Abstract
Upon microbial challenge, organs at various anatomic sites of the body employ different innate immune mechanisms to defend against potential infections. Accordingly, microbial pathogens evolved to subvert these organ-specific host immune mechanisms to survive and grow in infected organs. Francisella tularensis is a bacterium capable of infecting multiple organs and thus encounters a myriad of organ-specific defense mechanisms. This suggests that F. tularensis may possess specific factors that aid in evasion of these innate immune defenses. We carried out a microarray-based, negative-selection screen in an intranasal model of Francisella novicida infection to identify Francisella genes that contribute to bacterial growth specifically in the lungs of mice. Genes in the bacterial tryptophan biosynthetic pathway were identified as being important for F. novicida growth specifically in the lungs. In addition, a host tryptophan-catabolizing enzyme, indoleamine 2,3-dioxygenase 1 (IDO1), is induced specifically in the lungs of mice infected with F. novicida or Streptococcus pneumoniae. Furthermore, the attenuation of F. novicida tryptophan mutant bacteria was rescued in the lungs of IDO1−/− mice. IDO1 is a lung-specific innate immune mechanism that controls pulmonary Francisella infections.
Organs at different anatomic sites of the body have different physiological functions and are exposed to vastly different microbial and environmental challenges on a daily basis (32). As a result, depending on its relative sterility, each organ senses impending danger of infection differently and employs unique innate immune responses in an organ-specific homeostatic manner to effectively fight off any potential infection without compromising organ physiology and function. For instance, a sterile organ like the spleen is likely to induce a strong proinflammatory and bactericidal defense to maintain organ sterility, while such a defense would not be induced in a nonsterile organ like the colon. Instead, innate immune responses in the colon support a peaceful coexistence with the gut microflora rather than achieving sterility. To gain a better understanding of tissue-specific innate immune defense mechanisms, we have used Francisella tularensis as a model bacterial pathogen that is capable of entering and surviving in many different host tissues and organs.
Francisella tularensis is a facultative, intracellular Gram-negative bacterium that causes the highly debilitating zoonotic disease tularemia (28). There are currently 4 known subspecies of F. tularensis that cause disease of different severities. The most virulent subspecies of Francisella is F. tularensis subsp. tularensis, which is found predominantly in North America and has an infectious dose of less than 10 CFU in humans (33, 34, 37). It is also associated with lethal pulmonary infections. Francisella novicida is also found primarily in North America but rarely causes disease in immunocompetent individuals (16). However, F. novicida shares the same families of virulence genes as F. tularensis, causes a similar disease in mice (27), and is more genetically amenable than F. tularensis, thus making F. novicida infection of mice a good experimental model for the study of Francisella pathogenesis.
Francisella infects mammalian hosts via extremely diverse routes of entry, colonizing and replicating in different organs. Tularemia occurs in several forms, depending on the initial route of infection. The most common form of tularemia is ulceroglandular tularemia, which occurs when the bacterium enters the skin subcutaneously (9). Other infection routes include inoculation via the conjunctiva, which leads to oculoglandular tularemia (9, 38), and the ingestion of contaminated food and water, which leads to oropharyngeal or gastrointestinal tularemia (9). The most acute and fatal form of tularemia is pneumonic tularemia, which is caused by inhalation of the bacterium into the lungs. Pneumonic tularemia can also occur as a result of complications from the above forms of tularemia (9, 12), when the bacteria spread systemically from the initial peripheral site of infection to the lungs.
From the perspective of the fitness of a microbe, it is advantageous for a microbe to infect its host via multiple entry routes and infect multiple organs. Thus, it is plausible that F. tularensis, a microbe that infects a range of different organs, has to deal with a diverse repertoire of different innate immune responses launched in these various organs during infection. We utilized a genome-wide genetic screen in F. novicida as a tool to identify tissue-specific interactions between the host and pathogen.
Genetic screens are very effective tools that have been successfully used for the large-scale identification of virulence factors of many bacterial pathogens in vivo (6, 19, 44). Here, we utilize a microarray-based, negative-selection technique called transposon site hybridization (TraSH) (44) in an intranasal (i.n.) model of Francisella infection to identify Francisella factors important for bacterial growth and/or survival in the lungs. The screen identified almost all of the known Francisella virulence genes, including the Francisella pathogenicity island (FPI) genes. We also identified novel genes not previously known to be important for bacterial growth and/or survival in the lungs.
