Abstract
Alternative splicing and posttranslational modifications (PTMs) are major sources of protein diversity in eukaryotic proteomes. The SR protein SF2/ASF is an oncoprotein that functions in pre-mRNA splicing, with additional roles in other posttranscriptional and translational events. Functional studies of SR protein PTMs have focused exclusively on the reversible phosphorylation of Ser residues in the C-terminal RS domain. We confirmed that human SF2/ASF is methylated at residues R93, R97, and R109, which were identified in a global proteomic analysis of Arg methylation, and further investigated whether these methylated residues regulate the properties of SF2/ASF. We show that the three arginines additively control the subcellular localization of SF2/ASF and that both the positive charge and the methylation state are important. Mutations that block methylation and remove the positive charge result in the cytoplasmic accumulation of SF2/ASF. The consequent decrease in nuclear SF2/ASF levels prevents it from modulating the alternative splicing of target genes, results in higher translation stimulation, and abrogates the enhancement of nonsense-mediated mRNA decay. This study addresses the mechanisms by which Arg methylation and the associated positive charge regulate the activities of SF2/ASF and emphasizes the significance of localization control for an oncoprotein with multiple functions in different cellular compartments.
Pre-mRNA splicing is a required step for the expression of most human genes. Alternative splicing plays a key role in regulating gene expression and is a major source of protein isoform diversity. SR proteins are a family of closely related and highly expressed eukaryotic RNA-binding proteins that regulate both general and alternative splicing (22) and have additional roles in other aspects of gene expression (41). SF2/ASF, a prototypical member of the SR protein family, has a modular structure with two RNA recognition motifs (RRMs), which recognize a 7-nucleotide exonic splicing enhancer (ESE) motif on pre-mRNA, and an RS domain with numerous repeats of serine-arginine dipeptides (Fig. 1 A). SF2/ASF is required for general splicing, though sometimes interchangeably with other SR proteins (33), and also plays a role in the regulation of alternative splicing in a concentration-dependent manner, in part through the recognition of ESEs (16). In addition to its role as a broad-specificity splicing regulator, SF2/ASF strongly enhances nonsense-mediated mRNA decay (NMD) upon overexpression (65), stimulates translation both in vivo and in vitro in an enhancer-dependent manner (50), and functions as an adaptor protein for mRNA export (26). Some of these additional functions of SF2/ASF reflect its ability to shuttle between the nucleus and the cytoplasm (13). Reflecting its multifaceted role as a regulator in various cellular processes, SF2/ASF is a potent oncoprotein whose overexpression by as little as 2-fold is sufficient to transform immortal rodent fibroblasts (30). Conversely, the knockdown of SF2/ASF induces G2 cell cycle arrest and apoptosis (39).
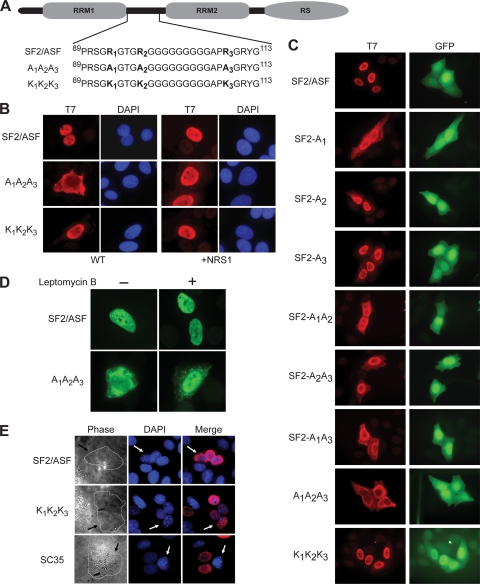
FIG. 1.
Localization and shuttling activity of wild-type and mutant SF2/ASF. (A) Modular structure of SF2/ASF and mutants in the inter-RRM linker region. The three Arg residues in boldface type are methylated, and we mutated each one to Ala or Lys. (B) Indirect immunofluorescence of HeLa cells transfected with wild-type or mutant T7-tagged SF2/ASF cDNA, with or without fusion to a nuclear retention signal peptide from SC35 (NRS1) (17). (C) Indirect immunofluorescence of HeLa cells cotransfected with GFP and wild-type or mutant T7-tagged SF2/ASF. (D) Indirect immunofluorescence of HeLa cells transfected with T7-tagged SF2/ASF or the A1A2A3 mutant with or without prior treatment with 10 ng/ml leptomycin B for 12 h. (E) Heterokaryon assay (13, 49). We transfected HeLa cells with T7-tagged SF2/ASF, K1K2K3, or SC35 as a control. After 24 h, we treated the cells with cycloheximide, fused them with mouse NIH 3T3 cells using polyethylene glycol, and incubated them for 2 h. After fixation, we determined the localization of the transiently expressed proteins in the heterokaryons by indirect immunofluorescence. The panels in the right column show the merged images of T7 and DAPI signals. The arrows indicate mouse nuclei within human-mouse heterokaryons, distinguished from human nuclei by their characteristic DAPI staining (middle column). The panels in the left column show phase-contrast images of the same heterokaryons.
One of the ways through which the various activities of SF2/ASF could be regulated is by posttranslational modifications (PTMs). Like alternative splicing, PTMs are an integral part of gene regulation and provide another way of creating protein isoform diversity, with widespread functional consequences. Studies of PTMs in SR proteins have focused mostly on reversible phosphorylation at multiple Ser residues within the C-terminal RS domain, as regulated by a combination of phosphatases and kinases (46). More recently, three methylated Arg residues (R93, R97, and R109) in human SF2/ASF were identified in a global analysis of protein methylation (48) (Fig. 1A). Arginine methylation is widespread in eukaryotes. In the budding yeast Saccharomyces cerevisiae, the protein Npl3p, which resembles both hnRNPs and SR proteins in primary sequence and domain structure (7), is involved in mRNA export (38, 54) and undergoes Arg methylation at RGG motifs in its C-terminal RGG/RS domain, catalyzed by Hmt1p (53). A lack of Arg methylation causes defects in the export of Npl3p from the nucleus (52), and the hypermethylation of RGG motifs blocks its nuclear import (63). In mammals, the methylation of RGG motifs in the C-terminal Gly-rich domain of the splicing factor hnRNP A2 promotes its nuclear localization (47). The methylated Arg residues in SF2/ASF are part of Arg/Gly-rich motifs, which are very similar to those in Npl3p and hnRNP A2, although these motifs differ in their locations in SF2/ASF. Indeed, it was shown previously that SF2/ASF and its close paralog SRp30c are methylated by the human homolog of Hmt1p, PRMT1 (11, 21). PRMT1 is a type I methyltransferase that catalyzes the formation of monomethyl and asymmetric dimethyl Arg on proteins (59, 60) preferentially at RGG/GRG and related motifs (28, 31).
To investigate the functional relevance of the modified Arg residues of SF2/ASF, we mutated these residues, singly or in combination, to Ala or Lys. After an extensive analysis of the various functions of SF2/ASF, we report that the three methylated Arg residues in the inter-RRM linker region control SF2/ASF's subcellular localization, which in turn determines the protein's activity in enhancing NMD and regulating the alternative splicing of endogenous pre-mRNA targets.
MATERIALS AND METHODS
Plasmids.
