Abstract
Maternally expressed gene 3 (MEG3) is a noncoding RNA highly expressed in the normal human brain and pituitary. Expression of MEG3 is lost in gonadotroph-derived clinically nonfunctioning pituitary adenomas. Meg3 knockout mice were generated to identify targets and potential functions of this gene in embryonic development and tumorigenesis. Gene expression profiles were compared in the brains of Meg3-null embryos and wild-type littermate controls using microarray analysis. Microarray data were analyzed with GeneSifter, which uses Kyoto Encyclopedia of Genes and Genomes pathways and Gene Ontology classifications to identify signaling cascades and functional categories of interest within the dataset. Differences were found in signaling pathways and ontologies related to angiogenesis between wild-type and knockout embryos. Quantitative RT-PCR and immunohistological staining showed increased expression of some Vascular Endothelial Growth Factor pathway genes and increased cortical microvessel density in the Meg3-null embryos. In conclusion, Meg3 may play an important role in control of vascularization in the brain and may function as a tumor suppressor in part by inhibiting angiogenesis.
Loss of Meg3 results in increased angiogenesis in the developing mouse brain.
Clinically nonfunctioning pituitary adenomas are common intracranial tumors (1) and often grow to sufficient size to cause mass effect-related symptoms, of which visual field impairment, headaches, and hypopituitarism are the most common (2,3,4). The pathogenesis of these tumors is largely unknown. Our group has shown that this tumor type, unlike other types of pituitary tumors, has lost expression of maternally expressed gene 3 (MEG3), a gene which encodes a noncoding RNA instead of a protein. This gene is highly expressed in the normal human brain and pituitary, including gonadotroph cells from which clinically nonfunctioning tumors are mainly derived (5).
MEG3 RNA has an antiproliferative function (5,6). It activates the tumor suppressor p53 and induces expression of p53 target genes in vitro (6). Loss of expression in clinically nonfunctioning pituitary tumors is associated with hypermethylation of its promoter, an enhancer region (7), and/or the intergenic differentially methylated region for imprinting control located 13 kb upstream of the MEG3 gene (8). Hypermethylation of this intergenic differentially methylated region has also been observed in neuroblastomas, Wilms' tumor (9), and multiple myeloma (10). These results suggest that MEG3 functions as a tumor suppressor in both the pituitary and other cell and tissue types. The murine homolog of the human MEG3 gene, Meg3/Gtl2 (gene trap locus 2), is highly expressed in many regions of the developing and adult mouse brain, including the neocortex, hypothalamus, hippocampus, and amygdala (11,12,13). We generated Meg3 knockout mice and compared gene expression profiles in the brains of Meg3-null embryos with wild-type littermates using microarray analysis to identify its target genes and potential functions in embryonic development and tumorigenesis. This analysis identified differences in signaling pathways and ontologies related to angiogenesis, brain development, postnatal brain function, and tumorigenesis in the brains of Meg3-null embryos compared with those of wild-type littermates. We further confirmed increased expression of angiogenesis-related genes with loss of Meg3 by quantitative RT-PCR and increased microvessel formation in the brains of Meg3-null embryos by immunohistological staining.
Materials and Methods
Embryos
Meg3 knockout mice were generated by deletion of the first five exons and approximately 300 bp of the adjacent upstream promoter region of the Meg3 gene using gene targeting techniques (Zhou, Y., P. Cheunsuchon, Y. Nakayama, M. W. Lawler, Y. Zhong, K. A. Rice, L. Zhang, X. Zhang, F. E. Gordon, H. G. W. Lidou, R. Bronson, and A. Klibanski, submitted for publication). Brains from embryonic day (E)18.5 Meg3-null embryos and wild-type littermate controls were excised, placed in RNAlater (QIAGEN, Valencia, CA), and stored at −20 C until RNA was extracted. This work was approved by the Subcommittee on Research Animal Care of Massachusetts General Hospital, and all experiments using mice were performed according to Massachusetts General Hospital Animal Care and Use Guidelines.
RNA extraction and GeneChip expression assay
Total RNA was extracted from the brains of eight littermate-matched pairs of wild-type and Meg3-null embryos using an RNeasy Lipid Tissue kit (QIAGEN). RNA was used for GeneChip analysis at the Harvard Medical School Biopolymers Facility (Boston, MA) with Mouse 430 2.0 chips (Affymetrix, Santa Clara, CA). This facility used Affymetrix kits and standard protocols for all steps of this assay. Spots were scanned with an Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE).
Normalization
Raw data were normalized using a previously described process, which has quality control and data transformation steps (14). The quality control process excluded chips with an unacceptable degree of variation from further analysis, leaving six wild-type and seven Meg3-null samples in the final dataset. Raw data were normalized by median scaling of P or present calls (P-calls), which are regions of successful hybridization between the spot on the chip and the cRNA in the sample.
GeneSifter
This program extracts patterns of gene expression from Affymetrix gene expression data and uses Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and Z-score reports to summarize the biological significance of a gene list. All analyses were based on the GeneSifter software (Geospiza, Inc., Seattle, WA), which was current as of August 7, 2008. This program uses a t test-based algorithm with a Bonferroni false-discovery correction to identify significant genes. Results were further screened by a quality cut-off of 0.5 and a fold change of at least 1.2. GeneSifter assigns a quality score of 1 to P-calls and 0 to A, or absent calls (i.e. either very little or no expression of a particular gene or a hybridization error for that particular probe set). The program also assigns a score of 0.5 to M, or marginal calls, which indicates either mean low expression or a hybridization error. A quality cut-off of 0.5 selects for genes mostly characterized by P-calls, filtering out the majority of poorer quality expression data characterized by M or absent calls. A fold change cut-off of 1.2 removes genes that do not change between wild-type and knockout embryos from further consideration, while still allowing for the detection of subtle expression changes. Genes on the Mouse 430 2.0 chips that passed these quality control filters and had an uncorrected P < 0.05 were grouped into two sets of categories by the program.