By comparing the genes that are important for Francisella growth and/or survival in the lungs with the genes identified previously for spleen colonization (44), we identified the Francisella tryptophan biosynthetic pathway as being important for bacterial survival and/or growth specifically in the lungs. We demonstrated that the host enzyme that is responsible for catalyzing the first rate-limiting step of tryptophan degradation, indoleamine 2,3-dioxygenase 1 (IDO1), is induced specifically in the lungs during Francisella infection, suggesting that the host innate immune response to F. novicida restricts the availability of this essential amino acid. Indeed, the F. novicida tryptophan mutant bacteria survived better in the lungs of mice that lack IDO1 compared to wild-type mice. These findings suggest that IDO1 acts as an organ-specific, host innate immune mechanism that defends against a highly virulent bacterial pathogen.
MATERIALS AND METHODS
Bacteria.
Wild-type F. novicida strain U112 was described previously (12). All bacterial strains were grown overnight with aeration in either modified tryptic soy broth (TSB; Difco/BD, Sparks, MD) and supplemented with 0.2% l-cysteine or in minimal Chamberlain's defined medium (CDM) (5).
Transposon mutant library.
We utilized a Tn5-based transposon mutant library in which each gene in the Francisella genome has been mutated via the insertion of a kanamycin resistance cassette with outward-facing T7 promoters at each end, as previously described (44). We also obtained a fully sequenced Francisella novicida transposon mutant library from the Biodefense and Emerging Infections Research resources repository, where each gene in the Francisella genome has been mutated via the insertion of a kanamycin resistance cassette.
Mice and Francisella infections.
Female C57BL/6J or IDO1−/− mice (The Jackson Laboratory, Bar Harbor, ME) between 5 and 7 weeks of age were kept under specific-pathogen-free conditions in filter-top cages at the animal facility at Stanford University and provided with sterile food and water ad libitum. All experimental studies were carried out in accordance with the Institutional Animal Care and Use Committee guidelines. Mice were inoculated with the indicated bacterial dosage via the appropriate route of infection. For intranasal infection with the transposon mutant library, mice were inoculated with 5 × 106 CFU in 30 μl of sterile phosphate-buffered saline (PBS). For the intranasal single and competition infections, mice were inoculated with 1 × 103 CFU or 3 × 104 CFU, respectively, in 30 μl of sterile PBS. For intraperitoneal (i.p.) and intradermal (i.d.) infections, mice were inoculated with either 1 × 104 CFU in 200 μl of sterile PBS or 1 × 105 CFU in 100 μl of sterile PBS, respectively. The indicated organs were harvested at the specified time point postinfection and homogenized, and dilutions were plated on supplemented Mueller-Hinton (MH) agar plates containing kanamycin (infections with library) or MH plates with or without the appropriate antibiotic (single infections and competition experiments). Plates were grown overnight at 37°C with 5% CO2, and colonies were enumerated. After infections with the library, ∼105 CFU were collected in sterile PBS for isolation of DNA. Competitive index (CI) values were calculated by using the formula CI = (mutant CFU in output/wild-type CFU in output)/(mutant CFU in input/wild-type CFU in input).
Microarrays.
Genomic DNA was purified from bacterial pellets using the DNeasy blood and tissue kit (Qiagen). Each DNA sample was divided in 2 and digested separately with BfaI and RsaI (NEB, Ipswich, MA). The digested DNA was used as the template for in vitro transcription with the AmpliScribe T7-Flash transcription kit (Epicentre) following the manufacturer's protocol, except that 2 μg of digested DNA was used, and the reaction was allowed to proceed for 12 to 16 h. Purified RNA was used in a reverse transcription (RT) reaction using SuperScript II (−) (Invitrogen, Carlsbad, CA) and random hexamers as primers. cDNA was labeled with amino-allyl dUTP by using the Klenow (exo-) enzyme (NEB, Ipswich, MA). The single-stranded DNAs (ssDNA) containing amino-allyl dUTP from the mouse output or the library input pools were labeled with Cy5 and Cy3, respectively, before hybridization to our Francisella microarray as described previously (44). All raw data sets are freely available for download from the GEO database.
Data analysis.
