Abstract
Streptococcus pyogenes, a multiple-auxotrophic human pathogen, regulates virulence gene expression according to nutritional availability during various stages in the infection process or in different infection sites. We discovered that CvfA influenced the expression of virulence genes according to growth phase and nutritional status. The influence of CvfA in C medium, rich in peptides and poor in carbohydrates, was most pronounced at the stationary phase. Under these conditions, up to 30% of the transcriptome exhibited altered expression; the levels of expression of multiple virulence genes were altered, including the genes encoding streptokinase, CAMP factor, streptolysin O, M protein (more abundant in the CvfA− mutant), SpeB, mitogenic factor, and streptolysin S (less abundant). The increase of carbohydrates or peptides in media restored the levels of expression of the virulence genes in the CvfA− mutant to wild-type levels (emm, ska, and cfa by carbohydrates; speB by peptides). Even though the regulation of gene expression dependent on nutritional stress is commonly linked to the stringent response, the levels of ppGpp were not altered by deletion of cvfA. Instead, CvfA interacted with enolase, implying that CvfA, a putative RNase, controls the transcript decay rates of virulence factors or their regulators according to nutritional status. The virulence of CvfA− mutants was highly attenuated in murine models, indicating that CvfA-mediated gene regulation is necessary for the pathogenesis of S. pyogenes. Taken together, the CvfA-enolase complex in S. pyogenes is involved in the regulation of virulence gene expression by controlling RNA degradation according to nutritional stress.
Streptococcus pyogenes is a strict human pathogen that causes a variety of diseases ranging from mild superficial infections, such as impetigo and pharyngitis, to life-threatening systemic diseases, such as toxic shock and necrotizing fasciitis. The primary infection sites of S. pyogenes are the upper respiratory mucosal epithelium and the superficial layers of the epidermis. On rare occasions, S. pyogenes causes severe invasive diseases by penetrating the bloodstream or deep tissue. Some infection sites, such as blood, provide a rich source of nutrients, but others, such as plasma and the interstitial tissue fluid, are not nutritionally rich in terms of free amino acids, inorganic phosphate, and glucose (36). Nutrients would be more limited when S. pyogenes comes into contact with the skin or throat as the primary infection site. Because each of the infection sites provides a unique nutritional environment, S. pyogenes must alter its virulence gene expression pattern by sensing nutrient availability in order to obtain more nutrients in each infection site.
The major regulatory molecules responding to nutrient starvation, especially amino acid starvation, are guanosine nucleotides, mainly guanosine tetraphosphate (ppGpp). Under conditions of nutritional stress, accumulation of ppGpp initiates a global change in cellular metabolism, a phenomenon termed the stringent response, which includes the downregulation of nucleic acid and protein syntheses, and the simultaneous upregulation of protein degradation and amino acid synthesis (27). Many virulence genes mostly involved in host protein degradation and peptide uptake in S. pyogenes are upregulated during amino acid starvation (36). This upregulation, however, is independent of RelA, which is the only enzyme to synthesize or hydrolyze guanosine tetraphosphate (ppGpp) in S. pyogenes. This implies that a new pathway to regulate virulence gene expression other than ppGpp-mediated stringent response under nutrient starvation conditions exists in S. pyogenes. Pathogens like S. pyogenes, which is a multiple-amino-acid auxotroph, are likely to use a strategy different from that of nonpathogenic microorganisms to survive under nutrient-limited conditions. In addition to applying the general stringent response, which downregulates metabolic processes, S. pyogenes appears to adopt an active strategy by increasing the expression of virulence factors, such as the major secreted protease SpeB to obtain essential amino acids in tissues (8, 14, 21, 44).
During the study of a transposon-generated CvfA (SPy_1633) null mutant, we discovered that CvfA regulated the expression of virulence factors according to growth phase and nutritional status. The cvfA (conserved virulence factor A) gene was previously identified as a gene affecting the virulence of Staphylococcus aureus through a silkworm infection model and a mouse intraperitoneal infection model (17). CvfA has been studied in only two bacteria, Staphylococcus aureus and Bacillus subtilis. CvfA has three distinctive domains: a transmembrane domain at the N terminus, a KH (ribonucleoprotein K homology) domain in the middle, which has RNA binding activity (42), and an HD (His-Asp-containing phosphohydrolase) domain in the C-terminal region, known to have metal-dependent phosphohydrolase activity (2). The CvfA ortholog in B. subtilis, which has been named YmdA (16), Rny (10), or RNase Y (35), is located on the cell membrane, probably through its N-terminal transmembrane domain (16). The KH and HD domains are required for virulence of S. aureus in the silkworm model, indicating that these motifs have an important role in CvfA function (17, 29).
Previously, on the basis of the existence of its domains, CvfA was postulated to be an RNase (1), and recent studies verified that CvfA is an endoribonuclease. YmdA, the CvfA ortholog in B. subtilis, is responsible for the endonucleolytic processing of the gapA operon mRNA, which encodes six genes in the glycolytic pathway (cggR, gapA, pgk, tpiA, pgm, and eno) (10). YmdA cleaves the gapA operon mRNA between cggR and gapA. YmdA also cleaves the 5′ monophosphorylated S-adenosylmethionine (SAM)-dependent riboswitches, and the depletion of YmdA increases the half-life of bulk mRNA more than 2-fold, indicating that YmdA is important for the initiation of mRNA decay as well as for riboswitch RNA turnover in B. subtilis (35). In addition, YmdA interacts with glycolytic enzymes phosphofructokinase and enolase (10). We also verified the interaction between CvfA and enolase in S. pyogenes. Taken together, a protein complex containing CvfA and enolase regulates the expression of virulence genes, and the regulation is dependent on the nutritional status of S. pyogenes. Most previously known regulatory mechanisms for the expression of virulence factors in S. pyogenes are mediated by transcriptional regulators (19). However, the CvfA-involved regulatory system appears to use a new mechanism to regulate the expression of virulence factors at the posttranscriptional level.
MATERIALS AND METHODS
Bacterial strains and media.
All experiments and strain construction employed Streptococcus pyogenes HSC5 (15). Molecular cloning experiments utilized Escherichia coli DH5α which was cultured in Luria-Bertani broth (34). Routine culture of S. pyogenes employed Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (Difco) (THY medium) with incubation at 37°C in sealed tubes without agitation. Unless indicated otherwise, growth of S. pyogenes for SpeB activity assay, Western blotting, and RNA preparation for microarray and real-time PCR used C medium (25). To produce solid media, Bacto agar (Difco) was added to a final concentration of 1.4% (wt/vol). Cultures on solid media were incubated under the anaerobic conditions produced using a commercial gas generator (GasPak) (catalogue no. 260678; BBL). A chemically defined S. pyogenes medium (CDM) (43) was modified by supplementation with 0.1% yeast extract (CDMY) to improve growth of culture inoculated with washed cells (23). To increase the amount of nitrogen source in the CDMY medium, dialyzed peptides (1%, wt/vol) were added to CDMY medium containing 0.05% glucose. Bacto proteose peptone no. 3 (BD Diagnostic Systems) was dialyzed at 4°C for 48 h with 10,000-molecular-weight cutoff (MWCO) dialysis tubing (SnakeSkin dialysis tubing; Pierce). Double deionized water (5 liters) was replaced every 12 h during dialysis. When appropriate, antibiotics were added to the media at the following concentrations if they are not specified; spectinomycin, 100 μg/ml for E. coli and S. pyogenes; kanamycin, 50 μg/ml for E. coli and 500 μg/ml for S. pyogenes.
Manipulation of DNA.
Plasmid DNA was isolated by standard techniques and used to transform E. coli by the method of Lederberg and Cohen (20) and to transform S. pyogenes by electroporation as described previously (6). Restriction endonucleases, ligases, and polymerases were used according to the recommendations of the manufacturers. Chromosomal DNA was purified from S. pyogenes as described previously (6). When required, DNA fragments were purified using MinElute gel extraction kit (Qiagen) following electrophoresis through an agarose gel.
Strain construction.
