Abstract
Eukaryotic endocytosis involves multivesicular bodies formation, which is driven by endosomal sorting complexes required for transport (ESCRT). Here, we showed the presence and expression of homologous ESCRT genes in Entamoeba histolytica. We cloned and expressed the Ehvps4 gene, an ESCRT member, to obtain the recombinant EhVps4 and generate specific antibodies, which immunodetected EhVps4 in cytoplasm of trophozoites. Bioinformatics and biochemical studies evidenced that rEhVps4 is an ATPase, whose activity depends on the conserved E211 residue. Next, we generated trophozoites overexpressing EhVps4 and mutant EhVps4-E211Q FLAG-tagged proteins. The EhVps4-FLAG was located in cytosol and at plasma membrane, whereas the EhVps4-E211Q-FLAG was detected as abundant cytoplasmic dots in trophozoites. Erythrophagocytosis, cytopathic activity, and hepatic damage in hamsters were not improved in trophozoites overexpressing EhVps4-FLAG. In contrast, EhVps4-E211Q-FLAG protein overexpression impaired these properties. The localization of EhVps4-FLAG around ingested erythrocytes, together with our previous results, strengthens the role for EhVps4 in E. histolytica phagocytosis and virulence.
1. Introduction
Entamoeba histolytica is the enteric protozoan parasite responsible for human amoebiasis that affects 50 million people around the world, causing colitis and liver abscesses [1]. In this organism, phagocytosis and vesicular trafficking play a critical role in ingestion and degradation of host cells and microorganisms. Vacuolar protein sorting (Vps) factors and some proteins that participate in vesicle transport in eukaryotes have been identified in E. histolytica [2–4].
In eukaryotic cells, endocytosis consists in phagocytosis, micropinocytosis and pinocytosis. Particularly, phagocytosis involves the ingestion of particles of varying size into phagosomes, which sequentially fuse with early and late endosomes forming multivesicular bodies (MVB), as well as with lysosomes to form phagolysosomes [5]. Additionally, MVB are critical for cell receptors down-regulation, retroviral budding and other processes [6–8]. MVB formation is driven by the assembly of endosomal sorting complexes required for transport (ESCRT), which result from the interaction of different class E Vps proteins [9, 10].
The MVB-sorting process initiates with the association of Vps27 and Hse1 proteins to form the ESCRT-0 complex, which is targeted to endosomal membrane domains that bind ubiquitinated cargo proteins (reviewed in [6, 7]). Then, ESCRT-0 recruits ESCRT-I formed by Vps23, Vps28, and Vps37 proteins, as well as an additional subunit called Mvb12 [11, 12]. Later, ESCRT-II, composed by Vps22, Vps25, and Vps36 proteins, is activated by ESCRT-I [13]. Ubiquitinated cargo proteins are recognized by ESCRT-I via Vps23 and by ESCRT-II via Vps36. Then, ESCRT-III, formed by Vps20, Vps32, Vps2, and Vps24, interacts with ESCRT-II components to complete ESCRT formation [14]. ESCRT-III is required for cargo molecules concentration into MVB vesicles and it coordinates the association of Bro1 protein and Doa4-deubiquitinating enzyme [15–17]. Finally, Vps4 protein catalyzes the ATP-dependent dissociation of ESCRT complexes from endosomes to initiate new rounds of vesicle formation and cargo molecules transport [6, 7]. Particularly, it has been reported that substitution of the conserved E amino acid residue by a Q residue in the Vps4 ATPase motif impairs ATP hydrolysis activity [18, 19], resulting in an inefficient protein transport from endosomal compartments to vacuoles or lysosomes [20–22], which evidenced the critical role of Vps4 in protein transport and vesicle trafficking.
In E. histolytica, experimental evidence suggests that MVB-like structures are formed by fusion of inward budding membranes with phagosomes [23]. Ubiquitin, deubiquitinating enzymes and a SNF7 homologue (vesicular trafficking protein) have been identified in isolated phagosomes by proteomic analysis, suggesting that a mechanism similar to MVB formation could be present in E. histolytica [23–25]. Additionally, the Bro1 domain-containing EhADH112 protein, which forms part of the E. histolytica EhCPADH complex involved in parasite virulence, is located in MVB-like structures in trophozoites [26, 27]. However, molecular mechanisms regulating MVB formation in E. histolytica remain poorly understood. Here, by in silico analysis from parasite genome databases, we identified 20 ESCRT protein-encoding genes in E. histolytica and showed that most of them were transcribed in trophozoites. Since Vps4 has been described as a key molecule to complete the disassembly of ESCRT and associated factors in other systems, we initiated the study of ESCRT machinery in E. histolytica by cloning and characterizing the EhVps4 protein. Biochemical assays showed that EhVps4 exhibits ATPase activity in vitro. Interestingly, by using trophozoites overexpressing the wild type and a mutant version of EhVps4, we provide data supporting a role for this protein in phagocytosis and virulence.
2. Material and Methods
2.1. In Silico Identification of Putative ESCRT Genes in E. histolytica
Sequence similarity searches for ESCRT genes were performed in E. histolytica genome database (http://pathema.jcvi.org/cgi-bin/Entamoeba/PathemaHomePage.cgi) by BLAST using human and yeast ESCRT protein sequences as queries. Putative E. histolytica ESCRT homologous proteins were selected using the following criteria: (i) at least 20% identity and 35% similarity to the query sequence; (ii) e-value lower than 0.002; and (iii) absence of stop codons in the coding sequence. Predicted amino acids sequences were aligned by ClustalW software (http://www.ebi.ac.uk/clustalw/). Functional and structural domains were predicted using PROSITE (http://www.expasy.org/tools/scanPROSITE/) and Pfam (http://www.sanger.ac.uk/Software/Pfam/) databases. Phylogenetic relationships among putative ESCRT proteins from E. histolytica and other organisms were analyzed using the Neighbor-Joining distance method [28] as implemented in the MEGA package version 3.1 [29]. Phylogenetic trees were generated for each putative E. histolytica ESCRT protein aligned with homologues from different species. Robustness of phylogenetic inferences was tested by bootstrapping method, involving 1000 replications of the data based on the criteria of 50% majority-rule consensus.
For 3D modeling of MIT and AAA domains of E. histolytica EhVps4, predicted tertiary structures were obtained with the Phyre server (http://www.sbg.bio.ic.ac.uk/phyre/), using crystal data from yeast Vps4 MIT (2v6xA) and AAA (2qpaB) domains as templates.
2.2. E. histolytica Cultures
Trophozoites of E. histolytica clone A (strain HM1: IMSS) were axenically cultured in TYI-S-33 medium at 37°C and harvested during exponential growth phase [30]. Medium for transfected trophozoites was supplemented with 40 μg/mL G418 (Gibco). Cell viability was monitored by microscopy using Trypan blue dye exclusion test.
2.3. Semi-Quantitative RT-PCR Assays
Using Trizol reagent (Invitrogen), total RNA was extracted from 106 trophozoites grown in TYI-S-33 medium or 5 minutes after red blood cells (RBC) ingestion.Semiquantitave RT-PCR was performed using 1 μg of DNAse I-treated total RNA that was reverse transcribed using Superscript II (Invitrogen) for 2 hours at 42°C. Control samples without Superscript-II were included in all experiments. cDNA samples were subjected to PCR amplification using specific internal primers for distinct E. histolytica putative ESCRT genes(see Table T1 in Supplementary material available on line at doi: 10.1155/2010/890674).Briefly, PCR consisted in an initial denaturation step at 94°C for 5 minutes followed by 25 cycles of 35 s at 94°C, 30 s at Tm calculated for each gene(Supplementary data Table T1),1 minute 30 s at 72°C and a final extension step at 72°C for 7 minutes. As a control, we amplified a Eh25S rRNA gene internal sequence which was used to normalize densitometric data. Products were separated by 1% agarose gel electrophoresis, stained with ethidium bromide and visualized by UV light in a Gel Doc 1000 apparatus (BioRad). Densitometric analysis was performed using the Quantity One software. Three independent experiments were done by duplicate. Statistical significance was determined by T Student test [31].