Quantitative RT-PCR
Quantitative RT-PCR was used to confirm the differences in gene expression between the wild-type and Meg3-null embryos identified by microarray analysis. RNA was extracted from the brains of E18.5 embryos collected from eight Meg3-null embryos and seven wild-type littermates of six litters, different from those used for microarray analysis. Reverse transcriptions were carried out with 1 μg of RNA using the Improm-II RT kit (Promega, Madison, WI). Quantitative PCR was carried out with the resulting cDNA using SYBR Green (Applied Biosystems, Foster City, CA). PCRs were performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) with the following reaction conditions: 50 C for 2 min, 95 C for 10 min, 40 cycles of 95 C for 15 sec, various annealing temperatures (see below) for 1 min, and 72 C for 1 min. Quantitative PCRs were immediately followed by a melt curve of 95 C for 15 sec, 60 C for 1 min, 95 C for 15 sec, and 60 C for 15 sec to confirm the PCR specificities. Primers and annealing temperatures are listed in Table 1. PCRs were performed in triplicate, and the expression levels of Meg3 and other genes were normalized with the expression levels of Gapdh. Normalized Δ threshold count (Ct) values were used to evaluate the significance of differences in gene expression between groups by a one-tailed t test. The statistical significance of changes in Vegfr2 expression between wild-type and Meg3-null embryos was assessed by a two-tailed t test, because the direction of change in expression of this gene was not predicted by the array.
Table 1.
Primers used for quantitative RT-PCR
| Gene name | Gene symbol | Forward primer | Reverse primer | Annealing temperature (C) |
|---|---|---|---|---|
| Delta-like 4 | Dll4 | GTTGCCCTTCAATTTCACCT | AGCCTTGGATGATGATTTGG | 55 |
| Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA | 58 |
| Hairy and enhancer of split 1 | Hes1 | CCAGCCAGTGTCAACACGA | AATGCCGGGAGCTATCTTTCT | 58 |
| IQ motif containing GTPase activating protein 1 | Iqgap1 | GATGTGACCCCTGAACAAGC | CTCTGCGTTCTCGTCACC | 58 |
| Neuropilin 1 | Nrp1 | CGTGGAAGTAATTGATGGGGAG | CATAGCGGATGGAAAACCCTG | 56 |
| Vascular endothelial growth factor A | Vegfa | CTTGTTCAGAGCGGAGAAAGC | ACATCTGCAAGTACGTTCGTT | 57 |
| Vascular endothelial growth factor receptor 1 | Vegfr1 | TGGACCCAGATGAAGTTCCC | GCGATTTGCCTAGTTTCAGTCT | 57 |
| Vascular endothelial growth factor receptor 2 | Vegfr2 | AGCACTGGTCCTATGGGTTG | GGTTCTGCCATTTGATCCA | 55 |
| Wiskott-Aldrich syndrome-like (human) | Wasl | GGAACTGTATGTGGGCAAAGAAGT | GACAAGTATCTCCAGCAAAGGTATG | 58 |
Immunohistological staining
Blood vessel density in the brains of mouse embryos was evaluated by immunohistological staining with two markers: platelet/endothelial cell adhesion molecule 1 (Pecam1) and vascular endothelial growth factor receptor 2 (Vegfr2). The former mediates angiogenesis through homophilic and heterophilic cell adhesion (15), and the latter is the primary receptor for vascular endothelial growth factor A (Vegfa) (16,17). Brains were excised from E18.5 Meg3-null embryos and wild-type littermate controls from litters different from the ones used for microarray analysis. One hemisphere was excised from each brain for RNA extraction as described above. The remaining hemisphere and cerebellum were fixed in 10% normal buffered formalin (Fisher Scientific, Waltham, MA) for 2–5 d before paraffin embedding. Five-micrometer tissue sections from cortical areas were prepared, because the large area of this region allowed for examination of multiple fields. For Pecam1 staining, paraffin sections were baked for 1 h at 60 C before deparaffinization with xylene. Antigen retrieval was performed by heating slides in boiling high pH target retrieval solution (Dako Cytomaton, Glostrup, Denmark) in a microwave oven for 20 min at low power and cooling for 20 min at 4 C. For Vegfr2 staining, antigen retrieval was performed after deparaffinization with xylene by heating slides in sodium citrate solution (pH 6.0) in a microwave oven for 15 min and cooling for 20 min at 4 C. Slides were blocked for 30 min in 10% normal serum (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in PBS with 1% BSA (Sigma, St. Louis, MO), followed by incubation with the primary antibody (anti-Pecam1 antibody, 1:500 dilution; Santa Cruz Biotechnology, Inc.; and anti-Vegfr2 antibody, 1:100 dilution; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. After washing with PBS, the slides were incubated with biotinylated secondary antibody (1:250 dilution for Pecam1 and 1:500 dilution for Vegfr2; Santa Cruz Biotechnology, Inc.) for 30 min. Slides were then incubated in ABC reagent (Vector Laboratories, Burlingame, CA) for 30 min followed by incubation in 3, 3-diaminobenzidine (Vector Laboratories) and counterstaining with hematoxylin.
Statistical analysis
Microvessel number in three equivalent cortical fields per brain section was counted blindly and independently by two investigators at ×200 magnification (field area, 0.17 mm2). Counts were divided by 0.17 to get the microvessel density per cortical field, and the mean cortical microvessel density was calculated before sample identification codes were released. Repeated measures ANOVA was used to compare mean cortical microvessel density between wild-type and Meg3-null embryos.