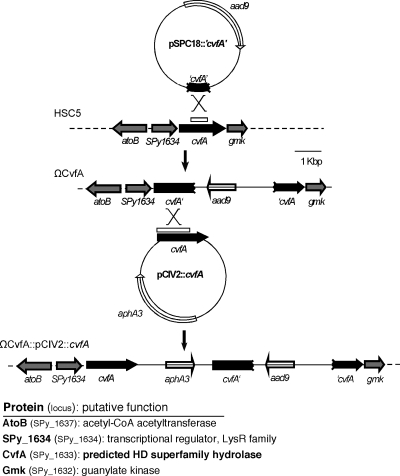
The ΩCvfA mutant strain contains insertional disruption of cvfA and was derived from S. pyogenes HSC5 as follows. An internal region of cvfA was amplified by PCR, inserted into the suicide vector pSPC18 (26), and then used to transform strain HSC5 to resistance to spectinomycin. To ensure that mutant phenotypes were specifically due to gene disruption and not due to a nonspecific effect of integration of pSPC18 into the chromosome, S. pyogenes HSC5Spc was included in selected analyses as a wild-type control strain for pSPC18 insertion (9). For cis complementation of the disrupted cvfA gene in the ΩCvfA mutant strain, the ΩCvfA::pCIV2::cvfA strain was constructed as follows. cvfA containing a putative promoter region was amplified by PCR, inserted into the suicide vector pCIV2 (32) and then used to transform the ΩCvfA strain. Suicide vectors, such as pSPC18 and pCIV2, do not have a replication origin of Streptococcus, so the plasmid can be maintained in Streptococcus only by integrating into the chromosome through homologous recombination. Details on each integrating plasmid and the sequences of the primers used for amplification of DNA segments are listed in Table 1. The gene disruption or reconstruction by homologous recombination was confirmed by PCR using appropriate primers.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | recA1 endA1 hsdR17 | 34 |
| CF1648 | Wild-type MG1655 | 46 |
| CF1693 | CF1648 ΔrelA251::kan ΔspoT207::cat | 46 |
| S. pyogenes | ||
| HSC5 | Wild type | 15 |
| HSC5Spc | Wild-type control strain for pSPC18 insertion; pSPC18 was inserted downstream of recF in the chromosome of strain HSC5 without disrupting any gene or operon | 9 |
| ΩCvfA strain | HSC5 mutant with cvfA disruption created by pSPC18::′cvfA′a | This study |
| ΩCvfA::pCIV2::cvfA strain | ΩCvfA strain with pCIV2::cvfA insertion in the chromosome; the disrupted cvfA gene in the ΩCvfA strain was restored by inserting an intact copy of cvfAa | This study |
| ΩCvfA(pRopB-HA) strain | ΩCvfA strain transformed with pRopB-HA | This study |
| Plasmids | ||
| pSPC18 | pUC18-based streptococcal integration vector containing aad9 (spectinomycin resistance gene from Enterococcus faecalis) | 26 |
| pCIV2 | pUC18-based streptococcal integration vector containing aphA3 (kanamycin resistance gene) | 32 |
| pSPC18::′cvfA′ | pSPC18 containing a 0.78-kbp internal fragment of cvfA generated by PCR with the CvfA-f primer (TCCCCGGGAATCTAAACACATTGATGAGCGGC) and the CvfA-r primer (CGCGGATCCTAATTCTCCATTGACTCATTACG)b | This study |
| pCIV2::cvfA | pCIV2 containing an intact copy of cvfA (1.80 kbp) generated by PCR with the CvfAcomp-f primer (ACATGCATGCGAAGCCTACATCATGGACGAC) and the CvfAcomp-r primer (GGGGTACCCTACTTGGCATAATCAACCG)b | This study |
| pRopB-HA | pABG5 derivative containing HA-tagged ropB | 24 |
| pSPC18::′cvfA-6xHis | pSPC18 containing a DNA sequence of the C-terminal part of CvfA fused to the 6× His tag sequence (0.75 kbp) generated by PCR with the CvfA6xΗisBam-f primer (TTTGGATCCTCTCTGATTGCTGATGGTCG) and the CvfA6xHisBam-r primer (TTTGGATCCTCAATGATGATGATGATGATGCTTGGCATAATCAACCGCTCTC)b | This study |
| pCIV2::′eno-HA | pCIV2 containing a DNA sequence of the C-terminal part of enolase (SPy_0731) fused to the HA tag sequence (0.75 kbp) generated by PCR with the EnoSal-f primer (AAAGTCGACAGGTGACGAAGGTGGATTTG) and the EnoHASal-r primer (AAAGTCGACCTAAGCGTAATCTGGAACATCGTAGCTGGTTTTTTTTAAGTTATAGAATGATTTGATAC)b | This study |
See Fig. 1.
Restriction sites embedded into primer sequences are underlined.
RNA isolation and real-time RT-PCR.
RNA isolation from S. pyogenes cultures and real-time reverse transcription-PCR (RT-PCR) were conducted as described elsewhere (4). The primers for real-time PCR were described elsewhere (9), except for the primers for gmk expression analysis (GGGTTGATTATTTCTTCCGC and AACATAAGTTAAAGGGGTGCC). The level of expression of the recA gene was used as an internal control to normalize the levels of expression of specific transcripts in different samples (4). Data reported represent the mean derived from at least 2 RNA preparations derived from a minimum of 2 independent experiments, each performed on a different day, and each data point was determined at least twice.
Microarray analysis.
Microarray analysis was conducted as described elsewhere (8). Briefly, an antisense oligonucleotide array based on the published genome of S. pyogenes SF370 (12) was made with a single oligonucleotide (60- to 80-mer) designed to hybridize to the putative transcripts from each annotated open reading frame (ORF). Synthesis and labeling of cDNA used a commercial kit (3DNA Array 900; Genisphere), and the signal of cyanine 3 (Cy3) or cyanine 5 (Cy5) was measured using a laser scanner (ScanArray Express HT; Perkin-Elmer). The fluorescence values from each array were normalized by the LOWESS (Locally Weighted Scatter plot Smoother) function of the laser scanner. Average values were calculated from a data set consisting of 18 total microarray hybridizations that were derived from RNA samples prepared from 2 independent experiments, each of which was conducted on a different day. The data were tested for significance computationally using significance analysis of microarrays (SAM) software (http://www-stat.stanford.edu/∼tibs/SAM/) (40) with the following settings: log2-based transformation of fold change values and a one-class response format. Genes considered differentially expressed fulfilled the following criteria: the estimated false discovery rate at the 90th percentile was ≤0.1%, and the fold change value was at least ±2.0.
Analysis of protein expression.
Use of protease indicator medium, Western blot analysis against SpeB, and SpeB cysteine protease activity were all performed as described previously (25). M protein in cell wall fractions was analyzed by Western blotting as described previously (9).
Streptokinase activity assay.
Streptokinase activity in S. pyogenes culture supernatants was determined by measuring the activity of plasmin activated by streptokinase as described elsewhere (39). The cysteine protease inhibitor E-64 (final concentration, 10 μM; Sigma) was added to S. pyogenes cultures to prevent degradation of streptokinase by SpeB. Briefly, 20 μl of filter-sterilized culture supernatant, 50 μl of 50 mM Tris buffer (pH 8.0), and 50 μl of human plasminogen (20 μg/ml) (catalogue no. P-5661; Sigma) were mixed and incubated for 15 min at 37°C. Then, 50 μl of a chromogenic plasmin substrate (H-D-valyl-leucyl-lysine-p-nitroaniline) (1 mg/ml) (catalogue no. V7127; Sigma) was added and further incubated for 30 min at 37°C. After the final incubation, absorbance at 405 nm was measured. Tests were performed in triplicate.
CAMP test.
CAMP factor activity of S. pyogenes strains was tested as described elsewhere (13). In brief, S. pyogenes strains were streaked perpendicular to, but not touching, a streak of S. aureus ATCC 25923, which produces a β-toxin. THY medium or C-medium agar plates containing 5% (vol/vol) sheep blood were used. After anaerobic incubation at 30°C overnight, the plates were inspected for CAMP hemolysis.
ppGpp measurements.
The assay to detect ppGpp was performed as described previously (28, 36). Briefly, S. pyogenes was grown in CDM supplemented with 0.1% (wt/vol) yeast extract and 26 mM MOPS (morpholinepropanesulfonic acid) at 37°C to the mid-exponential phase (optical density at 600 nm [OD600] of 0.35). The cells were washed with phosphate-buffered saline (PBS) buffer, suspended in 1 ml of low-phosphate medium containing 0.07% glucose for carbon source starvation or 1.0% (wt/vol) glucose, and incubated with 150 mCi carrier-free 32Pi for 90 min at 37°C. To determine nucleotide patterns, the cells were resuspended in 200 μl of ice-cold 13 M formic acid. After three freeze-thaw cycles, the samples were centrifuged, and the supernatant fluids were subjected to one-dimensional polyethyleneimine (PEI) thin-layer chromatography (TLC) developed with 1.5 M potassium phosphate solution (pH 3.4). The assay of ppGpp in E. coli control strains was performed as described elsewhere (11).
Formaldehyde cross-linking to examine the interaction between CvfA and enolase.
To test whether CvfA interacts with enolase (SPy_0731), CvfA was labeled with the 6× His tag, and enolase was labeled with hemagglutinin (HA) tag through homologous recombination. The C-terminal regions of CvfA and enolase were amplified with the proper primers (Table 1). The primers annealing to the sense strand (the primers binding to the 3′ end) included the sequence of each tag. Each amplified PCR product was inserted into a suicide vector (pSPC18 for the PCR product of CvfA C-terminal sequence fused to the 6× His tag sequence and pCIV2 for the PCR product of enolase C-terminal sequence fused to the HA tag sequence) and transferred to S. pyogenes HSC5 sequentially by electroporation. CvfA was tagged first, and then enolase was tagged. The strain with both tags was selected through antibiotic resistances conferred by each suicide vector (spectinomycin resistance from pSPC18 and kanamycin resistance from pCIV2), and proper insertions of the plasmids were examined through Western blotting using antibody against the His tag (anti-His tag antibody) or the HA tag (anti-HA tag antibody).
After both proteins were tagged, we performed protein cross-linking using formaldehyde, a cleavable and membrane-permeable cross-linker. The strain that has both proteins tagged was grown in 1 liter of C medium supplemented with 1% (wt/vol) glucose to the stationary phase (OD600 of 1.2) and washed with PBS buffer. Then, the cells were suspended in PBS to an optical density at 600 nm of 0.5, and formaldehyde was added to a final concentration of 0.5% (wt/vol). After the cells were incubated for 20 min at room temperature, the formaldehyde cross-linking was quenched by adding glycine to a final concentration of 125 mM, and the cells were extensively washed with PBS buffer to remove residual formaldehyde. The cells were then lysed with PlyC, which is the lysin produced by streptococcal phage C1, in 30% (wt/vol) sucrose solution supplemented with protease inhibitors (complete, mini, EDTA-free protease inhibitors; Roche Applied Science). PlyC cleaves the cell wall of S. pyogenes efficiently (31) and makes protoplasts in 30% sucrose solution. The collected protoplasts were lysed with a bead beater (FastPrep; MP Biomedicals), and the cell membrane was collected by ultracentrifugation (100,000 rpm for 12 h). CvfA, which is a membrane-anchored protein, was released from the membrane by dissolving the membrane with 4 ml of column binding buffer (50 mM sodium phosphate [pH 7.0], 300 mM NaCl, 5 mM imidazole) containing 1% (vol/vol) Nonidet P-40 (NP-40), a nonionic detergent. The His-tagged CvfA in the dissolved membrane fraction was then purified with a His tag affinity column containing cobalt resin (Talon His tag affinity resin; Clontech). After the column was washed with a washing buffer (50 mM sodium phosphate [pH 7.0], 300 mM NaCl, 10 mM imidazole, 1% [vol/vol] NP-40), the proteins bound to the resin were eluted with 1.0 ml of an elution buffer (50 mM sodium phosphate [pH 7.0], 300 mM NaCl, 250 mM imidazole) containing 1% (wt/vol) CHAPS {[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate}, a dialyzable detergent. It was necessary to add a detergent to the washing and the elution buffer to prevent aggregation of CvfA inside the affinity column. The eluted sample was dialyzed to remove salts and detergents and concentrated to 100 μl using a vacuum evaporator. Ten microliters of each sample was loaded onto a 8% polyacrylamide gel for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blotting was performed using anti-His tag antibody or anti-HA tag antibody. Before Western blotting, the purified proteins were incubated under cross-linker-cleavable or -noncleavable conditions; boiling for 20 min cleaves protein cross-linking by formaldehyde.