Plasmid pSP64-HβΔ6 (34) and the duplicated 5′-splice-site (5′ss) version pSP64-5′ssD-wt (65) were described previously. Plasmid pUCβ128SV has a full-length β-globin gene driven by its own promoter and the simian virus 40 (SV40) enhancer (14). The mutant version pUCβ128SV-T39 and the wild-type and mutant duplicated 5′ss versions, pUC-5′ssD-wt and pUC-5′SSD-mt, respectively, were described previously (65). Mammalian expression plasmid pCGT-SF2/ASF encodes a T7-tagged version of human SF2/ASF (12). We used site-directed mutagenesis to create the various T7-tagged pCGT versions of single-, double-, or triple-missense mutants in which the Arg residues at positions 93, 97, and 109 were changed to either Ala or Lys (see Table S1 in the supplemental material). Using pCGT-A1A2A3 and pCGT-K1K2K3 as templates, we made pCGT-SM and pCGT-KM by site-directed mutagenesis, changing the Gly residues in the RGG/GRG/RGR motifs to Ala. We replaced a ClaI-BstBI fragment in plasmid pCGT-SF2/ASF-NRS1 (17) with the corresponding fragments from pCGT-A1A2A3 and pCGT-K1K2K3 to create pCGT-A1A2A3-NRS1 and pCGT-K1K2K3-NRS1, respectively. We replaced the SacII-BamHI fragment in pET9c-SF2/ASF(R/S) (35) with the corresponding fragment from pCGT-A1A2A3 and pCGT-K1K2K3 to create pET9c-A1A2A3 and pET9c-K1K2K3, respectively, for bacterial expression and purification. We added a C-terminal 6×His tag to the cDNAs of wild-type SF2/ASF (wt-SF2/ASF), A1A2A3, and K1K2K3 by PCR using their pCGT versions as templates. We then individually subcloned these amplicons as XbaI-BamHI fragments into the mammalian expression vector pTT3 (20). Similarly, we inserted the amplified cDNAs of A1A2A3 and K1K2K3 with an N-terminal T7 tag as BamHI-EcoRI fragments into the retroviral vectors pBABE-puro and pWZL-hygro. Both pBABE-T7SF2/ASF and pWZL-T7SF2/ASF were described previously (30). Plasmids pLCS-EDA and pLCS-EDAmt, used for translation assays involving transient transfection, as well as constructs used for in vitro translation, pLuc-3×EDA and pLuc-3×EDAmt, were generous gifts from Javier Cáceres and were described previously (50).
Expression and purification of recombinant proteins.
We purified recombinant untagged SF2/ASF as described previously (35). Briefly, we expressed wild-type or mutant SF2/ASF cDNAs in Escherichia coli cells by using pET9c-SF2/ASF(R/S)-type plasmids, followed by a series of purification steps, including cesium-chloride gradient centrifugation to remove nucleic acids, selective precipitation in low salt, urea denaturation, ion-exchange chromatography, and the subsequent refolding of the protein. We purified C-terminal 6×His-tagged SF2/ASF expressed in human 293-EBNA1 (293E) cells grown in suspension using a variation of a method reported previously (20). We transfected 293E cells with wild-type or mutant pTT3-SF2/ASF-His and harvested the cells 3 to 4 days after transfection. We resuspended the cells in lysis buffer (50 mM Tris-HCl [pH 8.0], 1 M NaCl, 10 mM imidazole, 20 mM β-mercaptoethanol, 0.1% [vol/vol] Triton X-100) and centrifuged them after sonication to remove insoluble material. We then precipitated most of the cellular proteins by using ammonium sulfate at 40% saturation at 4°C and removed them by centrifugation. We then purified the His-tagged proteins from the supernatant by nickel-agarose affinity chromatography.
Cell culture and transient and stable expression of proteins.
We grew HeLa and IMR90 cells in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C with 5% CO2. We grew 293E suspension cultures in Joklik's modified Eagle's medium (JMEM) (Invitrogen) supplemented with 5% calf serum (Invitrogen). We transfected HeLa cells with Fugene 6 reagent (Roche Diagnostics) according to the manufacturer's recommendations, unless otherwise stated. We transfected 293E cells using linear polyethylenimine (PEI 25000; Polysciences Inc.). We prepared a transfection mix consisting of 1 μg plasmid DNA, 2 μg PEI, and 50 μl of JMEM for every 1 ml of cell culture to be transfected. We then incubated the mix at room temperature for 10 to 15 min and added it to the suspension culture, which we maintained at a density of 2 × 105 cells/ml. For stable expression, we generated retroviruses by cotransfecting pBABE or pWZL constructs of wild-type or mutant SF2/ASF, along with a vesicular stomatitis G protein (VSV-G) expression construct, into Phoenix cells (a gift from Scott Lowe), an ecotropic packaging cell line expressing essential viral proteins (23). We used the resulting viruses to infect primary human lung fibroblasts (IMR90) to establish stable cell lines overexpressing wild-type or mutant SF2/ASF, as described previously (30).
Indirect immunofluorescence (IF).
We determined the subcellular localizations of transiently expressed wild-type or mutant versions of SF2/ASF by indirect immunofluorescence using the N-terminal T7 epitope tag, as described previously (17). Briefly, we transfected HeLa cells grown on coverslips in six-well plates with 1 μg of wild-type or mutant pCGT-SF2/ASF. We fixed the cells with 4% (vol/vol) paraformaldehyde in phosphate-buffered saline (PBS) 36 h after transfection for 30 min, followed by incubation for 5 min in 0.2% (vol/vol) Triton X-100 to permeabilize the cells. We then incubated the cells with anti-T7 monoclonal antibody (MAb) (1:1,000; Novagen) for 1 h, washed the cells with PBS, and incubated them with Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (1:1,000; Invitrogen) for 1 h. We mounted the coverslips onto slides using ProLong Gold Antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) and imaged the cells with a fluorescent microscope (Axioskop 2 Plus; Carl Zeiss) equipped with a camera (AxioCam; Carl Zeiss).
Shuttling assays.
We transfected HeLa cells grown on coverslips with T7-tagged wild-type or A1A2A3 mutant SF2/ASF, as described above. Twenty-four hours after transfection, we incubated the cells in tissue culture medium supplemented with 10 ng/ml leptomycin B (Calbiochem) for an additional 12 h. We then fixed and permeabilized the cells and analyzed them by IF, as described above.
The heterokaryon assay to assess the shuttling activity of nuclear proteins was described previously (13, 49). In short, to achieve a high transfection efficiency, we electroporated 1 × 106 HeLa cells with 4 μg of either pCGT7-SF2/ASF or pCGT-K1K2K3 and plated the cells onto coverslips; we included pCGT-SC35 as a negative control. We then coincubated the cells with an excess of untransfected mouse NIH 3T3 cells for 3 h in medium supplemented with 50 μg/ml cycloheximide; we then increased the concentration of cycloheximide to 100 μg/ml, with additional incubation for 30 min. We fused the cells using polyethylene glycol (PEG) and further incubated the resulting heterokaryons for 2 h in medium containing 100 μg/ml cycloheximide. We visualized the cells by IF using T7 antibody, as described above. We distinguished mouse nuclei (NIH 3T3) from human nuclei (HeLa) in the heterokaryons by the former's characteristic punctate DAPI staining pattern.
Dual-luciferase assay and cellular fractionation.
We transfected 40 to 50% confluent HeLa cells grown in six-well plates with 0.4 μg pLCS reporter (50), 0.2 μg TK-Renilla luciferase (Promega), and 1.0 μg of wild-type or mutant pCGT-SF2/ASF. We lysed the cells by using passive lysis buffer (Promega) at 48 h posttransfection and assayed the levels of firefly and Renilla luciferase using a dual-luciferase assay kit (Promega), measuring the luminescence with a Monolight 2010 apparatus (Analytical Luminescence Lab). For cellular fractionation, we harvested the cells 48 h after transfection by trypsinization, resuspended them in PBS, and used 1/10 of the cell suspension for the luciferase assay. We collected the remaining cells by centrifugation, resuspended them in a hypotonic buffer (10 mM HEPES [pH 8.0], 10 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, 0.5% [vol/vol] Nonidet P-40), incubated them on ice for 5 min, and lysed them by gentle pipetting. We centrifuged the lysate with intact nuclei and collected the supernatant as the cytosolic fraction. We washed the nuclear pellet with PBS and extracted the nuclear proteins by resuspending the pellet in Laemmli buffer. We analyzed both nuclear and cytoplasmic fractions by Western blotting.
In vitro translation.
In vitro translation reaction mixtures consisted of 200 ng of in vitro-transcribed pLuc reporter mRNA, 200 ng of recombinant wild-type or mutant SF2/ASF, and translation-competent HeLa cell extracts (6), incubated for 30 min at 37°C, as described previously (50). We stopped the reactions by dilution with 100 μl of passive lysis buffer (Promega) and measured luminescence as described above by using luciferase-activating reagent (Promega).
In vitro splicing.