2.4. Cloning and Sequencing of Ehvps4 and Ehvps4-E211Q Genes
The full-length Ehvps4 gene (1260 bp) reported at locus EHI_118900 in E. histolytica Pathema database was PCR amplified from E. histolytica genomic DNA using the Ehvps4-forward (5′-CCCCCGGATCCATGACATCGTTACTTGATAAAGG-3′) and Ehvps4-reverse (5′-CCCCCCTCGAGTTATCCATCTTGTCCAAATTGTTC-3′) primers in the presence of 0.2 U Pfx DNA polymerase (Invitrogen). Briefly, PCR consisted in an initial denaturation step at 94°C for 5 minutes followed by 28 cycles (30 s at 94°C, 35 s at 55°C, and 1 minute 30 s at 72°C) and a final extension step at 72°C for 7 minutes. The PCR product was cloned into TOPO vector (Invitrogen) yielding the TOPO-Ehvps4 plasmid. Using the QuikChange mutagenesis kit (Stratagene), we generated a point mutation in the EhVps4 ATPase domain at amino acid 211 to replace glutamic acid (E) by glutamine (Q) to obtain the TOPO Ehvps4-E211Q plasmid. Both constructions were confirmed by automated DNA sequencing. Then, Ehvps4 and Ehvps4-E211Q genes were PCR amplified from TOPO plasmids and subcloned in the pGEX-6P1 expression vector (Amersham Biosciences) to generate the recombinant pGEX-6P1-Ehvps4 and pGEX-6P1-Ehvps4-E211Q plasmids, respectively. Constructions were confirmed by automated DNA sequencing.
2.5. Expression and Purification of Recombinant EhVps4-GST and EhVps4-E211Q-GST Proteins
Escherichia coli BL21 (DE3) pLysS (Invitrogen) bacteria were transformed with pGEX-6P1-Ehvps4 or pGEX-6P1-Ehvps4-E211Q plasmids to produce the GST-tagged rEhVps4 and rEhVps4E211Q proteins. Bacteria were grown at 37°C in 2-TY medium containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. rEhVps4-GST expression was induced by 1 mM isopropyl beta-D-thiogalacto pyranoside (IPTG) for 3 hours at 37°C. Cells were harvested, resuspended in ice cold PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 pH 7.3) and lysed by sonication at 4°C in the presence of lisozyme (1 mg/mL). rEhVps4-GST and rEhVps4-E211Q-GST polypeptides were purified near to homogeneity through glutathione affinity chromatography, according to manufacturer recommendations (Amersham Biosciences). To further use the recombinant protein as antigen, the GST tag (25 kDa) was removed from rEhVps4-GST protein by incubation with 2 U PreScission Protease (Amersham Biosciences) during 12 hours at 4°C. Cleaved rEhVps4 protein was dialyzed into PBS pH 7.4. Identity and integrity of purified rEhVps4-GST, rEhVps4-E211Q-GST and rEhVps4 proteins were confirmed by 10% SDS-PAGE and Western blot assays using anti-GST, (GE-Healthcare, 1 : 5000 dilution) and anti-rEhVps4 (1 : 15000 dilution) antibodies, respectively, and the ECL-Plus Western blotting detection system (Amersham Biosciences).
2.6. Generation of Polyclonal Antibodies against EhVps4
Purified rEhVps4 without GST tag was submitted to preparative 10% SDS-PAGE and electroeluted from Coomassie stained gels. rEhVps4 (200 μg) in complete Freund's adjuvant (Sigma) was subcutaneously inoculated into a New Zealand male rabbit, and then, three doses of 100 μg rEhVps4 in incomplete Freund's adjuvant were injected at 15 days intervals. Rabbit was bled to obtain polyclonal serum.
2.7. ATPase Activity Assay
The ATPase activity assay procedure was based on that previously described [32] using the PiPer Phosphate Assay kit (Invitrogen), with minimal modifications. Briefly, Amplex red was diluted in DMSO to a 10 mM final concentration. Maltose phosphorylase, maltose, glucose oxidase, and horseradish peroxidase were diluted in enzyme buffer (0.1 M Tris-HCl, pH 7.5) to a final concentration of 200 U/mL, 40 mM, 200 U/mL, and 100 U/mL, respectively. Assay buffer (100 mM Tris, 20 mM KCl, and 6 mM MgCl2, pH 7.4) was added to a 96-well plate. The volume corresponding to 3.5 μg and 5 μg of purified and dialyzed rEhVps4-GST, rEhVps4-E211Q-GST, and rGST proteins was added to wells and enzyme buffer was added up to 27 μL. 50 μL of working solution (2 U/mL glucose oxidase, 4 U/mL maltose phosphorylase, 0.4 mM maltose, 100 μM amplex red, 0.4 U/mL HRP) were added to wells. Controls consisting in enzyme buffer or enzyme buffer with working solution were included. Fresh ATP was diluted in assay buffer to a concentration of 2.5 mM and added to each well to a final volume of 100 μL. Plates were mixed by pipetting and shacked for 30 s to ensure homogeneity. Then, reactions were kept at 25°C for 30 minutes and protected from the light. The absorbance was measured at 562 nm at 30 minutes and data were documented. The purified rGST protein was used as negative control in ATPase assays. Assays were performed twice by triplicate.
2.8. Plasmids Construction for Transfection Assays
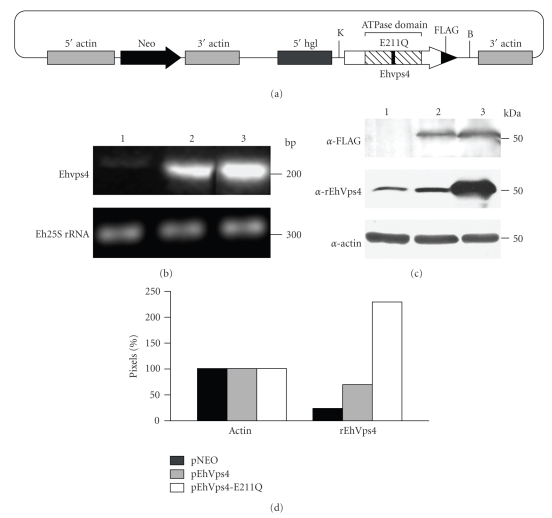
The mutant Ehvps4-E211Q gene and the wild type Ehvps4 gene were PCR amplified from TOPO Ehvps4-E211Q and TOPO-Ehvps4, respectively, using the Ehvps4-S-KpnI (5′-CCCCCGGTACCATGACATCGTTACTTGATAAAGG-3′) and Ehvps4-AS-FLAG-BamHI (5′-CCCCCGGATCCTTACTTATCGTCGTCATCCTTGTAATCTCCATCTTGTCCAAATTGTTC-3′) primers and cloned into pNEO vector for further transfection assays [33]. The underlined nucleotide sequence corresponds to the FLAG epitope (1 kDa) [34] that was added to the carboxy terminus of recombinant proteins to allow their specific immunodetection in transfected trophozoites. The resulting plasmids named pEhVps4-E211Q and pEhVps4 were confirmed by automated DNA sequencing.
2.9. In Vitro and In Vivo Virulence of Transfected Cells
Trophozoites were transfected with 200 μg of pEhVps4-E211Q, pEhVps4 and control pNEO plasmids by electroporation as described [33]. Briefly, amoebae (107) were washed twice in cold PBS and once in cold complete cytomix buffer. Electroporation was performed with the Bio-Rad Gene Pulser using 1200 V/cm and 25 μF, with a constant time of 0.4 ms. Electroporated cells were transferred into fresh culture medium for 48 hours before selecting them with 10 μg/mL G418.
