Abstract
Comparative genomic studies have identified many proteins that are found only in various Chlamydiae species and exhibit no significant sequence similarity to any protein in organisms that do not belong to this group. The CT670 protein of Chlamydia trachomatis is one of the proteins whose genes are in one of the type III secretion gene clusters but whose cellular functions are not known. CT670 shares several characteristics with the YscO protein of Yersinia pestis, including the neighboring genes, size, charge, and secondary structure, but the structures and/or functions of these proteins remain to be determined. Although a BLAST search with CT670 did not identify YscO as a related protein, our analysis indicated that these two proteins exhibit significant sequence similarity. In this paper, we report that the CT670 crystal, solved at a resolution of 2 Å, consists of a single coiled coil containing just two long helices. Gel filtration and analytical ultracentrifugation studies showed that in solution CT670 exists in both monomeric and dimeric forms and that the monomer predominates at lower protein concentrations. We examined the interaction of CT670 with many type III secretion system-related proteins (viz., CT091, CT665, CT666, CT667, CT668, CT669, CT671, CT672, and CT673) by performing bacterial two-hybrid assays. In these experiments, CT670 was found to interact only with the CT671 protein (YscP homolog), whose gene is immediately downstream of ct670. A specific interaction between CT670 and CT671 was also observed when affinity chromatography pull-down experiments were performed. These results suggest that CT670 and CT671 are putative homologs of the YcoO and YscP proteins, respectively, and that they likely form a chaperone-effector pair.
Chlamydiae are obligate intracellular bacteria that infect a variety of eukaryotes, including humans, animals, insects, and free-living amoebae (8, 31, 51). They are highly pathogenic and cause genital tract, ocular, and respiratory infections in humans (9, 55). A key characteristic of Chlamydiae is their biphasic developmental cycle, in which the bacteria alternate between two morphologies: the elementary body and the reticulate body (1, 31). The Chlamydiae species, like many other Gram-negative pathogenic bacteria, contain a type III secretion (T3S) system, which plays a major role in their pathogenicity (4, 7, 44). This key system aids pathogenicity by exporting bacterial proteins, termed effectors, into the host cell via a syringe-like nanomachine called an injectisome (4, 7). Once secreted into the host cell, these effectors may manipulate host cell functions to the advantage of the pathogen (44, 54).
Because of the unique chlamydial developmental cycle currently there are no tractable methods for genetic manipulation of these organisms (25, 49). Detailed bioinformatic investigations have identified approximately 200 proteins unique to various taxonomic levels of the Chlamydiae phylum (19, 21). Included in this pool of proteins are proteins whose genes occur in chlamydial T3S system loci (23, 44). Most of the Chlamydiae-specific genes in these loci encode proteins whose functions are unknown or putative and are referred to as Chlamydiae-specific putative T3S-related proteins. These genes include the ct670 and ct671 genes, which are downstream of the gene for the ATPase of the T3S system (ct669) and upstream of yscQ (ct672). CT670 is a Chlamydiales-specific protein with no known homologs (based on BLAST searches) in other species outside this group (21). However, based on similarities in size, charge distribution, and predicted secondary structure, CT670 is thought to be the Chlamydia trachomatis equivalent of YscO, a mobile core component of the T3S system in Yersinia (40). Previous studies of CPn0706, the Chlamydophila pneumoniae homolog of CT670, indicated that this protein was localized in the inclusion in infected cells, but it was not detected in the inclusion membrane or host cytosol, suggesting that it is not a secreted protein (24). Moreover, CT670 is not expected to be a type III secreted effector on the basis of the results obtained by a computational approach that was used to predict type III secreted effectors by comparison of sequences to sequences of known effectors (47). CT671 is a Chlamydiaceae-specific protein with no known homologs in any other species outside this family (21), but it is predicted to be a homolog of YscP, which is the molecular ruler and substrate specificity switch in Yersinia species (2). This protein is secreted by a heterologous T3S system and was predicted to be a T3S effector using the computational approach mentioned above (47, 53). Furthermore, the CCA00037 protein, the CT671 homolog in Chlamydophila caviae, has been visualized in the host cytosol of C. caviae-infected cells using specific antibodies, which provided evidence that it is secreted (53). Although CT670 and CT671 do not appear to be related to YscO and YscP, respectively, on the basis of the results of BLAST searches, it is possible that they have similar roles in the chlamydial T3S system based on their genetic neighborhood and other characteristics noted above. Further, because of their Chlamydiae specificity, they may provide a unique characteristic of the chlamydial T3S system. To examine this possibility, detailed investigations of the three-dimensional (3D) structure of CT670, its self-association properties, and its binding partners were carried out in the present study. Here we report elucidation of the CT670 crystal structure at a resolution of 2.0 Å. CT670 crystallized as a monomer with an elongated two-helix coiled coil. Using analytical ultracentrifugation, CT670 was found to be mostly monomeric in solution, but a dimeric form was also detected. Furthermore, by performing protein-protein interaction studies involving bacterial two-hybrid assays, as well as biochemical experiments, we obtained evidence showing that CT670 interacts specifically with CT671, which would be expected if these proteins have functions similar to those of YscO and YscP, respectively.
MATERIALS AND METHODS
Chlamydia growth and DNA extraction.
McCoy cells were grown at 37°C in minimal essential medium (MEM) supplemented with 10% (vol/vol) fetal calf serum in a humidified 5% CO2-95% air incubator. The cells were infected with C. trachomatis serotype L2 in this medium supplemented with 2 mM l-glutamine and 1 μg/ml cycloheximide. After 48 to 72 h, infected cells were harvested by vigorous shaking with glass beads (17). Total genomic DNA was extracted from C. trachomatis-infected McCoy cells by the phenol-chloroform method and was precipitated with ethanol (37). Using this DNA, different C. trachomatis genes were PCR amplified using custom-synthesized oligonucleotide primers and cloned in the vectors indicated below.
Size exclusion chromatography and analytical centrifugation.
Full-length ct670 was cloned into the expression plasmid p15TvLic (Clinical Genomics Centre), which produced an N-terminal His6-CT670 protein fusion. Escherichia coli BL21 cells transformed with this plasmid were induced in early log phase with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 to 5 h at 37°C. Cell extracts were obtained by sonication, and the recombinant protein was purified from cleared cell lysates by nickel affinity chromatography using Ni-ProBond resin (Invitrogen). The protein was concentrated to obtain a concentration of 10 to 15 mg/ml in a buffer containing 250 mM NaCl, 20 mM Tris (pH 7.9), and 1 mM MgCl2 (column buffer) using an Amicon ultracentrifugation filter. Size exclusion chromatography was carried out using a Superdex 200 (16/600) column, and the peak protein fractions were analyzed by 12% SDS-PAGE and visualized by staining with Coomassie blue. Sedimentation equilibrium experiments using analytical ultracentrifugation were carried out as described by Fraser et al. (18) using a Beckman Optima XL-A analytical ultracentrifuge with an An-60 Ti rotor. Data were collected in duplicate for two separately prepared samples of purified protein. For the first data set, three concentrations of CT670 (∼9, 28, and 44 μM; 1 μM was equivalent to 0.022 mg/ml) were analyzed by using centrifugation at 10,000, 15,000, and 20,000 rpm at 4°C. For the second data set, three concentrations of CT670 (∼9, 27, and 44 μM; 0.11, 0.31, and 0.51 mg/ml) were analyzed by using centrifugation at 7,280 × g, 16,380 × g, and 29,120 × g at 4°C. Data were analyzed using Origin 6.0 software. Th solvent density and the partial specific volume were calculated using the SEDNTERP program.