We prepared radiolabeled pre-mRNAs by the in vitro transcription of plasmids pSP64-HβΔ6 and pSP64-5′ssD-wt linearized with BamHI, with SP6 RNA polymerase, in the presence of [α-32P]UTP. We incubated the pre-mRNAs under splicing conditions in HeLa nuclear or cytoplasmic S100 extracts, as described previously (43, 44). We extracted the RNAs and analyzed them by denaturing PAGE and phosphorimager analysis on a Fujifilm FLA-5100 instrument (Fuji Medical Systems USA, Inc.). We quantified the band intensities using Multi Gauge software, version 2.3 (Fujifilm).
RPA.
We cotransfected 40 to 50% confluent HeLa cells grown in 10-cm plates with 0.2 μg green fluorescent protein (GFP) plasmid, 0.5 μg wild-type or mutant pCGT-SF2/ASF plasmid, and 2.5 μg wild-type β-globin or mutant (T-39) NMD reporter plasmids. To test both NMD and 5′ss selection, we cotransfected 0.2 μg GFP plasmid, 0.7 μg SF2/ASF plasmid, and 2.1 μg 5′ssD-wt or 5′ssD-mt plasmid. We isolated total RNA 36 h after transfection by ultracentrifugation of the cell lysates layered on a 5.7 M CsCl cushion at 20°C and carried out an RNase protection assay (RPA) as described previously (25, 65). We analyzed the radioactive protected fragments by denaturing PAGE and phosphorimaging, as described above.
Reverse transcription (RT)-PCR.
We extracted total RNA from IMR90 cells stably expressing wild-type or mutant SF2/ASF using Trizol reagent (Invitrogen) and reverse transcribed 2 μg from each RNA sample using Superscript-II (Invitrogen). We amplified the cDNAs corresponding to transcripts of endogenous target genes of SF2/ASF by PCR using Taq Gold polymerase (Invitrogen) with specific primers and conditions described previously (30).
Antibodies and Western blotting.
We lysed cells in Laemmli sample buffer and analyzed the proteins by Western blotting with various primary antibodies specific for SF2/ASF (MAb AK96), SRp55 (MAb 9-1-56), Myc (MAb 9E10), T7 (Novagen), β-catenin (Transduction Labs), and caspase-3 (a gift from Yuri Lazebnik). The secondary antibody was goat anti-mouse IgG conjugated to Alexa Fluor 532 (Invitrogen). After washing, we scanned the nitrocellulose membranes (Whatman) on a Fujifilm FLA-5100 instrument equipped with a 532-nm laser and quantified fluorescence band intensities as described above.
MS.
Wild-type or mutant versions of SF2/ASF were purified from 293E cells as described above and analyzed by mass spectrometry (MS). Briefly, we separated the purified proteins by SDS-PAGE and stained them with GelCode blue (Pierce). We digested and processed the bands for analysis as described previously (2). We analyzed the tryptic peptides either by liquid chromatography (LC)-tandem MS (MS/MS) on a linear trap quadrupole mass spectrometer (Thermo) or by targeted fragmentation on a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometer (Applied Biosystems) (8). We analyzed the LC-MS/MS runs using the X!Tandem search engine (19) with the following parameters: variable modifications included mono-, di-, and trimethylation of Lys and mono- or dimethylation of Arg residues. For targeted fragmentation, we first analyzed the precursor mass list using either the Profound or Peptidemap software package (http://prowl.rockefeller.edu/). We then selected peaks with precursor masses consistent with methylated peptides for fragmentation and analyzed the resulting spectra using MS-Product (http://prospector.ucsf.edu/). We resolved ambiguities by comparisons with theoretical spectra derived from all possible methylation patterns.
RESULTS
SF2/ASF is methylated at Arg residues 93, 97, and 109.
We expressed C-terminal 6×His-tagged SF2/ASF in human 293-EBNA1 (293E) cells, purified it, and analyzed it by MS. We verified the results by MS/MS analysis, which confirmed that SF2/ASF is methylated at Arg residues 93, 97, and 109 (data not shown), consistent with previous observations from global proteomic studies (48).
The methylated arginines affect the nucleocytoplasmic distribution of SF2/ASF.
Based on studies of Npl3p in yeast and of other RNA-binding proteins, we hypothesized that the sites of Arg methylation in SF2/ASF (Fig. 1A) might influence its localization. We anticipated that changes in the distribution of SF2/ASF between the nucleus and cytoplasm may affect the alternative splicing of target genes as well as its other functions. To investigate the functional significance of the modifications, we mutated these sites singly or in combination to Ala to prevent methylation as well as abolish the positive charge. Similarly, as a control, we also mutated these residues to Lys to retain the positive charge but presumably prevent methylation. We determined the subcellular localization of wild-type SF2/ASF (wt-SF2/ASF) and the missense mutants by indirect immunofluorescence (IF) via an N-terminal T7 epitope tag after the transient transfection of the cDNAs into HeLa cells. We observed that the triple-Ala mutant A1A2A3 was predominantly cytoplasmic, in contrast to the corresponding triple-Lys substitution mutant K1K2K3 or wt-SF2/ASF, which localized to nuclear speckles (Fig. 1B). The single- or double-Ala mutants displayed intermediate phenotypes (Fig. 1C).
To test whether the A1A2A3 mutant could still shuttle, we treated transiently transfected HeLa cells with a nuclear export inhibitor, leptomycin B. The accumulation of T7 signal in the nucleus upon treatment with the inhibitor indicates that A1A2A3 shuttles between the nuclear and cytoplasmic compartments (Fig. 1D). We also tested the shuttling of the nuclear protein K1K2K3 by using a heterokaryon assay and found it to be the same as that of wt-SF2/ASF (Fig. 1E). The heterokaryon assay (49), which is used to test the shuttling activity of nuclear proteins, could not be used in the case of the A1A2A3 mutant, as it showed predominant cytoplasmic localization.
Reduced activity of the A1A2A3 mutant in general and alternative splicing in vitro.
To analyze the effect of mutations in the inter-RRM linker on the splicing activity of these proteins, we purified untagged recombinant SF2/ASF and the A1A2A3 and K1K2K3 versions to homogeneity from E. coli cells. We also purified posttranslationally modified versions of these proteins after expression in 293E cells with a C-terminal 6×His tag (Fig. 2A). We then tested the purified proteins for their ability to complement HeLa S100 extracts to carry out the general splicing of various pre-mRNAs. We observed that the K1K2K3 mutant was as active as wt-SF2/ASF, but the A1A2A3 mutant was approximately 4-fold less active with β-globin pre-mRNA (Fig. 2B and D and see Fig. S1 in the supplemental material). We obtained similar results with other substrates, including Ftz, AdML, IgM-M1M2, IgM-C1C2, and δ-crystallin (data not shown).
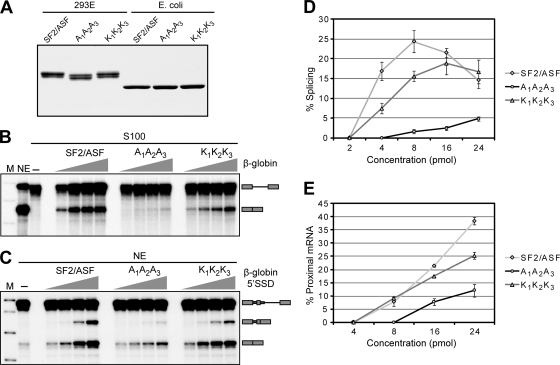
FIG. 2.
