) and (d) are metaphase and anaphase images of the same lacO array in a cell depleted of Cdc6. (e) Solid line represents eleven recoil events in cells with replicated chromosomes. The distance between the lacO spot and spindle pole decreases rapidly over time. Dotted line represents lacO spot to spindle pole distance in cells with unreplicated chromosomes. Tracks of unreplicated lacO array to pole distances taken while spindle is elongating as in cells with replicated chromosomes. Unreplicated telomeres show no rapid movement towards either pole. (f) Composite graph of spindle elongation from cells with replicated chromosomes (solid line). Dotted line is a composite graph of spindle elongation in cells with unreplicated chromosomes. Spindle elongation rate is greater in these cells than both wild type and sgo1Δ cells (Tables 4 and 5)
Table 5.
Rate of telomere–proximal lacO array recoil and spindle elongation in cells with replicated and unreplicated chromosomes
| Genotype | Carbon source | Rate of recoil (μm/min) | Rate of spindle pole separation (μm/min) | Number |
|---|---|---|---|---|
| CDC6 | Glucose | 3.52±2.26 | 0.58±0.19 | 11 |
| GAL-Ub-CDC6 | Glucose | – | 3.46±1.38 | 10 |
Difference in spindle elongation rates between these two types of cells is statistically significant (p<0.001)
Mechanical linkage between sister chromatids restrains spindle elongation
The stretching observed during rDNA array segregation in anaphase suggests that the chromatid arm is under tension. This tension indicates that the mechanical links between sister chromatid arms oppose the outward force of the spindle. To examine the effect of sister chromatid arm cohesion on spindle elongation, we quantitated and compared the average spindle elongation rate of wild-type, sgo1Δ, and CDC6-depleted cells. We found that the rate of spindle elongation during anaphase in sgo1Δ cells is approximately 1.6 times that in wild type (0.92±0.23 versus 0.58±0.19 μm/min). In the complete absence of sister–sister links (following CDC6 depletion), spindles elongate to greater than 5 μm in length at an average rate of 3.46 μm/min (Fig. 4f; Table 5). This rate is approximately six times faster than the fast phase of wild-type spindle elongation and almost four times faster than sgo1Δ spindle elongation (Tables 4 and 5). Additionally, we do not see the stereotypical two phases of spindle elongation (Pearson et al. 2001) in sgo1Δ or CDC6-depleted cells. We conclude that the spatially regulated release of intersister links acts as a governor, controlling the rate of spindle elongation. Furthermore, a subset of the intersister links depends on SGO1.
Discussion
Visibly distinct sister chromatid arms have been observed in many eukaryotes prior to anaphase onset. Termed individualization in metazoans, sister chromatid arm separation prior to anaphase onset is a result of the removal of cohesin in prophase (Waizenegger et al. 2000; Sumara et al. 2000, 2002; Gimenez-Abian et al. 2004). Despite the appearance of separated sister chromatid arms, there remain physical linkages throughout anaphase (Paliulis and Nicklas 2004). The data herein demonstrate that yeast sister rDNA arrays, loci located on chromatid arms, can also be visibly distinguished, but are mechanically linked in vivo. These residual linkages are partially dependent upon the protector of cohesin, SGO1, and serve a mechanical role in anaphase spindle elongation.
The ribosomal DNA locus is organized into visible rods following replication in budding yeast (Figs. 1 and 5). These rods are 1–2 μm in length in metaphase, reflecting a 300–500-fold compaction of B-form rDNA (Table 1). The strands stretch 2.5 times their metaphase length as they reach the anaphase spindle and segregate to opposite poles. While resolvable sister loci are not observed outside rDNA prior to anaphase onset, a similar cruciform-like segregation trajectory of chromosome arm loci is observed as well as rapid recoil of these loci to the poles. Thus, both rDNA and chromosome arms are under tension as they approach the anaphase spindle, indicative of persistent mechanical linkages. These linkages are partially dependent upon the “protector” of cohesin, SGO1. The rDNA loci no longer appear as two visible rods in the absence of SGO1. The inability to observe two strands could result from the lack of separation or from twists and/or entanglements between sister rDNA arrays (Fig. 5). The observation that sister telomeres appear as two distinct spots in sgo1Δ mutants indicates that these sister loci are separated in these mutants. Consistent with this, sister telomeres exhibit reduced recoil during anaphase in sgo1Δ cells. Thus, SGO1 not only plays a role in sister chromatid cohesion but also provides a scaffold for organizing sister chromatid arms into separated rods prior to anaphase.