In vitro virulence of nontransfected and transfected trophozoites (105) was measured on MDCK cells (6 × 104) as described [35, 36]. Rate of erythrophagocytosis was evaluated using trophozoites and RBC in a 1 : 100 relation, at 5, 10, and 15 minutes [37].
In vivo virulence of transfected trophozoites was evaluated as described [38]. Briefly, under sterile conditions and intraperitoneal anaesthesia with 0.2 mg Anesthesal (Pfizer), three groups of six hamsters each were laparotomized and intraportally challenged with pEhVps4-E211Q, pEhVps4, or pNEO transfected trophozoites (2.5 × 106) in a volume of 0.1 mL PBS using a 29-gauge needle. Seven days after challenge, animals were anaesthetized and livers were removed to evaluate hepatic damage.
2.10. Immunodetection of EhVps4 in Trophozoites
Western blot assays were performed using 30 μg of total proteins from nontransfected or pNEO, pEhVps4, and pEhVps4-E211Q transfected trophozoites. We used anti-FLAG monoclonal (1 : 700) or anti-rEhVps4 polyclonal (1 : 15000) antibodies and rabbit anti-mouse and goat anti-rabbit IgG horseradish peroxidase-labeled secondary antibodies (Zymed; 1 : 10000), respectively. Immunodetected proteins were revealed with the ECL Plus Western blotting system (Amersham Biosciences). As an internal control, we used monoclonal antibodies raised against E. histolytica actin (1 : 1000) and goat anti-mouse IgG horseradish peroxidase-labeled secondary antibodies (Zymed; 1 : 10000).
For confocal microscopy assays, nontransfected and transfected trophozoites were grown on coverslips, fixed with 4% paraformaldehyde at 37°C for 1 hour, permeabilized with Triton X-100 and blocked with 1% bovine serum albumin in PBS. Then, nontransfected and transfected cells were incubated with polyclonal anti-rEhVps4 (1 : 5000) or monoclonal anti-FLAG primary antibodies (Sigma; 1 : 500), respectively, at 37°C for 2 hours, followed by incubation with anti-rabbit or anti-mouse fluoresceinated (Zymed; 1 : 100) monoclonal antibodies, at 37°C for 1 hour. Next, trophozoites were washed three times with PBS 1X at room temperature and DNA was counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) (5 μg/mL) for 5 minutes. To immunolocalize EhVps4 during erytrophagocytosis, pEhVps4 transfected cells were incubated with RBC (1 : 20) for 10 minutes. Ingested RBC were stained with diaminobenzidine (0.84 mM 3,3′-diaminobenzidine, 0.048% H2O2 and 50 mM Tris-HCl, pH 9.5) for 5 minutes and cells were incubated with polyclonal anti-rEhVps4 (1 : 5000) to be processed for immunofluorescence as described above. Light optical sections (1.5 μm) were obtained through a Leica inverted microscope attached to a laser confocal scanning system TCS-SP2 (Leica).
3. Results
3.1. E. histolytica Genome Contains Homologous ESCRT Genes That Are Expressed in Trophozoites
To investigate the presence of ESCRT-related genes in E. histolytica, we surveyed the parasite genome sequence at Pathema database using the amino acid sequences of yeast and human ESCRT proteins as queries. We detected a set of 20 putative ESCRT protein-encoding genes, which exhibited significant e-values (1.1e − 114 to 0.00032) and high similarity (20 to 62%) to yeast and human ESCRT (ESCRT 0-III and associated factors) orthologues (see Table 1). However, we did not find homologues for yeast Mvb12, and Bul1p protein involved in protein ubiquitination, neither human Vps37-B and Vps37-C proteins. Phylogenetic inference revealed that E. histolytica ESCRT predicted proteins are closely related to homologous proteins from other protozoa, such as Giardia lamblia, Tetrahymena thermophila, Trichomonas vaginalis, Plasmodium vivax, Cryptosporidium homini, and Trypanosoma cruzi (data not shown). Bioinformatics analysis showed that E. histolytica ESCRT predicted proteins have the most characteristic functional domains (see Supplementary data Figure S1) and tertiary structure folding described for yeast and human ESCRT factors (data not shown).
Table 1.
Comparison of Entamoeba histolytica, Homo sapiens, and Saccharomyces cerevisiae ESCRT machineries.
| Entamoeba histolytica | Homo sapiens | Saccharomyces cerevisiae | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative complex | Predicted protein | Accesion numbera | Protein | Accesion numberb | e-value | S (%) | I (%) | Protein | Accesion numbera | e-value | S (%) | I (%) |
| ESCRT-0 complex | EhHse1 | EHI_091530 | STAM1 | Q92783 | 9e − 08 | 72 | 50 | Hse1p | P38753 | 2e − 10 | 70 | 62 |
| EhVps27 | EHI_117910 | HRS | O14964 | — | — | — | Vps27p | P40343 | 1e − 11 | 61 | 49 | |
| ESCRT-I complex | EhVps23 | EHI_135460 | TSG101 | Q99816 | 1.4e − 10 | 51 | 33 | Vps23p | P25604 | 0.0052 | 31 | 25 |
| EhVps28 | EHI_108630 | hVps28 | Q9UK41 | — | — | — | Vps28p | Q02767 | 0.00057 | 45 | 28 | |
| EhVps37A | EHI_077870 | hVps37A | Q6NW27 | 0.00022 | 55 | 24 | Vps37p | Q99176 | 0.00084 | 46 | 21 | |
| EhVps37D | EHI_060400 | hVps37D | Q6P2C3 | 3.8e − 06 | 50 | 35 | nd | — | — | — | — | |
| nd | — | nd | — | — | — | — | Mvb12 | P42939 | — | — | — | |
| ESCRT-II complex | EhVps22 | EHI_131120 | EAP30 | Q96H20 | 9e − 22 | 47 | 30 | Vps22p | Q12483 | 3.4e − 15 | 48 | 25 |
| EhVps25 | EHI_137860 | EAP20 | Q9BRG1 | 9e − 08 | 52 | 28 | Vps25p | P47142 | 0.00038 | 46 | 19 | |
| EhVps36 | EHI_045320 | EAP45 | Q86VN1 | 1.8e − 15 | 49 | 25 | Vps36p | Q06696 | 1.1e − 09 | 48 | 26 | |
| ESCRT-III complex | EhVps2 | EHI_194400 | CHMP2A | O43633 | 8.9e − 24 | 55 | 29 | Vps2p | P36108 | 2e − 05 | 50 | 29 |
| CHMP2B | Q9UQN3 | 7.9e − 16 | 48 | 25 | — | — | — | — | — | |||
| EhVps20 | EHI_066730 | CHMP6 | Q96FZ7 | — | — | — | Vps20p | Q04272 | 0.00015 | 54 | 26 | |
| EhVps24 | EHI_048690 | CHMP3 | Q9Y3E7 | — | — | — | Vps24p | P36095 | 2.2e − 05 | 48 | 22 | |
| EhVps32 | EHI_169820 | CHMP4A | Q9BY43 | 0.0012 | 48 | 24 | Vps32p | P39929 | 2.5e − 12 | 48 | 25 | |
| CHMP4B | Q9H444 | 9.6e − 07 | 43 | 20 | nd | — | — | — | — | |||
| CHMP4C | Q96CF2 | 1.4e − 06 | 43 | 20 | nd | — | — | — | — | |||
| Vps4 | EhVps4 | EHI_118900 | hVps4A | Q9UN37 | 1.1e − 114 | 69 | 52 | Vps4p | P52917 | 3e − 114 | 78 | 60 |
| hVps4B | O75351 | 3e − 114 | 78 | 60 | — | — | — | — | — | |||
| Other MVB proteins | EhVta1 | EHI_010040 | hVta1 | Q9NP79 | 2.8e − 05 | 44 | 23 | Vta1p | Q06263 | — | — | — |
| EhVps46 | EHI_093850 | CHMP1A | Q9HD42 | — | — | — | Vps46p | P69771 | 0.00032 | 48 | 18 | |
| CHMP1B | Q7LBR1 | — | — | — | nd | — | — | — | — | |||
| EhVps60 | EHI_114790 | CHMP5 | Q9NZZ3 | 5.6e − 15 | 45 | 28 | Vps60p | Q03390 | 9e − 06 | 46 | 19 | |
| EhDoa4 | EHI_012290 | UBP4 | Q13107 | 1e − 77 | 58 | 36 | Doa4p | P32571 | 2e − 27 | 50 | 31 | |
| EhADH112 | EHI_181220 | ALIX | Q9UKL5 | 1e − 21 | 40 | 20 | Rim20p | Q12033 | 2e − 12 | 38 | 21 | |
| BRO1p | P48582 | 0.003 | 50 | 32 | ||||||||
| Ubiquitination components | nd | — | nd | — | — | — | — | Bul1p | P48524 | — | — | — |
| EhRsp5 | EHI_011530 | NEED4 | P46934 | 1.6e − 50 | 53 | 33 | Rsp5p | P39940 | 7.8e − 57 | 57 | 36 | |
aPathema Entamoeba Bioinformatics Resource Center. bSwiss-Prot/TrEMBL databases. nd: not determined. —: Denote nonsignificant similarity/identity and e-values. S: Similarity. I: Identity.
