Abstract
A fundamental issue in cell biology is how migratory cell behaviors are controlled by dynamically regulated cell adhesion. Vertebrate neural crest (NC) cells rapidly alter cadherin expression and localization at the cell surface during migration. Secreted Wnts induce some of these changes in NC adhesion and also promote specification of NC-derived pigment cells. Here, we show that the zebrafish transcription factor Ovo1 is a Wnt target gene that controls migration of pigment precursors by regulating the intracellular movements of N-cadherin (Ncad). Ovo1 genetically interacts with Ncad and its depletion causes Ncad to accumulate inside cells. Ovo1-deficient embryos strongly upregulate factors involved in intracellular trafficking, including several rab GTPases, known to modulate cellular localization of cadherins. Surprisingly, NC cells express high levels of many of these rab genes in the early embryo, chemical inhibitors of Rab functions rescue NC development in Ovo1-deficient embryos and overexpression of a Rab-interacting protein leads to similar defects in NC migration. These results suggest that Ovo proteins link Wnt signaling to intracellular trafficking pathways that localize Ncad in NC cells and allow them to migrate. Similar processes probably occur in other cell types in which Wnt signaling promotes migration.
Keywords: Neural crest, N-cadherin, Wnt, Zebrafish
INTRODUCTION
Migratory cells must dynamically regulate adhesion as they move. Neural crest (NC) cells in vertebrate embryos are highly migratory progenitor cells that form diverse cell types including cartilage and bone of the skull, sensory neurons and glia of the peripheral nervous system, and pigment cells of the skin (Le Douarin et al., 2004). These cells actively regulate expression of cadherins on their surfaces as they delaminate and migrate from the dorsolateral edges of the neural tube (Monier-Gavelle and Duband, 1995; Nakagawa and Takeichi, 1995; Nakagawa and Takeichi, 1998). Members of the Wnt family of secreted signals induce NC cells to initiate migration but their roles in cadherin regulation and morphogenesis are largely unknown.
NC precursors in the neural ectoderm initially express both Cadherin 6b (Cad6b) and N-cadherin (Ncad; Cdh2 – Zebrafish Information Network), but downregulate both and upregulate Cadherin7 (Cad7) as they undergo an epithelial-mesenchymal transition (EMT) and begin to migrate (Nakagawa and Takeichi, 1995; Akitaya and Bronner-Fraser, 1992). Functions for individual cadherins in this process remain unclear. For example, either increasing or decreasing Ncad protein levels disrupts NC cell migration in avian embryos (Nakagawa and Takeichi, 1998). Also, overexpressing full-length Ncad inhibits NC migration, whereas a truncated version translocates to the nucleus and promotes cell cycle progression and EMT in response to Bmp signaling (Shoval et al., 2007). Ncad−/− mutant NC cells in mice migrate in inappropriate directions (Xu et al., 2001), whereas defects in neural tube closure (but not in NC) have been reported in Ncad−/− mutant zebrafish (Lele et al., 2002).
NC cells establish transient cell-cell contacts during migration, possibly by regulating Ncad localization to their filopodial tips (Monier-Gavelle and Duband, 1995). But how is this localization regulated? Members of the Rab family of GTPases are attractive candidates. Rab11-mediated recycling of Drosophila Ecad regulates cell intercalation during tracheal morphogenesis in response to Wnt signaling (Shaye et al., 2008). Interestingly, this regulation of cell adhesion is mediated by Spalt, a Wnt-responsive transcription factor (Shaye et al., 2008). These studies suggest roles for intracellular trafficking in cadherin localization and cell migration through tissue-specific transcriptional control in response to Wnt signaling.
Wnt signaling not only induces EMT in NC cells but also specifies NC lineages that give rise to pigment cells (Dorsky et al., 1998). In zebrafish, single NC cells labeled adjacent to the neural tube give rise to clones of progeny that are lineally restricted to single cell types (e.g. skeletal, neural, pigment), suggesting that fates are specified prior to migration (Schilling and Kimmel, 1994; Raible et al., 1994). Overexpression of a truncated, dominant-negative form of Tcf3 (dnTcf3) in individual NC cells inhibits pigment cell formation, whereas an activated form of β-catenin (βcat; Ctnnb – Zebrafish Information Network) promotes pigment cell fates (Dorsky et al., 1998). Wnt signaling also directly promotes mitfa expression, a key regulator of melanocytes (Dorsky et al., 2000). Wnt3a added to chick NC explants increases melanocyte number (Jin et al., 2001), whereas Wnt1−/−/Wnt3a−/− double-mutant mice form fewer melanoblasts and NC cells in general (Ikeya et al., 1997). Interestingly, Wnt signals regulate NC cell EMT, at least in the trunk, by controlling cyclin D expression and cell cycle progression (Burstyn-Cohen and Kalcheim, 2002; Burstyn-Cohen et al., 2004). Wnts also activate snail transcription factors, well-known transcriptional repressors of E-cadherin (Ecad; Cdh1 – Zebrafish Information Network) and Cad6b, providing a possible direct link between Wnt functions in cell fate determination and adhesion (Aybar et al., 2003; Cano et al., 2000; Taneyhill et al., 2007; Vallin et al., 2001).
In addition to snail, Wnt signaling also regulates expression of other zinc finger transcription factors, including members of the evolutionarily conserved family of Ovo genes (Dai et al., 1998; Mackay et al., 2006; Mevel-Ninio et al., 1991; Oliver et al., 1994). Drosophila ovo is a direct Wnt target involved in germline development and epidermal appendage formation (Khila et al., 2003; Mevel-Ninio et al., 1991; Payre et al., 1999). Mice have three Ovo-like (Ovol) genes, Ovol1-3, and Ovol1 is a Wnt target in the mammalian germline and epidermis (Dai et al., 1998; Li et al., 2002). By contrast, Ovol2 is required for neurulation and cranial NC migration; a subset of NC cells remains attached to the neural tube in Ovol2 mutant mice (Mackay et al., 2006). However, whether or not this reflects a role for Ovo downstream of Wnt proteins or in controlling NC cell adhesion and fate remains unclear.
Here we report the characterization of zebrafish ovo1 and show that it is a Wnt target that controls NC migration, in part by regulating intracellular trafficking of Ncad. Depletion of Ovo1 in vivo causes a subset of NC cells to cluster in the dorsal midline above the neural tube. Ovo1 genetically interacts with Ncad and promotes its membrane localization. One possible mechanism is through inhibition of rab expression. rab overexpression accounts for at least part of the NC defects in Ovo1 morphants as (1) a chemical inhibitor of Rab function rescues the morphant phenotype, and (2) misexpression of a rab11-interacting factor causes similar NC migration defects. These data suggest that Ovo1 links Wnt signaling to changes in NC cell adhesion by regulating Ncad localization to the membrane, which has important consequences for subsequent NC cell fates.
MATERIALS AND METHODS
Morpholinos and mRNA injections
Antisense morpholino oligonucleotides (MO) targeting the Ovo1 translation start site (Ovo1 ATG MO), Ovo1 5′ untranslated region (Ovo1 5′UTR MO) and Rab11fip2 exon2 splice-donor site (Fip2 MO) were purchased from Gene Tools and dissolved in 1× Danieau's buffer for injection (see Table S2 in the supplementary material). The Ncad MO was described previously (Lele et al., 2002). To test the efficacy of Ovo1 MOs, the target sequences were fused to Gfp, subcloned into pCS2+ to synthesize capped mRNA (mMessage mMachine Kit, Ambion), and co-injected with or without Ovo1 MO into 1- to 4-cell-stage embryos and assayed for Gfp expression. Ovo1 5′UTR MO was most effective and used for all subsequent experiments (Ovo1 MO). Fip2 MO efficacy was assayed by RT-PCR to detect alternatively spliced products. For single MO experiments, 3 ng of Ovo1 MO, 2 ng p53 MO, 0.75-3 ng Fip2 MO and 1 nl of a 50 μM solution of Ncad MO were injected into 1- to 4-cell-stage embryos. For genetic interactions studies, the amount of each MO injected was decreased by half. For rescue experiments, the full-length open reading frame (ORF) of ovo1 was cloned into the pEGFP:C3 vector and subcloned into pCS2+ to generate a gfp:ovo1 fusion construct. gfp:ovo1 mRNA (85 pg) was injected at the 1-cell stage.