Results
GeneSifter data analysis
GeneSifter grouped genes that passed both quality and expression filters into signaling pathways using KEGG criteria (18) and ontologies according to GO classifications (19). Fourteen KEGG pathways identified by GeneSifter were considered noteworthy because expression of at least three genes changed between wild-type and Meg3-null embryos and Z > 1.75 (Table 2). Several of these pathways are important for brain development, postnatal brain function, and/or angiogenesis. The calcium, Notch, and Wnt signaling pathways, which are important for brain development, were enriched in the Meg3-null group (Z-up = 2.15, Z-up = 2.02, and Z-down = 1.9, respectively). Three pathways important for postnatal brain function were enriched in the brains of Meg3-null embryos: long-term potentiation, GnRH, and calcium signaling (Z-up = 1.96, Z-up = 2.05, and Z-up = 2.15, respectively). Adherens junctions (Z-up = 3.36) and VEGF (Z-up = 1.89) signaling pathways are also enriched in Meg3-null embryos and are important for angiogenesis, in addition to the Notch pathway.
Table 2.
KEGG pathways with a noteworthy degree of enrichment upon loss of Meg3
| KEGG pathway | No. of genes with increased expression in KO embryos | No. of genes with decreased expression in KO embryos | Z-score (up) | Z-score (down) |
|---|---|---|---|---|
| Adherens junction | 6 | 1 | 3.36 | <1.75 |
| Glycerophospholipid metabolism | 5 | 0 | 3.19 | <1.75 |
| Taste transduction | 3 | 0 | 2.86 | <1.75 |
| Glycerolipid metabolism | 3 | 0 | 2.17 | <1.75 |
| Calcium signaling pathway | 8 | 1 | 2.15 | <1.75 |
| Long-term potentiation | 4 | 0 | 2.05 | <1.75 |
| Notch signaling pathway | 3 | 0 | 2.02 | <1.75 |
| Type 2 diabetes mellitus | 3 | 0 | 1.98 | <1.75 |
| GnRH signaling pathway | 5 | 0 | 1.96 | <1.75 |
| VEGF signaling pathway | 4 | 0 | 1.89 | <1.75 |
| MAPK signaling pathway | 10 | 2 | 1.81 | <1.75 |
| Inositol phosphate metabolism | 3 | 0 | 1.76 | <1.75 |
| Wnt signaling pathway | 6 | 2 | <1.75 | 1.9 |
| Starch and sucrose metabolism | 2 | 1 | <1.75 | 1.78 |
Pathways identified using the KEGG function in GeneSifter with a Z-score of at least 1.75 and three or more genes affected by loss of Meg3. KO, Knockout.
Changes in the VEGF signaling pathway between wild-type and Meg3-null brains were also indentified in GeneSifter with GO. Ontologies were considered to be important if Z > 2, and expression of three or more genes changed between groups. A graphic representation of the more common biological process and molecular function ontologies is available as Supplemental Fig. 1, A and B, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org. Some of the ontologies most relevant to development, tumorigenesis, and postembryonic physiological processes are listed in Table 3. Ontologies related to angiogenesis include VEGF signaling (Z-up = 4.19), and blood vessel development (Z-up = 3.33 and Z-down = 3.88). Homophilic cell adhesion (Z-up = 2.75), GTPase activator activity (Z-up = 2.60), and actin cytoskeletal organization and biogenesis (Z-up = 2.24) are important for cell migration during both angiogenesis and axonal guidance. A related ontology, axon extension (Z-up = 3.21), also changes significantly with loss of Meg3. Ontologies important for postembryonic brain function that changed significantly between groups included neuropeptide signaling (Z-up = 3.95), circadian rhythm (Z-up = 2.92), and voltage-gated channel activity (Z-up = 2.68).
Table 3.
Ontologies indentified by GeneSifter that change with loss of Meg3
| Ontology | No. of genes with increased expression in KO embryos | No. of genes with decreased expression in KO embryos | Z-score (up) | Z-score (down) |
|---|---|---|---|---|
| VEGF signaling | 3 | 0 | 4.19 | <2 |
| Neuropeptide signaling | 3 | 2 | 3.95 | <2 |
| Blood vessel patterning | 3 | 1 | 3.33 | 3.88 |
| Axon extension | 3 | 0 | 3.21 | <2 |
| Circadian rhythm | 3 | 0 | 2.92 | <2 |
| Homophilic cell adhesion | 6 | 0 | 2.75 | <2 |
| Voltage-gated channel activity | 10 | 0 | 2.68 | <2 |
| GTPase activator activity | 10 | 0 | 2.60 | <2 |
| Transcription | 67 | 5 | 2.54 | <2 |
| Calcium binding | 30 | 1 | 2.40 | <2 |
| Gated channel activity | 13 | 0 | 2.40 | <2 |
| Kinase activity | 32 | 3 | 2.31 | <2 |
| DNA repair | 10 | 2 | 2.25 | <2 |
| Regulation of neuronal action potential | 3 | 0 | 2.25 | <2 |
| Actin cytoskeletal organization and biogenesis | 9 | 0 | 2.24 | <2 |
| Embryonic pattern specification | 3 | 1 | <2 | 2.50 |
| Postembryonic development | 2 | 1 | <2 | 2.30 |
| Cellular lipid metabolic process | 14 | 5 | <2 | 3.08 |
Biological process or molecular function ontologies with a Z-score >2 and three or more genes that change with loss of Meg3 were considered significant. A subset of these ontologies that are relevant for development, tumorigenesis, and/or physiological functions are listed here. KO, Knockout.