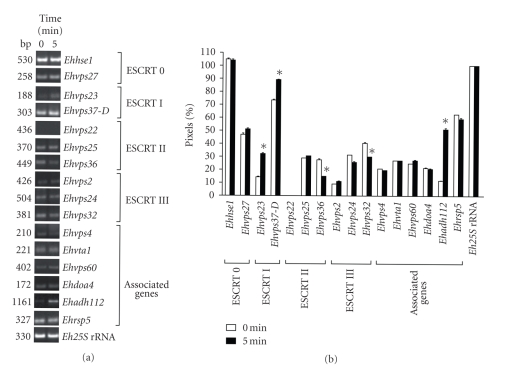
To determine whether the putative E. histolytica ESCRT genes were expressed, we performed RT-PCR assays for the 16 most conserved genes. In basal culture conditions, 15 genes were transcribed, but the Ehvps22 transcript was not detected in these experiments (see Figures 1(a) and 1(b)). We also performed RT-PCR assays using RNA obtained from trophozoites incubated with RBC for 5 minutes to investigate whether erythrophagocytosis has an effect on ESCRT gene expression (see Figures 1(a) and 1(b)). In these assays, we found that all genes were expressed during erythrophagocytosis at similar levels as in basal conditions. Again, we did not detect the Ehvps22 gene expression. Interestingly, Ehvps23 and Ehadh112 were 2- and 3-fold up-regulated, respectively. By densitometric analysis, the band given by the amplified Eh25S rRNA product in both conditions was taken as 100% and used to normalize ESCRT genes data.
Figure 1.
mRNA expression profile of E. histolytica putative ESCRT machinery genes. (a) RT-PCR products obtained from 1 μg of total RNA from trophozoites growing in TYI-S-33 medium (0 minute) or after 5 minutes of erythrophagocytosis. (b) Densitometric analysis of RT-PCR products in (a). Pixels corresponding to Eh25S rRNA amplified product were taken as 100% in each lane. Data represent the mean of three independent experiments performed by duplicate for each gene. Asterisk, genes whose transcription is significantly changed according to T Student test.
3.2. The Predicted EhVps4 Protein Conserves the Typical Architecture of Vps4 Homologues
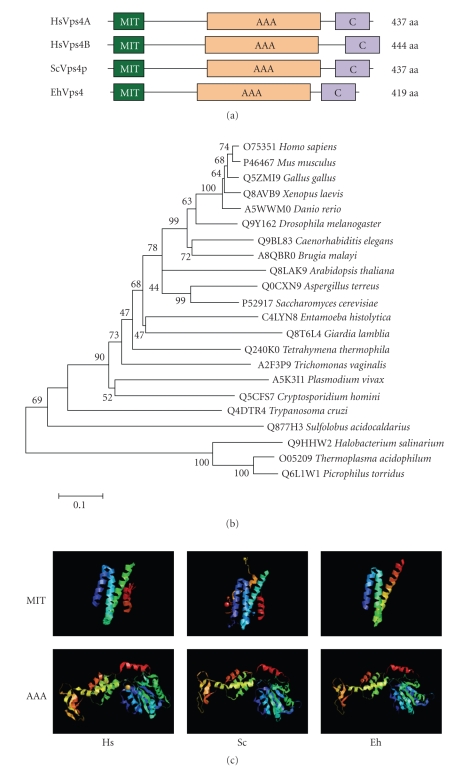
Since Vps4 orthologues have been described as key molecules for the dissociation of ESCRT, we first focused on the characterization of the E. histolytica EhVps4 protein. The predicted EhVps4 contains the N-terminal MIT (microtubule interacting and transport), AAA (ATPase associated with a variety of activities) and Vps4 C-terminal domains (see Figure 2(a)) that are characteristic and essential for Vps4 proteins biological functions [39, 40]. Phylogenetic trees revealed that EhVps4 is more related to protozoan Vps4 than orthologous proteins from higher eukaryotes (see Figure 2(b)). Interestingly, 3D modeling of EhVps4, using the crystal structure of MIT and AAA domains from yeast Vps4p and human Vps4B as templates (see Figure 2(c)), showed that EhVps4 region spanning the N-terminal MIT and AAA ATPase domains exhibited a similar folding with the conserved Vps4 α-helices.
Figure 2.
Comparison of EhVps4 with other Vps4 proteins. (a) Schematic representation of Vps4 proteins from human, yeast and E. histolytica. Conserved MIT, AAA, and Vps4 C-terminal (C) domains were predicted using PROSITE and Pfam programs. (b) Phylogenetic relationships between EhVps4 and homologous proteins from other organisms. The phylogenetic tree of Vps4 homologues was created with the MEGA 3.1 program using the Neighbor Joining algorithm based on ClustalW alignments of complete amino acids sequences. Numbers on tree nodes represent the bootstrap proportions (%) of 1000 replications. The UniProt KnowledgeBase database accession number for each protein is indicated before the organism. (c) Comparison of 3D structures of MIT and AAA domains from human (Hs), yeast (Sc) and E. histolytica (Eh) Vps4 homologues. 3D modeling of EhVps4 MIT and AAA domains was obtained using crystal data from yeast Vps4p MIT (2v6xA) and AAA (2qpaB) domains as template, respectively.
3.3. Expression and Immunodetection of EhVps4 Protein in Trophozoites
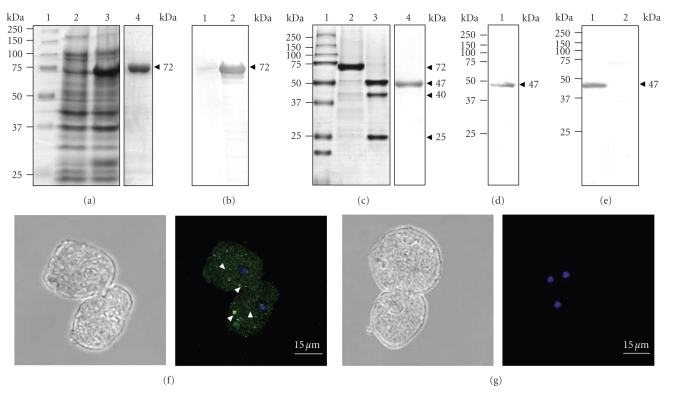
We produced and purified the recombinant EhVps4 protein as a GST-tagged fusion polypeptide (rEhVps4-GST) in E. coli (see Figure 3(a), lanes 3, 4). By Western blot assays using commercial anti-GST monoclonal antibodies, we immunodetected the purified rEhVps4-GST as a 72 kDa protein in IPTG-induced bacteria (see Figure 3(b), lane 2) but not in protein extracts from noninduced bacteria (see Figure 3(b), lane 1). The rEhVps4-GST was digested with the PreScission Protease to obtain the rEhVps4 untagged protein (47 kDa) (see Figure 3(c), lanes 2 and 3), which was also purified (see Figure 3(c), lane 4) and used to generate rabbit polyclonal antibodies. Anti-rEhVps4 serum recognized the untagged rEhVps4 protein, confirming the antibodies specificity (see Figure 3(d)). In addition, the same antibodies detected a single 47 kDa band in E. histolytica protein extracts (see Figure 3(e), lane 1), which corresponds to the expected molecular weight for the predicted EhVps4 polypeptide amino acid sequence. Antibodies did not detect any other AAA ATPases predicted in the E. histolytica genome [41]. Pre-immune serum, used as a control, did not recognize any band in trophozoite extracts (see Figure 3(e), lane 2). Through immunofluorescence and laser confocal microscopy assays, the specific rabbit polyclonal antibodies revealed EhVps4 as abundant small dots dispersed in the cytosol (see Figure 3(f)).