Normalized data were downloaded from the Stanford Microarray Database according to the median log2 Cy5/Cy3 ratio (logRAT2N). Filters for feature quality, including a Cy3 net median intensity of ≥150 and regression correlation of >0.6, were applied. To compare data from separate intranasal infection experiments, each experimental sample was zero transformed to the input/input control. Features (spots) missing values for ≥30% of the arrays were removed from the data set. The data sets were analyzed with the SAM (Significance Analysis of Microarray) program, by using the two-class analysis option to identify features that consistently deviated from the input and samples across all arrays with a false discovery rate of 1.1%.
Targeted mutagenesis.
Mutant and complemented strains were constructed as described by using the primers listed in Table S2 in the supplemental material. Primers (Table S2) were used to amplify sequences flanking genes of interest (primers F1/Inv1 and Inv2/R1) as well as a kanamycin resistance cassette (primers Kan-F/R). The PCR products were then put together using a sewing-PCR approach (primers F1/R1). Resulting constructs were chemically transformed into U112, and Kan-resistant colonies representing strains with double-crossover events were selected. Integration of the Kan cassette into the correct region of the genome was confirmed by sequencing using the Check F/R primers (Table S2). For the FPI mutant, PCR analysis was performed to verify that the strain did not contain DNA carrying portions of the iglA, iglC, cds1, cds2, pdpB, and pdpD genes as previously described (44). Constructs for complemented strains were engineered similarly, but with a chloramphenicol cassette, and the resulting constructs were chemically transformed into the corresponding mutant strain of interest. Allelic replacement was confirmed by chloramphenicol resistance and Kan sensitivity as well as by sequencing with the Check F/R primers.
Macrophage infections.
For replication experiments, 2.5 × 105 macrophages were seeded per well in 24-well plates and allowed to adhere overnight at 37°C with 5% CO2. The following day, bacteria were added to the cells at a multiplicity of infection (MOI) of 10:1, and plates were centrifuged at 19,000 rpm for 15 min at room temperature to spin the bacteria onto the cells and synchronize the infection. After infection, macrophages were lysed with 1% saponin (Sigma, St. Louis, MO) at 30 min (to measure bacterial entry) and at 8 h (to measure bacterial replication) and plated on MH agar plates for CFU enumeration.
RNA isolation, RT-PCR, and qRT-PCR analysis.
To isolate RNA, 1 ml of Trizol reagent (Invitrogen) was added to 30 mg of the various mouse organs or to 1 × 1010 bacterial cells. RNA was isolated using the RNeasy minikit (Qiagen). Reverse transcription (RT)-PCR was performed using the Superscript II reverse transcriptase enzyme (Invitrogen) and Phusion hot start high-fidelity DNA polymerase (Finnzymes). Quantitative real-time RT-PCR (qRT-PCR) was performed on a real-time detection system (iCycler; Bio-Rad Laboratories). For the IDO1 and tryptophan 2,3-dioxygenase (TDO) genes, rTth enzyme (Applied Biosystems), SYBR green, and the respective primers (see Table S2 in the supplemental material) were used. While for the IDO2 gene, the mRNA was reverse transcribed into cDNA using Superscript III reverse transcriptase (Invitrogen) before quantitative real-time PCR was performed on the iCycler using Platinum SYBR green qPCR Supermix-UDG (Invitrogen) and primers listed in Table S2. All gene-specific transcripts were normalized to the amount of β-actin mRNA to obtain relative quantities of message.
Growth curves.
Overnight bacterial cultures were subcultured the following day in the indicated media at an optical density of 0.04. At each hour postsubculturing, the optical density of the bacterial culture was determined by measuring the absorbance at 600 nm.
Statistical analysis.
All statistical analyses were performed using either the Student's t test or analysis of variance (ANOVA) with a Dunnett's posttest as indicated. For competition experiments, log10 values of the CI were compared with the corresponding values for the htpX control mutant.
RESULTS
Genome-wide in vivo negative-selection screens identify genes required for F. novicida survival and/or growth in the lungs.
We used a comprehensive F. novicida transposon insertion library, made up of 12,600 F. novicida mutants as previously described (44), to screen for genes important for Francisella growth and/or survival in the lungs of mice after intranasal infection. Two separate experiments were performed in which groups of 5 mice were infected via the intranasal route with the transposon insertion library. At 48 h postinfection, the bacteria surviving in the lungs were pooled and compared with bacteria in the input sample taken at the time of infection. A rigorous statistical program, Significance Analysis of Microarrays (SAM), was used to analyze the microarray data. We identified transposon insertion mutants representing 82 genes as present in the input pool but absent from the lungs of infected mice in both experiments (see Table S1 in the supplemental material). These genes are referred to as negatively selected genes.