Elevated expression of angiogenesis-related genes in Meg3-deficient brains validated by quantitative RT-PCR
Both KEGG and GO in GeneSifter found changes in signaling pathways and/or ontologies related to brain development, postnatal brain function, and angiogenesis that changed significantly with loss of Meg3. In validating changes of gene expression according to microarray analysis, we focused on genes involved in angiogenesis, because angiogenesis is a critical process for tumorigenesis. Gene expression changes related to this process have been reported in clinically nonfunctioning pituitary adenomas compared with normal tissue (17,20) and hormone-secreting tumors (21). Expression changes of genes in the VEGF pathway, including Vegfa, Vegfr1, Neuropilin 1 (Nrp1), IQ motif containing GTPase activating protein 1 (Iqgap1), and Wiskott-Aldrich syndrome-like (Wasl), were examined by quantitative RT-PCR in eight Meg3-null embryos and seven wild-type littermates from six litters, which had not been used in microarray analysis. The first three genes initiate the VEGF signaling pathway (17,22). Nrp1 binds to Vegfa and acts as a coreceptor for Vegfr2 during angiogenesis (23). Iqgap1 and Wasl are cytoskeletal remodeling proteins activated by VEGF signaling during blood vessel development (24). Change in Vegfr2 expression between wild-type and Meg3 knockout embryos was also measured because of its importance in VEGF signaling (17). In addition, Delta-like 4 (Dll4) and hairy and enhancer of split 1 (Hes1) were chosen to validate changes in the Notch signaling pathway, which is important for brain development (25,26,27). Dll4 is a major ligand of the Notch 1 and 4 receptors (25), and Hes1 is a basic helix-loop-helix repressor that is expressed in response to Notch signaling (28,29,30).
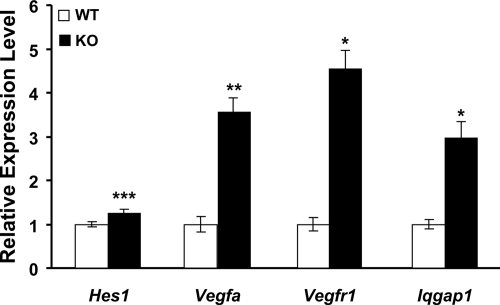
Significant changes in gene expression were found by quantitative RT-PCR for four genes indicated by microarray analysis. Vegfa expression was 3.56-fold higher in the brains of Meg3-null embryos compared with wild-type embryos (P < 0.01); and levels of one of its main receptors, Vegfr1, increased 4.55-fold in Meg3-null embryos compared with wild-type (P < 0.05). Iqgap1 expression was also significantly higher in the brains of Meg3-null embryos (2.97-fold, P < 0.05). Levels of Hes1 showed a subtle but highly significant (P < 0.005) increase in the brains of Meg3-null embryos compared with wild-type controls (Fig. 1). Expression of Dll4 increased 4.44-fold with loss of Meg3 (P = 0.06), and levels of Wasl increased 2.08-fold in Meg3-null embryos relative to wild-type (P = 0.07). In addition, a 4.33-fold increase in Vegfr2 expression was observed with loss of Meg3 (P = 0.18).
Figure 1.
Gene expression measured by quantitative RT-PCR. Expression of Hes1, Vegfa, Vegfr1, and Iqgap1 was determined by quantitative RT-PCR using RNA extracted from the brains of E18.5 embryos collected from eight Meg3-null embryos and seven wild-type (WT) littermates of six litters. KO, Knockout. Values are the mean fold change of quantitative RT-PCR results relative to wild-type. ***, P ≤ 0.005; **, P ≤ 0.01; *, P ≤ 0.05.
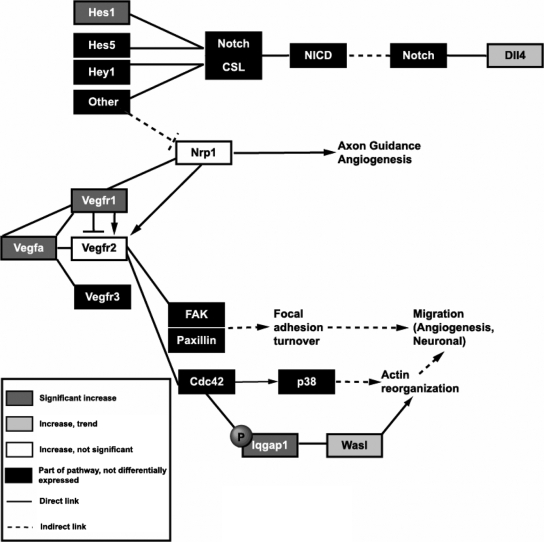
Figure 2 is a simplified schematic representation of the VEGF and Notch signaling pathways, including the major genes and the interaction between these factors, which were affected by loss of Meg3 according to our microarray analysis and quantitative RT-PCR results. Dark gray boxes represent genes (Hes1, Vegfa, Vegfr1, and Iqgap1) with increased expression in Meg3-null brains as predicted by microarray analysis and validated by quantitative RT-PCR. Light gray boxes represent genes (Dll4 and Wasl) with increased expression in Meg3-null brains as predicted by microarray analysis and determined by quantitative RT-PCR to approach statistical significance. White boxes represent genes (Nrp1 and Vegfr2) whose increased expression in Meg3-null brains was not statistically significant. Black boxes refer to genes not differentially expressed on the array.
Figure 2.
Simplified schematic representation of VEGF and Notch signaling cascades showing genes with expression change on the microarray and validated by quantitative RT-PCR. Genes whose increased expression predicted by the microarray analysis was validated by quantitative RT-PCR are represented by dark gray boxes. Light gray boxes denote genes whose increase predicted by microarray analysis approached statistical significance in quantitative RT-PCR assays. Nrp1, represented by a white box, displayed the same direction of change predicted by microarray analysis but did not reach statistical significance. Vegfr2 was not predicted to increase by microarray analysis but was chosen because of the importance of this gene in VEGF signaling. This gene is also denoted by a white box, indicating that the observed increase in expression of this gene was not statistically significant. Black boxes represent other genes that are part of the pathway but not predicted to change with loss of Meg3 by microarray analysis. Solid lines refer to direct interactions and/or effects, and dashed lines refer to indirect action. Arrows denote activation, and perpendicular lines denote repression. FAK, Focal adhesion kinase; Cdc42, cell division control protein 42 homolog; Hey1, Hes-related with YRPW motif 1; CSL, notch-activated coactivator complex of CBF1/Su(H)/Lag2; NICD, Notch intracellular domain.