To study Ncad subcellular localization, 1-cell-stage embryos were injected with 5-50 pg of linearized ncad:gfp plasmid (Jontes et al., 2004) and then photographed at 8-10 hours post-fertilization (hpf) with a Zeiss META 510 confocal microscope.
For rab11fip2 overexpression studies, the full-length ORF was cloned into XhoI sites of pCS2+ (see Table S3 in the supplementary material) and verified by sequencing with T7 and SP6. rab11fip2 mRNA was co-injected with 100 pg mCherry mRNA as a tracer into sox10(7.2):gfp transgenic embryos and photographed using a Zeiss Axioplan 2 microscope.
Heat-shock experiments
To test responses of genes (ovo1, rab3c, rab12, rab11fip2, sec6) to Wnt signaling, heterozygous Tg(hs:dnTcf-GFP)w26+/− males were outcrossed to wild-type females (Lewis et al., 2004). Transgenic embryos at 13 hpf were heat-shocked for 1 hour at 37°C and immediately transferred to embryo medium at room temperature. Gfp-positive (dnTcf3 transgenics) and Gfp-negative (wild type) controls were immersed in Trizol reagent 30 minutes post-heat-shock for ovo1 expression studies or 2 hours post-heat-shock for rab3c, rab12, rab11fip2 and sec6 expression studies, and mRNA was isolated for qPCR (see below).
For rescue experiments, fertilized eggs of Tg(hs:dnTcf-GFP)w26 heterozygotes (in similar wild-type outcrosses) were injected with mRNA encoding a gfp:ovo1 fusion protein and similarly heat-shocked at 10 hpf. Embryos were fixed 30 minutes after heat-shock and processed for gene expression by in situ hybridization.
Cell transplantation
Cells were grafted at gastrula stages from sox10(7.2):gfp transgenic donors into non-transgenic hosts. Cells were co-transplanted from control donors (injected with 2 ng of p53 MO to prevent MO-induced apoptosis) and rab11fip2-overexpressing donors were co-injected with 1.5 ng Ovo1 MO and 2 ng p53 MO (Fig. 7F,G) into wild-type hosts, or in some cases rab11fib2 mRNA, Ovo1 MO and p53 MO-injected cells were grafted alone (Fig. 7H-M). Embryos were selected based on NC-specific sox10(7.2):gfp expression and photographed at 24 hpf.
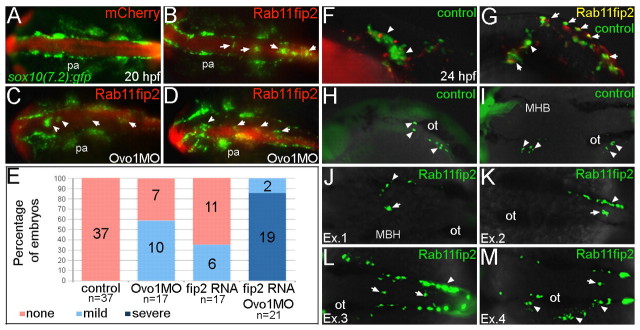
Fig. 7.
Overexpression of rab11fip2 disrupts NC cell migration and exacerbates the Ovo1 morphant phenotype. (A-E) Rab11fip2 and mCherry mRNAs were injected into sox10:gfp transgenics, in which NC cells fluoresce green in living embryos. Dorsal views, anterior to the left at 24 hpf. (A) Controls injected with mCherry mRNA alone show bilateral sox10:gfp+ cells. (B) By contrast, Gfp-positive cells aggregate in the dorsal midline (arrows) following co-injection of Rab11fip2 and mCherry mRNA. (C,D) Larger aggregates (>5 cells, arrowheads) form over the midbrain in embryos co-injected with subthreshold levels of Rabfip2 mRNA and Ovo1 MO. (E) Quantitation showing proportions of wild-type (red), mild (pale blue; <5 cells per aggregate located over the hindbrain) and severe (dark blue; >5 cells per aggregate located over both the midbrain and hindbrain) NC defects. (F-M) Cell transplantation of rab11fip2 mRNA-injected cells into wild-type hosts. (F,G) Co-transplantation of sox10:gfp, Rab11fip2, mCherry mRNA-injected (green and red) and sox10:gfp uninjected control cells (green) into wild-type hosts; lateral views. Rab11fip2-overexpressing cells remain dorsally located (arrows in G). (H-M) Transplantation of control sox10:gfp+ (H,I) or Rab11fip2 mRNA-injected sox10:gfp+ cells (J-M) into uninjected hosts; lateral view (H), dorsal views (I-M). Many Rab11fip2 overexpressing NC cells remain in the dorsal midline [arrows; examples (Ex) 1-4]. Arrowheads in F-M indicate NC cells that have migrated normally. MHB, mid-hindbrain boundary; ot, otic vesicle; pa, pharyngeal arches.
RNA in situ hybridization and immunohistochemistry
Wholemount RNA in situ hybridization was performed as previously described (Thisse et al., 1993). ovo1 (GenBank accession number CN021344) was subcloned into EcoRI and NotI sites of pCS2+ and T3 RNA polymerase was used for probe synthesis. Probes used for NC cell analysis included sox10, mitfa, gch, dlx2a, foxd3 and mbp (Knight et al., 2003). rab3c, sec6 and rab11fip2 were TOPO-cloned using primers listed in Table S3 in the supplementary material. For rab12, the predicted ORF was directionally cloned into pCS2+ and T7 RNA polymerase was used for probe synthesis.
To detect Ncad:Gfp, we performed wholemount immunohistochemistry using an anti-Gfp primary antibody (1:1000, Abcam) followed by an anti-rabbit IgG secondary antibody conjugated to Alexa Fluor488 (1:1000, Molecular Probes Invitrogen). Briefly, embryos were washed in PBT (phosphate buffer with 0.1% Tween 20) after overnight fixation in 4% paraformaldehyde and blocked with 5% goat serum for at least 1 hour at room temperature. Incubations with primary and secondary antibodies were performed overnight at 4°C with extensive washing in PBT in between.
Genotyping of pacpar2.10 mutants
To test genetic interactions between ncad and either ovo1 or rab11fip2, parachutepar2.10 (pacpar2.10; ncad) heterozygotes were first outcrossed to sox10(7.2):gfp transgenics, to label the NC cells in live embryos, and injected with subthreshold amounts of Ovo1 MO or rab11fip2 sense RNA. Individual embryos were separated based on phenotype and mRNA was extracted for subsequent RT-PCR genotyping as previously reported (Lele et al., 2002) using pacpar2.10 sense and antisense primers (Table S3 in the supplementary material). Briefly, a 266 bp ncad transcript appears in wild-type siblings, whereas pacpar2.10 heterozygous mutants exhibit additional larger and smaller bands (see Fig. S4 in the supplementary material).
Microarrays
Total RNA from control and Ovo1 morphant, sox10(7.2):gfp transgenic embryos was isolated using Trizol reagent (Gibco/BRL) from 12 hpf embryos, when morphants first show NC defects. Experiments were performed in triplicate. RNA samples were processed as per manufacturer's instructions (Affymetrix GeneChip Expression Analysis Technical Manual, Affymetric, Santa Clara, CA, USA). cRNA was hybridized to probe sets present on an Affymetrix GeneChip Zebrafish Genome Array at the MicroArray Facility at UCI. The results were quantified and analyzed using Expression Console ver.1.1 software (Affymetrix) using the PLIER algorithm default values. Microarray gene expression was further analyzed using the DNASTAR ArrayStar program, Version 2.0.0, build 61 (Madison, WI, USA), selecting for genes with a 2-fold or greater difference between samples. Microarray data are available at GEO with accession GSE21529.
Quantitative real time PCR
Total RNA was isolated from embryos using Trizol reagent (Gibco/BRL). First strand cDNA synthesis was performed on 1 μg of total RNA using oligo dT primers and Superscript III reverse transcriptase (Invitrogen). Quantitative real time RT-PCR (qPCR) was performed using the SYBR Green PCR mix (Roche Applied Science) in a DNA Engine Opticon continuous fluorescence detection system (MJ Research) using the qPCR primer sets in Table S3 in the supplementary material.