Murine subcutaneous infection model.
The virulence of S. pyogenes strains was examined using a murine subcutaneous infection model described previously (5). Briefly, 6- to 8-week-old female SKH1 hairless mice (Charles River Labs) received subcutaneous injections of 1 × 107 CFU in 100-μl volume into the right flank. Each study group consisted of 10 mice, and 5 mice were housed together. The area of the draining ulcer that formed was documented every 24 h by digital photography, and the precise area of each ulcer was calculated from the digital record using Meta-Morph image analysis software (version 4.6; Universal Imaging Corp.). Any differences in the areas of ulcers between experimental groups were tested for significance by the Mann-Whitney U test, and the null hypothesis was rejected when the P value was <0.05. The pathology of mouse ulcers caused by S. pyogenes was examined as follows. The lesions that developed on the right flanks of mice injected with 1 × 107 CFU of S. pyogenes subcutaneously were removed at 72 h postinjection using a sterile scalpel after euthanasia. Each lesion removed was fixed by immersion in 10 ml of a commercial tissue fixative (Histochoice; Sigma). The fixed samples were routinely processed and then embedded in paraffin, and 5-μm-thick sections were prepared which were dewaxed and rehydrated by standard methods and then stained with hematoxylin and eosin. The pathology of stained samples was observed and documented with a microscope equipped with digital photography (Leica microscope model DMIRB).
Microarray data accession number.
The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE15938.
RESULTS
Transposon mutagenesis identified the cvfA gene.
We employed Tn5-based transposome mutagenesis (9) to search for genes regulating the expression of virulence factors. A transposon insertion library created by transposome mutagenesis was examined for mutants that displayed reduced activity of the secreted SpeB cysteine protease following overnight culture on a protease indicator medium. Sequencing of the transposon insertion loci of mutants identified the Spy_1633 gene, also known as cvfA. In the transposon-generated cvfA mutant, transposon insertion occurred at the 3′ side of the cvfA gene (between TA and C of the 517th codon of tyrosine; the total number of codons in cvfA is 535). To confirm the results of transposon mutagenesis, we created a CvfA null mutant by directed insertional mutation using a streptococcal suicide vector (Fig. 1). An internal fragment of cvfA amplified through PCR was inserted into pSPC18, a streptococcal suicide vector that has a spectinomycin resistance gene (aad9) (26), and the construct was transferred to S. pyogenes HSC5, the wild-type strain, by electroporation. Strain HSC5Spc, which has pSPC18 inserted in the chromosome without disrupting any gene or operon, was used as a wild-type control for pSPC18 insertion in the chromosome (9). Like the transposon-generated mutant, the CvfA null mutant (ΩCvfA strain) also showed great reduction of SpeB production (Fig. 2). The growth rate of the CvfA null mutant was the same as that of the wild-type control in laboratory media.
FIG. 1.
Construction of the CvfA− mutant and its cis-complemented strain. The integrational plasmid (pSPC18::′cvfA′) shown at the top was used to disrupt the cvfA locus in S. pyogenes HSC5. The broken ends of the plasmid-encoded box indicate that the cvfA gene segment lacks the 5′ and 3′ ends of the corresponding chromosomal open reading frame, and the region is indicated by the open box above the cvfA gene. The other integrational plasmid (pCIV2::cvfA) shown in the middle was used to complement the disrupted cvfA in the CvfA− mutant. Chromosomal structures resulting from the single recombination event are indicated below the solid vertical arrows. The open reading frames labeled aad9 and aphA3 encode a spectinomycin-resistant determinant and a kanamycin-resistant determinant, respectively. acetyl-CoA, acetyl coenzyme A.
FIG. 2.
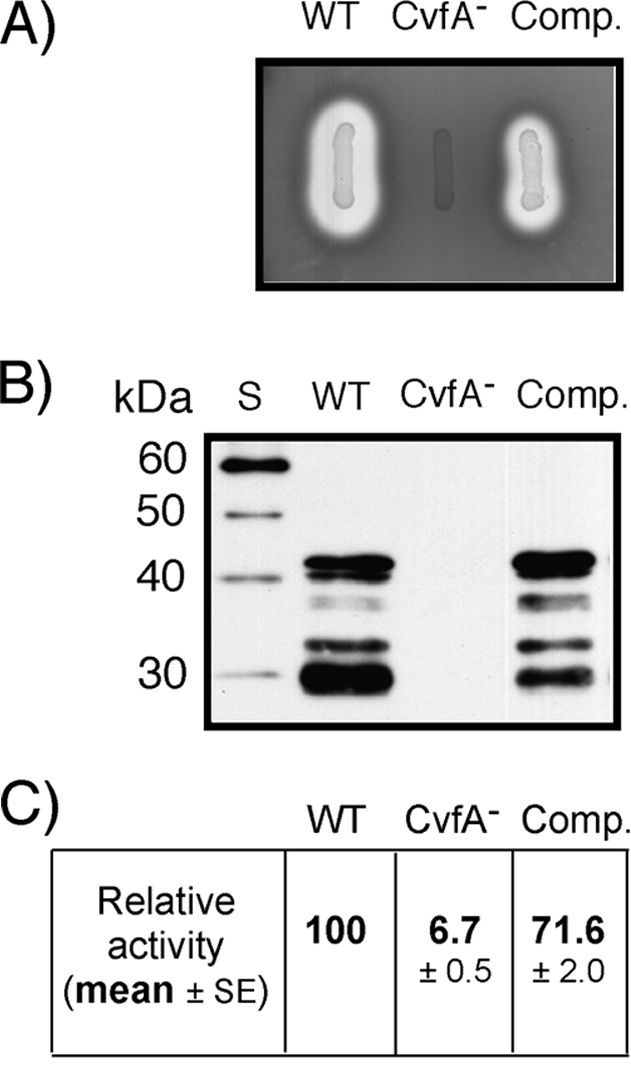
Contribution of cvfA to SpeB expression. (A) Protease production on indicator medium. The wild-type (WT) S. pyogenes strain (HSC5spc), CvfA− mutant strain (ΩCvfA strain), and CvfA− complemented strain (Comp.) (ΩCvfA::pCIV2::cvfA) were transferred to protease indicator medium and examined following overnight incubation. SpeB activity accounts for more than 95% activity of the secreted proteases by S. pyogenes. A zone of clearing resulted from protease activity. (B) Western blot analysis against SpeB. The SpeB cysteine protease in supernatants of 12-h cultures of strains are shown. Note that multiple proteolytic cleavages are required for activation of the 46-kDa SpeB zymogen to the active, 28-kDa SpeB. Lane S, protein size markers. (C) Quantitative assessment of protease activity. The SpeB cysteine protease activities in supernatants of 12-h cultures are shown relative to that of the wild-type strain. The data are the means ± standard errors of the means (SEs) derived from at least three independent experiments.
To examine whether the disruption of cvfA influences the transcription of downstream genes, we determined the level of expression of the first gene downstream, gmk, the direction of which is the same as that of cvfA. When the expression of gmk was examined with real-time RT-PCR, the ratio of expression of the gmk gene in the CvfA− mutant to the expression in the wild-type control was 1.24 ± 0.16 (mean ± standard deviation), indicating that cvfA disruption did not affect the expression of downstream genes (no polar effect).
To perform a complementation test for the disruption of cvfA, a copy of intact cvfA was introduced into the chromosome of the CvfA− mutant. The cvfA gene and its putative promoter region were amplified and inserted into pCIV2, a streptococcal suicide vector containing a kanamycin resistance gene (aphA3) (32), and transferred to the CvfA− mutant through electroporation. The introduction of an intact copy of cvfA through homologous recombination between the cvfA gene inserted into the second pCIV2 plasmid and the 5′ side of the disrupted cvfA on the chromosome of the ΩCvfA strain was confirmed by PCR using appropriate primers (Fig. 1). The complemented strain, the ΩCvfA::pCIV2::cvfA strain, exhibited the same phenotypic characteristics as those of the wild type, as shown in Fig. 2 (SpeB activity) and Fig. 4 (M-protein production, streptokinase activity, and CAMP factor activity in C medium). Thus, the cis complementation of cvfA reverted the phenotype of the CvfA− mutant to that of the wild type, indicating only cvfA disruption was responsible for the phenotype of the transposon-generated cvfA mutant.
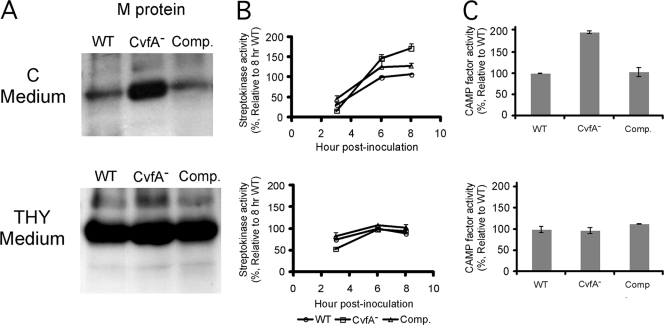
FIG. 4.