Increased microvessel formation in the brain of Meg3 knockout embryos
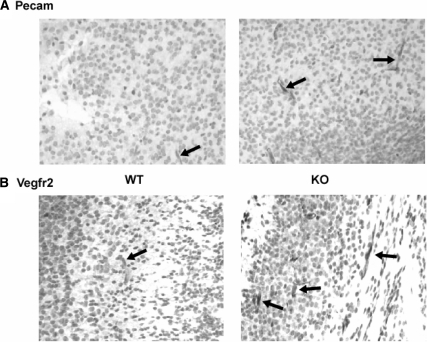
The results from both microarray analysis and quantitative RT-PCR suggest an increase in angiogenesis in the brains of Meg3-null embryos. Microvessel density was measured in equivalent cortical sections from Meg3-null embryos and litter-matched controls to confirm this finding. These sections were stained for Pecam1, a widely used endothelial cell marker (15), and Vegfr2, the main Vegfa receptor (16). Representative cortical sections from wild-type and Meg3-null brains stained with Pecam1 and Vegfr2 are shown in Fig. 3. As shown in Tables 4 and 5, average Pecam1-positive microvessels per mm2 in Meg3-null embryos increased by 30.38% compared with that in wild-type embryos, and Vegfr2 staining in Meg3-null embryos increased by 23.95% compared with that in wild type (P < 0.05 for both markers by repeated measures ANOVA). These data indicate that loss of Meg3 had a positive effect on brain blood vessel development.
Figure 3.
Representative cortical sections of wild-type (WT) and Meg3-null embryos with Pecam1 and Vegfr2 staining. Pecam1 (A) and Vegfr2 (B) staining of a representative cortical section in a wild-type (left) and a littermate-matched Meg3-null (right) embryo at ×200 magnification. KO, Knockout.
Table 4.
Changes in microvessel density visualized by Pecam1 staining with loss of Meg3
| Litter | Average microvessel density (WT) | Average microvessel density (KO) | Percent change |
|---|---|---|---|
| 1 | 72.10 | 123.95 | 41.83 |
| 2 | 75.52 | 121.90 | 38.05 |
| 3 | 78.92 | 113.06 | 30.20 |
| 4 | 67.27 | 81.73 | 17.69 |
| 5 | 72.91 | 86.09 | 15.31 |
| All | 73.34 | 105.35 | 30.38 |
Microvessels were visualized in 5-μm cortical sections from seven Meg3-null embryos and nine wild-type (WT) litter mate controls from five litters by Pecam staining. Microvessel number in three cortical fields per embryo was counted blindly and independently before sample identification codes were released. KO, Knockout.
Table 5.
Changes in microvessel density visualized by Vegfr2 staining with loss of Meg3
| Litter | Average microvessel density (WT) | Average microvessel density (KO) | Percent change |
|---|---|---|---|
| 1 | 57.89 | 121.93 | 52.52 |
| 2 | 43.23 | 82.49 | 47.60 |
| 3 | 99.06 | 128.25 | 22.76 |
| 4 | 87.46 | 110.59 | 20.92 |
| 5 | 87.99 | 108.47 | 18.88 |
| 6 | 101.11 | 109.18 | 7.39 |
| All | 83.77 | 110.15 | 23.95 |
Microvessels were visualized in 5-μm cortical sections from nine Meg3-null embryos and nine wild-type (WT) litter mate controls from six litters by VEGFR2 staining. Microvessel number in three cortical fields per embryo was counted blindly and independently before sample identification codes were released. KO, Knockout.
Discussion
MEG3 is a maternally expressed noncoding RNA gene whose expression is lost in clinically nonfunctioning pituitary adenomas (5,8,31). Previous studies from our lab indicate that this gene regulates p53 function and inhibits cell proliferation in an retinoblastoma protein (Rb) dependent manner (6). Results from this study suggest that Meg3 may regulate Vegf-mediated angiogenesis. Loss of Meg3 leads to up-regulation of signaling pathways related to angiogenesis. The brains of Meg3-null embryos have increased expression of genes in the VEGF (Vegfa, Vegfr1, Iqgap1, and Wasl) and Notch (Hes1 and Dll4) signaling cascades and increased cortical microvessel density compared with that of wild-type controls.
The results from microarray analysis also suggest that Meg3 is important for postembryonic brain function, cell differentiation, and axonal guidance in addition to angiogenesis. Long-term potentiation, calcium, and GnRH signaling pathways are important for postembryonic brain function (32,33), and loss of Meg3 can lead to changes in these signaling pathways according to the results from the microarray analysis. Enrichment of the calcium signaling KEGG pathway and axon extension, homophilic cell adhesion, GTPase activator activity, and actin cytoskeletal organization and biogenesis ontologies in our microarray data suggests that Meg3 regulates axonal guidance. Microarray analysis also predicted a change in Notch signaling with loss of Meg3, which is necessary for cell differentiation in both the brain and pituitary (34,35). Quantitative RT-PCR showed a significant increase in Hes1 levels in Meg3−/− embryos compared with wild-type. Hes1 is a downstream target gene of the Notch signaling pathway, which is essential for proper development and localization of α-glycoprotein subunit-positive and adrenocorticotropic hormone-producing cells in the anterior pituitary (34). All these results suggest that Meg3 is involved in a variety of physiological functions and plays an important role in different biological processes.