Crystallization.
For crystallization experiments, E. coli BL21(DE3)-RIPL (Stratagene) cells expressing His6-CT670 were grown in SeMet high-yield growth medium (Shanghai Medicilon) and induced with IPTG as described above. The His6-CT670 protein was purified as described above, and its His6 tag was removed by cleaving it with TEV protease and then running the sample once through an Ni-Probond column and collecting the unbound protein. The protein purified in this way was dialyzed against a buffer containing 10 mM HEPES (pH 7.5), 300 mM NaCl, and 0.5 mM Tris-(2-carboxyethyl) phosphine and concentrated to obtain a concentration of 10.0 mg/ml. Crystallization was performed with this protein at room temperature (21°C) using sitting-drop vapor diffusion with an optimized sparse matrix crystallization screen. The crystal used for multiwavelength anomalous dispersion (MAD) data collection at beamline 19-ID (Tables 1 and 2) was obtained using a crystallization liquor containing 0.2 M ammonium dihydrogen phosphate, 20% polyethylene glycol 3350 (PEG 3350), 1.5% (4s)-2-methyl-2,4-pentanediol (MPD), 20 mM magnesium acetate, and 10 mM sodium cacodylate (pH 6.5). The crystal used for data collection at a single wavelength on beamline 23-IDG was obtained using a similar crystallization liquor containing 0.2 M ammonium dihydrogen phosphate, 20% PEG 3350, 1% PEG 4000, 1 mM magnesium sulfate, and 25 mM morpholineethanesulfonic acid (MES) (pH 5.6).
TABLE 1.
Data collection and phasing statistics for CT670a
| Parameter | Single-wavelength anomalous dispersion (peak) | Multiwavelength anomalous dispersion |
|
|---|---|---|---|
| Peak | Inflection | ||
| Cell dimensions | |||
| a, b, c (Å) | 83.1, 23.2, 105.6 | 84.3, 23.5, 105.6 | 84.3, 23.6, 105.6 |
| α, β, γ (°) | 90, 107.5, 90 | 90, 107.7, 90 | 90, 107.7, 90 |
| Wavelength (Å) | 0.97872 | 0.97921 | 0.97942 |
| Resolution (Å) | 50.0-2.0 | 50.0-2.0 | 50.0-2.0 |
| No. of unique reflections | 13,438 (1,231) | 10,832 (460) | 11,202 (555) |
| Rmerge (%)b | 6.7 (19.1) | 6.6 (27.6) | 7.3 (29.6) |
| I/σI | 25.8 (6.3) | 43.6 (4.3) | 29.7 (3.2) |
| Completeness (%) | 99.0 (91.6) | 81.3 (34.7) | 85.8 (42.6) |
| Redundancy | 5.9 (4.6) | 8.6 (6.4) | 6.1 (4.0) |
The beamlines used for single-wavelength anomalous dispersion and multiwavelength anomalous dispersion were APS 23-IDG and APS 19-ID, respectively, and the space group for both types of dispersion was C2. The values in parentheses are the values for the highest-resolution shell.
Rmerge=∑hkl|I−〈I〉|/∑hklI.
TABLE 2.
Refinement statistics for CT670
| Parameter | Value |
|---|---|
| Resolution (Å) | 27.4-2.0 |
| No. of reflections | 12,778 |
| Rwork (%)a | 24.0 |
| Rfree (%)b | 28.5 |
| No. of atoms | |
| Protein | 1,403 |
| Water | 51 |
| B-factors (Å2) | |
| Protein | 24.0 |
| Water | 23.8 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.016 |
| Bond angles (°) | 1.535 |
| Ramachandran plot | |
| % in most favored regions | 98.6 |
| % in additionally allowed regions | 1.4 |
| % in generously allowed or disallowed regions | 0.0 |
Rwork=Σ|Fobs−Fcalc|/Σ|Fobs|, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
Rfree was calculated using 5% of the total reflections randomly chosen and excluded from the refinement.
Data collection, determination of the structure, and refinement.
The structure of CT670 was solved using the multiwavelength anomalous dispersion (MAD) method utilizing the anomalous signal from selenium atoms. A two-wavelength data set of the Se-Met derivative crystal was collected at a resolution of 2.0 Å on beamline 19-ID (Structural Biology Centre, Advanced Photon Source, Argonne National Laboratories), and data were integrated and scaled using the HKL2000 software package (38). Using data collected at the peak and inflection regions of the selenium K absorption edge, the positions of all four anomalous scatterers were determined using SHELXD (48) and data up to a resolution of 2.7 Å, followed by heavy-atom refinement and maximum-likelihood-based phasing utilized in the autoSHARP program suite (3, 11). Phase improvement by density modification (DM) generated an interpretable experimental electron density map at a resolution of 2.15 Å (the values of merit before and after DM were 0.481 and 0.791, respectively), which allowed approximately 70% of the asymmetric unit to be traced for an initial model using ARP/warp (43). The model was subsequently improved through alternate cycles of manual building using COOT (13) and restrained refinement against a maximum likelihood target with 5% of the reflections randomly excluded as an Rfree test set. All refinement steps were performed using REFMAC in the CCP4 program suite (6). The final model was refined against a fully complete data set collected at a resolution of 2.0 Å with a second crystal at beamline 23-IDG (GM/CA-CAT; Advanced Photon Source, Argonne National Laboratories). During refinement, the C-terminal residues Arg-163 to Asn-168 were omitted because of poor electron density. The final model comprised one molecule of CT670 and 51 solvent molecules refined to an Rwork of 24.0% and an Rfree of 28.5%, including TLS parameterization (58, 59). A Ramachandran plot generated by PROCHECK (33) showed excellent stereochemistry with no outliers and all residues in the most favored and additional allowed regions. Data collection and refinement statistics are summarized in Tables 1 and 2.
Bacterial two-hybrid assay.