Biochemical assay to confirm the results of transcript analysis. The wild-type (WT) strain (HSC5spc), CvfA− mutant strain (ΩCvfA strain), and CvfA− complemented strain (Comp.) (ΩCvfA::pCIV2:: cvfA) were used. The CvfA− mutant exhibits a phenotype different from that of the wild type only in C medium, not in THY medium. (A) Western blot against M protein. The amounts of M protein in cell wall fractions were compared by Western blotting. The cysteine protease inhibitor E-64 (final concentration, 10 μM) was added to S. pyogenes culture to prevent the degradation of M protein by SpeB. (B) Streptokinase activity. The activities of streptokinase in the supernatants of cultures 3, 6, and 8 h postinoculation were analyzed and expressed relative to the activity in supernatant of the wild-type culture 8 h postinoculation. The data are the means and standard errors of the means derived from at least three independent experiments. The cysteine protease inhibitor E-64 (final concentration, 10 μM) was added to S. pyogenes culture to prevent the degradation of streptokinase by SpeB. (C) CAMP test. CAMP factor activity was measured by streaking S. pyogenes strains perpendicular to a streak of S. aureus producing β-toxin on agar plates containing C medium or THY medium and 5% (vol/vol) sheep blood. CAMP factor activity was expressed relative to the activity of the wild type. The CAMP factor activities were compared by measuring the area of CAMP hemolysis zone using ImageJ software (NIH) after the plates were scanned. The data are the means and standard errors of the means derived from at least two independent experiments.
The effect of cvfA on virulence gene expression was most pronounced during stationary phase.
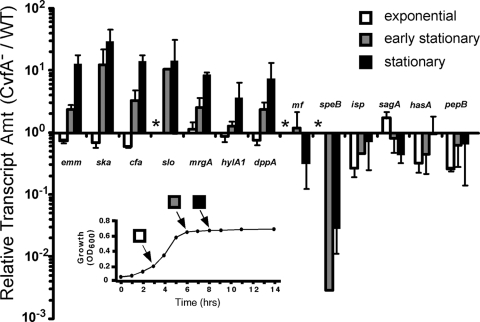
The virulence gene expression of the CvfA− mutant was examined at different growth stages with real-time RT-PCR and compared with that of S. pyogenes HSC5spc, the wild-type control strain. RNA was extracted from the cells grown for 3, 6, or 8 h in C medium (25). Each time point represents exponential, early stationary, and stationary growth of cells (Fig. 3). As the length of time of cultivation increased, the differences in gene expression of the CvfA− mutant and the wild type increased (Fig. 3). The exponentially growing cells at 3 h postinoculation (3 hpi) did not show much difference in the virulence gene transcript amount, the early-stationary-phase cells (6 hpi) started to show differences in the transcript amount, and cells at the stationary phase (8 hpi) showed the biggest differences in the transcript amounts of the CvfA− mutant and the wild type. At the stationary growth phase (8 hpi), the expression of the genes encoding streptokinase (ska), M protein (emm), CAMP factor (cfa), streptolysin O (slo), dipeptide ABC transporter substrate-binding protein (dppA), and the MrgA protein (mrgA) were upregulated, and the expression of the cysteine protease SpeB gene (speB) was significantly downregulated in the CvfA− mutant compared to the wild type (Fig. 3). Thus, CvfA downregulates the expression of ska, emm, cfa, slo, dppA, and mrgA and upregulates the expression of speB at the stationary phase in wild-type cells in C medium. Taken together, CvfA differentially regulates the expression of virulence factors mostly during stationary phase.
FIG. 3.
Disruption of cvfA results in altered patterns of virulence gene expression at stationary phase. The relative levels of the transcripts of virulence genes in the CvfA− mutant (ΩCvfA strain) were determined over the course of growth in C medium using real-time RT-PCR and expressed as changes relative to the levels in the wild-type (WT) strain. Levels that were not determined are indicated by an asterisk. The growth pattern of S. pyogenes is shown in the graph in the inset, and the growth stage of each sampling time is shown as indicated. The samples were taken at exponential growth phase at 3 h postinoculation (hpi), early stationary phase at 6 hpi, and stationary phase at 8 hpi. For clarity, only the growth pattern of the wild-type strain is shown, as the growth characteristics of the two strains were indistinguishable under these conditions. Amt, amount.
Microarray analysis to determine the effect of CvfA on global gene expression at stationary phase.
To gain insight into the expression of genes affected by CvfA, global transcriptome profiling between the CvfA− mutant and the wild type was compared through DNA microarray analysis. The design, construction, and validation of the microarray are described in detail elsewhere (8). Differential expression profiles were developed by comparing stationary-phase cells (8 hpi) in which the effect of CvfA on virulence gene expression was most pronounced (Fig. 3). This analysis revealed large differences in expression between the CvfA− mutant and the wild type. Approximately 14% of the total number of genes in the genome were significantly upregulated (219 genes), and 15% were significantly downregulated (228 genes) in the CvfA− mutant compared to the wild type (see Tables S1 and S2 in the supplemental material). To gain insight into the overall change, genes whose expression was altered by CvfA were assigned a code according to the COG (clusters of orthologous groups) database (Table 2). Each code of the COG system represents a basic cellular process, and open reading frames (ORFs) are assigned to one of the codes based on predicted function by orthologous relationships (38). In the analysis according to this system, the genes in the categories of transport and metabolism of most of the cellular components, including amino acids, carbohydrates, and lipids, were upregulated in the CvfA− mutant compared to the wild type (Table 2). Also, the genes in the category of energy production and conversion were upregulated. On the other hand, the genes in the transcription category, which includes the genes of transcriptional regulators, were downregulated. A number of recognized virulence factors were differentially expressed in the CvfA− mutant than in the wild type, and this result was confirmed by real-time RT-PCR (Table 3). The streptokinase (ska), exotoxin G (speG), CAMP factor (cfa), streptolysin O (slo), and M protein (emm) gene were upregulated, and the SpeB (speB), mitogenic factor (mf), and streptolysin S (sagA) genes were downregulated.
TABLE 2.
Genes that were significantly upregulated or downregulated in the CvfA− mutant compared to the wild-type control strain
| Code | Categorya (no. of genes in categoryb) | CvfA− strain vs. WT strain |
||
|---|---|---|---|---|
| No. of upregulated genesc (%d) | No. of downregulated genesc (%d) | Ratio (no. of upregulated genes/no. of downregulated genes) | ||
| J | Translation (140) | 36 (25.7) | 29 (20.7) | 1.2 |
| K | Transcription (93) | 9 (9.7) | 22 (23.7) | 0.4 |
| L | Replication, recombination, and repair (95) | 13 (13.7) | 18 (18.9) | 0.7 |
| C | Energy production and conversion (57) | 17 (29.8) | 7 (12.3) | 2.4 |
| E | Amino acid transport and metabolism (99) | 19 (19.2) | 8 (8.1) | 2.4 |
| F | Nucleotide transport and metabolism (59) | 6 (10.2) | 7 (11.9) | 0.9 |
| G | Carbohydrate transport and metabolism (124) | 25 (20.2) | 16 (12.9) | 1.6 |
| H | Coenzyme transport and metabolism (34) | 4 (11.8) | 5 (14.7) | 0.8 |
| I | Lipid transport and metabolism (45) | 8 (17.8) | 3 (6.7) | 2.7 |
| Q | Secondary metabolite biosynthesis, transport, and catabolism (11) | 2 (18.2) | 1 (9.1) | 2.0 |
| D | Cell cycle control, mitosis, and meiosis (16) | 2 (12.5) | 3 (18.8) | 0.7 |
| M | Cell wall/membrane biogenesis (64) | 6 (9.4) | 7 (10.9) | 0.9 |
| O | Posttranslational modification, protein turnover, chaperones (50) | 7 (14.0) | 6 (12.0) | 1.2 |
| P | Inorganic ion transport and metabolism (58) | 8 (13.8) | 7 (12.1) | 1.1 |
| T | Signal transduction mechanisms (37) | 3 (8.1) | 7 (18.9) | 0.4 |
| U | Intracellular trafficking and secretion (14) | 4 (28.6) | 1 (7.1) | 4.0 |
| V | Defense mechanisms (36) | 2 (5.6) | 9 (25.0) | 0.2 |
| R | General function prediction only (137) | 17 (12.4) | 20 (14.6) | 0.9 |
| S | Function unknown (126) | 7 (5.6) | 25 (19.8) | 0.3 |
| Not in COGs (222) | 24 (10.8) | 27 (12.2) | 0.9 | |
| Sum (1,517) | 219 (14.4) | 228 (15.0) | ||
Genes were categorized according to a functional classification, COGs (clusters of orthologous groups) (38).
Structural RNA genes (tRNA and rRNA) and phage genes were not included.
Number of genes displaying statistically significant differential expression fulfilling the following criteria: average fold change of at least ±2.0, and FDR (false discovery rate) in SAM (significance analysis of microarrays) analysis (40) with 90th percentile of ≤ 0.1%.
Number of statistically significant genes to the total number of genes assigned to the category.
TABLE 3.
Virulence factors significantly upregulated or downregulated in the CvfA− mutant compared to the wild-type control strain
| Locusa | Geneb | Gene productb | Fold change by microarrayc | Fold change by real-time PCRd |
|---|---|---|---|---|
| SPy-1979 | ska | Streptokinase A precursor | 9.28 | 21.8 ± 4.6 |
| SPy-0212 | speG | Exotoxin G precursor | 6.24 | ND |
| SPy-1273 | cfa | CAMP factor | 5.29 | 14.2 ± 4.0 |
| SPy-0167 | slo | Streptolysin O precursor | 3.82 | 4.8 ± 0.8 |
| SPy-0378 | hlyX | Putative hemolysin | 3.76 | ND |
| SPy-2018 | emm | M protein (type 1) | 3.74 | 14.5 ± 7.3 |
| SPy-1054 | Putative collagen-like protein | 3.65 | ND | |
| SPy-0591 | Putative protease | 2.63 | ND | |
| SPy-1896 | ropA | Transcription regulator (trigger factor, prolyl isomerase) | 2.48 | ND |
| SPy-0165 | nga or spn | Nicotine adenine dinucleotide glycohydrolase precursor | 2.43 | ND |
| SPy-2025 | isp | Immunogenic secreted protein precursor | −2.53 | −1.1 ± 3.1 |
| SPy-2043 | mf | Mitogenic factor | −3.17 | −3.6 ± 0.2 |
| SPy-0738 | sagA | Streptolysin S-associated protein | −4.36 | −2.3 ± 0.6 |
| SPy-2039 | speB | Pyrogenic exotoxin B | −130.54 | −76.7 ± 84.3 |
SPy numbers are assigned on the basis of the genome of serotype M1 group A streptococcus (GAS) strain SF370 (12).