Most importantly, our microarray analysis predicted that loss of Meg3 leads to changes in signaling pathways and ontologies important for angiogenesis. Adherens junctions, Notch, and VEGF signaling are all KEGG pathways related to blood vessel development that changed with loss of Meg3. Adherens junctions stabilize interendothelial cell connections in nascent blood vessels by forming connections between the extracellular matrix and structural proteins within the cell (36). Notch signaling is necessary for the stability of newly formed blood vessels (26) and mediates specification of tip cells that direct endothelial cell migration (25,37). VEGF signaling mediates proliferation (17), survival (17), and migration (24,38,39) of endothelial cells during angiogenesis, which are essential to the formation of new blood vessels. We chose to focus most of our validation on VEGF signaling because of its importance in both angiogenesis and tumorigenesis. In particular, changes in this pathway have been observed in clinically nonfunctioning pituitary adenomas, which do not express MEG3, compared with normal tissue (20) and hormone-secreting tumors (21), which express MEG3. Quantitative RT-PCR validation of our microarray findings confirmed that loss of Meg3 leads to increased expression of Vegfa, Vegfr1, Iqgap1, and Wasl, which are part of the VEGF pathway and of importance in angiogenesis. Vegfa is the most angiogenic of the four Vegfs (Vegfa–d) (16,40). It binds to three Vegfrs, Vegfr1–3 (41,42,43), and Vegf coreceptors, such as Nrp1 (23), and mediates formation of F-actin-containing stress fibers during migration through phosphorylation of focal adhesion kinase and Paxillin (39,44). Vegfa signaling also promotes activation of the small GTPases Ras-related C3 botulinum toxin substrate 1 and cell division control protein 42 homolog, which activate Iqgap1 (38,45). Activated Iqgap1 promotes loss of intercellular contacts (46) and Wasl-mediated cytoskeletal remodeling (24), both of which are essential for endothelial cell migration.
Visualization of blood vessels using Pecam1 and Vegfr2 staining indicated significantly greater cortical microvessel density in the brains of Meg3-null embryos compared with wild-type controls, supporting both the microarray and quantitative RT-PCR findings. RT-PCR confirmed significant increases in expression of both Vegfa and Vegfr1 with loss of Meg3. Vegfa positively regulates blood vessel development (16), whereas Vegfr1 has both a positive (47,48,49) and negative (17,50,51) role in angiogenesis. Some reports indicate that Vegfr1 represses Vegfr2 function (50) by acting as a “sink” for available Vegfa (17). However, Vegfr1 knockout embryos have vastly decreased blood vessel branching despite normal endothelial cell proliferation and differentiation (47), suggesting that this protein also has a positive role in blood vessel formation. A recent study showed that VEGF signals through the serine-threonine kinase Akt/ERK pathway to inhibit constitutive ubiquitination and induce rapid Vegfr1 accumulation in endothelial cells. Surprisingly, Vegfr1 is primarily localized in the nucleus of endothelial cells. In contrast, VEGF signals through the c-Jun N-terminal kinase/c-Jun pathway to induce endocytosis, nuclear translocation, and down-regulation of Vegfr2 via ubiquitination. VEGFR1 signaling is required for endothelial-cell survival, whereas VEGFR2 regulates capillary tube formation (49). Therefore, it is possible that local factors and other components in the VEGF signaling pathway are involved in determining the functions of VEGF receptors. In our study, we found a significant increase in Vegfr2 staining in the brains of Meg3-null embryos, in addition to increased expression of Vegfa. Furthermore, immunohistochemical staining clearly demonstrated an increase in microvessel density associated with the loss of Meg3. Taken together, our model system suggests that Meg3 functions to suppress VEGF signaling and angiogenesis.
At this moment, there is very little known about the mechanisms for the actions of MEG3, which encodes a noncoding RNA instead of a protein. We have previously shown that MEG3 RNA stimulates p53-mediated transcriptional activation; p53 is known to negatively regulate VEGFA transcription through binding to the transcription factor Sp1 sites on the VEGFA promoter (52,53,54). Therefore, it is possible that loss of MEG3 leads to reduced p53 activity and increased transcription of VEGFA. Recently, it has been shown that nutlin-3, an activator of the p53 pathway, functions to inhibit angiogenesis (55). Interestingly, both MEG3 and nutlin-3 activate GDP-15, a p53 downstream target with tumor suppressive function (6,55). Therefore, activation of the p53 network could be one of the possible mechanisms for the antiangiogenic effects of MEG3.
It has been reported that clinically nonfunctioning pituitary adenomas are more vascularized than hormonally functioning tumors (21). In addition, these tumors do not have centrally necrotic regions despite their large size (56), which is indicative of active angiogenesis. Based on these findings, together with results from our studies, we hypothesize that human MEG3 acts as a tumor suppressor, one of whose functions includes regulation of angiogenesis. One possible mechanism by which MEG3 regulates angiogenesis is the negative regulation of VEGF pathway genes. We hypothesize that loss of expression of this noncoding RNA gene in clinically nonfunctioning pituitary adenomas could enhance blood vessel development and allow for continual growth of these tumors. In addition, changes in other signaling pathways with loss of MEG3 may contribute to the development of nonfunctioning pituitary adenomas in humans.
Supplementary Material
Acknowledgments
We thank Dr. Hang Lee for his assistance with statistical analysis and Dr. Roderick Bronson for his assistance with the mouse brain histology.
Footnotes
This work was supported in part by National Institutes of Health Grants DK40947, UL1 RR025758, and F32DK09218, the Guthart Family Foundation, and the Jarislowsky Foundation.
Disclosure Summary: The authors have nothing to disclose.