All samples were quantified by the comparative cycle threshold (Ct) method for relative quantification of gene expression, normalized to ef1a (Livak and Schmittgen, 2001). Differences between two groups were analyzed using a two-tailed Student's t-test assuming unequal variances.
Chemical treatments
For Brefeldin A (BFA) experiments, sox10(7.2):gfp transgenic and/or Ovo1 morphant embryos at 10-11 hpf were treated with 1 μM BFA (initially dissolved in 100% ethanol) in embryo medium (4% dimethyl sulfoxide, DMSO) overnight at 28.5°C. For Ncad:Gfp localization studies, morphant embryos at 6 hpf were similarly treated with BFA for 3 hours prior to fixation at 9 hpf. Control embryos were treated identically, but with 4% DMSO alone in embryo medium.
Confocal imaging and movies
For analysis of NC cell migration, NC cells were labeled by the sox10(7.2):gfp transgene starting at 12-14 hpf. Transgenic embryos were manually dechorionated, anesthetized with ethyl-m-aminobenzoate methane sulfanate and mounted in 1% agarose in embryo medium on a coverslip. Approximately 70 μm z-stacks at 4 μm intervals were captured using a Zeiss LSM510 META confocal fluorescence microscope. Movies were assembled using ImageJ software and converting to Quicktime at 4 frames per second.
RESULTS
Identification and expression of zebrafish ovo genes
In Ovol2 mutant mice, a subset of cranial NC cells fails to migrate (Mackay et al., 2006). To investigate the roles of ovo genes in NC development, we isolated two zebrafish ovo orthologs, ovo1 and ovo3, by sequence similarity to mammalian Ovol1 (see Fig. S1A in the supplementary material). Both Ovo proteins contain the four C2H2 zinc finger motifs and the nuclear localization signal (NLS; see Fig. S1A in the supplementary material, underlined black and blue, respectively) characteristic of Ovo transcription factors, as well as the 10 amino acid SNAG domain, thought to act as a transcriptional repressor, at the N-terminus (see Fig. S1A in the supplementary material, red box). Phylogenetic analyses suggest that zebrafish ovo1 is an ortholog of murine Ovol1, with which it shares partial gene synteny (see Fig. S1B in the supplementary material). By contrast, ovo3 is more distantly related and conserved in pufferfish, Fugu rubripes, whereas a clear ortholog of Ovol2 has not been identified. Consistent with a function as a transcription factor, fusions of ovo1 or ovo3 to egfp (ovo:egfp) injected into zebrafish embryos localized to cell nuclei (see Fig. S1C,C′ in the supplementary material). Although we focus here on a functional analysis of ovo1, we obtained largely similar results for ovo3.
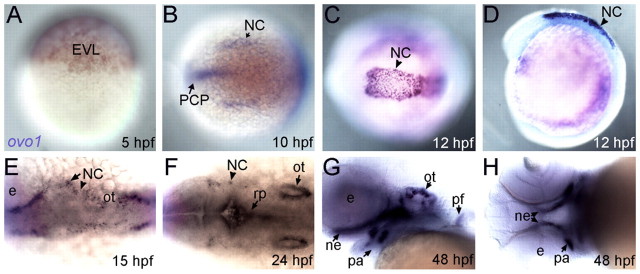
Zebrafish ovo1 expression was first detected by in situ hybridization after the mid-blastula transition in the enveloping layer (EVL; Fig. 1A), although earlier maternal expression was detected by RT-PCR (see Fig. S2A in the supplementary material). Expression was later restricted to premigratory neural crest (NC) at 10-12 hours post-fertilization (hpf) as well as prechordal plate mesoderm (PCP; Fig. 1B-D). By 15-24 hpf, ovo1 expression was detected in subsets of migratory NC cells (Fig. 1E,F), otic placodes and in the roof plate of the neural tube (Fig. 1F). By 48 hpf, expression was detected in the pharyngeal arches, particularly in the first and second arches and presumptive neurocranium (Fig. 1G,H). One consistent and potentially interesting feature of ovo1 mRNA expression was its dynamic localization within cells, not only in the perinuclear cytoplasm, as seen for most mRNAs (Fig. 1A,B,G,H; arrows), but also in punctate foci within cell nuclei (Fig. 1C,D; arrowheads).
Fig. 1.
Expression profile of ovo1. Wholemount in situ hybridization. (A) Enveloping layer (EVL) expression at 5 hours post-fertilization (hpf). (B) Neural crest (NC) and prechordal plate (PCP) expression at 10 hpf (dorsal view, anterior to the left). (C,D) Expression in premigratory cranial NC cells at 12 hpf (C, dorsal view; D, lateral view). (E,F) Expression in migrating cranial NC cells at 15 and 24 hpf (dorsal views). Additional expression at 24 hpf in otic vesicles (ot) and roof plate (rp; F, arrows). (G,H) Expression at 48 hpf in pharyngeal arches (pa), pectoral fins (pf), otic vesicles and neurocranium (ne; G, lateral view; H, ventral view). e, eye.
Lineage-specific requirements for Ovo in neural crest development
To investigate the functions of Ovo genes in zebrafish we designed antisense morpholino oligonucleotides (MOs) to create functional knockouts or ‘knockdowns’. Using the mouse Ovo-like proteins to deduce the translational start site, we targeted a translation-blocking MO to the 5′UTR of Ovo1. To test MO efficacy, we fused gfp to the 3′ end of the Ovo1 MO target site (ovo1 5′utr:gfp) and co-injected sense mRNA derived from this construct (500 pg) with 3 ng of the Ovo1 MO, which completely eliminated Gfp expression (see Fig. S2B,C in the supplementary material). To test MO specificity, a second MO was targeted to the translational start site, which caused an identical phenotype (data not shown).
In wild-type embryos, sox10 expression marks migrating NC in stripes on either side of the neural tube at 24 hpf (Fig. 2A; see also Movie 1 in the supplementary material). Injection of 3 ng Ovo1 MO caused groups of sox10+ cells to accumulate in the dorsal midline (Fig. 2B; see Movie 2 in the supplementary material). To determine the identities of these midline sox10+ cells, we examined markers of different NC lineages. Both mitfa, which marks melanocyte precursors (Fig. 2C,D), and gch, which marks xanthophore precursors (Fig. 2E,F) (Lister et al., 2006), were detected in the dorsal midline at 28 hpf in Ovo1 MO-injected embryos (morphants) in similar patterns to sox10. However, strikingly, we did not detect defects in patterns of dlx2 (putative skeletal precursors; Fig. 2G,H) or foxd3 expression (Fig. 2I,J) in Ovo1 morphants, suggesting that defects are restricted to pigment cell lineages. Consistent with these early defects, later numbers of NC-derived melanocytes were strongly reduced in morphants (see Fig. S3A-D in the supplementary material; arrows), whereas jaw morphology appeared normal (see Fig. S3C,D in the supplementary material; arrowheads).
Fig. 2.