The BacterioMatch II two-hybrid system (Stratagene) was used to examine protein-protein interactions in vivo in E. coli cells (12). Two-hybrid analyses were carried out by closely following the protocol provided by the manufacturer. Plasmid constructs were created by cloning the target and bait proteins in the pTRG and pBT vectors using an in-fusion dry-down PCR cloning kit (Clontech). All constructs were verified by sequencing. In a typical experiment, validation reporter strain XL1-blue MRF′ Kan cells were cotransformed with 50 ng each of pBT and pTRG containing the bait and target proteins, respectively. After incubation for 90 min at 37°C with shaking in LB medium, the cells were washed three times with M9 medium containing His drop-out broth (containing various other amino acids but not histidine) and then incubated at 37°C for 2 h. One hundred microliters of transformed cells was plated in triplicate on M9 His drop-out agar (i.e., agar lacking histidine) containing 5 mM 3-amino-1,2,4-triazole (3-AT), 34 μg/ml chloramphenicol, and 12.5 μg/ml tetracycline and also on agar plates containing the antibiotics mentioned above but no 3-AT. pBT-LGF2, pTRG-Gal11P, and empty pBT and pTRG constructs were used as positive and negative controls. The plates were incubated at 37°C for 24 h and then moved to room temperature until colonies appeared in the positive controls (the colonies indicating a positive interaction usually appeared between 48 and 72 h). The positive interactions were confirmed by streaking colonies on plates containing both 3-AT and streptomycin (10 μg/ml). The interaction of CT670 with CT671 and CT091 was also examined using another bacterial two-hybrid system in which the genes for the bait and target proteins were fused to different fragments of the green fluorescent protein (GFP) in the plasmid vectors pET11-a-link-NGF and pMRBAD-link-CGFP (57). Consequently, any interaction between the bait and target proteins led to reconstitution of the GFP, which was readily detected due to its fluorescence (57).
Strep-tag pull-down assay.
Full-length ct671 was cloned into the expression plasmid pTriEx (p52b+) (Novagen), which produced an N-terminal Strep-tag II-CT671 fusion protein (36). Full-length ct670 was cloned into the expression plasmid pCDFDuet-1 (Novagen), which produced an N-terminal His6-CT670 fusion protein and allowed cotransformation with the other pET vectors. After cotransformation of E. coli BL21 cells with both of these plasmids, the cells were induced and collected as described above. A control experiment with cells expressing only His6-CT670 was also carried out in parallel. Each cell pellet was suspended in Strep-tactin wash buffer (150 mM NaCl [pH 8.0], 100 mM Tris, 1 mM EDTA), and cell extract was obtained by sonication. The cleared cell lysates were applied to Strep-tactin superflow agarose resin (Novagen), and after extensive washing with the wash buffer (150 mM NaCl, 100 mM Tris-HCl [pH 8.0], 1 mM EDTA), they were eluted with the buffer described above containing 2.5 mM desthiobiotin. Fractions (0.5 ml) were collected, analyzed by 15% SDS-PAGE, and visualized by Coomassie blue staining. The proteins were also transferred to a nitrocellulose membrane for Western blot analysis with rabbit polyclonal antibodies to either CT670 or CT671. After the blots were reacted with anti-rabbit antibody conjugated to horseradish peroxidase, they were developed with horseradish peroxidase substrate.
Protein structure accession number.
The coordinates of the final CT670 structure model have been deposited in the Protein Data Bank under accession code 3K29.
RESULTS
CT670 structure.
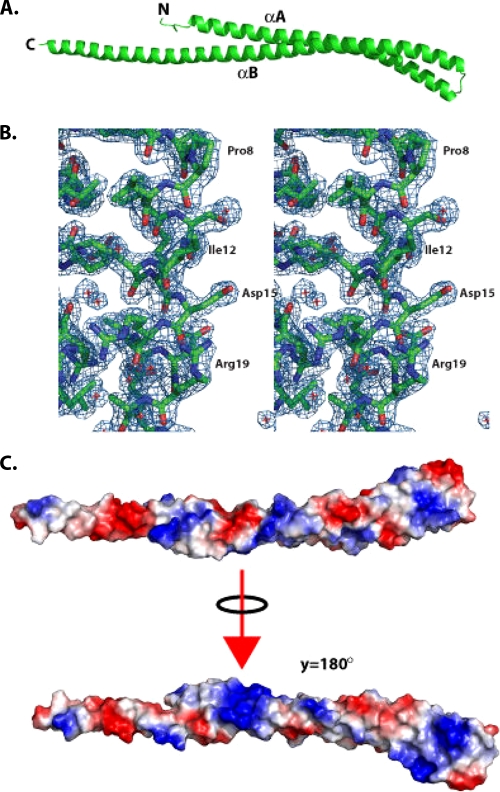
Full-length CT670 crystallizes as a monomer with a single molecule in the asymmetric unit which encompasses residues 1 to 162 (of 168 residues) (Fig. 1A). An electron density map of a small region of the molecule near the N terminus is shown in Fig. 1B, which indicates the quality of the data used to deduce the structure. The last 6 residues yielded no traceable electron density and are presumed to be disordered. Overall, the structure is a very elongated two-helix coiled coil with approximate dimensions of 144 by 19 by 12 Å. As shown in Fig. 1A, helices A and B, comprising residues 7 to 66 and 69 to 161, respectively, are joined by a four-residue turn defined by residues 66 to 69 (sequence GTTS). CT670 is highly enriched in charged residues, and these residues cluster on the outside of the structure; hydrophobic residues are located principally between the helices. The distribution of the charged residues is such that when the electrostatic potential along the molecular surface is calculated, bands of electropositive and electronegative potential lie along the length of the structure (Fig. 1C). A search for proteins that are structurally similar to CT670 using the DALI server revealed significant structural similarity to a number of protein domains (Z scores between 7 and 10) with a coiled-coil motif (26). However, coiled-coil domains are ubiquitous and play important roles in proteins that are not involved in the type III secretion system, so the relationship of these structural neighbors to CT670 is not obvious. For example, the top two hits for CT670 were hits for the colicin E3 (PDB code 2B5U; Z score, 10.9) and CheY (PDB code 1KMI; Z score, 8.7) proteins, which are involved in cell killing and chemotaxis, respectively, and have no obvious relationship to the type III secretion system. However, in a more general sense, elongated coiled-coil domains can function as adaptors for other proteins by providing a binding platform for multiple partners.
FIG. 1.
Structure of CT670. (A) Ribbon diagram of CT670, with the two helices and N and C termini labeled. (B) Electron density obtained from a 2Fo-Fc map generated from the final model, contoured at 1σ. A region representing the N terminus of helix αA is shown, and selected residues on one face of helix αA are labeled for clarity. (C) Electrostatic potential mapped on the molecular surface of the CT670 monomer, shown in two orientations, the first corresponding to the orientation shown in panel A and the second corresponding to a 180° rotation about the y axis relative to the first orientation.
Self-association properties of CT670.
The ability of CT670 to form coiled coils was investigated using the algorithm MultiCoil, which predicts two- and three-stranded coiled coils (60). CT670 showed a high probability (>60%) of forming dimeric coiled coils over about 40 amino acids (from amino acid 80 to amino acid 120) around the midpoint of the primary sequence (results not shown). This prediction provided insight into possible self-association properties of CT670, and therefore the oligomeric state of CT670 in solution was investigated using size exclusion chromatography and analytical ultracentrifugation.