In vitro splicing activity of wild-type and mutant SF2/ASF. (A) Coomassie stain of an SDS-polyacrylamide gel showing the different versions of recombinant SF2/ASF purified from either 293E cells or E. coli cells. (B) S100 complementation assay to test the general splicing activity of wild-type or mutant SF2/ASF purified from 293E cells using a radiolabeled β-globin minigene pre-mRNA. We added increasing amounts of each protein (4, 8, 16, and 24 pmol) to otherwise identical reaction mixtures. The mobilities of pre-mRNA and mRNA are indicated on the right. M, molecular weight marker; NE, splicing in nuclear extract. (C) In vitro splicing assay to test the alternative splicing activity of wild-type or mutant SF2/ASF purified from 293E cells using a model β-globin pre-mRNA with a duplicated 5′ss derivative (65). The positions of the pre-mRNA and the two spliced mRNA isoforms are indicated on the right. We supplemented the splicing reaction mixtures of HeLa nuclear extracts with increasing amounts of each protein (4, 8, 16, and 24 pmol). (D) Graph showing quantification of phosphorimager signals from multiple experiments described above (B). We normalized the mRNA intensities to the corresponding pre-mRNA intensities in each lane and plotted them as a percentage of splicing as a function of the amount of protein added (n = 2 to 3). Error bars indicate standard deviations (SD). (E) Graph showing quantification of phosphorimager signals from multiple experiments described above (C). We divided the proximal-mRNA intensities by the sum of proximal- and distal-mRNA intensities in each lane and plotted them as a percentage of proximal mRNA as a function of the amount of protein added (n = 2). Error bars indicate SD.
To test alternative splicing activity in vitro, we used a model β-globin pre-mRNA with a duplicated 5′ss in intron 1 (65). Again, we observed that the K1K2K3 mutant was as efficient as wt-SF2/ASF in promoting proximal 5′ss use in HeLa nuclear extracts, whereas the A1A2A3 mutant was slightly less active (Fig. 2C and E). In all the assays, the trend was similar and did not vary when the proteins were obtained from bacteria or mammalian cells. However, the specific activity of SF2/ASF obtained from 293E cells was consistently higher than that from bacteria (data not shown), which may reflect differences in PTMs and/or in purification/renaturation protocols and solubility. The A1A2A3 protein, irrespective of its source, was slightly less soluble under splicing conditions, especially at higher concentrations, which may be one of the reasons why the triple-Ala mutant was less active in these cell-free assays.
The A1A2A3 mutant fails to modulate splicing of endogenous pre-mRNAs.
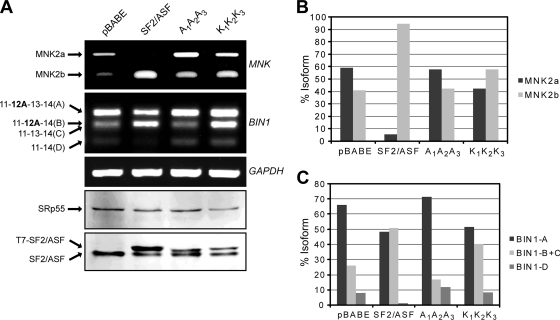
We recently identified BIN1 and MNK2 pre-mRNAs as endogenous targets of SF2/ASF (30). To test the effect of the mutant proteins on the alternative splicing of these transcripts, we stably expressed cDNAs encoding T7-tagged, wt-SF2/ASF or the mutant A1A2A3 and K1K2K3 versions in primary human lung fibroblasts (IMR90) using retroviral transduction. We analyzed total RNA extracted from transductant pools by RT-PCR to detect the isoforms of the above-described target genes. We observed that the overexpression of both wt-SF2/ASF and the K1K2K3 mutant promoted the inclusion of exon 12a in BIN1 and led to an increase in the levels of the MNK2 alternatively spliced isoform MNK2b; however, the K1K2K3 mutant was less active due to its lower level of expression. In contrast, the relative levels of alternatively spliced isoforms of the target genes upon the overexpression of the A1A2A3 mutant were similar to those of the empty vector control (Fig. 3).
FIG. 3.
Effects of wild-type or mutant SF2/ASF on alternative splicing of endogenous transcripts in IMR90 cells. (A) We extracted total RNA and protein from IMR90 cells stably overexpressing the indicated proteins after retroviral transduction (30). We analyzed total RNA by RT-PCR and detected the expressed T7-tagged proteins and endogenous SRp55 and SF2/ASF by Western blotting. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B and C) Graphs showing the levels of different mRNA isoforms of MNK2 (B) and BIN1 (C) after quantification of the corresponding lanes in panel A.
The triple-Ala mutant A1A2A3 fails to enhance NMD.
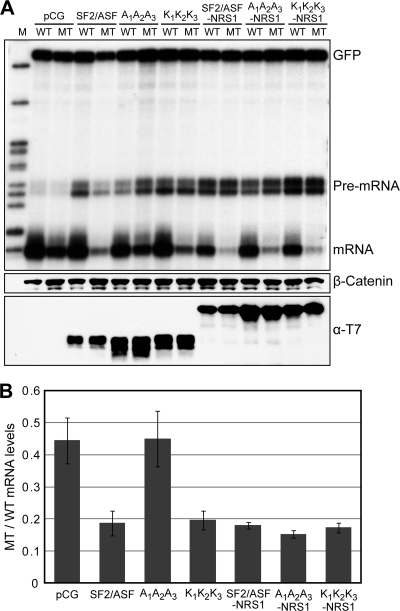
Splicing is a nuclear process (9), and a nuclear activity of SF2/ASF appears to be required for its stimulatory effect on NMD (65). Therefore, we next assayed the activities of the above-described SF2/ASF mutants to enhance NMD by using an RNase protection assay (RPA). We used a full-length β-globin gene and a mutant version with a nonsense mutation in codon 39 in exon 2 as reporters. The nonsense mutation introduces a premature termination codon (PTC), making the mRNA susceptible to NMD (Fig. 4A, left) (56, 64). RPA showed that upon overexpression, wt-SF2/ASF as well as the K1K2K3 version strongly enhanced NMD of the T39 (mut) mRNA, whereas the mislocalized A1A2A3 mutant was similar to the empty vector control (Fig. 4B and E). The single- and double-Ala mutants had intermediate effects (Fig. 4C and E).
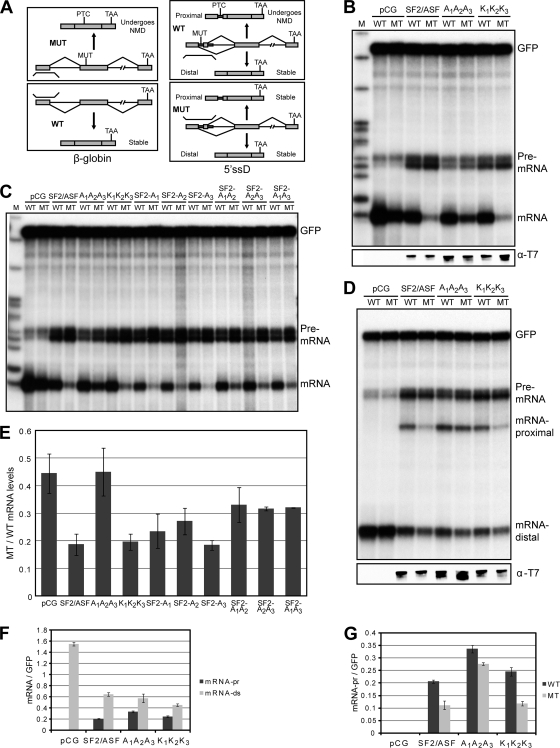
FIG. 4.
Effect of overexpression of wild-type or mutant SF2/ASF on NMD. (A) Schematic representation of the wild-type (WT) and mutant (MT) β-globin reporters (left), the wild-type and mutant β-globin reporters with a duplicated 5′ss (right), and their corresponding RPA probes. (B) We cotransfected a human β-globin gene (wild type) or a mutant version with an in-frame PTC with wild-type or mutant versions of SF2/ASF into HeLa cells. We extracted total RNA 36 h after transfection and analyzed it by RPA. We also cotransfected a GFP-expressing plasmid as an internal reference and used β-globin and GFP probes simultaneously for RPA. At the bottom, Western blotting with T7 antibody shows the expression of each tagged protein. (C) Effect of single- or double-Ala mutants of SF2/ASF on sensitivity to a PTC upon overexpression. We performed transient cotransfection and RPA as described above (B). (D) A cotransfected β-globin minigene (wild type) has a duplicated 5′ss and can be spliced via either the proximal or the distal 5′ss; the mutant version has an in-frame PTC such that the proximal mRNA is susceptible to NMD (65). We analyzed total RNA recovered from HeLa cells by RPA after transient cotransfection with wild-type or mutant versions of SF2/ASF. We cotransfected a GFP plasmid as a reference and used β-globin and GFP probes simultaneously. The bottom panel shows the expression of each tagged protein analyzed by Western blotting. (E) Quantification of phosphorimager signals from multiple experiments described above (B and C). We normalized the individual mRNA intensities to the corresponding GFP intensities in each lane. We plotted the ratios of normalized mutant mRNA to wild-type mRNA intensities to show the relative NMD enhancement upon the overexpression of each protein. (F) Quantification of phosphorimager signals from multiple experiments described above (D). The plot shows individual wild-type proximal- and wild-type distal-mRNA intensities normalized to the corresponding GFP intensities in each lane. (G) Same as panel F. We plotted the normalized intensities of both wild-type and mutant proximal-mRNA levels to show the NMD enhancement upon the overexpression of the depicted proteins (n = 2 to 5, and error bars show SD [E to G]).