Fig. 5.
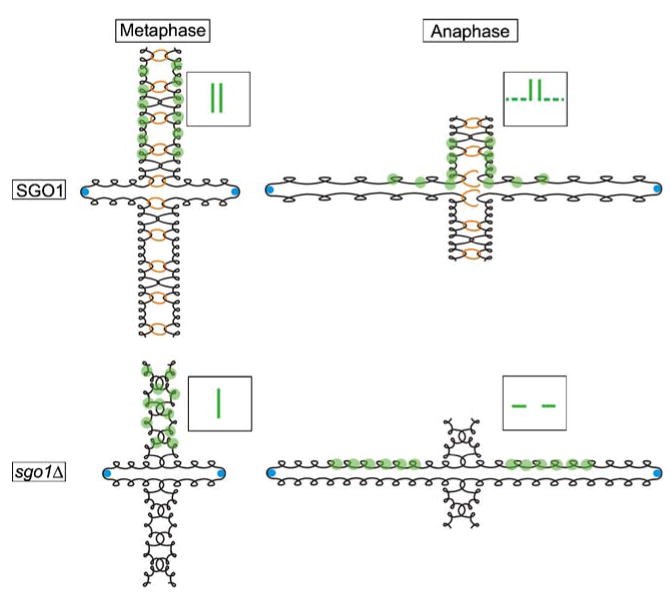
Chromosome morphology throughout mitosis. In the presence of SGO1, sister chromatid arms (black) are arranged as morphologically distinct and mechanically linked rods in metaphase. Blue circles represent the Cse4-containing nucleosome at the centromere. When bound by GFP (green circles), the chromatid arms appear as separated strands (SGO1/Metaphase, inset box). In anaphase, the elongating spindle applies an outward force on sister chromatids. SGO1-dependent links (orange rings) along with SGO1-independent links (depicted as DNA catenations) between sister chromatids participate in opposing the outward force of the spindle. The opposing forces introduce tension on the chromatid arm (SGO1/Anaphase, dashed line in inset box). The spatially regulated release of mechanical links between sister chromatid arms also causes them to adopt a cruciform-like structure during anaphase. In the absence of SGO1, sister chromatids fail to organize into morphologically distinct rods. When visualized using GFP, this unorganized state of sister chromatid arms appears as a single strand (sgo1Δ/Metaphase, inset box). During anaphase in sgo1Δ cells, a subset of mechanical links between sister chromatid arms are perturbed. This lessens the inward force opposing spindle elongation and sister chromatid arms are put under less tension
As the spindle elongates, mechanically linked sister arm loci follow a stereotypic trajectory to the spindle axis whereupon the chromatin stretches approximately 2.5-fold. Thus, residual linkages are retained until the arm becomes proximal to the spindle axis (Fig. 5). At this time, the linkages are removed, as evidence by the rapid recoil (greater than the rate of spindle elongation) of sister telomeres to the spindle poles. This step-wise spatially regulated removal of sister chromatid cohesion results in tension on chromatid arms. When the last mechanical link between sister chromatid arms is removed at the telomere, tension is relieved and the elastic properties of chromatin mediate a rapid migration of sister telomeres to their respective poles. The spatial regulation of sister chromatid arm cohesion dissolution is likely to contribute to the mechanisms that prevent entanglement of chromosome arms.