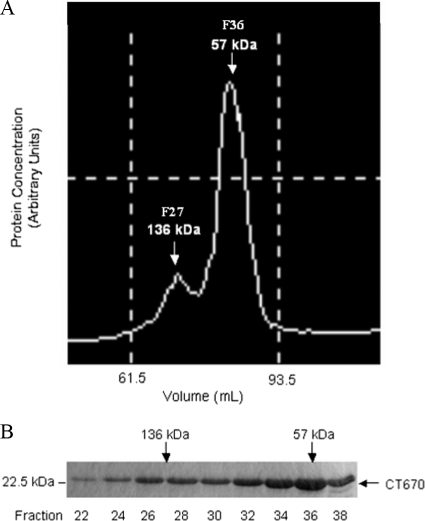
To obtain an initial estimate of the self-association properties of CT670, size exclusion chromatography using a Superdex 200 16/600 column was employed with nickel affinity-purified His6-CT670. Peak protein fractions from the column were visualized on an elution chromatogram (Fig. 2A) and were analyzed by SDS-PAGE (Fig. 2B). This experiment was carried out in triplicate with three independent purified preparations, and the protein always eluted in two peaks, as shown in the chromatogram in Fig. 2A. The majority of the protein eluted at an apparent molecular mass corresponding to the molecular mass of a CT670 dimer (∼57 kDa), and the minority eluted at an apparent molecular mass corresponding to the molecular mass of a hexamer (∼136 kDa). Caution should be employed when size exclusion chromatography is used to determine the oligomeric states of proteins, as it is highly dependent on protein shape and usually calibrated for folded proteins that are spherical (34). As shown previously, CT670 (Fig. 1A) is cigar shaped and far from spherical, and thus the chromatography results are probably not indicative of the actual molecular masses or self-association properties of this protein. However, the data do demonstrate that CT670 exists as two species in solution based on its consistent elution in two peaks.
FIG. 2.
Elution profile of CT670 resulting from size exclusion chromatography. (A) Chromatogram of CT670 elution from a size exclusion column. (B) Peak protein fractions from panel A analyzed by 12% SDS-PAGE. The apparent molecular mass of the eluted CT670 oligomer and fraction numbers are indicated.
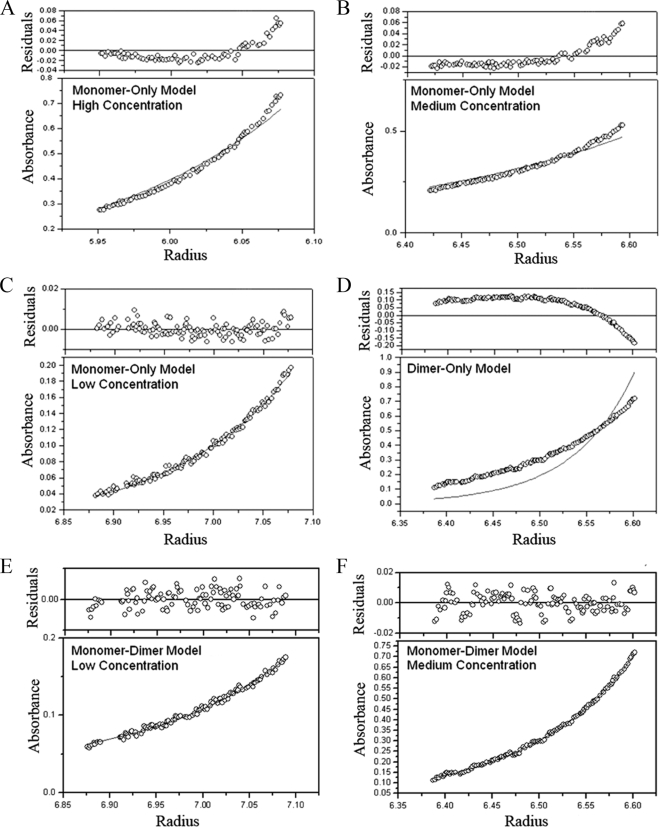
Analytical ultracentrifugation can also determine the oligomeric state of proteins by determining the molecular mass of the protein in solution. This method is superior to size exclusion chromatography because it is independent of the shape of the protein and dependent solely on the molecular mass (18). We carried out sedimentation equilibrium studies using two independently purified samples of His6-CT670, and each sample was tested at protein concentrations considered low, medium, and high (∼9, 27, and 44 μM). The data obtained from the sedimentation equilibrium runs were tested using several models for CT670 self-association. To obtain an initial estimate of the CT670 molecular mass, plots of apparent molecular mass versus protein concentration were generated, and the data trends predicted that CT670 may exist as a monomer at lower relative concentrations and have the potential to form up to a dimer at higher relative concentrations (data not shown). In addition, the average molecular masses of all species in the system were calculated by using Origin 6.0 software, and they ranged from 24.0 to 37.1 kDa. This is in accordance with a monomer and monomer-dimer system for His6-CT670. The observed average molecular masses did not support the hypothesis that there is a dimer-tetramer equilibrium.
Data sets were fitted to various molecular masses corresponding to a monomer, a dimer, a trimer, a tetramer, etc. or combinations of these forms for CT670, and the resulting plots were analyzed. The first systems analyzed for CT670 associations were monomer-only, dimer-only, and trimer-only models, where the molecular mass was fitted to the monomer size of His6-CT670 and two and three times this size (Fig. 3). As the plots show, at higher relative concentrations (Fig. 3A and B), the monomer-only model resulted in bad fits of the data, as indicated by poor overlap of the best-fit curve with data points in the lower absorbance plots, as well as by poor dispersion of residuals about zero in the upper residual plots. In addition, the residual distribution curve had an upward slope, indicating that the aggregation states present were higher than those predicted by the model. However, at low relative protein concentrations (Fig. 3C), the monomer-only model provided a good fit to the data, as indicated by good overlap of the best-fit curve with data points and residuals dispersed around zero. A representative plot of data fitted to twice the molecular mass of His6-CT670 for a dimer-only model is shown in Fig. 3D. The data fit this model poorly, and the downward slope of residuals indicated that the system was not ideal. Similar results were obtained for a trimer-only model (data not shown). Together, these data suggest that CT670 does not exist solely as one species in solution, except at low concentrations, where the monomer-only model fits well. Various monomer-n-mer models, including but not limited to monomer-dimer, monomer-trimer, monomer-dimer-trimer, and dimer-tetramer models, were analyzed next. Overall, the model that resulted in the best data fit was the monomer-dimer equilibrium model (Fig. 3E and F). This model is in accordance with the data reported above which showed that CT670 can exist in a monomer-dimer form. The association constant (Ka) was calculated using Origin 6.0 software for the CT670 monomer-dimer equilibrium. The Ka was 2.02 × 103 M−1, suggesting that there is weak CT670 self-association.
FIG. 3.