Figure 3.
Expression and immunodetection of EhVps4. (a) Expression and purification of recombinant EhVps4-GST polypeptide. Proteins were separated through 10% SDS-PAGE and gels were stained with Coomassie blue. Lane 1, molecular weight markers; lane 2, noninduced bacteria extract; lane 3, IPTG-induced bacteria extract; lane 4, affinity purified rEhVps4-GST from IPTG-induced bacteria extract. (b) Immunodetection of rEhVps4-GST. Western blot assays were performed using noninduced bacteria extract (lane 1) and IPTG-induced bacteria extract (lane 2), with anti-GST antibodies. (c) PreScission Protease digestion of rEhVps4-GST revealed by Coomassie blue stained gels. Lane 1, molecular weight markers; lane 2, purified rEhVps4-GST protein; lane 3, rEhVps4-GST digested with PreScission Protease; lane 4, purified rEhVps4 protein. (d) Immunodetection of purified rEhVps4 protein by Western blot assays using specific anti-rEhVps4 antibodies (lane 1). (e) Immunodetection of EhVps4 in total extracts of trophozoites using specific anti-rEhVps4 antibodies (lane 1) and preimmune serum (lane 2). Proteins are indicated by arrowheads. (f) and (g) Cellular localization of EhVps4 in trophozoites. Trophozoites of clone A were incubated with rabbit anti-rEhVps4 (f) or preimmune (g) serum, treated with FITC-labeled secondary antibodies, counterstained with DAPI and analyzed through confocal laser microscopy. Left, cells observed in phase contrast; right, merge (trophozoites observed in the green (FITC) and blue (DAPI) channels). Arrowheads, EhVps4 signal in small cytoplasmic dots.
3.4. ATPase Activity Assays
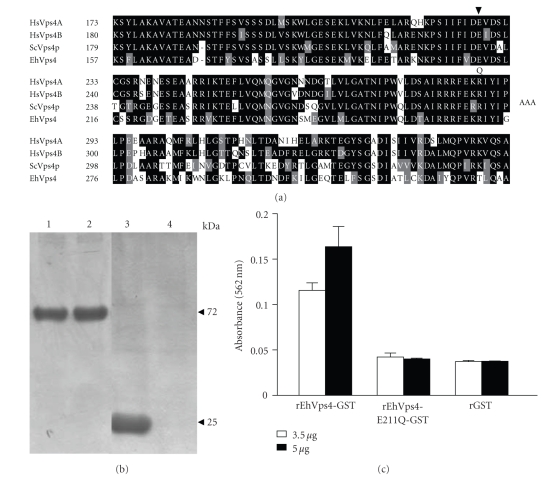
To investigate whether the recombinant protein exhibited ATPase activity we used the rEhVps4 purified protein and a mutant version (EhVps4-E211Q) in which we changed the E211 residue of the AAA motif, essential for enzyme activity, by a Q residue (see Figure 4(a)) [18, 42]. Reactions were carried out using the PiPer Phosphate Assay Kit (Invitrogen) in the presence of 500 μM ATP. Inorganic phosphate (Pi) release was measured spectrophotometrically at A562. At zero time, absorbance values of samples containing rEhVps4-GST, rEhVps4-E211Q-GST and control rGST purified proteins (see Figure 4(b)) were similar and they were taken as background. At 30 minutes, A562 values increased to 0.115 ± 0.008 and 0.162 ± 0.022, using 3.5 μg and 5 μg of rEhVps4-GST protein, respectively. No significant A562 increase was detected when we used rEhVps4-E211Q-GST or rGST proteins at 30 minutes (see Figure 4(c)). These results showed that rEhVps4-GST has ATPase activity and that the AAA motif is essential for ATP hydrolysis.
Figure 4.
Measurement of ATPase activity of recombinant EhVps4. (a) Multiple sequence alignment of Vps4 AAA domain from Homo sapiens (HsVps4A and HsVps4B), S. cerevisiae (ScVps4p) and E. histolytica (EhVps4). Black boxes, identical amino acid (aa); grey boxes, conserved substitutions; open boxes, different aa. Numbers at the left are relative to the position of the initial methionine in each protein. Arrowhead indicates the position of the conserved E residue. (b) Purification of recombinant proteins. Proteins were expressed in E. coli, purified through affinity chromatography, separated through 10% SDS-PAGE and stained with Coomassie blue. Lane 1, rEhVps4-GST; lane 2, rEhVps4-E211Q-GST; lane 3, rGST; lane 4, purified fraction from noninduced bacteria extract. (c) ATPase assay. ATPase activity of rEhVps4-GST and rEhVps4-E211Q-GST was monitored as described in Section 2.7. rGST was included as control. Data are the mean of three independent assays.
3.5. Generation of Trophozoites Overexpressing the EhVps4-FLAG and Mutant EhVps4-E211Q-FLAG Proteins
To investigate the role of EhVps4 in E. histolytica, we generated transfectant trophozoites that overexpress the EhVps4 and the mutant EhVps4-E211Q FLAG-tagged proteins (see Figure 5(a)). Trophozoites transfected with pNEO vector were used as control in these experiments. Viability of transfected trophozoites was up to 90% as determined by trypan blue exclusion assays.
Figure 5.
Expression of exogenous EhVps4 in transfected trophozoites. (a) Schematic representation of pEhVps4-E211Q construct. Mutant Ehvps4 gene was cloned into the KpnI and BamHI sites of pNEO vector, upstream the sequence coding for FLAG epitope. B, BamHI site; K, KpnI site. (b) 2% agarose gel with RT-PCR assays products. An internal fragment of the Ehvps4 gene (upper panel) or the Eh25S rRNA gene (bottom panel) was RT-PCR amplified using 1 μg of total RNA from trophozoites of clone A transfected with pNEO (lane 1), pEhVps4 (lane 2) or pEhVps4-E211Q (lane 3) plasmid. (c) Western blot assays. Anti-FLAG (upper panel), anti-rEhVps4 (middle panel) and anti-actin (lower panel) antibodies were used to analyze protein lysates (30 μg) from pNEO (lane 1), pEhVps4 (lane 2) and pEhVps4-E211Q (lane 3) transfected cells. (d) Densitometric analysis of bands corresponding to actin and EhVps4 in (c). Pixels corresponding to actin control were taken as 100% in each lane and used to normalize EhVps4 data.
RT-PCR assays, using specific primers for Ehvps4 gene, showed that trophozoites transfected with pEhVps4 or pEhVps4-E211Q plasmids expressed a higher amount of the Ehvps4 transcript in comparison to pNEO transfected cells (see Figure 5(b), upper panel, lanes 1, 2, and 3). As control, we amplified the Eh25S rRNA transcript, which showed minimal changes among the different transfectants studied (see Figure 5(b), lower panel).