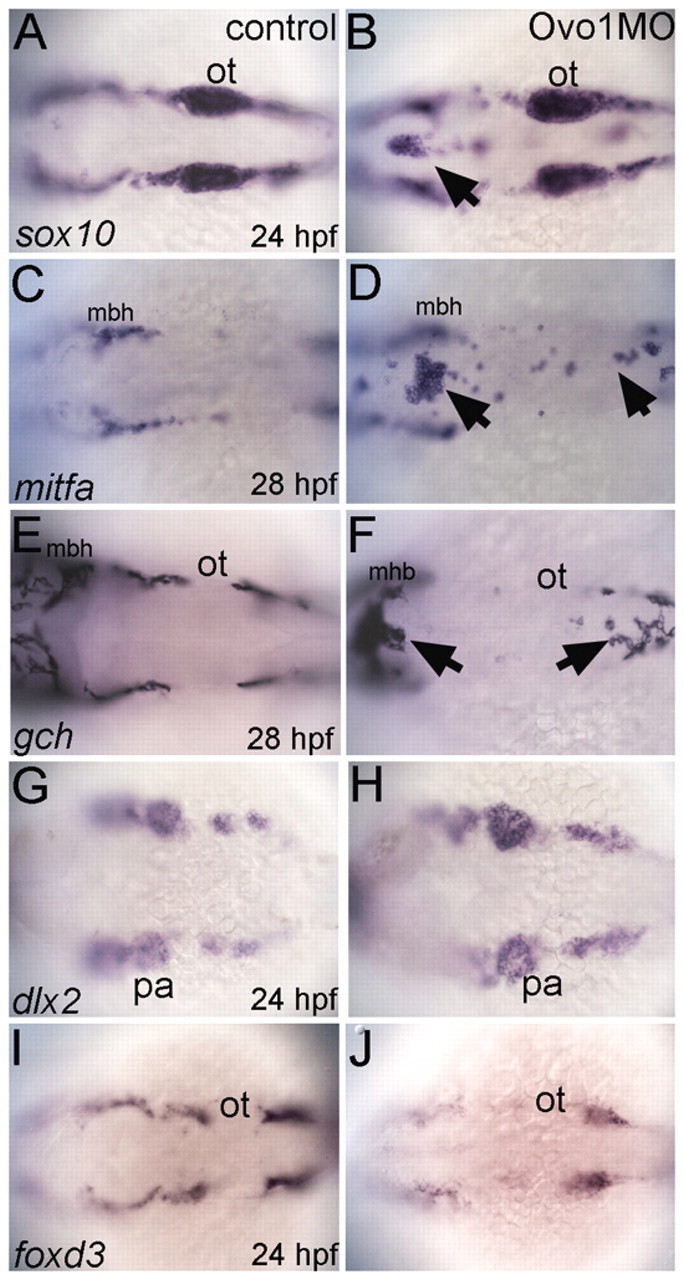
Ovo1 is required for the migration of NC-derived pigment precursors. Dorsal views, anterior to the left. (A,C,E,G,I) Controls; (B,D,F,H,J) Ovo1 morphants. Embryos were processed by wholemount in situ hybridization for expression of sox10 (A,B), mitfa (C,D), gch (E,F), dlx2 (G,H) and foxd3 (I,J). NC cells migrate in bilateral streams in control embryos (A), but aggregate in the dorsal midline of Ovo1 morphants (B, arrow). Pigment precursors, which express mitfa and gch at 28 hpf, form ectopic clumps in the dorsal midline in Ovo1 morphants (D,F arrows) when compared with wild-type controls (C,E). However, skeletogenic and glial precursors, which express dlx2 and foxd3 at 24 hpf, respectively, do not form clumps in the midline in Ovo1 morphants (G-J). mbh, midbrain-hindbrain boundary; ot, otic vesicle; pa, pharyngeal arches.
Wnt signaling regulates ovo1 expression
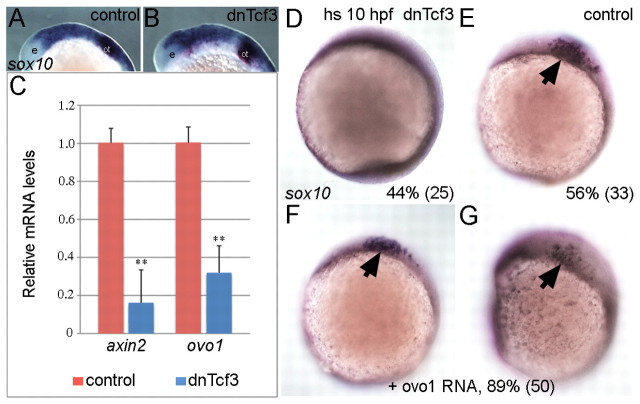
Pigment cell fate depends on Wnt signaling, and members of the Ovo family are direct Wnt targets in both flies and mice (Li et al., 2002; Payre et al., 1999). Thus, Ovo1 could mediate Wnt signaling during pigment cell fate specification. To address this hypothesis, we analyzed ovo1 mRNA levels in response to altered Wnt signaling by real-time, quantitative PCR (qPCR). We inhibited Wnt signaling at the transcriptional level using a transgenic line that expresses a truncated, dominant-negative form of Tcf3 (dnTcf3) tagged with Gfp under the control of the heat-shock promoter Tg(hs:dnTcf-GFP)w26. Embryos were heat-shocked at 13 hpf, bypassing initial requirements for Wnt signaling in NC induction, and transgenic embryos were selected by Gfp expression. As expected, this late heat-shock did not affect initial NC induction, as sox10-expressing NC cells were present in both dnTcf3 overexpressing and control embryos (Fig. 3A,B). ovo1 RNA levels were strongly reduced in embryos overexpressing dnTcf3 when compared with heat-shocked wild-type controls, similar to axin2, a direct Wnt target (Fig. 3C). Thus, ovo1 is a (possibly direct) downstream target of Tcf.
Fig. 3.
ovo1 is induced by Wnt signaling and Ovo1 overexpression partially rescues loss of Wnt signaling in NC cells. (A,B) sox10 expression appears identical in controls and embryos overexpressing a heat-shock-inducible form of the dominant-negative form of Tcf3 (dnTcf3), heat-shocked at 12 hpf. e, eye; ot, otic vesicle. (C) Real-time PCR shows reduced ovo1 RNA levels in embryos overexpressing dnTcf3 (blue bars) when compared with wild-type siblings (red bars). Similar reductions occur for axin2 mRNA, a direct Wnt target. (D,E) Loss of sox10 expression in NC in dnTcf3-injected embryos heat-shocked at 10 hpf (D), compared with controls (E, arrow). Percentages in lower right corners indicate phenotype frequency with number of embryos in parentheses. (F,G) Rescue of sox10 expression in NC (arrows) in heat-shocked, dnTcf3-injected embryos overexpressing ovo1 (**, P≤0.01).
Based on these data, we reasoned that restoring ovo1 expression in embryos lacking Wnt signaling might rescue NC development. To address this question, we injected a gfp-tagged ovo1 (gfp:ovo1) mRNA into a pool of 50% heterozygous Tg(hs:dnTcf-GFP)w26 and 50% wild-type embryos and performed heat-shock at 10 hpf. This early heat-shock completely eliminated sox10-positive NC cells in 45% (25/59) of the uninjected embryos (Fig. 3D; presumably the transgenics) while the rest remained unaffected (n=59; Fig. 3E) (Lewis et al., 2004). By contrast, 90% (50/56) of heat-shocked embryos injected with gfp:ovo1 mRNA showed some sox10 expression (n=56; Fig. 3F,G). Taken together, these data suggest that zebrafish ovo1 mediates the effects of Wnts, at least in part, on NC formation and pigment cell development.
Genetic interactions between Ovo and N-cadherin
Wnt signaling modulates Ecad, Cad6b and Ncad to regulate NC cell adhesion during EMT and migration. We noticed that our Ovo1 morphants superficially resembled zebrafish ncad−/− mutants. Therefore, to elucidate the cellular mechanisms through which ovo1 genes exert their functions on NC, we focused on possible regulation of adhesion through Ncad.
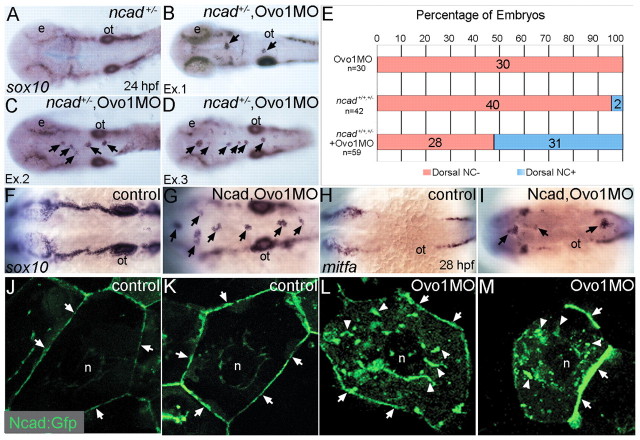
To test for genetic interactions between ovo1 and ncad, we injected subthreshold levels of Ovo1 MO into Ncad-deficient embryos. To reduce Ncad levels, we either used heterozygous ncad+/− mutants (genotyped as shown in Fig. S4 in the supplementary material) or injected subthreshold levels of Ncad MO, neither of which disrupted NC cells on their own (Fig. 4A,F). sox10-positive NC cells formed aggregates in the dorsal midlines of 52.5% (31/59) of ncad+/− mutants injected with Ovo1 MO (Fig. 4B-E) and 100% of Ncad/Ovo1 double-morphant embryos (Fig. 4G). Similar results were obtained for mitfa-positive pigment precursors (Fig. 4H,I). These results point to a genetic interaction between Ovo1 and Ncad and suggest that they act in a common pathway.