To assay both alternative splicing and NMD using RPA, we used a derivative of β-globin with a 5′-splice-site (5′ss) duplication in the first exon (5′ssD-wt) (65). Because of alternate 5′ss usage, the pre-mRNA gives rise to two distinct mRNAs: a longer proximal mRNA and a shorter distal mRNA. The mutant version of this gene (5′ssD-mt) carries a point mutation that introduces a PTC into the proximal, but not the distal, spliced mRNA (Fig. 4A, right). As reported previously (65), SF2/ASF promoted the selection of the proximal 5′ss and enhanced NMD of the resulting mRNA. In contrast, the A1A2A3 mutant efficiently promoted proximal splice-site selection but failed to enhance NMD; the K1K2K3 mutant, however, showed activity identical to that of wt-SF2/ASF (Fig. 4D, F, and G). Again, as described above, the single- and double-Ala mutants had intermediate effects (data not shown).
In contrast to the transduced IMR90 cells, which result in only about a 2-fold overexpression of SF2/ASF and the various mutants (Fig. 3A), these experiments involving transient cotransfection result in substantial overexpression in the cells that take up the DNA. Because the A1A2A3 mutant retains some ability to shuttle, the high levels of overexpression presumably allow enough of the A1A2A3 protein to transit through the nucleus to alter the splicing of β-globin pre-mRNA with a 5′ss duplication (Fig. 1D and 4D). In contrast, with more physiological levels of overexpression, the triple-Ala mutant fails to modulate the splicing of target genes (Fig. 3). Furthermore, the solubility and/or folding of the transiently overexpressed proteins is not likely to be affected in HeLa cells, which could explain the observed parity between the activities of A1A2A3 and wt-SF2/ASF in promoting the selection of the proximal 5′ss after cotransfection (Fig. 4D and F), in contrast to the cell-free assays (Fig. 2C and E).
A nuclear-retained version of the A1A2A3 protein promotes NMD.
Based on the above-described observations, it seemed plausible that the mislocalization of the A1A2A3 mutant was responsible for its loss of function in NMD. To test this hypothesis, we fused a nuclear retention signal (NRS) from the nonshuttling SR protein SC35 to the A1A2A3 and K1K2K3 versions of SF2/ASF at their respective C termini. A C-terminal fusion of the NRS sequence, in the case of wt-SF2/ASF, results in a loss of shuttling activity (17). Furthermore, this nuclear-retained version of SF2/ASF is active in splicing and NMD (65), as well as in autoregulation (55), but fails to enhance translation in cells (50). Immunofluorescence analysis confirmed that the fusion of the NRS sequence to the A1A2A3 mutant restored nuclear localization (Fig. 1B). Furthermore, when assayed for NMD, the NRS version of the A1A2A3 mutant was as efficient as wt-SF2/ASF in enhancing NMD, clearly indicating that nuclear localization is essential for this activity (Fig. 5).
FIG. 5.
Enhancement of NMD by SF2/ASF or mutants fused to a nuclear retention signal. We cotransfected T7-tagged, wt-SF2/ASF or the A1A2A3 and K1K2K3 mutants, with or without the NRS1 peptide from SC35 fused at the C terminus, with β-globin with or without a PTC. (A) RPA of total RNA after transient expression, as described in the legend of Fig. 4A. At the bottom are Western blots with anti-T7 antibody to detect the expressed tagged proteins and endogenous β-catenin as a reference. (B) Quantification of phosphorimager signals from multiple experiments described above (A). We normalized the individual mRNA intensities to GFP mRNA and plotted the ratios of normalized mutant to wild-type mRNA intensities as described in the legend of Fig. 4C (n = 2 to 5; error bars indicate SD).
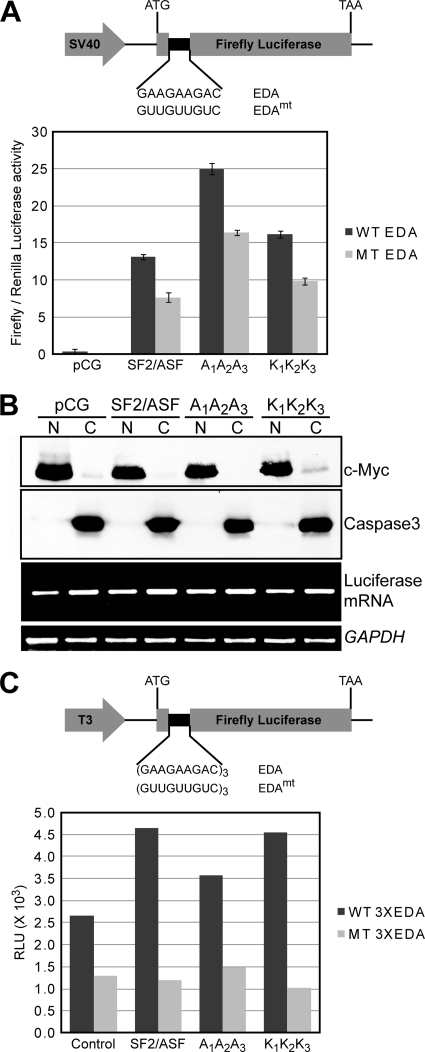
The A1A2A3 protein is more active than wt-SF2/ASF in enhancing translation in cells.
To test whether the triple-Ala mutant of SF2/ASF can enhance translation, we used previously described reporter constructs in which either a wild-type or a mutant ESE recognized by SF2/ASF is fused at the 5′ end of firefly luciferase (50). The cotransfection of the reporters along with wild-type or mutant versions of SF2/ASF and subsequent analysis of luciferase activity revealed that the A1A2A3 protein is twice as active as wt-SF2/ASF in promoting translation in an enhancer-dependent manner (Fig. 6A). The increase in luciferase activity in the cytoplasm upon the overexpression of A1A2A3 could be due to the following three reasons: increased translation-stimulatory activity due to mutation, greater accumulation of A1A2A3 in the cytoplasm, or increased levels of luciferase mRNA in the cytoplasm due to enhanced export. RT-PCR analysis of luciferase mRNA levels in nuclear and cytoplasmic fractions from the transfected HeLa cells revealed that A1A2A3 does not enhance mRNA export (Fig. 6B). Furthermore, recombinant A1A2A3 was equally as active as wt-SF2/ASF in enhancing the translation of the in vitro-transcribed luciferase reporter in a cell-free assay with a translationally competent HeLa cell extract (Fig. 6C). We conclude that the apparent increased activity of the A1A2A3 mutant in enhancing translation in cells is due to its increased residence time in the cytoplasm.
FIG. 6.