Analyses of different self-association models for His6-CT670 by sedimentation equilibrium. The results for sedimentation equilibrium from 15 data sets obtained at different protein concentrations and sedimentation speeds (see Materials and Methods) were fitted to different models. The plots were generated using Origin 6.0. (A, B, and C) Plots for a monomer-only model for three protein concentrations (low, medium, and high) of CT670 from data obtained by centrifugation at 29,120 × g. (D) Data fit for a dimer-only model (at the medium protein concentration) for centrifugation at 29,120 × g. (E and F) Data fit for a monomer-dimer model for (E) the low relative protein concentration for centrifugation at 16,380 × g and (F) the medium relative protein concentration for centrifugation at 29,120 × g.
Sequence similarity of the CT670 and YscO proteins.
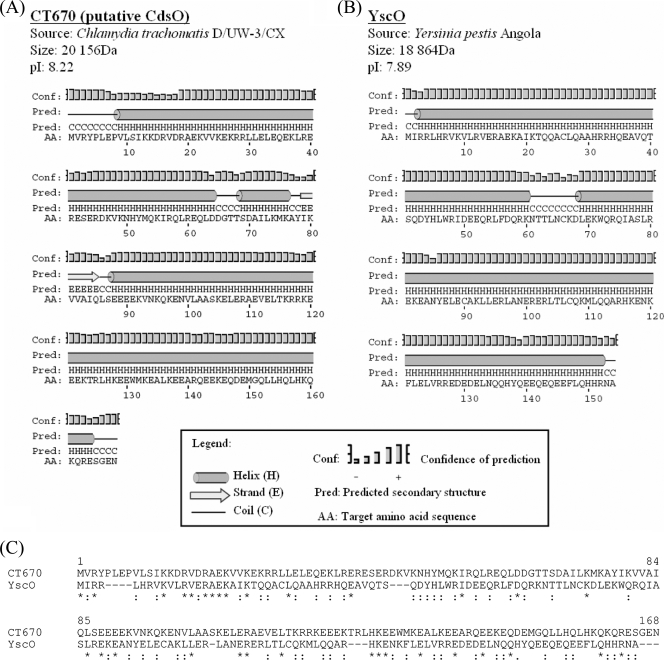
CT670 shares many characteristics with YscO, a T3S mobile core component in Yersinia (40). The genes for both of these proteins are immediately downstream of the ATPase gene (yscN) and shortly upstream of yscQ (with the yscP gene in between). Additionally, these two proteins have very similar sizes, charges, and secondary structures (Fig. 4A and B), supporting the view that they may be related. Although BLAST searches with CT670 did not detect YscO, in view of the observations described above sequence similarity between these two proteins was examined more closely by using additional methods. Figure 4C shows a sequence alignment of the CT670 and YscO proteins created using the ClustalX 1.83 program (29). As this figure shows, the two proteins can be aligned over their entire lengths with only a few small gaps. Large numbers of residues in these proteins, which are distributed over their entire lengths, either are identical (31/154) or involve conservative substitutions (48/154) (Fig. 4C). The significance of the sequence similarity of these two proteins was examined using the PCOMPARE program (39) from the PCGENE software package. We used this program in previous work to discover or demonstrate that the sequence similarities between the Hsp60 and Tcp-1 proteins and between the DnaK and MreB proteins were significant (20, 22). When this program was used, the amino acid sequence of one of the two proteins (YscO or CT670) was randomly shuffled (e.g., 50 to 100 times), and a mean alignment score for the shuffled sequences was calculated. This score was compared with the observed alignment score for the two proteins without any shuffling. An observed alignment score that differed from the mean alignment score for shuffled sequences by >3.0 standard deviations (i.e., z value) indicated that there is significant sequence similarity (39, 42). The analysis of the CT670 and YscO proteins by this method showed that the z value of the observed alignment score for these proteins for 100 randomized runs was 3.68, which provides evidence that the sequence similarity between these two proteins is significant.
FIG. 4.
Shared protein characteristics of CT670 and YscO. (A and B) Protein, source, size, pI, and predicted secondary structure for C. trachomatis D/UW-3/CX (A) and Y. pestis Angola (B). Secondary structure predictions were obtained with the PSIPRED algorithm using the primary protein sequences of CT670 and YscO (30). (C) Sequence alignment of CT670 and the YscO protein from Y. pestis. The alignment was constructed using the ClustalX 1.83 program. Identical residues are indicated by asterisks, and conservative substitutions are indicated by colons. The significance of sequence similarity between the two proteins was determined using the PCOMPARE program of the PCGENE software package (39) with the unitary matrix and a gap penalty value of 3. The YscO sequence was randomized 100 times. The normal score of this sequence alignment differed from the mean score of the randomized sequence alignment by 3.7 standard deviations, indicating that the observed sequence similarity of CT670 and YscO is significant and it is not due to the amino acid compositions of the proteins by chance.
Protein-protein interaction studies with CT670.
In previous studies, the YscO protein was reported to copurify with glutathione S-transferase (GST)-tagged YscP protein from Yersinia lysate (46). This finding led to the prediction that CT670, the putative YscO homolog, may interact with CT671, the putative YscP homolog, in C. trachomatis. However, CT670 has also been reported to interact with a number of other proteins of the type III secretion apparatus, including CT091 (the YscU homolog), proteins of the type III secretion ATPase (CT669), or other type III chaperones (46, 52). Hence, we examined the interaction of CT670 with a number of T3S-related proteins using a bacterial two-hybrid system (12). When this system is used, protein-protein interaction results in transcriptional activation of the HIS-3-aadA cassette. The product of the HIS-3 gene allows the cells to grow in the presence of 3-amino-1,2,4-triazole (3-AT), and the expression of the aadA gene (encoding adenyltransferase) confers resistance to streptomycin. 3-AT is a competitive inhibitor of the His3 enzyme; therefore, bacteria with low levels of His3 (leaky expression) are not able to grow on His drop-out medium (12). Protein interactions result in high-level expression of the His3 gene product, resulting in growth of bacteria on medium containing 3-AT. The positive interaction can be further confirmed by testing the growth of bacteria on plates containing both 3-AT and streptomycin. Using this system, it was determined that CT670 interacts specifically with CT671, whereas no interaction was observed with the predicted pilin protein (CT666) or its chaperones (CT665 and CT667), the type III secretion ATPase (CT669), and several proteins encoded in the same operon as CT670 (CT668, CT672, and CT673) (Table 3). In addition, YscU has also been reported to be a binding partner for YscO and YscP (46). However, no interaction between CT670 and CT091, the C. trachomatis equivalent of YscU, was observed (Table 3). These experiments were repeated at least twice, and the same results were obtained. We also confirmed the results of interaction of CT670 with CT671 and CT091 using another bacterial two-hybrid system, where the bait and the target proteins are fused to different fragments of GFP. The positive interaction in this system leads to reconstitution of GFP, which is readily detected due to fluorescence of the resulting cells or colonies (57). In these experiments, fluorescent colonies were observed only when the interaction of CT670 with CT671 was studied, whereas no fluorescence was observed when CT670 and CT091 were tested.
TABLE 3.