In addition to the stretching of chromosomes in anaphase, we have found that loss of SGO1-dependent mechanical linkages between sister chromatid arms leads to an increased rate of spindle elongation. In sgo1Δ mutants, the anaphase spindle elongates approximately two times the rate of wild-type spindle elongation. In cells containing unreplicated chromosomes, spindles elongate three to four times faster than sgo1Δ cells and six- to eightfold faster than wild-type cells. Taken together, these data suggest that the elongating (anaphase) spindle produces an outward force on sister chromatids that is opposed by mechanical links between sister chromatids. The SGO1-dependent linkages that are present after anaphase onset contribute to a restraining force that controls the spindle elongation. Also, the difference between the rate of spindle elongation in a sgo1Δ mutant versus a CDC6-depleted cell is strong evidence for the existence of SGO1-independent linkages between sister chromatids, a matter discussed below.
Cohesin is present at the rDNA locus (Laloraya et al. 2000) and is responsible for sister telomere cohesion (Antoniacci and Skibbens 2006). Additionally, cohesin can exchange between chromatin-bound and chromatin-unbound states (Ocampo-Hafalla et al. 2007). SGO1 has been shown to be a “protector” of cohesin at centromeres in meiosis and mitosis. Thus, it is possible that SGO1 functions to stabilize and/or protect the bound form of cohesin at the rDNA array and telomeres during mitosis. If SGO1 does function through the cohesin complex, then removal of SGO1-dependent linkages would depend on separase. Interestingly, separase localizes to the spindle axis during anaphase (Jensen et al. 2001). This would explain why sister arm loci are observed to juxtapose to the spindle axis prior to pole-ward migration. In zygotes, it is possible that separase localization along the spindle axis is biased toward the mother, which would contribute to an increase in daughter-bound rDNA array stretching compared to their mother-bound sisters.
Though SGO1-dependent linkages do contribute to sister rDNA array stretching and resisting spindle elongation in anaphase, we do find evidence that suggests that SGO1-independent mechanical linkages between sister chromatid arms also exist. In sgo1Δ mutants, daughter-bound sister rDNA arrays still show a mild stretching phenotype (Table 3). Also, though sgo1Δ mutants exhibit an increase in rate of spindle elongation presumably by disrupting a subset of mechanical links between sister chromatids, completely abolishing all mechanical links between sisters via CDC6 depletion increases the rate of spindle elongation to four times that in sgo1Δ mutants. These two observations are consistent with previous studies which have shown that intersister chromatid DNA catenations exist from replication to anaphase onset (DiNardo et al. 1984; Holm et al. 1988, Gimenez-Abian et al. 2002). Other studies have found that intersister catenations at the rDNA array are introduced by RNA polymerase I (Tomson et al. 2006). Some of these interchromatid links have been shown to be resolved by topoisomerase II in a process independent of the separase inhibitor PDS1 (Andrews et al. 2006). Cohesin-independent linkages at the rDNA array have been shown to be resolved by a CDC14-dependent localization of condensin (D'Amours et al. 2004; Sullivan et al. 2004; Wang et al. 2004). Thus, both cohesin-dependent and cohesin-independent mechanisms serve to mechanically link sister chromatids until anaphase onset.
Based on these observations, we put forth the model depicted in Fig. 5. Immediately after replication, sister chromatids are not resolved in the light microscope. During prometaphase and metaphase, sister arms are organized and can appear separated. SGO1 is responsible for protecting a subset of mechanical linkages at arm loci, possibly through cohesin. Based on the observations of rDNA herein and sister chromatid arms in mammalian cells, SGO1 may also contribute to a physical scaffold that separates and aligns chromosome arms into two distinct structures prior to anaphase onset. This scaffold functions to both organize and mechanically link sister chromatid arms. The function of such a scaffold may be critical in preventing the arm entanglement that would occur if all linkages between sister chromatids were simultaneously severed. The residual mechanical linkages provide a mechanism to prevent sister chromatid entanglements and to restrain spindle elongation. Thus, the temporal and spatial regulation of sister chromatid separation impacts both chromosome organization and spindle mechanics to ensure the fidelity of chromosome segregation in mitosis.