Effect of wild-type or mutant SF2/ASF on ESE-dependent stimulation of translation. (A) Diagram of the translation reporter with an in-frame fibronectin extradomain A (EDA) wild-type or mutant ESE upstream of the firefly luciferase open reading frame (ORF) (50) (top). We cotransfected HeLa cells with the reporter, wild-type or mutant T7-tagged SF2/ASF cDNA, and a Renilla luciferase reporter as a control for transfection efficiency. We harvested the cells after 48 h and analyzed them by a dual-luciferase reaction (DLR) assay. The bottom shows the ratios of firefly activity to Renilla luciferase activity (n = 4; error bars indicate SD). (B) Subcellular distribution of the luciferase reporter mRNAs upon the overexpression of wild-type or mutant SF2/ASF. We prepared nuclear and cytosolic fractions from aliquots of the transfected cells used for the DLR assay described above (A) and measured the levels of firefly reporter mRNA by RT-PCR as well as endogenous GAPDH mRNA as a reference (bottom two panels). Western blots show the levels of the nuclear c-Myc pro-tein and cytosolic caspase-3 protein to verify proper fractionation (top two panels). (C) Effect of wild-type or mutant SF2/ASF on stimulation of translation in vitro. At top is a diagram of the in vitro reporter (50). We incubated in vitro-transcribed reporter mRNAs (200 ng) with three copies of wild-type or mutant EDA ESE in a HeLa cell translation extract with or without 200 ng of recombinant wild-type or mutant SF2/ASF at 37°C for 30 min. The histogram shows the luciferase activity for each reaction. RLU, relative light units.
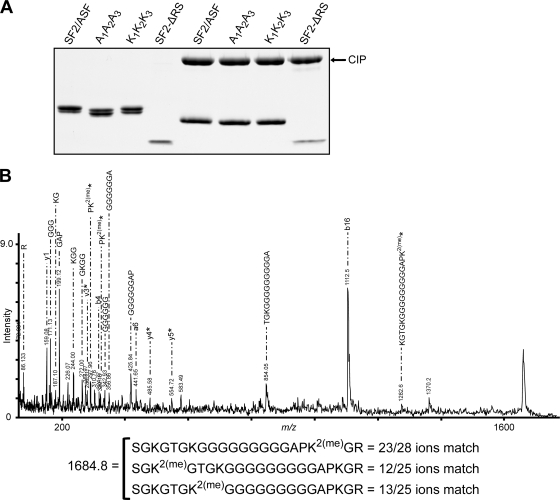
Tandem mass spectrometry reveals that the triple-Lys mutant is dimethylated.
Surprisingly, our results showed that in all the assays, the K1K2K3 mutant had effects that were very similar to those of wt-SF2/ASF. To address the possibility that the substituted Lys residues at these positions undergo methylation, we expressed the K1K2K3 mutant in 293E cells and analyzed the purified protein by mass spectrometry. MS/MS analysis of one of the peptides corresponding to the region of substitution revealed that the substituted Lys at position 109 was indeed dimethylated (Fig. 7). This unexpected finding points toward the possibility that both the methylation state and the positive charge at these positions could be responsible for the localization of SF2/ASF (see Discussion).
FIG. 7.
Tandem mass spectrometry of the K1K2K3 protein. (A) Coomassie-stained SDS-polyacrylamide gel showing the different versions of SF2/ASF purified from 293E cells before and after treatment with calf intestinal phosphatase (CIP). (B) We purified wild-type and mutant SF2/ASF expressed in 293E cells, treated them with calf intestinal phosphatase, separated them by SDS-PAGE, and stained them with GelCode blue. Next, we excised individual bands, digested them with trypsin, and analyzed them by MALDI-TOF MS. We selected peptides corresponding to the region of interest and analyzed them by MS/MS. The MS/MS spectrum shown is from one such peptide from the K1K2K3 protein with two methyl groups. The largest peak (b16 = 1,112.5) has a match only when one of the three Lys residues is dimethylated and not if two of them are monomethylated. The peptides marked with asterisks correspond to peptide mass peaks that can occur only when Lys at position 109 is dimethylated. The ion matches when one of the three Lys residues is dimethylated [K2(me)] are represented at the bottom, indicating that Lys109 is indeed dimethylated in this peptide.
To address the question of whether positive charge, methylation, or both are involved in controlling the localization of SF2/ASF, we mutated the Gly residues surrounding the methyl-arginine residues to Ala as well as Arg at position 111 to Lys, resulting in the SM mutant. As a control, we also made the corresponding mutations (Gly to Ala and Arg111 to Lys) in the K1K2K3 version, giving rise to the KM mutant (see Fig. S2A in the supplemental material). All the Gly-to-Ala changes disrupt the RGG/GRG motifs, whereas the Arg111-to-Lys substitution disrupts an RGR motif (59).
We hypothesized that if methylation is responsible for the mislocalization of SF2/ASF, and methylation at these positions is dependent upon RGG motifs, then replacing the glycines should result in a phenotype similar to that observed for the A1A2A3 mutant. Furthermore, if methylation of the substituted Lys in K1K2K3 also depends upon the introduced KGG and related motifs, which seems highly unlikely, the KM mutant should also behave like the triple-Ala mutant. Alternatively, if positive charge alone, and not methylation, is involved in the regulation of SF2/ASF localization, then both SM and KM mutants should behave like wt-SF2/ASF.
Both the SM and KM versions of SF2/ASF showed predominant nuclear localization, with only slight cytoplasmic staining (see Fig. S2B in the supplemental material). Both proteins showed NMD enhancement activities similar to that of wt-SF2/ASF (Fig. S2C). Although these results suggest that positive charge, and not methylation, controls SF2/ASF localization, they are based on the assumption that the RGG-type motifs are essential for the methylation of the Arg residues, which may not be the case, as methylation by PRMT1 is not restricted strictly to RGG and related motifs, and/or other methyltransferases may also modify SF2/ASF (see Discussion). Although the possibility still remains that both the Arg residues in SM and the Lys residues in KM undergo methylation, these results reaffirm the importance of positive charge in controlling the localization of SF2/ASF (see Discussion).
DISCUSSION
The coordinated and sequential events that lead to regulated gene expression in eukaryotes are extremely complex, spanning across cellular compartments and requiring numerous protein and RNA factors at various stages and at specific locations. When a single protein factor, such as SF2/ASF, is involved in multiple posttranscriptional events, it is essential that its movements between cellular compartments be tightly regulated. SF2/ASF is a shuttling protein that shows predominant nuclear localization in the steady state (13). Within the nucleus, SF2/ASF accumulates in nuclear speckles, and its recruitment to active sites of transcription is modulated by the phosphorylation of Ser residues in the RS domain and various protein-protein interactions (46). The phosphorylation of SF2/ASF also modulates its subcellular localization (32). The RS domain is required for the shuttling of SF2/ASF (13) and contributes to its nuclear localization (12).
Here we have demonstrated that additional signals that control the cellular localization of SF2/ASF are present in the linker between RRM1 and RRM2. The Arg residues in this linker region, in particular R93, R97, and R109, are methylated (48) and are important for correct localization, as we found that mutating these residues simultaneously to Ala resulted in cytoplasmic rather than nuclear accumulation.
The role of the SF2/ASF inter-RRM linker in RNA binding is unclear, as a structure of full-length SF2/ASF or its two RRMs in complex with RNA is lacking. Evidence from the structures of other two-RRM proteins, such as UP1, Sex-lethal, and nucleolin, indicates that the inter-RRM linkers, which are disordered when unbound, cooperate with the RRMs in binding the nucleic acid by providing increased affinity and specificity (42). In our study, the triple-Lys mutant was functionally indistinguishable from wt-SF2/ASF, and the triple-Ala mutant was as active as wt-SF2/ASF in promoting translation in vitro, suggesting that RNA binding was not affected by these substitutions.
The three guanidino nitrogen atoms in the Arg side chain can potentially form five hydrogen bonds (H bonds) with H-bond acceptors in RNA, resulting in a network of H-bond interactions, which are not possible with Lys, as it has a single terminal amino group (10, 15). For example, a short Arg-rich basic peptide from the HIV-1 Tat protein binds specifically to the transactivation-responsive region RNA, and a Lys substitution results in a loss of binding and transactivation (15).
The methylation of the two terminal amino groups in the Arg side chain does not alter the positive charge but increases the hydrophobicity, makes the side chain bulkier, and, most importantly, blocks any potential H-bond formation. This could provide a potential means of regulating protein-RNA as well as protein-protein interactions involving SF2/ASF such that the methylation of the Arg residues abolishes some interactions based on H bonding while leaving electrostatic interactions unaffected. For example, Pro-rich motifs in Sam68 interact with both SH3 and WW domains present in interacting partners; the methylation of RG repeats that flank the Pro-rich motifs reduces the binding of Sam68 to the SH3 domains of p59fyn and phospholipase C-γ1 without affecting binding to the WW domain of FBP30 (4).