Bacterial two-hybrid protein-protein interaction results
| Bait | Prey | Growth on medium without His containing 3-ATa | Growth on medium containing 3-AT and streptomycinb | Interaction |
|---|---|---|---|---|
| CT670 | CT665 | − | NA | − |
| CT670 | CT666 | − | NA | − |
| CT670 | CT667 | − | NA | − |
| CT670 | CT668 | − | NA | − |
| CT670 | CT669 | − | NA | − |
| CT670 | CT671 | + | +c | + |
| CT670 | CT672 | − | NA | − |
| CT670 | CT673 | − | NA | − |
| CT670 | CT091 | − | NAc |
−, no growth on medium lacking histidine in the presence of 3-AT, indicating that there is not an interaction between the bait and prey proteins: +, growth on this medium, indicating that there is a positive interaction.
Colonies which grew on medium without His containing 3-AT were also tested for growth in this medium in the presence of streptomycin. NA, not applicable (because no colonies grew on medium without His containing 3-AT, further testing on this medium was not possible).
The interaction was further tested and confirmed using another two-hybrid system based on reconstitution of GFP fluorescence in the case of the positive interaction (57).
The fact that CT670 showed positive interactions only with CT671 in both bacterial two-hybrid systems suggests that the results were not false-positive results. To further confirm this finding, ct671 was cloned in the pTriEx vector to produce an N-terminal fusion protein with the Strep-tag II peptide (Novagen), and the interaction of CT671 with CT670 lacking this tag was examined by coexpressing these two proteins in the same cells. The Strep-tag II peptide has a very high affinity for the streptavidin derivative Strep-tactin which is bound to agarose beads (Novagen) (36). When extracts from the coexpressed cells were passed over a Strep-tactin column, Strep-tagged CT671 bound to it, as expected, and it was the major protein in the elution fractions; most of the protein eluted in fractions 2 to 4, as visualized by SDS-PAGE (Fig. 5A). However, Western blot analyses of these fractions with antibodies to CT670 showed that a protein that migrated in the same position as CT670 and cross-reacted with CT670 was also present in these fractions (Fig. 5B). The elution (or reactivity) profile of CT670 was very similar to that of CT671 (Fig. 5A and B). It should be noted that the antibody to CT670 used in this experiment reacts specifically with CT670, and it shows no cross-reactivity with any other protein in cell extracts of E. coli BL21 cells (Fig. 5C). In control experiments, in which cell extracts of E. coli that expressed only His6-tagged CT670 were applied to the Strep-tactin column, no CT670 was detected in the eluted fractions (results not shown). This indicated that CT670 was copurified with CT671 and that the observed cross-reactive band was not due to a nonspecific reaction.
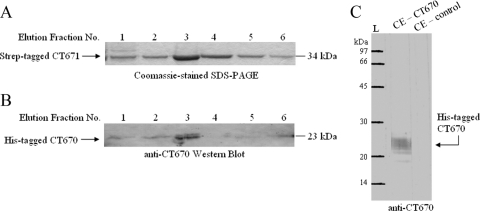
FIG. 5.
Strep-tag pull down assay of coexpressed Strep-tag II-CT671 and His6-CT670. (A) Coomassie blue-stained 12% SDS-PAGE gel of Strep-tagged CT671 elution fractions from Strep-tactin chromatography. CT671 eluted at 34 kDa. (B) Elution fractions from the assay were transferred to a nitrocellulose membrane and exposed to anti-CT670 antibodies for Western blot analysis as described in Materials and Methods. Antibodies recognized CT670 at 23 kDa in the elution fractions, indicating that it copurified with Strep-tagged CT671. In control experiments with cells expressing only His6-CT670, no binding to the Strep-tactin column was observed (not shown). (C) Cell extract (CE) from E. coil BL21 cells expressing CT670 or with no expression plasmid (control) was exposed to anti-CT670 antibodies. Antibodies recognized only the overexpressed CT670 protein.
DISCUSSION
In this paper we describe the crystal structure and several biochemical studies of the CT670 protein of C. trachomatis. The CT670 protein was identified in our recent work as a Chlamydiales-specific protein, as all significant BLAST hits with this protein were restricted to various Chlamydiae species (19, 21). Based on this observation and the fact that very little information regarding its function in Chlamydiae species is available, CT670 is annotated as a hypothetical protein in the NCBI database (32, 51). The presence of the CT670 gene in a cluster of genes for T3S system-related proteins provided the initial evidence that this protein plays some role in this system (23, 44). The genes for theT3S systems in various species are tightly clustered, and their genomic neighborhood is generally highly conserved (44). This indicated that CT670 (designated CdsO) might be a homolog of the YscO protein in Yersinia species (40). The sizes, charge distribution profiles, and predicted secondary structures of the CT670 and YscO proteins are similar (Fig. 4). Although BLAST searches with CT670 did not detect YscO, the results of our analysis presented here provide evidence that the sequence similarity between these two proteins is significant and that these proteins are related or homologous. At present, very little information regarding the cellular function of CdsO and YscO is available, and a protein with a homologous structure has not yet been solved and deposited in the Protein Data Bank (PDB).
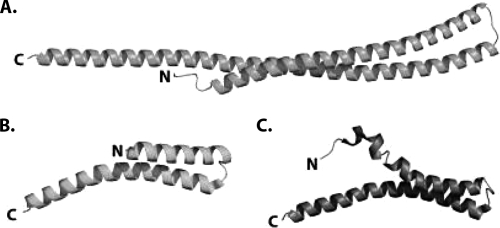
Because the structures of proteins are more conserved than their sequences (28), structure determination can provide more definitive information that establishes the functional relatedness of two proteins and also their possible cellular functions. In the present work, we solved the structure of the CT670 protein, which is the first structure of a YscO homolog determined for any species. The crystal structure of CT670 shows that this protein is an elongated two-helix coiled coil that is approximately 144 Å long in which helix A is 33 residues shorter than helix B (Fig. 1A). The helices are connected by a tight turn consisting of four residues (GTTS). The structure of YscO is not known. However, recently, preliminary X-ray diffraction data were collected for the FliJ protein (27), which is involved in the flagellar export system and shares many of the characteristics of the CdsO and YscO proteins (14, 18). When the structure of FliJ is known, it will be very interesting to determine how similar it is to CT670. We also note that the structure of CT670 also shows similarity to the structure of the T3S needle-forming protein (Fig. 6). The proteins whose structures have been solved include MxiH (PDB code 2CA5) of Shigella flexneri (10) and the PrgI protein (PDB code 2JOW) of Salmonella enterica serovar Typhimurium (56). These structures are also two-helix coiled coils in which the N-terminal helix is shorter than the C-terminal helix. However, these proteins are approximately 80 residues long; i.e., they are half the size of CT670, and therefore the helices are half the size of CT670 helices. However, it should be mentioned that although a search for structural neighbors of CT670 using the DALI server revealed structural similarity to a number of protein domains (Z scores between 7 to 10) with a coiled-coil motif, no structural similarity to any protein domain related to the type III secretion system was identified.
FIG. 6.