Fig. 4.
Interactions between Ovo1 and Ncad. (A-I) Ovo1 genetically interacts with Ncad. (A-G) In situ hybridization for sox10 expression. In both wild types and heterozygous ncad+/− mutants (A), sox10 expression marks bilateral stripes of migrating NC at 24 hpf. Dorsal views, anterior to the left. (B-D) In ncad+/− heterozygotes injected with subthreshold levels of Ovo1 MO, a subset of sox10-positive NC cells remains at the dorsal midline [arrows; examples (Ex) 1-3]. (E) Quantitation of wild-type (red bars) and ectopic dorsal (blue bars) sox10+ NC cells. In contrast to controls (F), ectopic sox10+ cells (G) cluster in the dorsal midline in embryos injected with subthreshold levels of both Ovo1 and Ncad MOs. (H,I) In situ hybridization for mitfa expression. In contrast to controls (H), ectopic cells at the dorsal midline express mitfa (I, arrows). (J-M) Ovo1 regulates Ncad localization. Confocal images of EVL cells in embryos injected with Ncad:Gfp. Ncad:Gfp localizes to the membrane (arrows) and perinuclear region (n, nucleus) of control cells (J,K), whereas Ncad:Gfp accumulates in the cytoplasm of Ovo1 morphant cells (L,M arrowheads). e, eye; n, nucleus; ot, otic vesicle.
We found significant reductions in ncad mRNA levels in Ovo1 morphant zebrafish by qPCR (see Fig. S5A in the supplementary material), but injection of small amounts (~300 pg) of ncad mRNA failed to rescue the morphant phenotype (see Fig. S5B-E in the supplementary material). However, this experiment was difficult to interpret because injecting larger amounts of ncad mRNA disrupted gastrulation and/or NC migration. These results suggest that reduced ncad expression alone cannot account for the Ovo1 loss-of-function phenotype.
Alternatively, Ovo1 might regulate post-translational modifications of Ncad such as its sub-cellular localization. Ncad localization to the tips of filopodia in migratory NC cells is necessary for their proper migration (Monier-Gavelle and Duband, 1995). To address this hypothesis, we used a Gfp-tagged, full-length Ncad (ncad:gfp) previously shown to localize to neuronal cell membranes (Jontes et al., 2004). Confocal images of live embryos at gastrula stages revealed ncad:gfp at the cell surfaces of epithelial cells of the enveloping layer (EVL; Fig. 4J,K; arrows). By contrast, Ovo1 morphant cells accumulated ncad:gfp in punctate structures within the cytoplasm (Fig. 4L,M; arrowheads).
Ovo1 regulates intracellular traffic
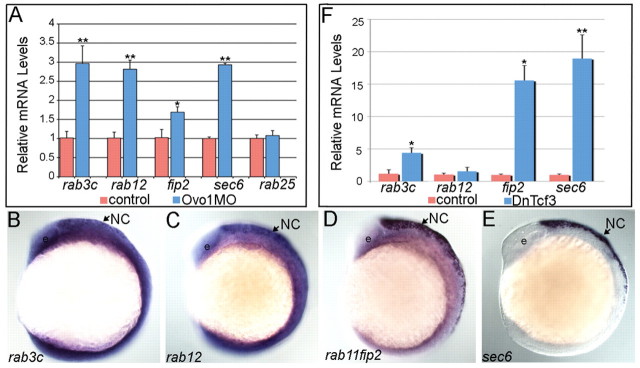
How does Ovo1 regulate Ncad localization? We performed a microarray analysis on Ovo1 morphants to identify genes transcriptionally regulated by Ovo1 (see Table S1 in the supplementary material). Interestingly, many of the upregulated genes in Ovo1 morphants encoded proteins involved in cell migration and intracellular vesicle transport. Genes encoding members of the Rab family of GTPases, Rab-interacting proteins, and other secretory pathway components were upregulated more than two-fold. We confirmed this with qPCR for rab3c, rab12, rab11fip2 and sec6, using rab25 as an internal control (Fig. 5A).
Fig. 5.
Ovo1 and Wnt signaling regulate expression of genes involved in intracellular trafficking. (A) Real-time PCR in Ovo1 morphants reveals elevated expression levels of rab3c, rab12, rab11fip2 and sec6 mRNA levels (blue bars), compared with controls (red bars), whereas rab25 levels do not change. (B-E) In situ hybridization for expression of rab3c (B), rab12 (C), rab11fip2 (D) and sec6 (E) in NC cells (arrows) at 13 hpf. Lateral views, anterior to the left. e, eye. (F) Inhibition of Wnt signaling using heat-shock-inducible dnTCF (Tg(hs:dnTcf-GFP)w26) increases rab3c, rab11fip2 (indicated as fip2) and sec6 expression (blue bars), but not rab12, compared with wild-type siblings (red bars). *, P≤0.05; **, P≤0.01.
Surprisingly, we found that rab3c, rab12, rab11fip2 and sec6 were all highly enriched in premigratory NC cells at 12 hpf, the same stage at which we performed the microarray and first detected the Ovo1 morphant phenotype (Fig. 5B-E). At earlier stages, expression of all four genes was ubiquitous throughout the embryo but later became enriched in the NC at 12 hpf and in the dorsal hindbrain and pharyngeal arches at 24-72 hpf (data not shown). Interestingly, we also found significant upregulation of rab3c, rab11fip2 and sec6 in embryos overexpressing the dnTcf3 transgene when compared with wild-type controls (Fig. 5F). These results suggest that both Wnt and Ovo1 regulate intracellular trafficking pathways, specifically in the NC.
In mice, Rab3c regulates neurotransmitter release, whereas Rab11fip2 belongs to the receptor recycling cascade (Schluter et al., 2004; Somsel Rodman and Wandinger-Ness, 2000) and sec6 influences both exocytosis and receptor recycling (Langevin et al., 2005). Based on their upregulation in Ovo1 morphants, we hypothesized that suppressing their expression, or the processes they regulate, should at least partially rescue the Ovo1 morphant phenotype. To address this, we used the chemical inhibitor Brefeldin A (BFA), a fungal metabolite that disrupts both endo- and exocytosis via regulation of Golgi complex integrity (Nebenfuhr et al., 2002).
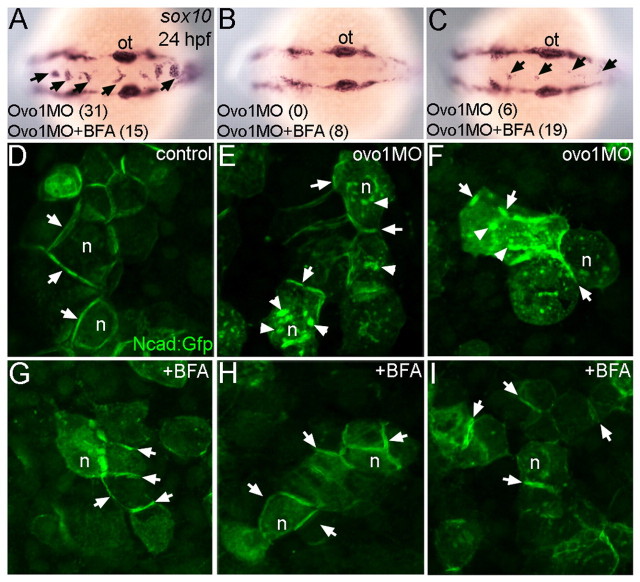
Large aggregates of sox10-positive NC cells (>5 cells per aggregate) accumulated in the dorsal midline in most Ovo1-deficient embryos (83.8%, n=37; Fig. 6A), while 15% showed a less severe NC defect (fewer than 5 cells per aggregate; 16.2%; Fig. 6C). Treatments of Ovo1 morphants with 1 μM BFA eliminated aggregates either entirely (19.0%, n=42; Fig. 6B) or partially (45.2%; Fig. 6C), indicating rescue. BFA treatments also restored the membrane localization of ncad:gfp in EVL cells in Ovo1-deficient embryos (Fig. 6H,I), similar to untreated controls or cells treated with BFA alone (Fig. 6D,G), eliminating the intracellular accumulations of ncad:gfp+ seen in Ovo1 morphant cells (Fig. 6E,F).
Fig. 6.