First Published Online April 14, 2010
Abbreviations: Dll4, Delta-like 4; E, embryonic day; GO, Gene Ontology; GTPase, enzymes with guanosine triphosphate (GTP) binding and hydrolysis activity; Hes1, hairy and enhancer of Split 1; Iqgap1, IQ motif containing GTPase activating protein 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; MEG3, maternally expressed gene 3; Nrp1, Neuropilin 1; P-calls, present calls; Pecam1, platelet/endothelial cell adhesion molecule 1; Vegfa, Vegf A; Vegfr2, vascular endothelial growth factor receptor 2; Wasl, Wiskott-Aldrich syndrome-like.
References
- Asa SL, Ezzat S 2002 The pathogenesis of pituitary tumours. Nat Rev Cancer 2:836–849 [DOI] [PubMed] [Google Scholar]
- Dekkers OM, Hammer S, de Keizer RJ, Roelfsema F, Schutte PJ, Smit JW, Romijn JA, Pereira AM 2007 The natural course of non-functioning pituitary macroadenomas. Eur J Endocrinol 156:217–224 [DOI] [PubMed] [Google Scholar]
- Dekkers OM, de Keizer RJ, Roelfsema F, Vd Klaauw AA, Honkoop PJ, van Dulken H, Smit JW, Romijn JA, Pereira AM 2007 Progressive improvement of impaired visual acuity during the first year after transsphenoidal surgery for non-functioning pituitary macroadenoma. Pituitary 10:61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colao A, Di Somma C, Pivonello R, Faggiano A, Lombardi G, Savastano S 2008 Medical therapy for clinically non-functioning pituitary adenomas. Endocr Relat Cancer 15:905–915 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A 2003 A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 88: 5119–5126 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A 2007 Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282:24731–24742 [DOI] [PubMed] [Google Scholar]
- Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A 2005 Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab 90:2179–2186 [DOI] [PubMed] [Google Scholar]
- Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A 2008 Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab 93:4119–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, Grundy R, Ferguson-Smith AC, Maher ER 2005 Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour. Br J Cancer 92:1574–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Dasoula A, Hatzimichael E, Georgiou I, Syrrou M, Bourantas KL 2008 Promoter hypermethylation of the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin Lymphoma Myeloma 8:171–175 [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Tevendale M, Knowles E, Takada S, Watkins M, Ferguson-Smith AC 2007 Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: implications for cis-acting control. Dev Biol 306:810–823 [DOI] [PubMed] [Google Scholar]
- McLaughlin D, Vidaki M, Renieri E, Karagogeos D 2006 Expression pattern of the maternally imprinted gene Gtl2 in the forebrain during embryonic development and adulthood. Gene Expr Patterns 6:394–399 [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A 1998 The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn 212:214–228 [DOI] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, Black PM, von Deimling A, Pomeroy SL, Golub TR, Louis DN 2003 Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res 63:1602–1607 [PubMed] [Google Scholar]
- DeLisser HM, Christofidou-Solomidou M, Strieter RM, Burdick MD, Robinson CS, Wexler RS, Kerr JS, Garlanda C, Merwin JR, Madri JA, Albelda SM 1997 Involvement of endothelial PECAM-1/CD31 in angiogenesis. Am J Pathol 151:671–677 [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C 2003 VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol 161:1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, LeCouter J 2003 The biology of VEGF and its receptors. Nat Med 9:669–676 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000 KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G 2000 Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe CJ, Boelaert K, Tannahill LA, Heaney AP, Stratford AL, Khaira JS, Hussain S, Sheppard MC, Franklyn JA, Gittoes NJ 2002 Vascular endothelial growth factor, its receptor KDR/Flk-1, and pituitary tumor transforming gene in pituitary tumors. J Clin Endocrinol Metab 87:4238–4244 [DOI] [PubMed] [Google Scholar]
- Niveiro M, Aranda FI, Peiró G, Alenda C, Picó A 2005 Immunohistochemical analysis of tumor angiogenic factors in human pituitary adenomas. Hum Pathol 36:1090–1095 [DOI] [PubMed] [Google Scholar]
- Vieira JM, Schwarz Q, Ruhrberg C 2007 Selective requirements for NRP1 ligands during neurovascular patterning. Development 134:1833–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Takagi H, Otani A, Koyama S, Kemmochi S, Uemura A, Honda Y 2002 Selective induction of neuropilin-1 by vascular endothelial growth factor (VEGF): a mechanism contributing to VEGF-induced angiogenesis. Proc Natl Acad Sci USA 99:383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, Kroschewski R 2007 IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem 282:426–435 [DOI] [PubMed] [Google Scholar]
- Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalén M, Gerhardt H, Betsholtz C 2007 Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445:776–780 [DOI] [PubMed] [Google Scholar]
- Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, Gerhardt H 2009 Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev Cell 16:70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CK, Li JL, Murga M, Harris AL, Tosato G 2006 Up-regulation of the Notch ligand Δ-like 4 inhibits VEGF-induced endothelial cell function. Blood 107:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R 2008 Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development 135:2531–2541 [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M 2007 Δ-Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res 35:4583–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R 2004 Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131:5539–5550 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F 2000 Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5:211–220 [DOI] [PubMed] [Google Scholar]