Assignment based on annotation of serotype M1 GAS strain SF370 ORFs (http://www.ncbi.nlm.nih.gov/genomes/altik.cgi?gi=178&db=Genome).
Average of a data set consisting of 18 total microarray hybridizations with 2 independently prepared RNA samples. The values here fulfilled the following criteria: the estimated false discovery rate at the 90th percentile in SAM (significance analysis of microarrays) analysis (40) was ≤0.1%, and the fold change value was at least ±2.0.
Fold change determined by real-time RT-PCR. Data show mean ± standard deviation derived from at least two independent experiments. ND, not determined.
The role of CvfA in influencing virulence gene expression was affected by growth media.
The activities or amounts of protein of three virulence factors (M protein, streptokinase, and CAMP factor), whose levels of expression were altered in the CvfA− mutant compared to the wild type, were examined to confirm the results of transcript analyses. As expected, when S. pyogenes was grown in C medium, which was used for real-time RT-PCR analysis, the amount of M protein, streptokinase activity, and CAMP factor activity of the CvfA− mutant were higher than those of the wild type, indicating that CvfA is involved in the downregulation of these virulence factors in the wild type in C medium (Fig. 4). However, when S. pyogenes was grown in THY medium, the amount of M protein, streptokinase activity, and CAMP factor activity of the CvfA− mutant were the same as those of the wild type, indicating that the CvfA protein does not downregulate these activities in THY medium (Fig. 4). Transcript analysis performed with real-time RT-PCR indicated that the altered phenotypic outcomes in different growth media originated at transcript level (Fig. 5A). The emm, ska, and cfa transcript ratios of the CvfA− mutant to the wild type (ΩCvfA/HSC5Spc) in C medium were much higher than the ratios in THY medium.
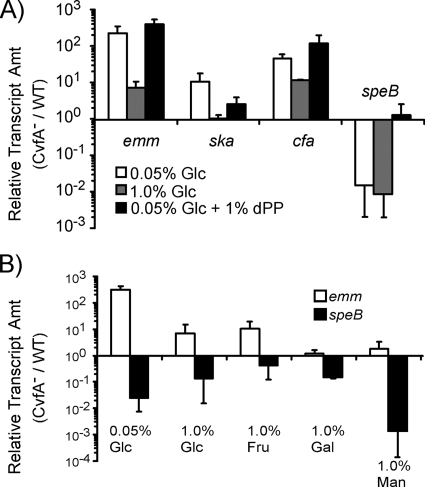
FIG. 5.
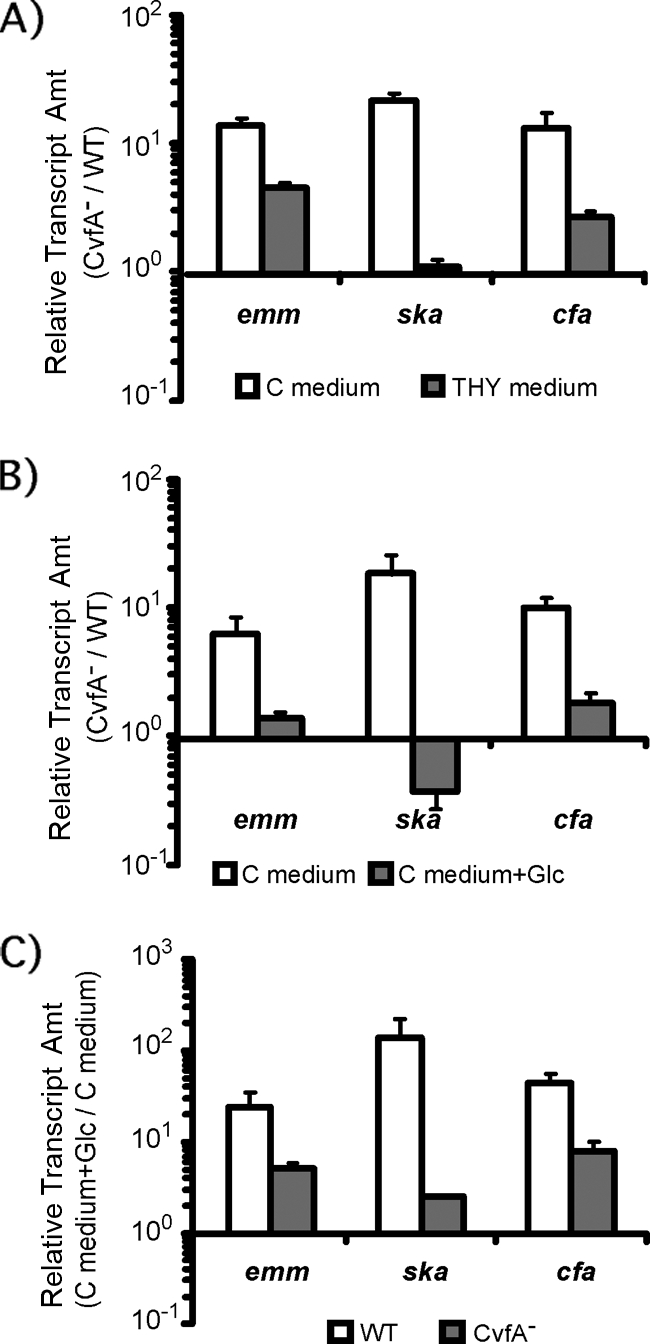
Glucose influences virulence gene expression through CvfA. The amounts (Amt) of transcripts of the genes encoding M protein (emm), streptokinase (ska), and CAMP factor (cfa) in S. pyogenes strains at the stationary phase (8 h postinoculation) were determined by real-time RT-PCR. The S. pyogenes strains used were the wild-type strain (WT) (HSC5spc) and the CvfA− mutant strain (ΩCvfA strain). (A) The ratio of emm, ska, or cfa transcript of the CvfA− mutant to that in the wild type (CvfA−/WT) for cells grown in C medium or THY medium is shown. The ratios became smaller in THY medium, which contains glucose, unlike C medium. (B) The ratio of emm, ska, or cfa transcript in the CvfA− mutant to that in the wild type (CvfA−/WT) for cells grown in C medium or C medium supplemented with 1% (wt/vol) glucose (C medium+Glc) is shown. The ratios became smaller when glucose was added to the medium. (C) The ratio of emm, ska, or cfa transcript for the wild-type or CvfA− mutant cells grown in C medium supplemented with 1% (wt/vol) glucose to transcript for cells grown in C medium (C medium+Glc/C medium) is shown. The ratios for the CvfA− mutant were smaller than those for the wild type.
Glucose supplementation to C medium made the phenotype of the CvfA− mutant similar to that of the wild type.
A major difference between THY medium and C medium is the amount of carbohydrates. C medium consists of 0.5% (wt/vol) proteose peptone no. 3 (Difco), 1.5% (wt/vol) yeast extract (Difco), 10 mM K2HPO4, 0.4 mM MgSO4, and 17 mM NaCl (pH 7.5). Unlike THY medium, C medium does not contain glucose, so it is rich in peptides and poor in carbohydrates. When glucose (1%, wt/vol) was added to C medium, the emm, ska, and cfa transcript ratios in the CvfA− mutant to the wild type (ΩCvfA/HSC5Spc) were greatly reduced, so the behavior of the CvfA− mutant in the glucose-supplemented C medium was similar to that in THY medium (Fig. 5B). This outcome resulted from decreased expression of most of these genes by glucose in the CvfA− mutant. In glucose-supplemented C medium, the expression of these virulence genes in both the wild type and the CvfA− mutant increased, but the degree of increase was much less in the CvfA− mutant than in the wild type: 5-fold less for emm, 55-fold less for ska, and 6-fold less for cfa (Fig. 5C). Thus, the expression of these virulence genes in the CvfA− mutant was less affected by the addition of glucose compared to the wild type. Overall, adding glucose to C medium reduced the difference in virulence gene expression between the CvfA− mutant and the wild type because the response of the CvfA− mutant to glucose was less sensitive than that of the wild type. This result suggests that a CvfA-involved regulatory system directly or indirectly senses the amount of glucose in medium and regulates the expression of some virulence genes by controlling the amount of their transcripts.
The expression of CvfA in the wild type in C medium with or without glucose did not change; the ratio of cvfA transcript of cells grown in glucose-supplemented C medium to that in C medium was 0.84 ± 0.08 (mean ± standard deviation), indicating that the amount of cvfA transcript itself was not affected by the addition of glucose to C medium.
Effect of the supplementation of peptides or sugars (monosaccharides) on virulence gene expression regulated by CvfA.
To test whether the addition of peptides to the medium affects the CvfA-involved regulation of virulence gene expression, the expression of emm, ska, cfa, and speB in the CvfA− mutant and the wild type in a chemically defined medium (43) supplemented with peptides (1%, wt/vol) was compared. In the chemically defined medium supplemented with peptides, the ratio of emm, ska, or cfa transcript of the CvfA− mutant to that in the wild type did not change (Fig. 6A). However, the speB transcript in the CvfA− mutant increased 100-fold up to the wild-type level in the peptide-supplemented chemically defined medium (Fig. 6A), indicating that the CvfA-involved regulatory system in the wild type downregulates the expression of speB by responding to the added peptides.
FIG. 6.