Materials and methods
The Saccharomyces cerevisiae strains used in these experiments are listed in Table 6. Strains with stable integrations were maintained in YPD (2% glucose, 2% peptone, and 1% yeast extract) at 32°C.
Table 6.
Strains table
| Strain Name | Relevant Genotype | Source or Reference |
|---|---|---|
| DCB190 | Mat a trp1Δ63, leu2Δ1, ura3-52, his3-Δ200, lys2-801, Cdc14-GFP∷KANr | This study |
| DCB194 | Mat a trp1Δ63, leu2Δ1, ura3-52, his3-Δ200, lys2-801, Cdc14-GFP∷KANr, sgo1∷NATr | This study |
| KBY9361 | Mat α trp1Δ63, leu2Δ1, ura3-52, his3-Δ200, lys2-801, Nuf2-GFP∷URA3 | This study |
| MH3341 | Mat a ura3-52, leu2Δ1, his3Δ-200, trp1-63, lys2-301, ade2-101, rDNA-5×LacO, ura3-52∷ pGalL-GFPLacI∷URA3 | Margaret Hoang/Douglas Koshland |
| MH3342 | Mat a ura3-52, leu2Δ1, his3Δ-200, trp1-63, lys2-301, ade2-101, rDNA-5×LacO, ura3-52∷ pGalL-GFPLacI∷URA3, Spc29-RFP∷Hbr | Margaret Hoang/Douglas Koshland and this study |
| MH3344 | Mat a ura3-52, leu2Δ1, his3Δ-200, trp1-63, lys2-301, ade2-101, rDNA-5×LacO, ura3-52∷ pGalL-GFPLacI∷URA3, sgo1∷NATr | Margaret Hoang/Douglas Koshland and this study |
| MH3346 | Mat a ura3-52, leu2Δ1, his3Δ-200, trp1-63, lys2-301, ade2-101, rDNA-5×LacO, ura3-52∷ pGalL-GFPLacI∷URA3, Spc29-RFP∷Hbr, sgo1∷NATr | Margaret Hoang/Douglas Koshland and this study |
| BDH2008 | Mat α trp1Δ63, leu2Δ1, ura3-52, his3-Δ200, lys2-801, Spc29-RFP∷Hbr | This study |
| BDH2009 | Mat α trp1Δ63, leu2Δ1, ura3-52, his3-Δ200, lys2-801, Spc29-RFP∷Hbr, sgo1∷NATr | This study |
| EYY1131 | Mat a ade1, met14, ura3-52, leu2-3,112, lys2delta∷lacI-GFP-NLS∷NATr, his3-11,15 TEL3∷ lacO∷LEU2 (pAFS102), SPC29-RFP∷Hbr, pRS313 | This study |
| EYY1152 | Mat a ade1, met14, ura3-52, leu2-3,112, lys2delta∷lacI-GFP-NLS-NATr, his3-11,15 TEL3∷ lacO∷LEU2 (pAFS102), SPC29-RFP∷Hbr, nat∷KANr, sgo1∷NATr, pRS313 | This study |
| EYY1171 | Mat a ade1, met14, ura3-52, leu2-3,112, lys2delta∷lacI-GFP-NLS-NATr, his3-11,15 TEL3∷ lacO∷LEU2 (pAFS102), LEU2∷KAN, pGAL-UB-CDC6∷LEU2, SPC29-RFP∷Hbr,r, pRS313 | This study |
Strain MH3341 was created by transforming YPH499 with p5lacOTtLSU and pCPIPpo (Lin and Vogt 1988) and plated on −URA −HIS +2% glucose to select for plasmids. Colonies were restruck on −URA −HIS +2% galactose to induce I-PpoI endonuclease to cleave within each rDNA repeat. Gal-resistant colonies were screened for gene conversion off p5lacOTtLSU by colony polymerase chain reaction (PCR). Southern blot analysis verified 5lacO construct within all rDNA repeats. Strains were cured of both plasmids by nonselective growth. Lastly, strains were transformed with linearized pMH4, selected on −ura +2% glucose and subsequently verified by Southern to target pGal-GFP-lacI (lacI dimerization form) to ura3–52 locus. p5lacOTtLSU was constructed in two steps: (1) PCR off pCM40 to produce 296-bp fragment with five lacO binding sites with ClaI linkers and (2) cloned PCR fragment into unique ClaI site of pRSTtLSU (Lin and Vogt 1998).