By Western blot assays, using the anti-FLAG antibodies, we detected the EhVps4-FLAG and EhVps4-E211Q-FLAG proteins in pEhVps4 and pEhVps4-E211Q transfectant cells, whereas no signal was detected in pNEO transfectantes, as expected (see Figure 5(c), upper panel). However, specific anti-rEhVps4 antibodies recognized endogenous EhVps4 protein in pNEO transfectants (see Figure 5(c), middle panel, lane 1), meanwhile signal increased 2.8- and 9.2-fold in pEhVps4 and pEhVps4-E211Q transfected cells, respectively (see Figure 5(c), middle panel, lanes 2, and 3). Anti-actin antibodies detected similar amounts of protein in the different transfectants (see Figure 5(c), lower panel). For densitometric analysis, actin band was taken as 100% in each lane and used to normalize EhVps4, EhVps4-FLAG, and EhVps4-E211Q-FLAG data (see Figure 5(d)).
3.6. Cellular Localization of EhVps4-FLAG and EhVps4-E211Q-FLAG Proteins in Transfected Trophozoites
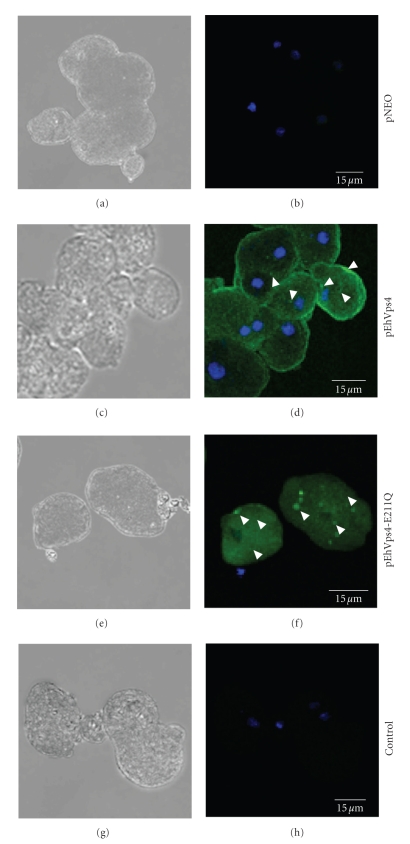
To investigate the cellular localization of exogenous wild type EhVps4 and mutant EhVps4-E211Q FLAG-tagged proteins in transfected trophozoites, we performed immunofluorescence assays using anti-FLAG antibodies (see Figure 6). In pEhVps4 transfected trophozoites, anti-FLAG antibodies detected the overexpressed exogenous protein diffuse in the cytoplasm and at the plasma membrane (see Figure 6(d)). In contrast, in mutant pEhVps4-E211Q transfected cells, signal was not observed at the plasma membrane, but it was present as cytoplasmic structures of varying sizes (see Figure 6(f)). As expected, no signal was detected by anti-FLAG antibodies in pNEO transfected cells (see Figure 6(b)). Control assays without anti-FLAG primary antibodies did not show any signal in transfected trophozoites (see Figure 6(h)).
Figure 6.
Cellular localization of exogenous EhVps4 in transfected trophozoites. Trophozoites transfected with pNEO ((a) and (b)), pEhVps4 ((c) and (d)) and pEhVps4-E211Q ((e) and (f)) were incubated with mouse anti-FLAG antibodies, treated with FITC-labeled secondary antibodies, counterstained with DAPI and analyzed through confocal laser microscopy. pEhVps4-E211Q transfected trophozoites incubated with FITC-labeled secondary antibodies were used as control ((g) and (h)). (a), (c), (e) and (g) Cells observed in phase contrast; (b), (d), (f) and (h) Merge, trophozoites observed in the green (FITC) and blue (DAPI) channels. Arrowheads, EhVps4 signal in plasma membrane and cytoplasmic dots.
3.7. Cytopathic Activity, Erythrophagocytosis and Liver Abscesses Formation Are Impaired in Trophozoites Overexpressing the Mutant EhVps4-E211Q-FLAG Protein
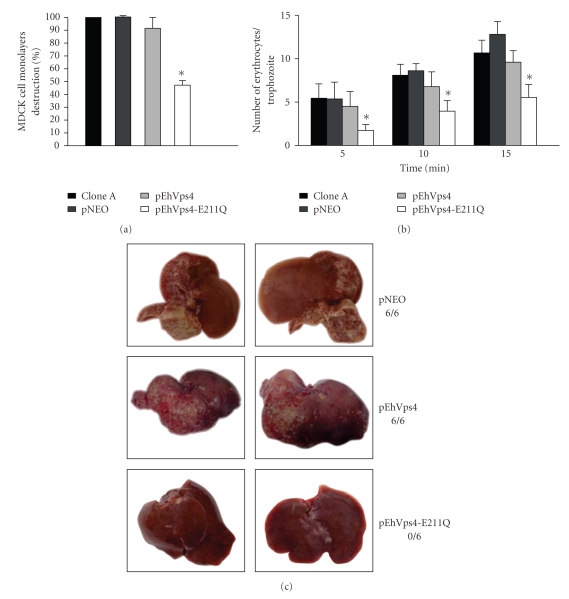
To initiate the phenotypical characterization of trophozoites transfected with pEhVps4 or pEhVps4-E211Q constructs, we first evaluated their cytopathic activity and rate of erythrophagocytosis. Nontransfected clone A and pNEO transfected cells were used as controls in these experiments. After 1 hour interaction of MDCK cells with trophozoites of clone A, pNEO or pEhVps4 transfected trophozoites, monolayers presented in mean 86.8 ± 13.2% destruction. Interestingly, pEhVps4-E211Q transfected cells were less efficient to destroy MDCK cell monolayers, since they only produced 46.4 ± 6.4% cell destruction (see Figure 7(a)).
Figure 7.
Virulence assays of transfected trophozoites. (a) Cytopathic activity. The destruction of MDCK cells monolayers by clone A trophozoites, pNEO, pEhVps4, or pEhVps4-E211Q transfected cells was determined as described [32, 33]. (b) Erythrophagocytosis. Rate of phagocytosis of clone A trophozoites, pEhVps4, pEhVps4-E211Q or pNEO transfected cells was evaluated at 5, 10, and 15 minutes [34]. Histograms show the mean count ± SD of three independent experiments by duplicate. (c) Hepatic damage in hamsters infected with transfected trophozoites. Three groups of six hamsters were infected with pNEO, pEhVps4, or pEhVps4-E211Q transfected trophozoites. After 7 days, animals were sacrificed and liver damage was recorded. Pictures were taken from the liver side where the abscesses appeared to be larger and are representative for each group. Upper panels, livers from hamsters infected with pNEO transfected trophozoites (control); middle panels, livers from hamsters infected with pEhVps4 transfected trophozoites; lower panels, livers from hamsters infected with pEhVps4-E211Q transfected trophozoites. 6/6, 6/6 and 0/6 denote number of infected animals/number of inoculated animals.
The rates of erythrophagocytosis of non-transfected clone A, pNEO and pEhVps4 transfected cells were similar, at each time tested. In contrast, pEhVps4-E211Q transfected trophozoites showed 60, 55%, and 57% decrease in erythrophagocytosis rate at 5, 10, and 15 minutes, respectively, when compared with the mean rate from the other trophozoites (see Figure 7(b)).
Then, we investigated the capacity of pEhVps4 and pEhVps4-E211Q transfected trophozoites to induce hepatic abscesses formation in hamsters. None of the six hamsters that were infected with pEhVps4-E211Q transfected trophozoites presented hepatic damage (see Figures 7(c)), whereas the six animals infected with pNEO and pEhVps4 transfected trophozoites developed extensive abscesses.