Effect of supplementing the medium with peptides or sugars on the expression of virulence genes regulated by CvfA. The wild-type (WT) strain (HSC5spc) and CvfA− mutant strain (ΩCvfA strain) were used. (A) Glucose influences the expression of emm, ska, and cfa, and peptides influence the expression of speB through CvfA. The amounts (Amt) of transcripts of the genes encoding M protein (emm), streptokinase (ska), CAMP factor (cfa), and streptococcal pyrogenic exotoxin B (speB) in the CvfA− mutant were determined by real-time RT-PCR and expressed relative to those of the wild type (CvfA−/WT). The cells were grown to the stationary phase (8 h postinoculation) in chemically defined medium containing glucose (0.05% or 1.0% [wt/vol]) as a carbohydrate source and/or dialyzed proteose peptone (1.0% [wt/vol]) as a supplemented peptide source. (B) Supplementation with fructose, galactose, or mannose gave results similar to the results of supplementation with glucose on the expression of the emm and speB genes regulated by CvfA. The amounts of transcripts of the genes encoding M protein (emm) and streptococcal pyrogenic exotoxin B (speB) in the CvfA− mutant were determined by real-time RT-PCR and expressed relative to those of the wild type (CvfA−/WT). The cells were grown to the stationary phase (8 h postinoculation) in chemically defined medium containing glucose (0.05% or 1.0% [wt/vol]), fructose (1.0% [wt/vol]), galactose (1.0% [wt/vol]), or mannose (1.0% [wt/vol]) as a carbohydrate source. Abbreviations: Glc, glucose; Gal, galactose; Fru, fructose; Man, mannose; dPP, dialyzed proteose peptone.
When the amount of glucose increased to 1.0% (wt/vol) in the chemically defined medium, glucose downregulated the expression of the emm, ska, and cfa virulence genes in the CvfA− mutant compared to the wild type, as in the glucose-supplemented C medium (Fig. 5B and 6A). However, the speB expression ratio was not changed by glucose supplementation (Fig. 6A).
We also tested the effects of other sugars on the expression of virulence genes in the CvfA− mutant and the wild type by examining the expression of the emm gene and the speB gene. When a monosaccharide other than glucose, such as fructose, galactose, or mannose, was added to the chemically defined medium, overall, the effects of these sugars on the expression of the emm and speB genes were similar to those of glucose (Fig. 6B). The addition of sugar to the medium made the ratio of emm transcript in the CvfA− mutant to that in the wild type smaller. The ratio was even smaller in media supplemented with galactose or mannose than in media supplemented with glucose or fructose. The addition of glucose, fructose, or galactose gave similar speB transcript ratios of the CvfA− mutant to the wild type, but adding mannose gave a smaller ratio than the other sugars. Mannose gave the largest effect among the sugars tested and showed the result that was most opposite the effect of peptide supplementation.
Taken together, these data suggest that the CvfA-involved regulatory system regulates the expression of two sets of virulence genes in different ways: the expression of a set of genes such as emm, ska, and cfa is affected by sugars (possibly carbon sources), and the expression of the other set of genes, including speB, is influenced more by peptides (possibly nitrogen sources). When sugar is depleted, the CvfA-involved regulatory system downregulates the expression of the emm, ska, and cfa genes. When peptides are depleted, the CvfA-involved regulatory system upregulates the expression of speB, but the levels of expression of the emm, ska, and cfa genes are not changed much. Thus, the CvfA-involved regulatory system seems to sense nutrients in the environment or sense cellular metabolic status to differentially regulate the expression of streptococcal virulence genes.
Overexpression of the transcriptional regulator RopB in the CvfA− mutant restored SpeB production to the wild-type level.
In S. pyogenes, transcription of speB is under the control of the transcriptional regulator RopB (30). To test whether CvfA influences SpeB production via the transcription regulator RopB, the CvfA− mutant was transformed with a plasmid that expresses high levels of a functional RopB protein that has been modified to include an influenza virus HA tag at its carboxy terminus (24). The CvfA− mutant in which RopB was overexpressed secreted the same level of SpeB protein as the wild type did (Fig. 7), indicating that CvfA influences SpeB expression via RopB.
FIG. 7.
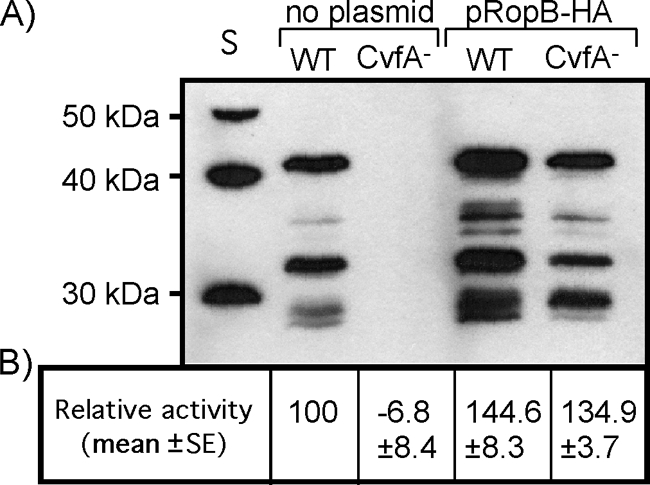
Overexpression of RopB restores SpeB expression in the CvfA− mutant. (A) Western blot analysis to detect SpeB in S. pyogenes strains containing a plasmid that overexpresses RopB (pRopB-HA) or in S. pyogenes strains without the plasmid. The wild-type (WT) S. pyogenes strain (HSC5spc) and CvfA− mutant strain (ΩCvfA strain) were used. Note that multiple proteolytic cleavages are required for activation of the 46-kDa SpeB zymogen to the 28-kDa active protease. S, protein size markers. (B) The SpeB cysteine protease activities in supernatants from 12-h cultures are shown relative to that of the wild-type strain.
The nutrient-dependent virulence gene regulation by the CvfA-involved regulatory system was not caused by the ppGpp-mediated stringent response.
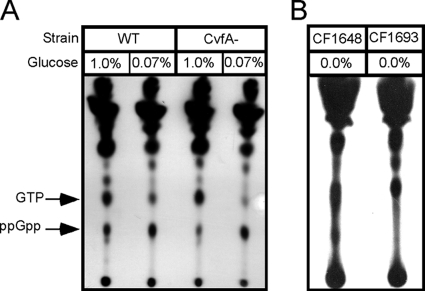
The nutrient-dependent virulence gene expression through CvfA was suggestive of the stringent response. The stringent response is triggered by the accumulation of guanosine nucleotides, mainly guanosine tetraphosphate (ppGpp) to conserve energy when microorganisms face nutrient starvation especially on glucose or amino acids. Also, CvfA contains an HD domain like the streptococcal RelA enzyme, a bifunctional ppGpp synthetase/hydrolase. Thus, we compared the patterns of ppGpp production in the CvfA− mutant and the wild type under glucose starvation conditions to investigate whether the phenotype of the CvfA− mutant is caused by the stringent response. However, the ppGpp accumulation patterns in the CvfA− mutant and the wild type under glucose starvation conditions were identical (Fig. 8), indicating that the ppGpp-mediated stringent response is not involved in the phenotype of the CvfA− mutant.
FIG. 8.
The ppGpp accumulation pattern of the CvfA− mutant under glucose starvation conditions is not different from that of the wild type. (A) ppGpp accumulation in S. pyogenes strains. The wild-type (WT) strain (HSC5spc) and CvfA− mutant strain (ΩCvfA strain) were used. The cells were labeled with [32P]H3PO4 in MOPS under glucose-rich (1.0%, wt/vol) or glucose-starved (0.07%) condition. Nucleotides were acid extracted, and 32Pi-labeled nucleotides were resolved by polyethyleneimine-coated TLC plates followed by autoradiography. (B) ppGpp accumulation in E. coli control strains under glucose-starved conditions. E. coli strains CF1648 (wild type) and CF1693 (RelA− SpoT−) were used.
CvfA interacts with enolase, an enzyme in the glycolysis pathway.
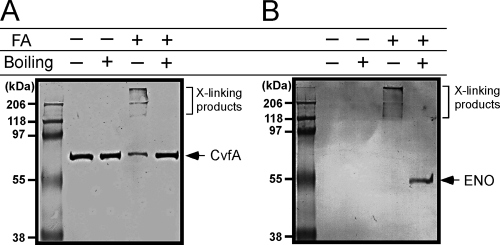
Recent studies indicated that the CvfA ortholog in B. subtilis has endoribonuclease activity and interacts with the RNA processing enzymes RNase J1 and polynucleotide phosphorylase and with the glycolytic enzymes phosphofructokinase and enolase (10, 35). Because the CvfA-involved regulation controls the expression of streptococcal genes according to nutritional status, we examined whether CvfA interacts with enolase (Spy_0731), one of the glycolytic enzymes interacting with the B. subtilis CvfA ortholog, in S. pyogenes. For the test, we tagged CvfA with the 6× His tag and enolase with the HA (hemagglutinin) tag at their C termini through homologous recombination. After we tagged CvfA with the 6× His tag, we analyzed the cysteine protease SpeB activity in culture supernatant to examine whether the tagged CvfA is functional, since the CvfA− ΩCvfA mutant strain showed prominent reduction in SpeB production (Fig. 2). The SpeB activity did not change after CvfA was tagged with 6× His (data not shown), indicating that the His-tagged CvfA was functional. Enolase in B. subtilis is essential even under conditions when no glucogenic activities are required (10). We could tag enolase in S. pyogenes with the HA tag, suggesting that the enolase tagged with the HA tag was functional. After both proteins were tagged, we performed protein cross-linking using formaldehyde, a cleavable and membrane-permeable cross-linker. CvfA is a membrane-anchored protein, so a detergent, ND-40, was used to purify CvfA, and this usage of a detergent made us employ a chemical cross-linking strategy for the protein interaction study. Cross-linking was performed with a 0.5% (wt/vol) formaldehyde solution for 20 min at room temperature. Then, CvfA tagged with the 6× His tag was purified with a His tag affinity resin, and the purified proteins were applied to Western blots to see whether enolase was copurified under this condition. The purified proteins were treated in a cross-linking cleavable or noncleavable condition before they were applied to Western blots. Protein cross-linking by formaldehyde can be cleaved by boiling samples for 20 min. In the noncleavable condition of formaldehyde cross-linking, three high-molecular-mass bands with a mass of more than 100 kDa were detected when anti-His tag antibody, which detects the His-tagged CvfA, was used. This indicates that CvfA (size, 60.3 kDa) is interacting with itself or other proteins inside cells (Fig. 9A). The three high-molecular-weight protein bands in the same sample disappeared in the cross-linking cleavable condition, indicating that the high-molecular-weight bands containing CvfA are products of formaldehyde cross-linking. When Western blotting was performed with the purified proteins in the noncleavable condition using anti-HA tag antibody, which detects the HA-tagged enolase, the same three high-molecular-weight protein bands were detected, and no enolase band was detected (Fig. 9B). In the cross-linking cleavable condition, only the protein band of enolase (47.4 kDa) was detected. These results indicate that there was no contamination by free enolase while His-tagged CvfA was purified, and all enolase in the purified sample was cross-linked with CvfA directly or indirectly. In conclusion, the interaction between CvfA and enolase was detected in S. pyogenes.