The negative-selection technique we used here identified all of the genes within the FPI, metabolic genes involved in amino acid transport and purine/biotin synthesis, and genes involved in the stress response (htpG), as well as genes involved in capsule biosynthesis and lipopolysaccharide O antigen biogenesis (Fig. 1; see Table S1 in the supplemental material). We had expected these genes to be negatively selected in vivo since they code for gene functions that are known to be required for Francisella growth and survival in vivo (3, 26), thus validating the robustness and sensitivity of this screen for identifying negatively selected genes in the lungs.
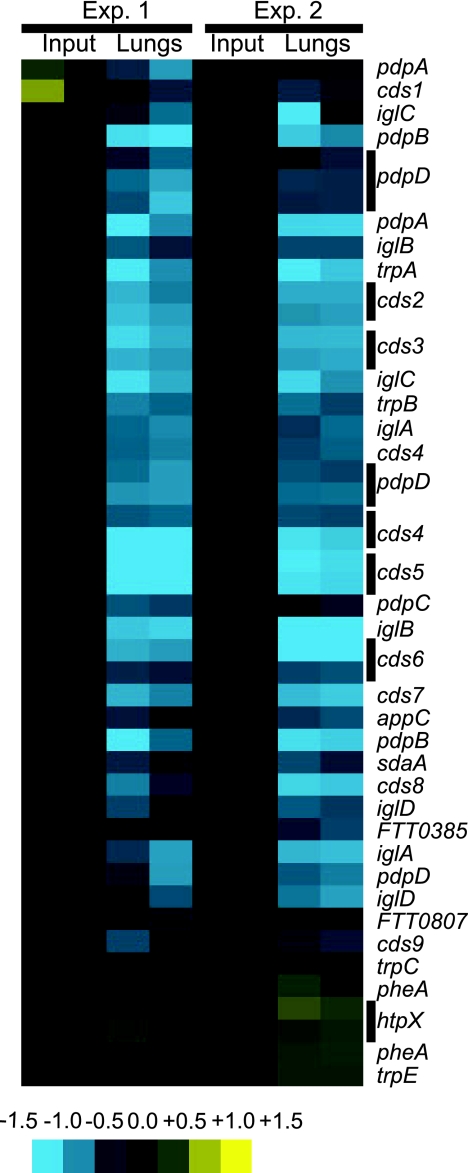
FIG. 1.
Array data for genes targeted in this study. Bacteria surviving in lung samples at 48 h postinfection from two groups of 5 mice each were collected. The transposon mutants present in each sample were identified by microarray analysis. Data for a subset of the negatively selected transposon insertion mutants is shown. Inputs are shown as controls. Yellow represents overabundance of signal compared to the input, and blue represents absence relative to the input. Multiple rows for a given gene represent multiple spots on the arrays. Data are representative of two independent experiments (Exp.).
We next compared our list of negatively selected genes identified in the lungs to a previously published list of genes that were identified as being important for F. novicida growth and/or survival in the spleen after an intraperitoneal (i.p.) route of infection (44). Among the 82 negatively selected genes in the lungs, there were 16 novel genes that were previously not identified in the screen for genes required for F. novicida survival in the spleen.
In vivo validation of negatively selected genes identified in the lungs.
We used SAM to determine which of the 16 novel genes were most significantly negatively selected and chose 6 of these to validate in vivo. We performed competition experiments in which mice were infected intranasally (i.n.) with a 1:1 mixture of the wild-type U112 strain and one of the mutant strains (Fig. 2). In this secondary screen, we used mutant strains from the fully sequenced Francisella transposon insertion mutant library obtained from the Biodefense and Emerging Infections Research resources repository (Manassas, VA), with the exception of the Francisella pathogenicity island deletion mutant strain (ΔFPI). Forty-eight hours after infection, lungs and spleens were harvested and the number of wild-type and mutant bacteria in each sample was enumerated by plating on MH agar plates with or without the appropriate antibiotic.
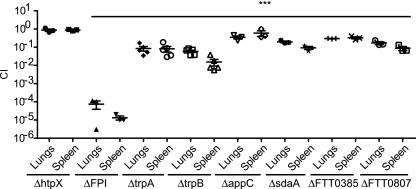
FIG. 2.