3.8. EhVps4 Was Detected around Ingested RBC
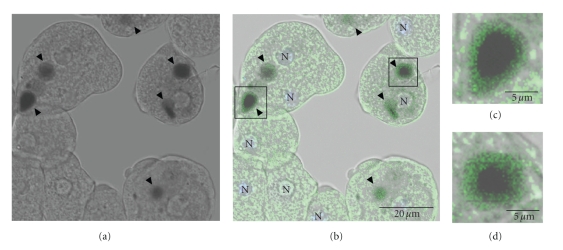
To determine the localization of EhVps4 in pEhVps4 transfected trophozoites during erythrophagocytosis, we performed immunofluorescence assays with anti-rEhVps4 serum. Antibodies detected EhVps4 as a signal surrounding ingested erythrocytes, suggesting that this protein may be involved in phagocytosis (see Figure 8).
Figure 8.
Cellular localization of EhVps4 protein during erythrophagocytosis. Trophozoites transfected with pEhVps4 were incubated with RBC, treated with diaminobenzidine, anti-rEhVps4 and FITC-labeled secondary antibodies, and analyzed through confocal laser microscopy. (a) Cells observed in phase contrast. (b) Trophozoites observed in the green (FITC) channel and phase contrast. (c) Magnification of diaminobenzidine stained RBC squared in (b). Arrowhead, EhVps4 signal around RBC. N, nuclei.
4. Discussion
In E. histolytica trophozoites, phagocytosis, and vesicular trafficking are important events for parasite nutrition and destruction of target cells. As in other eukaryotes, the endocytosis and phagocytosis pathways involve the formation of endosomes, MVB and lysosomes [5, 43]. Indeed, MVBlike structures have been detected in trophozoites [26, 27] and within isolated phagosomes [24], but proteins participating in their formation have not been identified.
Here, by mining the parasite genome sequence, we demonstrated the presence of 20 genes that encode putative ESCRT machinery components, which could participate in endosomal transport and MVB formation in E. histolytica. Most predicted proteins have the conserved functional domains and share high similarity and phylogenetic relationship with homologous proteins from other eukaryotes, strongly suggesting that they have similar functions. RT-PCR assays evidenced that 15 out of 16 ESCRT components tested are transcribed, which is consistent with the activity of endosome formation and vesicle trafficking exhibited by E. histolytica trophozoites (reviewed in [5, 43]). Except for the Ehadh112 and Ehvps23 genes, transcript amount was not significantly modified after 5 minutes of RBC interaction. Analysis of published E. histolytica microarrays data confirmed that most ESCRT genes are transcribed without any significant change under distinct experimental conditions [44, 45]. However, these experiments were not focused on phagocytosis. Additionally, in Dictyostelium discoideum, microarrays assays evidenced that expression of most genes involved in intracellular vesicle traffic is not significantly changed during phagocytosis [46]. In mouse macrophages, phagocytosis causes cellular redistribution of Hrs protein (the homologue of yeast Vps27p), but the protein amount remains unchanged [47]. It is possible that trophozoites could have enough ESCRT and associated proteins to perform erythrophagocytosis, if, as we think, these proteins are involved in phagocytosis in E. histolytica. However, changes in the gene expression of E. histolytica ESCRT members at shorter or larger times cannot be discarded.
In yeast and mammal cells, Vps4 is a key component for disassembly of ESCRT complexes from the endosomal membrane at the MVB invagination pathway [42, 48, 49]. We focused on EhVps4 protein to investigate its role in endocytosis, specifically in one of the “professional function” of E. histolytica trophozoites: phagocytosis. As in yeast, E. histolytica has only one vps4 gene, while higher eukaryotes have two vps4 genes [21, 50, 51]. By confocal microscopy, EhVps4 was immunodetected as abundant small dots dispersed in the cytosol. In yeast, mammals, plants, and the protozoan Leishmania major, similar structures have been identified as MVB formed during endocytosis and vesicular trafficking [22, 42, 51, 52].
Yeast Vps4p protein exhibits ATPase activity, which depends on the E233 residue present within the AAA domain [18, 42]. In this work, we showed that EhVps4 is an ATPase, which conserves the characteristic domains and folding of Vps4 homologues. This activity depends on the AAA domain since the mutant rEhVps4-E211Q-GST, in which E211 amino acid residue was substituted by Q residue, did not exhibit detectable ATP hydrolysis, as reported in yeast [18, 42].
We further investigated the biological relevance of EhVps4 by generating trophozoites that overexpress wild type EhVps4-FLAG and mutant EhVps4-E211Q-FLAG protein. As the endogenous protein, exogenous EhVps4-FLAG was located in abundant punctuate structures in the cytosol as reported in other organisms [42, 50, 52, 53]. However, overexpressed EhVps4-FLAG was also detected at plasma membrane. In transfected mammal cells that overexpress Sendai virus protein C and wild type Vps4, ALIX/AIP1 proteins are recruited at the plasma membrane to facilitate the budding of virus-like particles [54]. EhADH112, the E. histolytica homologue of ALIX, has been detected at the plasma membrane [55]. Experiments currently in progress will help us to define the role of Vps4 at the membrane of trophozoites.
Erythrophagocytosis and cytopathic activities were not improved in trophozoites overexpressing wild type EhVps4-FLAG, probably because other ESCRT proteins are limiting factors for cell destruction and RBC intake [42, 52, 56]. However, the dominant negative effect of mutant EhVps4-E211Q-FLAG protein in MDCK cells destruction, the rate of RBC intake, and liver abscesses formation in hamsters, suggest a role for EhVps4 in E. histolytica virulence properties. The localization of EhVps4-FLAG protein around ingested RBC strengthens this hypothesis.
In conclusion, we showed that E. histolytica has an ESCRT machinery, which is transcribed in trophozoites. Particularly, we presented evidence that the conserved EhVps4 is an ATPase that could participate in cytopathic activity and erythrophagocytosis, as well as in hepatic damage in hamster. Work currently in progress is focusing on the identification of cytoplasmic structures containing EhVps4, as well as its interaction with other components of the E. histolytica ESCRT machinery.
Supplementary Material
Schematic representation of ESCRT machinery in E. histolytica, human and yeast. Most known domains described in homologous ESCRT proteins are depicted for E. histolytica factors. EhHse1, EhVps27, EhVps23, EhVps28, EhVps46 and EhRsp5 proteins that lack several consensus domains were considered as bona fide ESCRT components, since BLAST analysis showed that they share high identity with homologous human and yeast proteins (see Table 1). The scale is shown at the bottom.
Acknowledgments
Monoclonal antiactin antibodies were gently provided by Dr. Manuel Hernández (CINVESTAV-IPN). The authors are grateful to M. Sc. Eduardo Carrillo (UACM) for laser confocal microscopy assistance. Their thanks are also to Alfredo Padilla (ICyTDF) for his help in the artwork. This work was supported by a Grant from CONACyT (Mexico) and by CYCYT Grant BFU2005-01970 (to O. Vincent).