Comparison of the structures of CT670 and the type III needle proteins MxiH and PrgI. (A) CT670 in the orientation shown in Fig. 1A. (B) MxiH (PDB code CA5) from S. flexneri. (C) PrgI (PDB code 2JOW) from S. enterica serovar Typhimurium. The structures are represented by ribbons, and the N and C termini are labeled.
We also describe here studies of the self-association properties of CT670. The analysis of the structure resulted in a strong prediction that this protein should form dimeric coiled coils (results not shown). This prediction was validated by the behavior of CT670 both in size exclusion chromatography and in analytical ultracentrifugation experiments. The results of these studies show that at low protein concentrations CT670 exists predominantly as a monomer, whereas at higher concentrations both monomeric and dimeric forms seem to coexist in solution (Fig. 3). It should be noted that CPn0706, the CT670 homolog of C. pneumoniae, has also been reported to exist as a monomer and a dimer as determined by nondenaturing SDS-PAGE (52).
Identification of proteins that interact with a protein can provide valuable information regarding the cellular function of the latter protein (12, 49, 50, 57). The YscO protein (CT670 homolog in Yersinia) has previously been shown to interact specifically with the neighboring protein YscP, indicating that these two proteins function together in the secretion of proteins by a T3S system (40, 41, 46). In the present work, we showed that CT670 interacts specifically with its immediate downstream neighbor, the CT671 protein, whereas no interaction was detected for several other T3S-related proteins, including the predicted pilin protein (CT666) and its chaperones (CT665 and CT667), the type III secretion ATPase (CT669), and several proteins that are part of the same operon as CT670 (CT668, CT672, and CT673). It should be noted that although a positive interaction between CPn0707 (C. pneumoniae homolog of CT670) and CPn0706 (C. pneumoniae homolog of CT669) has been reported (52), such an interaction was not observed in our work despite repeated attempts. The possibility that this interaction occurs specifically in C. pneumoniae cannot be excluded based on our experiments. It should also be noted that in our work CT670 did not interact with the YscU homolog CT091. YscU is an ∼350-residue protein which contains transmembrane helices throughout most of the protein and has a cytoplasmic C-terminal domain with autoproteolytic processing at N263 (45). Processing of this protein by autocleavage is necessary for assembly of the type III secretion apparatus, as mutants with mutations at this residue do not secrete effectors. The C-terminal domain of YscU was shown to interact with YscO as well as the ATPase regulator YscL (45), and in particular, an autocleavage-negative mutant G270N bound strongly to YscO; thus, a model was proposed in which YscU binds YscO during preprocessing, but YscU binds to the ATPase complex after processing (45). The possibility that some of these protein-protein interactions are also species specific cannot be excluded.
The interaction between CT670 and CT671 was observed in our work in two independent bacterial two-hybrid systems, as well as in affinity pull-down experiments, indicating that this interaction is specific and likely physiological. This is the first reported interaction between these proteins in Chlamydiales species. Based on the observation that YscO interacts with the neighboring protein YscP in Yersinia (46), the analogous positions of the CT670 and CT671 genes in various Chlamydiae genomes (23, 44), and the significant sequence similarity between CT670 and YscO, it is highly likely that CT670 (CdsO) and CT671 (CdsP) function in Chlamydiae like the YscO and YscP proteins (40, 41, 46). A number of observations indicate that the CT670 and CT671 proteins may constitute a chaperone-effector pair. First, typically, most T3S chaperones are dimers, and the ability of CT670 to dimerize suggests that it could act as a T3S chaperone (7, 35). Second, the genes encoding the T3S chaperones are located adjacent to or near the genes encoding their cognate effectors; the fact that ct670 and ct671 are direct neighbors in the same operon is also consistent with this role (23, 44). Additionally, the T3S chaperones are generally not secreted but rather aid in the secretion of their effectors. Previous studies of CT670 and its C. pneumoniae homolog, CPn0706, indicated that these proteins are not secreted (24, 47). However, CT671 was shown to be secreted by a heterologous T3S system, and its homolog was detected in the host cytosol of infected cells (53). Likewise, studies with the Yersinia homologs showed that YscP is a secreted protein, whereas whether YscO is secreted is debatable (40, 41). Specifically, YscP secretion is necessary for its role as the molecular ruler for T3S needle length (2). It is tempting to speculate that YscO or its chlamydial homolog CT670, by interacting with YscP (or CT671) and acting as a chaperone, may aid in the secretion of the latter protein during needle assembly. However, it is important to confirm the chaperone and effector roles of CT670 and CT671 by performing more detailed cell biological studies, as has been done for several Chlamydiae proteins (5, 50, 54).
As noted above, CT670 and YscO also share several features with the FliJ protein. FliJ is reported to act as a chaperone escort protein in the flagellar system, binding to flagellar subunit chaperones when they are free of their cargo (15). Recently, this role was investigated for YscO and InvI, which are orthologs of FliJ in the T3S system. It was found that YscO and InvI recognized the translocon chaperones SycD and SicA, respectively, by using GST pull-down assays (14). These chaperones bind the translocator pore proteins YopB/D and SipC, respectively (14). These data supported the hypothesis that the YscO and InvI proteins have a FliJ-like chaperone escort protein role. The C. trachomatis homologs of SycD are Scc2 and Scc3, which are the proposed chaperones of the translocated T3S pore proteins CopB1/D1 and CopB2/D2, respectively (16). Given that CT670 is homologous to YscO, it is possible that it may also act as a chaperone escort protein for the translocon chaperones Scc2 and Scc3. Thus, it should be interesting to investigate the possible interactions between CT670 and these proteins.
In summary, the findings presented here for the crystal structure, self-association properties, and interacting partners for CT670 provided insights into the possible function of this protein in the T3S system. In the future, as structural information for FliJ, YscO, and other related proteins become available, structural comparisons should lead to more definitive insights into whether CT670 functions as a chaperone for CT671 and/or as a chaperone escort protein for the translocon chaperones Scc2 and Scc3. The specific interaction between CT670 and CT671 observed in these studies further corroborates the predicted homology between these proteins and the Yersinia T3S proteins YscO and YscP, respectively. This interaction also suggests that it may be possible to determine a crystal structure for the CT670-CT671 protein complex, which would be very valuable for understanding how these proteins interact and their cellular functions.