Because the motif recognized by PRMT1 is not limited strictly to RGG and related sequences (60), it is possible that other Arg residues of SF2/ASF are also modified. One precedent is the methylation of R3 in histone H4 by PRMT1, in which the methylated Arg is not part of an Arg/Gly-rich region (58). As in the case of histones, the methylation state in the SF2/ASF linker region may control various protein-protein interactions, either directly or by influencing other modifications of SF2/ASF, such as the phosphorylation of the RS domain. Such regulation via Arg methylation, in conjunction with phosphorylation-dephosphorylation cycles of the RS domain, could play a role in the localization and trafficking of SF2/ASF between cellular compartments. A precedent for this type of cross talk was observed previously for Npl3p of budding yeast (63). The phosphorylation of the RS domain is essential for the interaction of SF2/ASF with transportin-SR2 (TRN-SR2), which acts as a receptor for the nuclear import of SR proteins (36). Furthermore, the phosphorylation state of SF2/ASF influences its activity as an adaptor protein for Tip-associated protein (TAP)-mediated mRNA export (27, 37). However, when we analyzed the interactions of wt-SF2/ASF and its mutant versions (A1A2A3 and K1K2K3) with either TAP or TRN-SR2, we observed that all versions of SF2/ASF interacted similarly with these two proteins, arguing against the possibility that these three Arg residues, or their methylation states, affect these interactions (data not shown).
In addition to changing the properties of binding sites and affecting other modifications, methylated arginines are also involved directly in protein-protein interactions. The Tudor domain of the SMN protein interacts directly with symmetric dimethyl Arg residues in proteins with this modification (18). However, proteins and their respective domains that may bind to asymmetric dimethyl Arg residues in SF2/ASF, and other proteins with the same modification, are yet to be discovered.
The triple-Ala mutant of SF2/ASF was unable to enhance NMD and failed to modulate the alternative splicing of endogenous target pre-mRNAs when modestly overexpressed. Furthermore, due to the accumulation of the protein in the cytoplasm, A1A2A3 was more efficient at enhancing the translation of a luciferase reporter than wt-SF2/ASF. When we restored the nuclear localization of the A1A2A3 protein by the C-terminal fusion of a nuclear retention sequence from the nonshuttling SR protein SC35 (17), the resulting A1A2A3-NRS1 protein was as effective in promoting NMD as wt-SF2/ASF, demonstrating that the effects observed with A1A2A3 were due to mislocalization. However, we note that at high levels of overexpression, A1A2A3 was as effective as SF2/ASF in promoting the selection of the proximal 5′ss of the β-globin model pre-mRNA with a duplicated 5′ss, most likely due to its ability to shuttle, resulting in enough protein in the nucleus to modulate alternative splicing. However, it is interesting that even at high levels of overexpression and with the ability to shuttle, A1A2A3 was inactive in promoting NMD. The ability of SF2/ASF to regulate splicing in a concentration-dependent manner is well documented (16), whereas the precise mechanisms underlying the effect of SF2/ASF in NMD (65) remain largely unknown, although it was shown recently that the transient overexpression of SF2/ASF promotes an increase in the efficiency of the pioneer round of translation (51). Nonetheless, we have conclusively confirmed the initial observation (65) that the nuclear localization of SF2/ASF is essential for its activity in enhancing NMD.
Although the detailed mechanisms through which SF2/ASF promotes transformation are understood only in part (30), an improper cellular localization of SF2/ASF may be one of the ways through which it exerts its oncogenic activity, especially due to its regulatory roles in multiple posttranscriptional events in both the nucleus and the cytoplasm. For example, in sputum, an increase in the cytoplasmic levels of another splicing factor, hnRNP A2, serves as a powerful predictor of lung cancer almost a year prior to clinical detection (57), suggesting that such a change may be a prerequisite for the transformation of lung epithelial cells.
Intriguingly, the K1K2K3 mutant was functionally indistinguishable from wt-SF2/ASF, pointing toward the importance of charge at these positions as opposed to the methylation state per se. Studies of the yeast SR-like protein Npl3p (52, 63); other shuttling RNA-binding proteins in mammals, such as some hnRNPs (24, 40, 47); and the transcription factors TAF15 and Ewing's sarcoma oncoprotein (EWS) (3, 29) have shown that the Arg methylation of RGG motifs is involved in controlling the nucleocytoplasmic distribution of these proteins. However, no obvious defect in the nuclear export or localization of Npl3p was observed upon the replacement of Arg residues in the RGG motifs with Lys (45, 61). A similar observation was made with the yeast protein Hrp1p, which resembles hnRNPs and also contains RGG motifs; but in this case, additional changes of the Arg residues to Glu or Gln resulted in cytoplasmic localization, most likely through impaired nuclear import (61). In mammals, EWS interacts with components of both the transcriptional machinery, via its N-terminal transactivation domain, as well as the splicing machinery, via its C-terminal domain, and is thought to couple transcription and splicing (62). EWS is methylated at two RGG boxes, RGG2 and RGG3, which are required for its nuclear localization (3, 5). However, only the replacement of Arg residues in the RGG boxes with Ala, and not Lys, altered the transcriptional activity of EWS (1).
Our observation that at least one of the substituted lysines (R109K) in the K1K2K3 version of SF2/ASF was dimethylated suggests that both the methylation state and the positive charge at these positions may contribute to the localization of SF2/ASF. Although it is possible that naturally occurring Lys residues in SF2/ASF are methylated, we had not expected to find methylation of the substituted Lys residues. Also, in the absence of quantitative mass spectrometry data, we do not know what fraction of the total K1K2K3 protein expressed in 293E cells underwent this modification, so it is possible that dimethylation of the substituted Lys109 is present in only a small fraction of the protein.
Experiments involving the SM and the KM mutants (see Fig. S2 in the supplemental material), which had activities similar to those of wt-SF2/ASF, did not conclusively solve the conundrum of charge versus methylation due to the possibility that even in the absence of the glycines in the RGG/GRG motifs, the Arg residues in the SM mutant might still be methylated by PRMT1 and/or other methyltransferases. Furthermore, the treatment of HeLa cells with the specific PRMT1 inhibitor AMI-1 did not alter the localization of either endogenous SF2/ASF or transfected wt-SF2/ASF and K1K2K3 proteins, consistent with the importance of positive charge (data not shown). However, PRMT1 methylates many proteins in the cell; therefore, blocking its activity may affect various cellular processes, including nucleocytoplasmic trafficking, which could account for the observed negative result. Another possibility is that methylation of the three arginines in SF2/ASF may also involve other methyltransferases, which may be active when PRMT1 is inhibited.
In summary, our findings underscore the importance of the proper localization of SF2/ASF for its activity in key nuclear and cytoplasmic processes. We have further identified the signals that control the distribution of SF2/ASF between nucleus and the cytoplasm and also generated mutants of SF2/ASF with a partial loss of function. Such mutants will prove useful in future studies to dissect the mechanisms through which SF2/ASF affects various normal cellular processes as well as oncogenic transformation.
Supplementary Material
Acknowledgments
We thank Javier Cáceres for generously providing the pLCS and pLuc constructs, Yuri Lazebnik for caspase-3 antibody, and Scott Lowe for Phoenix cells. We thank Rolf Sternglanz and Rui-Ming Xu for helpful discussions and Xavier Roca and Yimin Hua for critical reading of the manuscript.
This work was supported by grant CA13106 from the National Cancer Institute.