References
- 1.Abd-Alla MD, Jackson TFHG, Reddy S, Ravdin JI. Diagnosis of invasive amebiasis by enzyme-linked immunosorbent assay of saliva to detect amebic lectin antigen and anti-lectin immunoglobulin G antibodies. Journal of Clinical Microbiology. 2000;38(6):2344–2347. doi: 10.1128/jcm.38.6.2344-2347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez MA, García-Pérez RM, García-Rivera G, et al. An Entamoeba histolytica Rab-like encoding gene and protein: function and cellular location. Molecular and Biochemical Parasitology. 2000;108(2):199–206. doi: 10.1016/s0166-6851(00)00216-4. [DOI] [PubMed] [Google Scholar]
- 3.Marion S, Laurent C, Guillén N. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cellular Microbiology. 2005;7(10):1504–1518. doi: 10.1111/j.1462-5822.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- 4.Nakada-Tsukui K, Saito-Nakano Y, Ali V, Nozaki T. A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica. Molecular Biology of the Cell. 2005;16(11):5294–5303. doi: 10.1091/mbc.E05-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Souza W, Sant’Anna C, Cunha-e-Silva NL. Electron microscopy and cytochemistry analysis of the endocytic pathway of pathogenic protozoa. Progress in Histochemistry and Cytochemistry. 2009;44(2):67–124. doi: 10.1016/j.proghi.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annual Review of Biophysics and Biomolecular Structure. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nature Reviews Molecular Cell Biology. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 8.Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? Journal of Cell Science. 2009;122(13):2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- 9.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Molecular Biology of the Cell. 1992;3(12):1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley JH. The ESCRT machinery at a glance. Journal of Cell Science. 2009;122(13):2163–2166. doi: 10.1242/jcs.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106(2):145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 12.Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Molecular Biology of the Cell. 2007;18(2):636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Developmental Cell. 2002;3(2):283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 14.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Developmental Cell. 2002;3(2):271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 15.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. Journal of Cell Science. 2003;116(10):1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Developmental Cell. 2005;8(6):937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. The Journal of Cell Biology. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. The EMBO Journal. 1997;16(8):1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nature Reviews Molecular Cell Biology. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 20.Finken-Eigen M, Rohricht RA, Kohrer K. The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Current Genetics. 1997;31(6):469–480. doi: 10.1007/s002940050232. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimori T, Yamagata F, Yamamoto A, et al. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Molecular Biology of the Cell. 2000;11(2):747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas TJ, Sliwinski MK, Martínez DE, et al. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. The Plant Cell. 2007;19(4):1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada M, Nozaki T. New insights into molecular mechanisms of phagocytosis in Entamoeba histolytica by proteomic analysis. Archives of Medical Research. 2006;37(2):244–252. doi: 10.1016/j.arcmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Okada M, Huston CD, Mann BJ, Petri WA, Jr., Kita K, Nozaki T. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryotic Cell. 2005;4(4):827–831. doi: 10.1128/EC.4.4.827-831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada M, Huston CD, Oue M, et al. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Molecular and Biochemical Parasitology. 2006;145(2):171–183. doi: 10.1016/j.molbiopara.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Bañuelos C, López-Reyes I, García-Rivera G, González-Robles A, Orozco E. The presence of a Snf7-like protein strenghtens a role for EhADH in the Entamoeba histolytica multivesicular bodies pathway. In: Boeree MJ, editor. In: Proceedings of the 5th European Congress on Tropical Medicine and International Health, vol. 978; May 2007; Amsterdam, The Netherlands. pp. 31–35. PP-292. [Google Scholar]
- 27.Bañuelos C, García-Rivera G, López-Reyes I, Orozco E. Functional characterization of EhADH112: an Entamoeba histolytica Bro1 domain-containing protein. Experimental Parasitology. 2005;110(3):292–297. doi: 10.1016/j.exppara.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 30.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1978;72(4):431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 31.Student. The probable error of a mean. Biometrika. 1908;6:1–25. [Google Scholar]
- 32.Avila Ch, Kornilayev BA, Blagg BSJ. Development and optimization of a useful assay for determining Hsp90’s inherent ATPase activity. Bioorganic and Medicinal Chemistry. 2006;14(4):1134–1142. doi: 10.1016/j.bmc.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 33.Hamann L, Nickel R, Tannich E. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(19):8975–8979. doi: 10.1073/pnas.92.19.8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung M-K, Ha CW, Huh W-K. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast. 2008;25(4):301–311. doi: 10.1002/yea.1588. [DOI] [PubMed] [Google Scholar]
- 35.Orozco E, Martínez-Palomo A, López-Revilla R. An in vitro model for the quantitative study of the virulence of Entamoeba histolytica. Archivos de Investigación Médica. 1978;9(supplement 1):257–260. [PubMed] [Google Scholar]
- 36.Bracha R, Mirelman D. Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions, and metronidazole. The Journal of Experimental Medicine. 1984;160(2):353–368. doi: 10.1084/jem.160.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orozco E, Guarneros G, Martínez-Palomo A, Sánchez T. Entamoeba histolytica. Phagocytosis as a virulence factor. The Journal of Experimental Medicine. 1983;158(5):1511–1521. doi: 10.1084/jem.158.5.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutsumi V, Mena-López R, Anaya-Velázquez F, Martínez-Palomo A. Cellular bases of experimental amebic liver abscess formation. American Journal of Pathology. 1984;117(1):81–91. [PMC free article] [PubMed] [Google Scholar]
- 39.Hurley JH, Yang D. MIT domainia. Developmental Cell. 2008;14(1):6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Xiao J, Xia H, Zhou J, et al. Structural basis of Vta1 function in the multivesicular body sorting pathway. Developmental Cell. 2008;14(1):37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loftus B, Anderson I, Davies R, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 42.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. The EMBO Journal. 1998;17(11):2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meza I, Clarke M. Dynamics of endocytic traffic of Entamoeba histolytica revealed by confocal microscopy and flow cytometry. Cell Motility and the Cytoskeleton. 2004;59(4):215–226. doi: 10.1002/cm.20038. [DOI] [PubMed] [Google Scholar]
- 44.Santi-Rocca J, Weber C, Guigon G, Sismeiro O, Coppée J-Y, Guillén N. The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cellular Microbiology. 2008;10(1):202–217. doi: 10.1111/j.1462-5822.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- 45.Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cellular Microbiology. 2009;11(1):51–69. doi: 10.1111/j.1462-5822.2008.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sillo A, Bloomfield G, Balest A, et al. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics. 2008;9, article 291 doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vieira OV, Harrison RE, Scott CC, et al. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Molecular and Cellular Biology. 2004;24(10):4593–4604. doi: 10.1128/MCB.24.10.4593-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sachse M, Strous GJ, Klumperman J. ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. Journal of Cell Science. 2004;117(9):1699–1708. doi: 10.1242/jcs.00998. [DOI] [PubMed] [Google Scholar]
- 49.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Molecular Biology of the Cell. 2000;11(1):227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheuring S, Rohricht RA, Schoning-Burkhardt B, et al. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. Journal of Molecular Biology. 2001;312(3):469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 51.Beyer A, Scheuring S, Muller S, Mincheva A, Lichter P, Kohrer K. Comparative sequence and expression analyses of four mammalian VPS4 genes. Gene. 2003;305(1):47–59. doi: 10.1016/s0378-1119(02)01205-2. [DOI] [PubMed] [Google Scholar]
- 52.Besteiro S, Williams RAM, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. The Journal of Biological Chemistry. 2006;281(16):11384–11396. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 53.Yang M, Coppens I, Wormsley S, Baevova P, Hoppe HC, Joiner KA. The Plasmodium falciparum Vps4 homolog mediates multivesicular body formation. Journal of Cell Science. 2004;117(17):3831–3838. doi: 10.1242/jcs.01237. [DOI] [PubMed] [Google Scholar]
- 54.Irie T, Nagata N, Yoshida T, Sakaguchi T. Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology. 2008;371(1):108–120. doi: 10.1016/j.virol.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 55.García-Rivera G, Rodríguez MA, Ocádiz R, et al. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein. Molecular Microbiology. 1999;33(3):556–568. doi: 10.1046/j.1365-2958.1999.01500.x. [DOI] [PubMed] [Google Scholar]
- 56.Fujita H, Yamanaka M, Imamura K, et al. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. Journal of Cell Science. 2003;116(2):401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of ESCRT machinery in E. histolytica, human and yeast. Most known domains described in homologous ESCRT proteins are depicted for E. histolytica factors. EhHse1, EhVps27, EhVps23, EhVps28, EhVps46 and EhRsp5 proteins that lack several consensus domains were considered as bona fide ESCRT components, since BLAST analysis showed that they share high identity with homologous human and yeast proteins (see Table 1). The scale is shown at the bottom.