FIG. 9.
CvfA interacts with enolase. Western blotting against CvfA (A) or enolase (B) after in vivo cross-linking with formaldehyde (FA). The S. pyogenes strain that has both 6× His-tagged CvfA and HA-tagged enolase was treated with formaldehyde (+), and CvfA was purified with His tag affinity resin. The presence of CvfA and enolase in the sample was then determined through Western blotting using anti-His tag antibody (A) or anti-HA tag antibody (B). Enolase was detected when CvfA was purified, and this copurification indicates the interaction between CvfA and enolase. The cross-linking between proteins by formaldehyde can be cleaved by boiling samples (+). The positions of cross-linking products (X-linking products), CvfA, and enolase (ENO) are shown to the right of the blots.
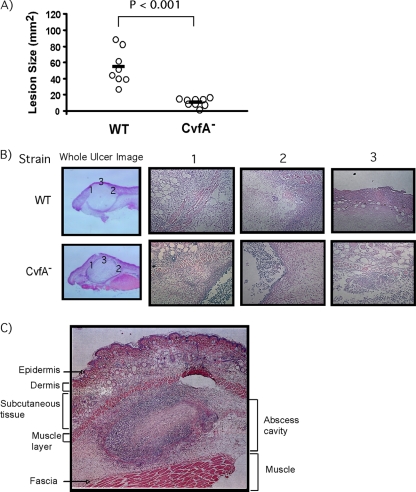
The CvfA− mutant is highly attenuated in a murine model of soft tissue infection.
To test whether cvfA disruption alters the virulence of S. pyogenes in a subcutaneous infection model, a dose (1 × 107 CFU) of the wild type or the CvfA− mutant was subcutaneously injected into SKH1 hairless mice. For the strain used in this study (HSC5), injection into the subcutaneous tissue of an SKH1 hairless mouse results in a local draining ulcer, whose area reaches the maximum value 3 days postinfection and which then goes on to heal over the next 10 to 14 days (5). For this study, 10 mice were injected with each S. pyogenes strain, and the sizes of the lesions were measured every day. The CvfA− mutant produced significant smaller lesions than the wild type did on day 3 postinjection (Fig. 10A). Histological examination of the lesions clearly showed that destruction of mouse tissue infected with the CvfA− mutant was much milder than that infected with the wild type (Fig. 10B), even though the sizes of abscess cavities caused by the injection of the wild type and the CvfA− mutant were the same. The subcutaneous tissue and the muscle layer right below the subcutaneous tissue (Fig. 10B1 and 3) showed less destruction by the CvfA− mutant infection than by the wild-type infection. Also, a clearer barrier between the ulcer and tissue was observed in the lesion caused by infection with the CvfA− mutant than in the lesion caused by infection with the wild-type strain (Fig. 10B2). Thus, cvfA disruption attenuated the virulence of S. pyogenes in a self-limited soft tissue infection model as well as in a lethal intraperitoneal systemic infection model (17).
FIG. 10.
The CvfA− mutant is highly attenuated. (A) The ability of the CvfA− mutant to cause disease in the murine subcutaneous infection model was determined. Virulence was evaluated on the basis of the area of the ulcer produced at the time when ulcer formation was maximal in the wild-type strain (3 days postinfection). Each symbol represents the area of the ulcer in an individual mouse infected with the wild type or the CvfA− mutant, and the solid bars indicate the mean values for the strains. The wild-type (WT) strain (HSC5spc) and CvfA− mutant strain (ΩCvfA strain) were used. The P value for the means determined by a Mann-Whitney U test statistic is indicated above the bracket. (B) Microscopic appearance of mouse ulcers caused by the wild type or the CvfA− mutant. The ulcers that developed were removed at 3 days postinfection, fixed and processed by standard methods, and stained with hematoxylin and eosin. Images were taken with a microscope equipped with digital photography. Three local images from each ulcer were taken, and each number above the images (1, 2, or 3) matches the number in the whole-ulcer image. (C) An example of microscopic appearance of the whole ulcer formed by S. pyogenes infection. Abscess cavity by S. pyogenes injection is formed between the muscle layer under the subcutaneous tissue and fascia.
DISCUSSION
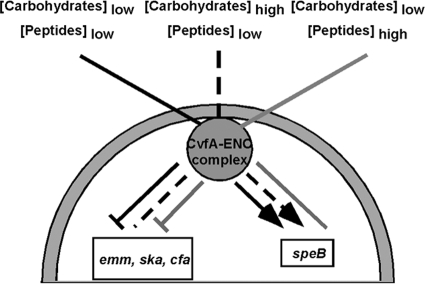
In this study, the effect of CvfA on the expression of S. pyogenes genes was examined under different conditions. Microarray analysis revealed that 29% of the total number of genes in the genome was significantly up- or downregulated by CvfA, indicating that CvfA extensively affects the expression of genes in S. pyogenes. The effect of CvfA on virulence gene expression was affected by growth phase and medium components. CvfA represses the expression of a set of virulence genes such as emm, ska, and cfa in medium containing minimal amounts of carbohydrates and activates speB gene expression in medium containing minimal amounts of peptides (Fig. 11). Thus, CvfA appears to respond to carbon and nitrogen sources to regulate the expression of S. pyogenes virulence genes.
FIG. 11.
Schematic representation of the role of the CvfA-enolase complex in the regulation of virulence gene expression in S. pyogenes. The large gray half circle represents an S. pyogenes cell, and a CvfA-enolase complex is attached to the membrane inside the cell. The lines from outside of the cell represent the status of the nutritional condition affecting CvfA-involved regulation (concentrations of carbohydrates or peptides), and the lines inside the cell indicate the effect of the CvfA-involved regulatory system on the expression of virulence genes. Each type of line (solid black, broken line, and solid gray) outside and inside the cell match each other. For the lines inside the cell, arrowheads indicate activation, flat ends reflect repression, and plain lines with no arrowhead or flat end indicate no effect by the CvfA-enolase complex. Overall, the CvfA-involved regulatory system represses the expression of emm, ska, and cfa in low-carbohydrate conditions and activates the expression of speB in low-peptide conditions.
The effect of CvfA on the expression of many virulence genes in S. pyogenes was altered depending on the cell growth phase (Fig. 3). Previously, it has been shown that the expression of most virulence factors of S. pyogenes is influenced by growth phase (21, 41). Growth phase status may be sensed as a result of the change of environment by metabolic by-products or nutrient consumption (36, 37), as cell cycle status (33), or as population densities (45). The growth-phase dependence of gene regulation by CvfA could be due to the lack of carbon or nitrogen sources at the stationary phase because the expression of virulence genes was also influenced by CvfA when carbohydrates or peptides are scarce.
Even though the fact that the CvfA-involved regulation of virulence gene expression is influenced by nutritional stress and cell growth phase implies the involvement of the ppGpp-mediated stringent response, this study showed that the regulation by the CvfA-involved regulatory system is not involved in the stringent response, because the pattern of ppGpp production in the CvfA− mutant and the wild type under glucose starvation conditions did not change (Fig. 8). Also, CcpA, the major carbon catabolite protein, seems not to be involved in the CvfA-involved regulatory system. The transcriptome analysis showed that the transcript amount of the ccpA gene in the CvfA− mutant was the same as that in the wild type, indicating that the CvfA-mediated regulatory system does not affect the expression of ccpA. Also, the expression of SpeB in a CcpA− mutant was almost the same as that in the wild type in C medium (18), but SpeB expression in the CvfA− mutant was negligible. Thus, the CvfA-mediated regulation of virulence gene expression in S. pyogenes seems to be a novel way to control gene expression under nutrient-deficient conditions. YmdA, the CvfA ortholog in B. subtilis, which shows 56% identity and 76% sequence similarity to S. pyogenes CvfA in amino acid level, is copurified with the glycolytic enzymes enolase and phosphofructokinase (PFKase) (10). We also verified in this study that CvfA interacts with enolase in S. pyogenes. Thus, the interactions between CvfA and the glycolytic enzymes might play a role in the regulation of gene expression according to the nutritional condition of cells by transmitting a signal from the glycolysis pathway to CvfA.
YmdA, the CvfA ortholog in B. subtilis, is responsible for the endonucleolytic processing of the gapA operon mRNA, which encodes six genes in the glycolytic pathway (cggR, gapA, pgk, tpiA, pgm, and eno) (10). YmdA cleaves the gapA operon between cggR and gapA. S. pyogenes, however, does not have the gapA operon. The glycolytic enzyme genes in the B. subtilis gapA operon are scattered throughout the genome of S. pyogenes. In our transcriptome analyses, however, the transcripts of many glycolytic enzymes are reduced by CvfA at the stationary phase: glucose-6-phosphate isomerase (Spy0215, 6.9-fold reduction), 6-phosphofructokinase (Spy1283, 2.3-fold reduction), enolase (Spy0731, 2.7-fold reduction), pyruvate kinase (Spy1282, 2.5-fold reduction), and l-lactate dehydrogenase (Spy1151, 2.2-fold reduction). Thus, this reduction might indicate that even though S. pyogenes does not have the gapA operon, CvfA in S. pyogenes still affects the glycolysis pathway by regulating the expression of glycolytic enzymes, and the CvfA-interacting enolase, a key glycolytic enzyme, might be involved in the regulation of expression of glycolytic enzymes.