Footnotes
Published ahead of print on 22 March 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alex, D., and K. A. Lee. 2005. RGG-boxes of the EWS oncoprotein repress a range of transcriptional activation domains. Nucleic Acids Res. 33:1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allemand, E., M. L. Hastings, M. V. Murray, M. P. Myers, and A. R. Krainer. 2007. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 14:630-638. [DOI] [PubMed] [Google Scholar]
- 3.Araya, N., H. Hiraga, K. Kako, Y. Arao, S. Kato, and A. Fukamizu. 2005. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem. Biophys. Res. Commun. 329:653-660. [DOI] [PubMed] [Google Scholar]
- 4.Bedford, M. T., A. Frankel, M. B. Yaffe, S. Clarke, P. Leder, and S. Richard. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030-16036. [DOI] [PubMed] [Google Scholar]
- 5.Belyanskaya, L. L., P. M. Gehrig, and H. Gehring. 2001. Exposure on cell surface and extensive arginine methylation of Ewing sarcoma (EWS) protein. J. Biol. Chem. 276:18681-18687. [DOI] [PubMed] [Google Scholar]
- 6.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney, E., S. Kumar, and A. R. Krainer. 1993. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21:5803-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bish, R. A., O. I. Fregoso, A. Piccini, and M. P. Myers. 2008. Conjugation of complex polyubiquitin chains to WRNIP1. J. Proteome Res. 7:3481-3489. [DOI] [PubMed] [Google Scholar]
- 9.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 10.Borders, C. L., Jr., J. A. Broadwater, P. A. Bekeny, J. E. Salmon, A. S. Lee, A. M. Eldridge, and V. B. Pett. 1994. A structural role for arginine in proteins: multiple hydrogen bonds to backbone carbonyl oxygens. Protein Sci. 3:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bressan, G. C., E. C. Moraes, A. O. Manfiolli, T. M. Kuniyoshi, D. O. Passos, M. D. Gomes, and J. Kobarg. 2009. Arginine methylation analysis of the splicing-associated SR protein SFRS9/SRP30C. Cell. Mol. Biol. Lett. 14:657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cáceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cáceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cáceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 15.Calnan, B. J., B. Tidor, S. Biancalana, D. Hudson, and A. D. Frankel. 1991. Arginine-mediated RNA recognition: the arginine fork. Science 252:1167-1171. [DOI] [PubMed] [Google Scholar]
- 16.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 17.Cazalla, D., J. Zhu, L. Manche, E. Huber, A. R. Krainer, and J. F. Cáceres. 2002. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 22:6871-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Côté, J., and S. Richard. 2005. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280:28476-28483. [DOI] [PubMed] [Google Scholar]
- 19.Craig, R., J. P. Cortens, and R. C. Beavis. 2004. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3:1234-1242. [DOI] [PubMed] [Google Scholar]
- 20.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulet, I., G. Gauvin, S. Boisvenue, and J. Côté. 2007. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282:33009-33021. [DOI] [PubMed] [Google Scholar]
- 22.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemann, M. T., J. T. Zilfou, Z. Zhao, D. J. Burgess, G. J. Hannon, and S. W. Lowe. 2004. Suppression of tumorigenesis by the p53 target PUMA. Proc. Natl. Acad. Sci. U. S. A. 101:9333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann, F., M. Bossert, A. Schwander, E. Akgun, and F. O. Fackelmayer. 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem. 279:48774-48779. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Y., and G. G. Carmichael. 1996. A suboptimal 5′ splice site is a cis-acting determinant of nuclear export of polyomavirus late mRNAs. Mol. Cell. Biol. 16:6046-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 27.Huang, Y., T. A. Yario, and J. A. Steitz. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U. S. A. 101:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyun, Y. L., D. B. Lew, S. H. Park, C. W. Kim, W. K. Paik, and S. Kim. 2000. Enzymic methylation of arginyl residues in -gly-arg-gly- peptides. Biochem. J. 348:573-578. [PMC free article] [PubMed] [Google Scholar]
- 29.Jobert, L., M. Argentini, and L. Tora. 2009. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 315:1273-1286. [DOI] [PubMed] [Google Scholar]
- 30.Karni, R., E. de Stanchina, S. W. Lowe, R. Sinha, D. Mu, and A. R. Krainer. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S., B. M. Merrill, R. Rajpurohit, A. Kumar, K. L. Stone, V. V. Papov, J. M. Schneiders, W. Szer, S. H. Wilson, W. K. Paik, and K. R. Williams. 1997. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry 36:5185-5192. [DOI] [PubMed] [Google Scholar]
- 32.Koizumi, J., Y. Okamoto, H. Onogi, A. Mayeda, A. R. Krainer, and M. Hagiwara. 1999. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem. 274:11125-11131. [DOI] [PubMed] [Google Scholar]
- 33.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 34.Krainer, A. R., T. Maniatis, B. Ruskin, and M. R. Green. 1984. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36:993-1005. [DOI] [PubMed] [Google Scholar]
- 35.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 36.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A. 98:10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, M. C., and W. Y. Tarn. 2004. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279:31745-31749. [DOI] [PubMed] [Google Scholar]
- 38.Lee, M. S., and P. A. Silver. 1997. RNA movement between the nucleus and the cytoplasm. Curr. Opin. Genet. Dev. 7:212-219. [DOI] [PubMed] [Google Scholar]
- 39.Li, X., J. Wang, and J. L. Manley. 2005. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 19:2705-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15:2800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, J. C., and J. F. Cáceres. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417:15-27. [DOI] [PubMed] [Google Scholar]
- 42.Maris, C., C. Dominguez, and F. H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118-2131. [DOI] [PubMed] [Google Scholar]
- 43.Mayeda, A., and A. R. Krainer. 1999. Mammalian in vitro splicing assays. Methods Mol. Biol. 118:315-321. [DOI] [PubMed] [Google Scholar]
- 44.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 45.McBride, A. E., J. T. Cook, E. A. Stemmler, K. L. Rutledge, K. A. McGrath, and J. A. Rubens. 2005. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 280:30888-30898. [DOI] [PubMed] [Google Scholar]
- 46.Misteli, T. 1999. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 9:R198-R200. [DOI] [PubMed] [Google Scholar]
- 47.Nichols, R. C., X. W. Wang, J. Tang, B. J. Hamilton, F. A. High, H. R. Herschman, and W. F. Rigby. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256:522-532. [DOI] [PubMed] [Google Scholar]
- 48.Ong, S. E., G. Mittler, and M. Mann. 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1:119-126. [DOI] [PubMed] [Google Scholar]
- 49.Piñol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 50.Sanford, J. R., N. K. Gray, K. Beckmann, and J. F. Cáceres. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, H., N. Hosoda, and L. E. Maquat. 2008. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell 29:255-262. [DOI] [PubMed] [Google Scholar]
- 52.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebel, C. W., and C. Guthrie. 1996. The essential yeast RNA binding protein Np13p is methylated. Proc. Natl. Acad. Sci. U. S. A. 93:13641-13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singleton, D. R., S. Chen, M. Hitomi, C. Kumagai, and A. M. Tartakoff. 1995. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J. Cell Sci. 108:265-272. [DOI] [PubMed] [Google Scholar]
- 55.Sun, S., Z. Zhang, R. Sinha, R. Karni, and A. R. Krainer. 2010. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17:306-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thermann, R., G. Neu-Yilik, A. Deters, U. Frede, K. Wehr, C. Hagemeier, M. W. Hentze, and A. E. Kulozik. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 17:3484-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tockman, M. S., Y. S. Erozan, P. Gupta, S. Piantadosi, J. L. Mulshine, and J. C. Ruckdeschel. 1994. The early detection of second primary lung cancers by sputum immunostaining. LCEWDG Investigators. Lung Cancer Early Detection Group. Chest 106:385S-390S. [DOI] [PubMed] [Google Scholar]
- 58.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 59.Weiss, V. H., A. E. McBride, M. A. Soriano, D. J. Filman, P. A. Silver, and J. M. Hogle. 2000. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 7:1165-1171. [DOI] [PubMed] [Google Scholar]
- 60.Wooderchak, W. L., T. Zang, Z. S. Zhou, M. Acuña, S. M. Tahara, and J. M. Hevel. 2008. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry 47:9456-9466. [DOI] [PubMed] [Google Scholar]
- 61.Xu, C., and M. F. Henry. 2004. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 24:10742-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, L., H. A. Chansky, and D. D. Hickstein. 2000. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J. Biol. Chem. 275:37612-37618. [DOI] [PubMed] [Google Scholar]
- 63.Yun, C. Y., and X. D. Fu. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150:707-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, J., X. Sun, Y. Qian, and L. E. Maquat. 1998. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4:801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Z., and A. R. Krainer. 2004. Involvement of SR proteins in mRNA surveillance. Mol. Cell 16:597-607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.