To induce lacI-GFP-NLS, cells were resuspended in synthetic media containing glucose (0.67% yeast nitrogen base, 2% glucose, and appropriate amino acids) and lacking histidine (SD-HIS) for ∼2 h before adding 20 mM 3-aminotriazole (Sigma-Aldrich) for 1 h. Induction steps were performed at 32°C. All cells were grown to midlogarithmic phase before preparation for imaging.
Depletion of CDC6 was achieved by arresting an asynchronous culture growing on SG-His using 0.2 μm/ ml nocodazole. After 1 h in SG-His plus nocodazole, cells were collected, washed, and resuspended in SD-His plus nocodazole. After 1 h, nocodazole was washed out and cells were resuspended in fresh SD-His. Cells were imaged 2 h after release. All growth carried out at 32°C.
MATa and MATα cells were grown early to midexponential phase in YPD at 32°C. Five hundred microliters of cells from each mating type was mixed, transferred to a 1-ml syringe, and collected on a 13-mm 0.45-μm membrane (Millipore, Billerica, MA, USA). In matings that required induction of pGalL-GFPlacI, all media postcollection contained galactose. Media used for other matings contained glucose. The membrane was placed on a 60×15-mm YPD plate with collected cells facing the plate. Cells were allowed to mate for 120–180 min at 32°C before imaging. Cells were liberated from the membrane by placing membrane in a 1.7-ml Eppendorf tube containing 100 μl of yeast complete media and vortexing.
Live-cell images of cells requiring induction of pGalL-GFPlacI were collected using cells immobilized on 25% gelatin slabs containing 2% galactose. All other cells were imaged on 25% gelatin slabs containing 2% glucose. Image acquisition was carried out using a TE2000 microscope (Nikon, East Rutherford, NJ, USA) with a 1.4 N.A. ×100 differential interference contrast (DIC) oil immersion lens. Images were acquired with an ORCA II ER CCD camera. MetaMorph 4.6 software (Molecular Devices, Downington, PA, USA) controlled the microscope as it executed an acquisition protocol taking five fluorescence images every minute at 0.5-μm axial steps and a single DIC image corresponding to the central fluorescence image. Exposure times ranged from 300–400 ms. For fast acquisition of telomere–proximal spot separation, single plane DIC and fluorescent images were taken at 10 s intervals using 300– 400-ms exposure times.
Bleaching of NET-GFP was done using a Spectra Physics Advantage 163C Air-cooled Ion Laser. Three 50-ms pulses of a focused beam were used to bleach a region of the fluorescent strand. The beam was filtered to allow only the 488 nm wavelength light to pass through to the sample. Immediately before and immediately following laser exposure, five plane fluorescence Z-series were taken to confirm the bleaching event.
Distances were measured using Measure Pixel tool in MetaMorph 4.6 software. To correct for random errors, each frame stack analysis was repeated three times. Data sets were exported into Microsoft Excel™ (Microsoft, Richmond, WA, USA) for analysis. Rapid telomere separation was defined as at least three consecutive time points of spot separation during which the distance between spots increased. Rapid separation ended with the first time point of decreased distance between spots or a sixfold decrease in rate. Linear regression plots were drawn through data points and the slopes were used to determine rates of separation. The average for all observed separation events was calculated. Distance between rDNA strands was determined using the Linescan tool in Metamorph 4.6. Linescans were drawn perpendicular to the rDNA filaments allowing for the identification of two fluorescence peaks. The distance between the peaks was then calculated.