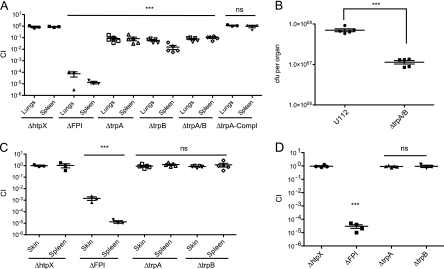
Mutants with mutations in negatively selected genes are attenuated in vivo and validate the results of our TraSH screen. Groups of 3 to 5 mice were intranasally infected with a 1:1 mixture of wild-type bacteria and the indicated bacterial mutant strain. The data represent the CI value for the CFU of mutant/wild-type bacteria in the lungs and spleens at 48 h after intranasal infection. Bars represent the geometric mean CI value for each group of mice. Data are representative of two independent experiments. Statistical significance is shown based on the ANOVA with a Dunnett's posttest compared to the corresponding CI values of the htpX mutant: ***, P < 0.05.
The ΔFPI strain was highly attenuated in the lungs and spleens of mice (Fig. 2) in our i.n. model of infection, similar to previously published data showing high levels of attenuation of the ΔFPI strain in the spleen following i.p. infection (44). In contrast, a mutant with a mutation in a heat shock protein gene that was not negatively selected, ΔhtpX, colonized the lungs and spleens similarly to wild-type bacteria. The FTT0092c (cytochrome oxidase bd-II, subunit I, pseudogene; appC), FTT0577 (l-serine dehydratase 1; sdaA), FTT0385 (hypothetical protein), and FTT0807 (conserved hypothetical membrane protein) mutant bacterial strains exhibited mild to moderate attenuation (∼2- to 15-fold) (Fig. 2). In contrast, the FTT1772c (trpA) and FTT1773c (trpB) mutants were strongly attenuated (∼10- to 100-fold) (Fig. 2). These results are in accordance with our microarray screen data and validate the utility of this genetic screen in the identification of genes that are negatively selected in the intranasal route of infection.
We decided to focus on the mutants (trpA and trpB mutations) with the most severe attenuation in the lungs for further characterization. Furthermore, these genes code for gene products that are involved in the same bacterial metabolic pathway, tryptophan biosynthesis.
F. novicida tryptophan auxotroph mutants do not grow in minimal medium.
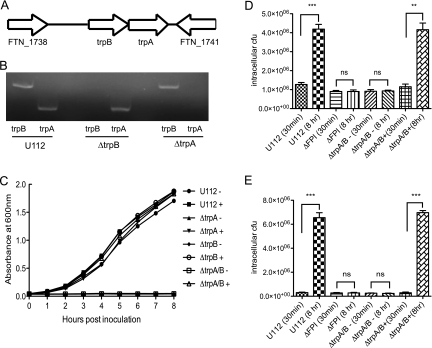
trpA and trpB encode the tryptophan synthase alpha and beta chains, respectively. These two chains make up the tetrameric α2β2 tryptophan synthase enzyme complex that catalyzes the final steps in tryptophan biosynthesis. The α subunits convert indole-3-glycerolphosphate to indole and glyceraldehyde 3-phosphate, while the β subunits catalyze the condensation of indole and serine to tryptophan (8, 25, 30). The trpA and trpB genes are positioned next to each other in the Francisella genome (Fig. 3A). The flanking genes FTN1738 and FTN1741 code for a putative metallocarboxypeptidase and hypothetical protein, respectively, and are not predicted to be involved in tryptophan biosynthesis (Fig. 3A). To test the role of tryptophan synthase in F. novicida growth and/or survival in vitro and in vivo, we made F. novicida mutant strains that lack either FTT1772c (ΔtrpA) or FTT1773c (ΔtrpB). We also constructed an F. novicida mutant (ΔtrpA/B) strain that lacks both the alpha and beta chains of the tryptophan synthase. The deletion of the targeted genes were confirmed by PCR and sequencing of the PCR product (data not shown). In addition, the ΔtrpA and ΔtrpB mutants are found not to have polar effects on each other's expression, as demonstrated by RT-PCR analysis (Fig. 3B). The targeted deletion mutants grew with similar kinetics to the wild-type U112 strain in rich broth (data not shown). To determine if tryptophan is an amino acid essential for Francisella growth, we tested the ability of the wild-type U112 and ΔtrpA, ΔtrpB, and ΔtrpA/B mutant bacterial strains to grow in Chamberlain's defined medium (CDM), a minimal growth medium that lacks tryptophan. The wild-type U112 strain grew in CDM with kinetics that are similar to previously published studies (Fig. 3C) (5, 40). In contrast, the F. novicida mutants that lack either one or both of the tryptophan synthase chains fail to grow (Fig. 3C). Not only does the addition of exogenous tryptophan to the medium restore the ability of the mutant strains to grow in minimal media, it also allows the wild-type U112 strain to replicate with slightly faster kinetics compared to those in the absence of tryptophan (Fig. 3C).