Inhibition of intracellular trafficking rescues the Ovo1 morphant phenotype. (A-C) In situ hybridization for sox10 at 24 hpf. Dorsal views, anterior to the left. Percentages of embryos with NC cell aggregates at the dorsal midline (arrows) with or without BFA treatment are indicated at the bottom of each panel. (A) A severe example with >5 cells per aggregate over both the midbrain and hindbrain. (C) A less severe example with <5 cells per aggregate exclusively located over the hindbrain. (D-I) Ncad:Gfp injections. Ncad:Gfp localizes to the membranes (arrows) of untreated (D) and BFA-treated control cells (G). In Ovo1 morphant cells, Ncad:Gfp also accumulates in the cytoplasm (E,F; arrowheads), but is largely restored to the membrane in BFA-treated Ovo1 morphant cells (H,I). n, nucleus; ot, otic vesicle.
Roles for rab3c, rab12 and rab11fip2 in intracellular trafficking have been studied in vitro but their functions during embryonic development, especially in the NC, have not been described. If elevated levels of one or more of these genes in NC are responsible for the Ovo1 loss-of-function phenotype, then we reasoned that we might be able to phenocopy the Ovo1 morphant phenotype by overexpressing them in early embryos. To address this hypothesis, we injected synthetic mRNA encoding rab11fip2 and constitutively active forms of rab3c and rab12 (carab3c and carab12, respectively), together with mCherry mRNA to mark injected cells, into transgenic embryos expressing Gfp in NC cells under the control of the sox10 promoter [sox10(7.2):gfp; Fig. 7]. Gfp-positive NC cells migrated normally both in control embryos injected with 100 pg of mCherry RNA alone (Fig. 7A) or with different combinations of carab3c and carab12 (data not shown). However, 250 pg of rab11fip2 alone caused the formation of NC cell aggregates in the dorsal neural tube in 35.3% of injected embryos (Fig. 7B,E) and, similar to Ovo1 morphants, these expressed mitfa (see Fig. S6A-C in the supplementary material). However, 90.5% of embryos co-injected with subthreshold levels of both rab11fip2 mRNA and Ovo1 MO had more severe NC cell aggregates (>5 cells per aggregate), especially at the level of the midbrain, than were observed in single-injected embryos (Fig. 7C-E; arrows), further indicating genetic interactions between the effects of reducing Ovo1 or increasing Rab11fip2 activity.
To corroborate that such defects in migration were autonomous to the NC, we transplanted cells at blastula stages from sox10(7.2):gfp transgenics into wild-type embryos. When Ovo1 morphant and wild-type cells were co-transplanted into the same hosts, subsets of rab11fip2 overexpressing, Ovo1 morphant NC cells (mCherry-positive) failed to migrate and accumulated in the dorsal neural tube (Fig. 7G; arrows), whereas wild-type NC cells (mCherry-negative) migrated normally (Fig. 7F, arrowheads). Similarly, transplantation of rab11fip2 overexpressing, Ovo1 morphant cells alone resulted in numerous aggregates (Fig. 7H-M; arrows). Taken together, these data suggest that Rab11fip2 modulates NC cell migration.
Is the role of Rab11fip2 in NC cell migration Ncad-dependent? To test this idea, we injected subthreshold levels of rab11fip2 RNA into heterozygous ncad+/− mutants (Fig. 8A,J). In contrast to ncad+/− alone, 43.1% (n=65) of embryos injected with rab11fip2 mRNA displayed ectopic aggregates of sox10-positive NC cells in the dorsal midline (Fig. 8B,C,J) that also expressed mitfa (Fig. 8G,H). Similar results were obtained when subthreshold levels of rab11fip2 mRNA and Ncad MO were co-injected (Fig. 8D-F,I,J), demonstrating that Rab11fip2 and Ncad interact genetically and might act in a common pathway.
Fig. 8.
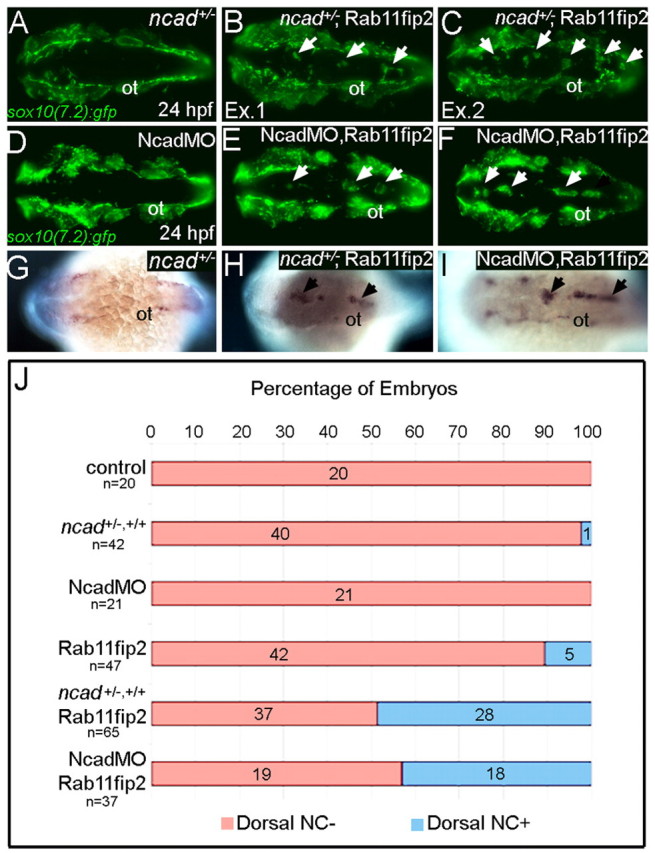
rab11fip2 genetically interacts with ncad to regulate NC migration. (A-F) Live images of tg:sox10(7.2):gfp labeling migratory NC cells in ncad+/− embryos (A-C) and single Ncad morphants (D-F). In contrast to controls (A,D), Gfp-positive NC cells remain in the dorsal neural tube of ncad+/− mutants [B,C; arrows; examples (Ex) 1 and 2] and Ncad morphants (E,F; arrows) injected with subthreshold levels of Rab11fip2 mRNA. (G-I) Similarly, ectopic mitfa-positive cells cluster in the dorsal midline (arrows) of ncad+/− mutants (H) and Ncad morphants (I) overexpressing Rab11fip2, but not in controls (G). (J) Quantitation of genetic interaction studies; numbers of embryos with or without ectopic sox10+ NC clusters in the dorsal midline (blue and red bars, respectively). ot, otic vesicle.
DISCUSSION
The NC is classically thought to be induced by Wnts and to downregulate Ecad and Ncad in order to exit the neuroepithelium and migrate (Nakagawa and Takeichi, 1995; Nakagawa and Takeichi, 1998). Wnt signaling is also required for NC to form pigment cells but no clear link exists between this and cadherin regulation. The role of Ncad in NC cells is also controversial, as both gain- and loss-of-function disrupt migration (Bronner-Fraser et al., 1992; Nakagawa and Takeichi, 1998; Shoval et al., 2007). Here, we help resolve this debate by suggesting that localization of Ncad to the cell surface is vital for its functions in NC migration and that this is regulated by Wnts. We show that the Wnt target gene, Ovo1, regulates the intracellular trafficking of Ncad, and provide the first evidence for NC-specific functions for intracellular trafficking components, such as rab11fip2, in migration (Fig. 9). This might be a more general mechanism by which Wnts control morphogenesis in many contexts.
Fig. 9.