The kymograph was created using MetaMorph 4.6 software. It was used to project all data points for an entire collected sequence of anaphase movements at 10 s intervals. Prior to kymograph creation, the spindle axis was aligned to a horizontal axis in each image of the time lapse image series using MATLAB-based (The Mathworks, Inc., Natick, MA, USA) software developed in house by Ajit Joglekar. A region 5 pixels wide was drawn through the long axis of the mitotic spindle and the brightest pixel was recorded on a single line. This was repeated for each time point and was displayed along the X-axis to show the entire time course. Schematics of chromosome segregation were created using CorelDRAW 11.
Supplementary Material
Acknowledgments
We thank Irem Unlu and Sena Özşeker for their hard work imaging rDNA-imbedded lacO arrays. We thank Jeff Molk, Julian Haase, Elaine Yeh, Gidi Shemer, Dr. Doug Koshland, and members of the Bloom laboratory for advice, assistance, and critical readings of the manuscript. This work was funded by the National Institute of Health RO1 Grant GM32238 (K. Bloom).
Footnotes
Communicated by F. Uhlmann
Electronic supplementary material The online version of this article (doi:10.1007/s00412-009-0224-6) contains supplementary material, which is available to authorized users.
Contributor Information
Benjamin D. Harrison, Department of Biology, University of North Carolina at Chapel Hill, CB# 3280, Coker Hall, Chapel Hill, NC 27599-3280, USA
Margaret L. Hoang, Department of Embryology, Carnegie Institution, Baltimore, MD 21218, USA
Kerry Bloom, Department of Biology, University of North Carolina at Chapel Hill, CB# 3280, Coker Hall, Chapel Hill, NC 27599-3280, USA, kerry_bloom@unc.edu.
References
- Andrews C, Vas A, Meier B, Gimenez-Abian JF, Diaz-Martin L, Green J, Erickson S, VanderWaal K, Hsu WS, Clarke D. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/ securin. Genes Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L, Skibbens R. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr Biol. 2006;16:902–906. doi: 10.1016/j.cub.2006.03.060. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. PNAS. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Clarke D, Gimenez-Martin G, Weingartner M, Gimenez-Abian MI, Carballo J, Diaz de la Espina S, Borge L, de la Torre C. DNA catenation that link sister chromatids until the onset of anaphase are maintained by a checkpoint mechanism. EJCB. 2002;81:9–16. doi: 10.1078/0171-9335-00226. [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. The Journal of Cell Biology. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1988;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Segal M, Clarke DJ, Reed SI. A novel role of the budding yeast Separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol. 2001;151:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis V, Galova M, Rabitsch K, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Vogt V. I-PpoI, the endonuclease encoded by the group I intron PpLSU3, is expressed from an RNA polymerase I transcript. Mol Cell Biol. 1998;18:5809–5817. doi: 10.1128/mcb.18.10.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F, Torres-Rosell J, Jarmuz A, Aragon L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci. 2007;120:4188–4196. doi: 10.1242/jcs.011528. [DOI] [PubMed] [Google Scholar]
- Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliulis LV, Nicklas RB. Micromanipulation of chromosomes reveals that cohesion release during cell division is gradual and does not require tension. Curr Biol. 2004;14:2124–2129. doi: 10.1016/j.cub.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Higuchi T, Katis V, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Geiffers C, Peters B, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Norbert R, Nigg E, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Tomson BN, D'Amours D, Adamson BS, Aragon L, Amon A. Ribosomal DNA transcription-dependent processes interfere with chromosome segregation. Mol Cell Biol. 2006;26:6239–6247. doi: 10.1128/MCB.00693-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wang BD, Yong-Gonzalez V, Strunnikov A. Cdc14/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