Acknowledgments
The work in the lab of R.S.G. was supported by a research grant from the Canadian Institute of Health Research. The work at the Ontario Centre for Structural Proteomics was funded in whole or in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272200700058C.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Abdelrahman, Y. M., and R. J. Belland. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Agrain, C., I. Sorg, C. Paroz, and G. R. Cornelis. 2005. Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57:1415-1427. [DOI] [PubMed] [Google Scholar]
- 3.Bricogne, G., C. Vonrhein, C. Flensburg, M. Schiltz, and W. Paciorek. 2003. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. Sect. D Biol. Crystallogr. 59:2023-2030. [DOI] [PubMed] [Google Scholar]
- 4.Coburn, B., I. Sekirov, and B. B. Finlay. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20:535-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchiaro, J. L., Y. Kumar, E. R. Fischer, T. Hackstadt, and R. H. Valdivia. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379-9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 8.Corsaro, D., and G. Greub. 2006. Pathogenic potential of novel chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clin. Microbiol. Rev. 19:283-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, D., R. P. Kandel, H. K. Adhikari, and T. Hessel. 2008. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. 5:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deane, J. E., P. Roversi, F. S. Cordes, S. Johnson, R. Kenjale, S. Daniell, F. Booy, W. D. Picking, W. L. Picking, A. J. Blocker, and S. M. Lea. 2006. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl. Acad. Sci. U. S. A. 103:12529-12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Fortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction method. Methods Enzymol. 276:472. [DOI] [PubMed] [Google Scholar]
- 12.Dove, S. L., and A. Hochschild. 2001. Bacterial two-hybrid analysis of interactions between region 4 of the sigma(70) subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J. Bacteriol. 183:6413-6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Evans, L. D., and C. Hughes. 2009. Selective binding of virulence type III export chaperones by FliJ escort orthologues InvI and YscO. FEMS Microbiol. Lett. 293:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, L. D., G. P. Stafford, S. Ahmed, G. M. Fraser, and C. Hughes. 2006. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. U. S. A. 103:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields, K. A., E. R. Fischer, D. J. Mead, and T. Hackstadt. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 187:6466-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields, P. I., and R. C. Barnes. 1992. The genus Chlamydia, p. 3691-3709. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, NY.
- 18.Fraser, G. M., B. Gonzalez-Pedrajo, J. R. Tame, and R. M. Macnab. 2003. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J. Bacteriol. 185:5546-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths, E., M. S. Ventresca, and R. S. Gupta. 2006. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, R. S. 1990. Sequence and structural homology between a mouse T-complex protein TCP-1 and the ‘chaperonin’ family of bacterial (GroEL, 60-65 kDa heat shock antigen) and eukaryotic proteins. Biochem. Int. 20:833-841. [PubMed] [Google Scholar]
- 21.Gupta, R. S., and E. Griffiths. 2006. Chlamydiae-specific proteins and indels: novel tools for studies. Trends Microbiol. 14:527-535. [DOI] [PubMed] [Google Scholar]
- 22.Gupta, R. S., and B. Singh. 1992. Cloning of the HSP70 gene from Halobacterium marismortui: relatedness of archaebacterial HSP70 to its eubacterial homologs and a model for the evolution of the HSP70 gene. J. Bacteriol. 174:4594-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hefty, P. S., and R. S. Stephens. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189:198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann, M., A. Schuhmacher, I. Muhldorfer, K. Melchers, C. Prothmann, and S. Dammeier. 2006. Identification and characterization of secreted effector proteins of Chlamydophila pneumoniae TW183. Res. Microbiol. 157:513-524. [DOI] [PubMed] [Google Scholar]
- 25.Heuer, D., C. Kneip, A. P. Maurer, and T. F. Meyer. 2007. Tackling the intractable—approaching the genetics of Chlamydiales. Int. J. Med. Microbiol. 297:569-576. [DOI] [PubMed] [Google Scholar]
- 26.Holm, L., and C. Sander. 1999. Protein folds and families: sequence and structure alignments. Nucleic Acids Res. 27:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibuki, T., M. Shimada, T. Minamino, K. Namba, and K. Imada. 2009. Crystallization and preliminary X-ray analysis of FliJ, a cytoplasmic component of the flagellar type III protein-export apparatus from Salmonella sp. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Illergard, K., D. H. Ardell, and A. Elofsson. 2009. Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins 77:499-508. [DOI] [PubMed] [Google Scholar]
- 29.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 30.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 31.Kalayoglu, M. V., and G. I. Byrne. 2001. Chlamydia, In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.7. Springer-Verlag, New York, NY. http://link.springer-ny.com/link/service/books/10125/.
- 32.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283. [Google Scholar]
- 34.Le Maire, M., E. Rivas, and J. V. Moller. 1980. Use of gel chromatography for determination of size and molecular weight of proteins: further caution. Anal. Biochem. 106:12-21. [DOI] [PubMed] [Google Scholar]
- 35.Letzelter, M., I. Sorg, L. J. Mota, S. Meyer, J. Stalder, M. Feldman, M. Kuhn, I. Callebaut, and G. R. Cornelis. 2006. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 25:3223-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier, T., N. Drapal, M. Thanbichler, and A. Bock. 1998. Strep-tag II affinity purification: an approach to study intermediates of metalloenzyme biosynthesis. Anal. Biochem. 259:68-73. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Minor, W., M. Cymborowski, Z. Otwinowski, and M. Chruszcz. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. Sect. D Biol. Crystallogr. 62:859-866. [DOI] [PubMed] [Google Scholar]
- 39.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 40.Payne, P. L., and S. C. Straley. 1998. YscO of Yersinia pestis is a mobile core component of the Yop secretion system. J. Bacteriol. 180:3882-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne, P. L., and S. C. Straley. 1999. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J. Bacteriol. 181:2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 44.Peters, J., D. P. Wilson, G. Myers, P. Timms, and P. M. Bavoil. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15:241-251. [DOI] [PubMed] [Google Scholar]
- 45.Riordan, K. E., and O. Schneewind. 2008. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol. Microbiol. 68:1485-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riordan, K. E., J. A. Sorg, B. J. Berube, and O. Schneewind. 2008. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J. Bacteriol. 190:6204-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samudrala, R., F. Heffron, and J. E. McDermott. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. Sect. D Biol. Crystallogr. 58:1772-1779. [DOI] [PubMed] [Google Scholar]
- 49.Sisko, J. L., K. Spaeth, Y. Kumar, and R. H. Valdivia. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 60:51-66. [DOI] [PubMed] [Google Scholar]
- 50.Spaeth, K. E., Y. S. Chen, and R. H. Valdivia. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 52.Stone, C. B., D. L. Johnson, D. C. Bulir, J. D. Gilchrist, and J. B. Mahony. 2008. Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J. Bacteriol. 190:6580-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subtil, A., C. Delevoye, M. E. Balana, L. Tastevin, S. Perrinet, and A. Dautry-Varsat. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 56:1636-1647. [DOI] [PubMed] [Google Scholar]
- 54.Valdivia, R. H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53-59. [DOI] [PubMed] [Google Scholar]
- 55.Vandahl, B. B., S. Birkelund, and G. Christiansen. 2004. Genome and proteome analysis of Chlamydia. Proteomics 4:2831-2842. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., A. N. Ouellette, C. W. Egan, T. Rathinavelan, W. Im, and R. N. De Guzman. 2007. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371:1304-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, C. G., T. J. Magliery, and L. Regan. 2004. Detecting protein-protein interactions with GFP-fragment reassembly. Nat. Methods 1:255-262. [DOI] [PubMed] [Google Scholar]
- 58.Winn, M. D., M. N. Isupov, and G. N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 57:122-133. [DOI] [PubMed] [Google Scholar]
- 59.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300-321. [DOI] [PubMed] [Google Scholar]
- 60.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]