The virulence of the CvfA− mutant was highly attenuated in the model of soft tissue infection, indicating that the expression of virulence genes in the CvfA− mutant was misregulated in soft tissue. Virulence gene expression in the CvfA− mutant was highly misregulated in stationary-phase cultures grown on C medium, but not in exponential-phase cultures grown on C medium or in cultures grown on THY medium, so it seems that the environment of stationary-phase culture in C medium is similar to the soft tissue environment. Loughman and Caparon employed the same soft tissue infection model and compared the gene expression of several S. pyogenes virulence factors in the tissue environment against the expression in different media and growth phases, and the gene expression profile of stationary-phase cultures grown on C medium was most similar to the profile in the tissue environment (21).
Our transcriptome analyses showed that in nutrient-limited conditions, CvfA downregulated the expression of most of the genes encoding cellular components in the categories of transport and metabolism and the genes in the category of energy production and conversion but upregulated the expression of some virulence factors, such as SpeB (protease), streptolysin S (hemolysin), and mitogenic factor (DNase). These virulence factors are able to destroy cells (streptolysin S) or degrade host cellular components (SpeB against proteins and mitogenic factor against nucleic acids). These data suggest that in nutritionally unfavorable conditions, the CvfA-involved regulatory system in S. pyogenes, a multiple-auxotrophic human pathogen, not only decreases cellular metabolism but also increases the expression of some virulence factors to obtain essential cellular components such as amino acids and nucleotides.
It has been shown that the CvfA orthologs in Bacillus subtilis and Staphylococcus aureus have endoribonuclease activity (29, 35). CvfA orthologs contain two functional domains, a KH domain, which has RNA binding activity (42), and an HD domain known to have metal-dependent phosphohydrolase activity (2). The HD domain in the S. aureus CvfA protein has manganese ion (Mn2+)-dependent phosphodiesterase activity, which is able to cleave phosphodiester bonds in nucleic acids and 2′,3′-cyclic nucleotides (29). KH domains consist of approximately 70 amino acids and bind RNA or single-stranded DNA (ssDNA). Proteins containing KH domains are found in archaea, bacteria, and eukaryota and involved in a myriad of different biological processes, including splicing, transcriptional regulation, and translational control (42). KH domains are classified into two types. Type I domains are found in multiple copies in eukaryotic proteins, whereas type II KH domains, like the one in CvfA, are typically found in a single copy in prokaryotic proteins. Unlike other RNA recognition motifs, which recognize diverse RNA lengths, the typical binding surface of KH domains is a cleft that can typically accommodate only four consecutive unpaired bases, so a KH domain usually has broad substrate specificity. Consistent with this notion, the depletion of the CvfA ortholog in B. subtilis increases the half-life of bulk mRNA more than 2-fold (35). However, even though CvfA is an endoribonuclease with broad substrate specificity, the specificity of the KH domain in CvfA does not seem to be totally nonspecific and bacteria seem to use the CvfA activity for specific cellular events. For example, the B. subtilis CvfA ortholog is involved in the processing of RNA of the glycolytic gapA operon, which cleaves between the cggR and gapA transcripts in order to separate their levels of expression (10). Also, the ortholog initiates the degradation of all 11 SAM-dependent riboswitches only when the transcripts containing the riboswitch are prematurely terminated by binding to SAM, but the riboswitches in antiterminated full-length mRNAs are not cleaved (35). This differential specificity indicates that the base-pairing status of the four bases recognized by CvfA can contribute to the substrate specificity of CvfA.
The E. coli RNA degradosome is composed of RNase E endoribonuclease, PNPase exoribonuclease, RhlB RNA helicase, and enolase glycolytic enzyme (for a review, see reference 7). B. subtilis CvfA also interacts with glycolytic enzymes enolase and PFKase and nucleases PNPase and RNase J1 (10). These interactions suggest that CvfA could be a member of the RNA degradosome in Firmicutes (10). Members of the phylum Firmicutes do not have an RNase E homolog, which is the major endoribonuclease in the E. coli RNA degradosome, so CvfA might be a functional homolog of E. coli RNase E (10). The role of enolase in the RNA degradosome is not known, but it has been speculated that it is a sensor for the cellular nutritional status (7), so the presence of enolase might explain the dependence of the activity of the CvfA-involved regulatory system on cellular nutritional status. Like CvfA, S. pyogenes PNPase is also involved in the regulation of expression of some virulence genes, indicating that RNA decay by RNases is involved in the regulation of virulence gene expression. S. pyogenes PNPase is a major factor to control the decay of the transcripts of the streptolysin S gene (sagA) and the streptodornase gene (sda) (3). The stability of those mRNAs was greater in a PnpA− mutant, while the stability of mRNAs of other virulence genes, such as the capsule synthetase gene (hasA) and the streptolysin O gene (slo), was not affected.
In the microarray analysis, 14% of the total number of genes in the genome were significantly upregulated (219 genes), and 15% were significantly downregulated (228 genes) in the CvfA− mutant compared to the wild type. A simple explanation for overexpressed transcripts in the CvfA− mutant is that CvfA degrades the RNA or their transcriptional activator mRNA. In the case of the transcripts whose amount was decreased in the CvfA− mutant, CvfA might degrade (i) the mRNA of the transcripts’ direct repressors, (ii) the mRNA of repressors of the transcripts’ transcription activators, or (iii) the mRNA of inhibitors to inhibit the transcripts’ activator activity. The transcription of SpeB, whose transcript was downregulated in the CvfA− mutant, is activated by the transcriptional regulator RopB. When we introduced the ropB gene into the CvfA− mutant using a multicopy plasmid, the introduction of ropB rescued the production of SpeB (Fig. 7), indicating that there was no or much less RopB available for the activation of speB transcription in the CvfA− mutant compared to that in the wild type. This result suggests that the mRNA of a transcriptional repressor or of a protein inhibitor binding to RopB might be less degraded in the CvfA− mutant. At this time, no transcriptional repressor of RopB is known, but a protein inhibitor of RopB exists. LacD.1, an aldolase enzyme that has been adapted to function exclusively as a regulatory component (22, 23), is known to inhibit speB transcription activation by RopB through direct binding to RopB (22).
Supplementary Material
Acknowledgments
We thank Doug Fix, John Martinko, and David Clark at Southern Illinois University for helpful discussions or critical reading of the manuscript. We gratefully acknowledge Michael Cashel at NIH for kindly providing E. coli strains CF1648 and CF1693 and Wess Warren, Seth Crosby, and Michael Heinz at the microarray core facility of Washington University for technical assistance on the microarray analysis.
Editor: A. Camilli
Footnotes
Published ahead of print on 12 April 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2001. A natural classification of ribonucleases. Methods Enzymol. 341:3-28. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, T. C., J. V. Bugrysheva, and J. R. Scott. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J. Bacteriol. 189:1866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221-234. [DOI] [PubMed] [Google Scholar]
- 5.Brenot, A., K. Y. King, B. Janowiak, O. Griffith, and M. G. Caparon. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 7.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61:71-87. [DOI] [PubMed] [Google Scholar]
- 8.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 9.Cho, K. H., and M. G. Caparon. 2008. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect. Immun. 76:3176-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Commichau, F. M., F. M. Rothe, C. Herzberg, E. Wagner, D. Hellwig, M. Lehnik-Habrink, E. Hammer, U. Volker, and J. Stulke. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell Proteomics 8:1350-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, B., and R. K. Bhadra. 2008. Molecular characterization of Vibrio cholerae ΔrelA ΔspoT double mutants. Arch. Microbiol. 189:227-238. [DOI] [PubMed] [Google Scholar]
- 12.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gase, K., J. J. Ferretti, C. Primeaux, and W. M. McShan. 1999. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 67:4725-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U. S. A. 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt, A., J. P. Rawlins, H. B. Thomaides, and J. Errington. 2006. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152:2895-2907. [DOI] [PubMed] [Google Scholar]
- 17.Kaito, C., K. Kurokawa, Y. Matsumoto, Y. Terao, S. Kawabata, S. Hamada, and K. Sekimizu. 2005. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56:934-944. [DOI] [PubMed] [Google Scholar]
- 18.Kietzman, C. C., and M. G. Caparon. 2010. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 20.Lederberg, E. M., and S. N. Cohen. 1974. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J. Bacteriol. 119:1072-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loughman, J. A., and M. Caparon. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loughman, J. A., and M. G. Caparon. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughman, J. A., and M. G. Caparon. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269-280. [DOI] [PubMed] [Google Scholar]
- 24.Loughman, J. A., and M. G. Caparon. 2007. Contribution of invariant residues to the function of Rgg family transcription regulators. J. Bacteriol. 189:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 27.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236-242. [DOI] [PubMed] [Google Scholar]
- 28.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata, M., C. Kaito, and K. Sekimizu. 2008. Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem. 283:2176-2184. [DOI] [PubMed] [Google Scholar]
- 30.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, D., R. Schuch, P. Chahales, S. Zhu, and V. A. Fischetti. 2006. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U. S. A. 103:10765-10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 33.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 34.Scott, J. R. 1974. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology 62:344-349. [DOI] [PubMed] [Google Scholar]
- 35.Shahbabian, K., A. Jamalli, L. Zig, and H. Putzer. 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28:3523-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 37.Steiner, K., and H. Malke. 2001. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tewodros, W., M. Norgren, and G. Kronvall. 1995. Streptokinase activity among group A streptococci in relation to streptokinase genotype, plasminogen binding, and disease manifestations. Microb. Pathog. 18:53-65. [DOI] [PubMed] [Google Scholar]
- 40.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unnikrishnan, M., J. Cohen, and S. Sriskandan. 1999. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect. Immun. 67:5495-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valverde, R., L. Edwards, and L. Regan. 2008. Structure and function of KH domains. FEBS J. 275:2712-2726. [DOI] [PubMed] [Google Scholar]
- 43.van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U. S. A. 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.