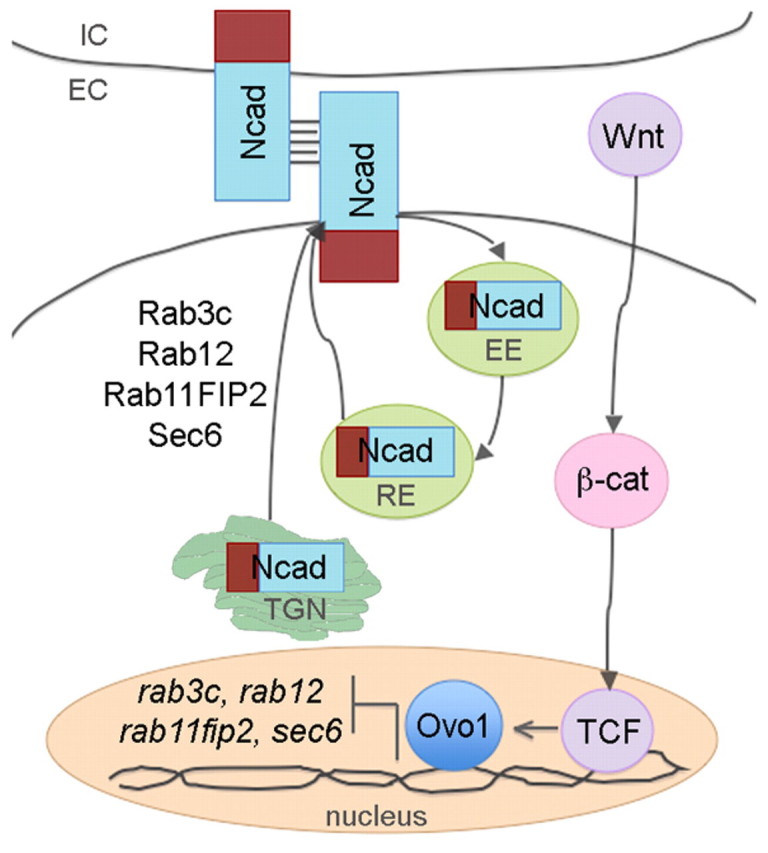
Model depicting regulation of Ncad trafficking in NC cells by Ovo1. Canonical Wnt/β-catenin signaling through Tcf transcription factors regulates ovo1 transcript levels in NC cells. Ovo1 normally represses the transcription of rab3c, rab12, rab11fip2 and sec6, which maintain the balance of intracellular and membrane-localized Ncad. Abnormally high levels of Rab3c, Rab12, Rab11fip2 and Sec6 in Ovo1-deficient NC promote hyperactive intracellular trafficking of Ncad; Ncad secretion from the trans-Golgi network (TGN) and recycling through the early and recycling endosomes (EE and RE) increases. Consequently, Ncad becomes depleted at the membrane. EC, extracellular space; IC, intracellular space.
Our model reconciles several observations (Fig. 9). It agrees with experimental evidence that Ovo1 and Ncad are both required for NC migration. It accommodates data showing that Wnt signaling regulates Ovo1 expression to control Ncad localization. Importantly, it helps explain how defects in Wnt and Ovo1 function disrupt the same subset of NC cells that form pigment cells. This might be the major mechanism by which Wnts control specification and migration of pigment cells and its key feature might be regulated cell adhesion.
Why is Ncad localization important for NC cell migration? These cells need to lose contact with their neighbors during EMT and later establish transient contacts while they pull apart from one another in a process called contact inhibition (Carmona-Fontaine et al., 2008). Ncad localizes to the filopodial tips of migrating NC cells (Monier-Gavelle and Duband, 1995) and, in Ncad−/− mutant mouse embryos, NC cells migrate in inappropriate directions (Xu et al., 2001). Thus, rapid turnover of Ncad in different regions of a cell membrane might help it establish transient contacts that allow it to move and guide its path.
Consistent with this idea, we show that many NC cells lose direction in Ovo1-deficient zebrafish embryos and this correlates with reductions in Ncad at the membrane. NC cells end up in ectopic clusters at the roof of the fourth ventricle of the hindbrain or otherwise attach to the neuroepithelium. Cells in Ovo1 morphants show aberrant cytoplasmic accumulation of ncad:gfp and rescue of NC cell migration correlates with restoration of cell surface Ncad.
How does Ovo1, a transcription factor, regulate the subcellular localization of Ncad? Our results suggest that among the key transcriptional targets are components of the secretory pathway (including rab3c, rab12, rab11fip2 and sec6). These are upregulated in Ovo1 morphants and blocking their functions (e.g. with BFA) rescues the morphant phenotype. Surprisingly, all four (rab3c, rab12, rab11fip2 and sec6) are also normally expressed at high levels, specifically in migrating NC cells. Previous studies have shown requirements for endocytosis and receptor recycling in NC cell motility in vitro (Monier-Gavelle and Duband, 1997). Our results in vivo suggest that rab3c, rab12, rab11fip2 and sec6 function in transport and/or removal of Ncad from the membrane (Fig. 9) and that this is crucial for proper NC migration. Interestingly, Wnt-dependent regulation of rab11 was recently shown to control tracheal cell intercalation in Drosophila; upregulation of rab11 in response to Wnt enhanced membrane recycling of Ecad and inhibited cell intercalation (Shaye et al., 2008). Similarly, we find that overexpression of rab11fip2 in zebrafish is sufficient to restrict the directional migration of NC cells. More than one Rab is probably involved as downregulation of rab11fip2 alone does not rescue the Ovo1 morphant phenotype (see Fig. S7A-D in the supplementary material).
Ovo1 most likely regulates Ncad transport through the Golgi network, as BFA treatments rescue both the morphant phenotype and the cytoplasmic localization of ncad:gfp. BFA causes the trans-Golgi network (TGN) to form a complex with endosomes through its effects on Arf1, a protein that recruits coat proteins of the Golgi complex (Nebenfuhr et al., 2002). Arf1 mutations in yeast disrupt both endo- and exocytic pathways (Gaynor et al., 1998). We find that, at certain concentrations, BFA restores Ncad to the membrane in Ovo1-deficient embryos, rescuing NC migration. BFA was previously shown to restore migration and Ncad localization in avian NC cells (Monier-Gavelle and Duband, 1997). Precisely which aspects of trafficking (endo- versus exocytosis, vesicle recycling, etc.) are most crucial in NC cell migration is an interesting subject for future studies using specific genetic or chemical interventions.
One striking similarity between Ovo1- and Wnt-deficient zebrafish embryos is their specific pigment defect. We hypothesize that the Ovo1- and Wnt-dependent Ncad localization that we have described here promotes migration of NC cells that will form pigment. There are several possible explanations for this lineage-specific requirement. Although widely expressed in the NC at premigratory stages, ovo1 localizes to a subset of NC cells by 15 hpf, which could be the pigment precursors that fail to migrate in morphants. Alternatively, Ovo1 deficiency might disrupt migration of a random subset of homogeneous NC cells, which subsequently adopt a pigment precursor identity. A subset of NC cells also fails to migrate in Ovol2−/− mutant mice, although it is not known if these are also pigment progenitors (Mackay et al., 2006). Intriguingly, NC that forms pigment migrates later than many other populations and this timing might be altered in Ovo mutants (Schilling and Kimmel, 1994). Unlike in mice, where NC cells fail to proliferate and die in Ovol2−/− mutants (Mackay et al., 2006), in zebrafish we find no changes in proliferation or survival (see Fig. S8A,B in the supplementary material) and cannot rescue defects by inhibiting apoptosis with a p53 MO (see Fig. S8C-J in the supplementary material). Both ectodermal specification and subsequent epidermal differentiation also appear normal (see Fig. S8K-N in the supplementary material). Thus, our results suggest that the effects of Ovo1 on pigment precursors are due to its roles in migration.
The NC has played an important role in the evolution of chordates and is one of the defining characteristics of vertebrate embryos (Gans and Northcutt, 1983). How did this cell population arise during evolution? NC cells originate, at least in part, from the non-neural ectoderm that later forms the epidermis, and relatives of many genes known to mark NC cells in vertebrates are expressed in this ectoderm in their invertebrate chordate relatives. For example, single dlx and tfap2a genes in amphioxus, AmphiDll and AmphiAp2, respectively, are expressed in non-neural ectoderm, suggesting ancestral functions in epidermal development (Holland et al., 1996; Meulemans and Bronner-Fraser, 2002). Drosophila ovo acts as a transcriptional switch in the epidermis integrating the Wingless (Wg) and Epidermal growth factor receptor (Der) signaling pathways to induce cytoskeletal changes associated with denticle production, including F-actin bundling and Apc2 localization (Delon et al., 2003; Payre et al., 1999). Similarly, Ovo proteins expressed in the epidermis might have been co-opted during chordate evolution to integrate signaling pathways that control cell shape and movement, leading to the advent of a novel migratory NC cell population.