- Ottem EN, Godwin JG, Petersen SL 2002 Glutamatergic signaling through the N-methyl-D-aspartate receptor directly activates medial subpopulations of luteinizing hormone-releasing hormone (LHRH) neurons, but does not appear to mediate the effects of estradiol on LHRH gene expression. Endocrinology 143:4837– 4845 [DOI] [PubMed] [Google Scholar]
- Krsmanovic LZ, Mores N, Navarro CE, Tomić M, Catt KJ 2001 Regulation of Ca2+-sensitive adenylyl cyclase in gonadotropin-releasing hormone neurons. Mol Endocrinol 15:429–440 [DOI] [PubMed] [Google Scholar]
- Himes AD, Raetzman LT 2009 Premature differentiation and aberrant movement of pituitary cells lacking both Hes1 and Prop1. Dev Biol 325:151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F 2008 Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135:3113–3124 [DOI] [PubMed] [Google Scholar]
- Wallez Y, Huber P 2008 Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim Biophys Acta 1778:794–809 [DOI] [PubMed] [Google Scholar]
- Phng LK, Gerhardt H 2009 Angiogenesis: a team effort coordinated by notch. Dev Cell 16:196–208 [DOI] [PubMed] [Google Scholar]
- Mataraza JM, Briggs MW, Li Z, Entwistle A, Ridley AJ, Sacks DB 2003 IQGAP1 promotes cell motility and invasion. J Biol Chem 278:41237–41245 [DOI] [PubMed] [Google Scholar]
- Le Boeuf F, Houle F, Huot J 2004 Regulation of vascular endothelial growth factor receptor 2-mediated phosphorylation of focal adhesion kinase by heat shock protein 90 and Src kinase activities. J Biol Chem 279:39175–39185 [DOI] [PubMed] [Google Scholar]
- Turner HE, Harris AL, Melmed S, Wass JA 2003 Angiogenesis in endocrine tumors. Endocr Rev 24:600–632 [DOI] [PubMed] [Google Scholar]
- de Vries C, Escobedo JA, Ueno H, Houck K, Ferrara N, Williams LT 1992 The fms-like tyrosine kinase, a receptor for vascular endothelial growth factor. Science 255:989–991 [DOI] [PubMed] [Google Scholar]
- Matthews W, Jordan CT, Gavin M, Jenkins NA, Copeland NG, Lemischka IR 1991 A receptor tyrosine kinase cDNA isolated from a population of enriched primitive hematopoietic cells and exhibiting close genetic linkage to c-kit. Proc Natl Acad Sci USA 88:9026–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T, Zarkada G, Wallgard E, Murtomäki A, Suchting S, Wirzenius M, Waltari M, Hellström M, Schomber T, Peltonen R, Freitas C, Duarte A, Isoniemi H, Laakkonen P, Christofori G, Ylä-Herttuala S, Shibuya M, Pytowski B, Eichmann A, Betsholtz C, Alitalo K 2008 Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 454:656–660 [DOI] [PubMed] [Google Scholar]
- Rousseau S, Houle F, Kotanides H, Witte L, Waltenberger J, Landry J, Huot J 2000 Vascular endothelial growth factor (VEGF)-driven actin-based motility is mediated by VEGFR2 and requires concerted activation of stress-activated protein kinase 2 (SAPK2/p38) and geldanamycin-sensitive phosphorylation of focal adhesion kinase. J Biol Chem 275:10661–10672 [DOI] [PubMed] [Google Scholar]
- Lamalice L, Houle F, Jourdan G, Huot J 2004 Phosphorylation of tyrosine 1214 on VEGFR2 is required for VEGF-induced activation of Cdc42 upstream of SAPK2/p38. Oncogene 23:434–445 [DOI] [PubMed] [Google Scholar]
- Yamaoka-Tojo M, Tojo T, Kim HW, Hilenski L, Patrushev NA, Zhang L, Fukai T, Ushio-Fukai M 2006 IQGAP1 mediates VE-cadherin-based cell-cell contacts and VEGF signaling at adherence junctions linked to angiogenesis. Arterioscler Thromb Vasc Biol 26:1991–1997 [DOI] [PubMed] [Google Scholar]
- Fong GH, Rossant J, Gertsenstein M, Breitman ML 1995 Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature 376:66–70 [DOI] [PubMed] [Google Scholar]
- Kappas NC, Zeng G, Chappell JC, Kearney JB, Hazarika S, Kallianos KG, Patterson C, Annex BH, Bautch VL 2008 The VEGF receptor Flt-1 spatially modulates Flk-1 signaling and blood vessel branching. J Cell Biol 181:847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Neiva KG, Lingen MW, Ellis LM, Nor JE 2010 VEGF-dependent tumor angiogenesis requires inverse and reciprocal regulation of VEGFR1 and VEGFR2. Cell Death Differ 17:499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H, Kowalski J, Yu L, Chen H, Pisabarro MT, Davis-Smyth T, Ferrara N 2000 A repressor sequence in the juxtamembrane domain of Flt-1 (VEGFR-1) constitutively inhibits vascular endothelial growth factor-dependent phosphatidylinositol 3′-kinase activation and endothelial cell migration. EMBO J 19:4064–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin CH 1994 Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem 269:26988–26995 [PubMed] [Google Scholar]
- Pal S, Datta K, Mukhopadhyay D 2001 Central role of p53 on regulation of vascular permeability factor/vascular endothelial growth factor (VPF/VEGF) expression in mammary carcinoma. Cancer Res 61:6952–6957 [PubMed] [Google Scholar]
- Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH 2008 The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol Cancer Ther 7:880–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wu J, Oyesanya RA, Lee Z, Mukherjee A, Fang X 2009 Sp-1 and c-Myc mediate lysophosphatidic acid-induced expression of vascular endothelial growth factor in ovarian cancer cells via a hypoxia-inducible factor-1-independent mechanism. Clin Cancer Res 15:492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchiero P, Corallini F, Gonelli A, Dell'Eva R, Vitale M, Capitani S, Albini A, Zauli G 2007 Antiangiogenic activity of the MDM2 antagonist nutlin-3. Circ Res 100:61–69 [DOI] [PubMed] [Google Scholar]
- Honegger J, Zimmermann S, Psaras T, Petrick M, Mittelbronn M, Ernemann U, Reincke M, Dietz K 2008 Growth modelling of non-functioning pituitary adenomas in patients referred for surgery. Eur J Endocrinol 158:287–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.