FIG. 3.
F. novicida requires tryptophan for growth in minimal medium and intracellular replication. (A) Diagram illustrating the genomic positions of trpA and trpB within the F. novicida genome. (B) RT-PCR analysis of the gene expression of the trpA and trpB genes in the indicated bacterial strains. (C) The indicated bacterial strains were inoculated in Chamberlain's defined medium in the absence (−) or presence (+) of exogenously added tryptophan (2 g/liter) and the optical density of the bacterial culture at 600 nm was determined at each hour postinoculation. Bone marrow-derived macrophages (D) or RAW macrophages (E) were infected with the indicated bacterial strains at a multiplicity of infection (MOI) of 10 in tryptophan-free Dulbecco's modified Eagle's medium (DMEM) supplemented with 1 mg/ml of bovine serum albumin (BSA) either in the absence (−) or presence (+) of exogenously added tryptophan (2 g/liter). Cells were lysed, and bacteria were collected at 30 min and 8 h after infection and plated for enumeration. Data are representative of two independent experiments. Statistical significance is shown based on the t test as compared to bacterial counts at 30 min postinfection: ***, P < 0.0005; **, P < 0.005; ns, not statistically significant.
F. novicida tryptophan auxotrophs do not replicate within macrophages.
Macrophages are one of the primary intracellular niches for Francisella survival in the host (28). To determine if the absence of tryptophan would affect the ability of F. novicida to replicate within macrophages, bone marrow-derived macrophages (BMDMs) were infected with the ΔtrpA/B mutant strain in tryptophan-free cell culture media. Bacterial uptake and replication were assayed at 30 min and 8 h postinfection, respectively. The ability of the ΔtrpA/B mutant strain to invade and replicate in macrophages was compared to the wild-type U112 and ΔFPI strains. The ΔtrpA/B mutant entered macrophages at similar levels as wild-type bacteria (Fig. 3D). The wild-type U112 strain replicated within macrophages in the absence of tryptophan in the media (Fig. 3D). However, the ΔtrpA/B mutant exhibited a strong intracellular replication defect in the absence of added tryptophan (Fig. 3D), similar to the ΔFPI strain (44). The intracellular replication defect was reversed by the addition of tryptophan to the cell culture media (Fig. 3D). The same results were obtained in RAW 264.7 macrophage infections (Fig. 3E).
The above results indicate that tryptophan is an essential amino acid to Francisella, where the presence of tryptophan is needed for normal bacterial replication both intracellularly and in broth.
Tryptophan biosynthesis genes, trpA and trpB, contribute to F. novicida growth specifically in the lung.
To test the ability of the ΔtrpA and ΔtrpB mutant bacterial strains to colonize the lungs of mice, we repeated the in vivo competition experiments with the defined tryptophan synthase deletion mutants. The ΔtrpA, ΔtrpB, and ΔtrpA/B mutant bacterial strains were outcompeted by wild-type F. novicida in the lungs and spleen after an i.n. route of inoculation (Fig. 4A). The attenuation of the ΔtrpA and ΔtrpB strains observed in the spleens of mice inoculated via the i.n. route (Fig. 4A) likely reflects the lower numbers of mutant bacteria in the lungs spreading to systemic tissues, such as the spleen. To ensure that the attenuation observed for the tryptophan biosynthesis mutants was due to the deletion of the targeted gene, we complemented the ΔtrpA strain with a wild-type copy of the gene. Complementation was done in cis, rather than in trans, to ensure that the complementing gene would be retained throughout the course of an in vivo infection in the absence of antibiotic selection. Virulence was restored to the ΔtrpA strain upon complementation because equal numbers of wild-type and complemented bacteria (competitive index of ∼1) were present in the lungs and spleens of i.n.-infected mice (Fig. 4A). In addition, we infected mice i.n. with either the wild-type strain or ΔtrpA/B mutant strain in single infections. The numbers of the tryptophan auxotroph bacteria recovered from the lungs were significantly lower (P < 0.0001) than the levels of wild-type bacteria at 48 h (Fig. 4B). Thus, the tryptophan auxotroph is attenuated in the lungs compared to wild-type bacteria even in the absence of competition.