Supplementary Material
Acknowledgements
We thank M. Hammerschmidt (ncad-MO), R. Brewster (ncad mutants), R. Dorsky (dntcf3 transgenics) and J. Jontes (ncad:gfp constructs) for reagents; Dr X. Dai for advice; Drs X. Dai, I. Blitz, K. Cho and M. Waterman for comments on the manuscript; and the Schilling laboratory for help with the manuscript and technical support. Funding for this study was provided by the NIH (R01 NS-41353 and R01 DE-13828) to T.F.S. and an NSF GRF to S.P. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.048439/-/DC1
References
- Akitaya T., Bronner-Fraser M. (1992). Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Dev. Dyn. 194, 12-20 [DOI] [PubMed] [Google Scholar]
- Aybar M. J., Nieto M. A., Mayor R. (2003). Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130, 483-494 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M., Wolf J. J., Murray B. A. (1992). Effects of antibodies against N-cadherin and N-CAM on the cranial neural crest and neural tube. Dev. Biol. 153, 291-301 [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T., Kalcheim C. (2002). Association between the cell cycle and neural crest delamination through specific regulation of G1/S transition. Dev. Cell 3, 383-395 [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T., Stanleigh J., Sela-Donenfeld D., Kalcheim C. (2004). Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development 131, 5327-5339 [DOI] [PubMed] [Google Scholar]
- Cano A., Perez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000). The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76-83 [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Matthews H. K., Kuriyama S., Moreno M., Dunn G. A., Parsons M., Stern C. D., Mayor R. (2008). Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Schonbaum C., Degenstein L., Bai W., Mahowald A., Fuchs E. (1998). The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12, 3452-3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon I., Chanut-Delalande H., Payre F. (2003). The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech. Dev. 120, 747-758 [DOI] [PubMed] [Google Scholar]
- Dorsky R. I., Moon R. T., Raible D. W. (1998). Control of neural crest cell fate by the Wnt signalling pathway. Nature 396, 370-373 [DOI] [PubMed] [Google Scholar]
- Dorsky R. I., Raible D. W., Moon R. T. (2000). Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 14, 158-162 [PMC free article] [PubMed] [Google Scholar]
- Gans C., Northcutt R. G. (1983). Neural crest and the origin of vertebrates: a new head. Science 220, 268-273 [DOI] [PubMed] [Google Scholar]
- Gaynor E. C., Chen C. Y., Emr S. D., Graham T. R. (1998). ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell 9, 653-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N. D., Panganiban G., Henyey E. L., Holland L. Z. (1996). Sequence and developmental expression of AmphiDll, an amphioxus Distal-less gene transcribed in the ectoderm, epidermis and nervous system: insights into evolution of craniate forebrain and neural crest. Development 122, 2911-2920 [DOI] [PubMed] [Google Scholar]
- Ikeya M., Lee S. M., Johnson J. E., McMahon A. P., Takada S. (1997). Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389, 966-970 [DOI] [PubMed] [Google Scholar]
- Jin E. J., Erickson C. A., Takada S., Burrus L. W. (2001). Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Dev. Biol. 233, 22-37 [DOI] [PubMed] [Google Scholar]
- Jontes J. D., Emond M. R., Smith S. J. (2004). In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J. Neurosci. 24, 9027-9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khila A., El Haidani A., Vincent A., Payre F., Souda S. I. (2003). The dual function of ovo/shavenbaby in germline and epidermis differentiation is conserved between Drosophila melanogaster and the olive fruit fly Bactrocera oleae. Insect Biochem. Mol. Biol. 33, 691-699 [DOI] [PubMed] [Google Scholar]
- Knight R. D., Nair S., Nelson S. S., Afshar A., Javidan Y., Geisler R., Rauch G. J., Schilling T. F. (2003). lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130, 5755-5768 [DOI] [PubMed] [Google Scholar]
- Langevin J., Morgan M. J., Sibarita J. B., Aresta S., Murthy M., Schwarz T., Camonis J., Bellaiche Y. (2005). Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev. Cell 9, 365-376 [DOI] [PubMed] [Google Scholar]
- Le Douarin N. M., Creuzet S., Couly G., Dupin E. (2004). Neural crest cell plasticity and its limits. Development 131, 4637-4650 [DOI] [PubMed] [Google Scholar]
- Lele Z., Folchert A., Concha M., Rauch G. J., Geisler R., Rosa F., Wilson S. W., Hammerschmidt M., Bally-Cuif L. (2002). parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281-3294 [DOI] [PubMed] [Google Scholar]
- Lewis J. L., Bonner J., Modrell M., Ragland J. W., Moon R. T., Dorsky R. I., Raible D. W. (2004). Reiterated Wnt signaling during zebrafish neural crest development. Development 131, 1299-1308 [DOI] [PubMed] [Google Scholar]
- Li B., Mackay D. R., Dai Q., Li T. W., Nair M., Fallahi M., Schonbaum C. P., Fantes J., Mahowald A. P., Waterman M. L., et al. (2002). The LEF1/beta - catenin complex activates movo1, a mouse homolog of Drosophila ovo required for epidermal appendage differentiation. Proc. Natl. Acad. Sci. USA 99, 6064-6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister J. A., Cooper C., Nguyen K., Modrell M., Grant K., Raible D. W. (2006). Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev. Biol. 290, 92-104 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402-408 [DOI] [PubMed] [Google Scholar]
- Mackay D. R., Hu M., Li B., Rheaume C., Dai X. (2006). The mouse Ovol2 gene is required for cranial neural tube development. Dev. Biol. 291, 38-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2002). Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development 129, 4953-4962 [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio M., Terracol R., Kafatos F. C. (1991). The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 10, 2259-2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier-Gavelle F., Duband J. L. (1995). Control of N-cadherin-mediated intercellular adhesion in migrating neural crest cells in vitro. J. Cell Sci. 108, 3839-3853 [DOI] [PubMed] [Google Scholar]
- Monier-Gavelle F., Duband J. L. (1997). Cross talk between adhesion molecules: control of N-cadherin activity by intracellular signals elicited by beta1 and beta3 integrins in migrating neural crest cells. J. Cell Biol. 137, 1663-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Takeichi M. (1995). Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development 121, 1321-1332 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takeichi M. (1998). Neural crest emigration from the neural tube depends on regulated cadherin expression. Development 125, 2963-2971 [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A., Ritzenthaler C., Robinson D. G. (2002). Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B., Singer J., Laget V., Pennetta G., Pauli D. (1994). Function of Drosophila ovo+ in germ-line sex determination depends on X-chromosome number. Development 120, 3185-3195 [DOI] [PubMed] [Google Scholar]
- Payre F., Vincent A., Carreno S. (1999). ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 400, 271-275 [DOI] [PubMed] [Google Scholar]
- Raible D. W., Eisen J. S. (1994). Restriction of neural crest cell fate in trunk of embryonic zebrafish. Development 120, 495-503 [DOI] [PubMed] [Google Scholar]
- Schilling T. F., Kimmel C. B. (1994). Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483-494 [DOI] [PubMed] [Google Scholar]
- Schluter O. M., Schmitz F., Jahn R., Rosenmund C., Sudhof T. C. (2004). A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629-6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Casanova J., Llimargas M. (2008). Modulation of intracellular trafficking regulates cell intercalation in the Drosophila trachea. Nat. Cell Biol. 10, 964-970 [DOI] [PubMed] [Google Scholar]
- Shoval I., Ludwig A., Kalcheim C. (2007). Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development 134, 491-501 [DOI] [PubMed] [Google Scholar]
- Somsel Rodman J., Wandinger-Ness A. (2000). Rab GTPases coordinate endocytosis. J. Cell Sci. 113, 183-192 [DOI] [PubMed] [Google Scholar]
- Taneyhill L. A., Coles E. G., Bronner-Fraser M. (2007). Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development 134, 1481-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B., Schilling T. F., Postlethwait J. H. (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203-1215 [DOI] [PubMed] [Google Scholar]
- Vallin J., Thuret R., Giacomello E., Faraldo M. M., Thiery J. P., Broders F. (2001). Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J. Biol. Chem. 276, 30350-30358 [DOI] [PubMed] [Google Scholar]
- Xu X., Li W. E., Huang G. Y., Meyer R., Chen T., Luo Y., Thomas M. P., Radice G. L., Lo C. W. (2001). Modulation of mouse neural crest cell motility by N-cadherin and connexin 43 gap junctions. J. Cell Biol. 154, 217-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.