FIG. 4.
trpA and trpB are lung-specific virulence factors. Groups of 3 to 5 mice were infected with a 1:1 mixture of wild-type bacteria and the indicated bacterial mutant strain (A, C, and D) or the indicated bacterial strains (B) either via the intranasal (A and B), intradermal (C), or intraperitoneal (D) route. The data represent the CI value for the CFU of mutant/wild-type bacteria (A, C, and D) or CFU counts of the indicated bacterial strain (B) in the lungs and spleens (A), lungs (B), skins and spleens (C), or spleens (D) at 48 h postinfection. Bars represent the geometric mean for each group of mice. The data are representative of two independent experiments. (A, C, and D) Statistical significance based on the ANOVA with a Dunnett's posttest as compared to the corresponding CI values of the htpX mutant: ***, P < 0.05; ns, not statistically significant. (B) Statistical analysis based on the t test: ***, P < 0.0001.
The analysis of our negative-selection screen results from the lungs (i.n. infection model) and from the spleen (i.p. infection model) (44) suggested that the tryptophan biosynthesis genes, trpA and trpB, contribute to growth and/or survival in the lungs of mice but are not necessary for growth and/or survival in the spleen of mice. To determine whether trpA or trpB contributed to growth and/or survival of F. novicida in the skin or spleen, competition experiments were performed in which mice were infected via the intraperitoneal (i.p.) or intradermal (i.d.) route. Competition experiments with the ΔFPI and ΔhtpX strains were included as controls. The ΔtrpA and ΔtrpB strains were not attenuated in the skin and spleen of mice that were infected via the i.d. or i.p. routes (Fig. 4C and D). In contrast, the ΔFPI mutant was strongly attenuated (>5 logs) in the lungs, skin, and spleen following all routes of inoculation (Fig. 4A, C, and D). Taken together, our results indicate that trpA and trpB contribute to growth and/or survival of F. novicida specifically in the lungs.
The tryptophan biosynthetic pathway is important for F. novicida growth and/or survival in the lungs.
For many organisms, tryptophan is an essential amino acid that cannot be synthesized and therefore needs to be taken in as part of its diet. However, plants and some microorganisms have the ability to synthesize tryptophan by encoding for a functional tryptophan biosynthetic pathway (genes trpA to trpG) within its genome. Larrson et al. previously reported the use of a computational method in the prediction of a functional tryptophan biosynthetic pathway within the Francisella genome (18). As mentioned earlier, while the trpA and trpB genes are located next to each other in the genome, genes trpC to trpG are located in other parts of the genome. However, genes trpC to trpG were not identified in our i.n. negative-selection screen. Two possible explanations include (i) the array data points for the other trp genes were weak and thus the stringent selection criteria used in the SAM analysis (false discovery rate of 1.1%) had filtered them out; or (ii) the required metabolic intermediates for tryptophan biosynthesis are present in the organs and tryptophan can still be synthesized in the absence of these genes.
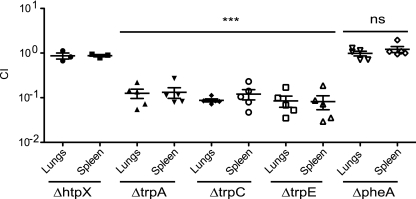
To differentiate between these 2 possibilities, targeted mutants in trpC and trpE, two additional genes in the tryptophan biosynthetic pathway, were made and tested in i.n. in vivo competition experiments. Both the ΔtrpC and ΔtrpE strains exhibited similar levels of attenuation to the ΔtrpA strain in the i.n. route of infection (Fig. 5). This suggests that all of the tryptophan biosynthesis genes are important for F. novicida growth and/or survival in the lungs, and it is highly likely that the stringent selection criteria used in the SAM analysis had filtered out weaker data points corresponding to the other trp genes.
FIG. 5.
The tryptophan biosynthetic pathway is important for Francisella virulence in the lungs. Groups of 5 mice were i.n. infected with a 1:1 mixture of wild-type bacteria and the indicated bacterial mutant strain. The data represent the CI value for the CFU of mutant/wild-type bacteria in the lungs and spleens at 48 h postinfection. Bars represent the geometric mean CI value for each group of mice. The data are representative of two independent experiments. Statistical significance is based on the ANOVA with a Dunnett's posttest as compared to the corresponding CI values of the htpX mutant: ***, P < 0.05; ns, not statistically significant.
We were interested in determining if tryptophan is a specific or general amino acid requirement for F. novicida virulence in the lung. We chose to target phenylalanine biosynthesis, as both tryptophan and phenylalanine are essential aromatic amino acids. A functional phenylalanine biosynthetic pathway has also been predicted to exist in the Francisella genome (18). We generated a targeted mutation in a gene essential for phenylalanine biosynthesis, FTT0575, pheA. The ΔpheA strain colonized the lungs and spleens of i.n.-infected mice to levels comparable to those in the wild-type U112 strain (Fig. 5). This provides evidence that the attenuation of the trp mutants in the lung is a specific and not general bacterial requirement for aromatic amino acids in the lungs.
IDO1 gene expression is induced in an organ-specific manner during infection with F. novicida or Streptococcus pneumoniae.
Why are the F. novicida tryptophan auxotrophs specifically attenuated in the intranasal route of infection? Since the tryptophan auxotrophs require tryptophan for replication in minimal media and in macrophages, we reasoned that the host could be differentially upregulating the expression of host tryptophan catabolism enzymes in the different organs, to place a selective pressure on the bacteria specifically in the lungs and deprive the trp mutant bacteria of tryptophan. Thus, we next focused our study on host enzymes that are involved in tryptophan catabolism, tryptophan 2,3-dioxygenase (TDO), indoleamine 2,3-dioxygenase 1 (IDO1) and IDO2.
TDO is involved in the catabolism of dietary l-tryptophan to n-formylkynurenine (31, 43). IDO1 is a host monomeric hemoprotein that is the first rate-limiting enzyme in the kynurenine pathway, catalyzing the breakdown of l-tryptophan to l-kynurenine and n-formylkynurenine (35, 41, 42, 45). IDO2 on the other hand, is a novel and relatively uncharacterized homolog of IDO1 (2).
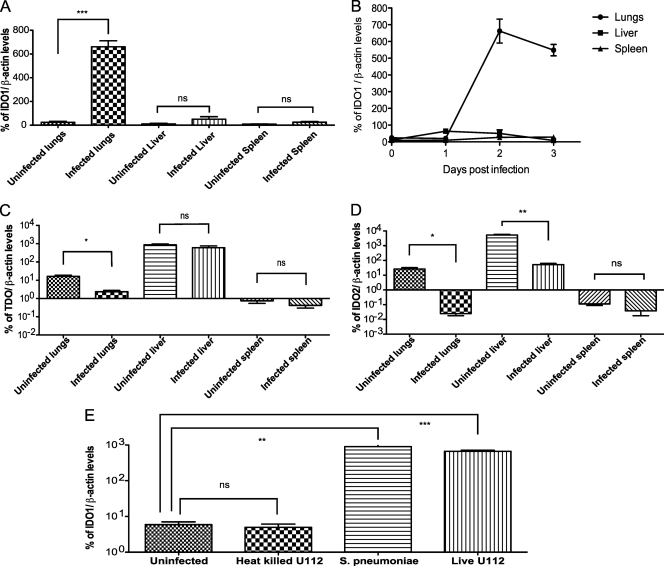
To determine if the expression of the TDO, IDO1, and IDO2 genes in the different organs was altered upon Francisella infection, mice were infected via the i.n., i.p., or i.d. routes of infection with the wild-type U112 strain. Different routes of infection were chosen in order to analyze gene expression in tissues that contained the same levels of bacteria per gram of tissue. Two days postinfection, infected lungs, livers, and spleens, as well as uninfected control organs, were harvested and the corresponding gene expression levels were determined via quantitative RT-PCR. IDO1 gene expression was highly induced in infected lungs and not in infected livers, spleens, or uninfected tissues (Fig. 6A). To further study the temporal distribution of IDO1 gene induction in infected organs, as well as to determine if IDO1 gene expression is induced in other infected organs at other time points, we determined the levels of IDO1 gene expression in infected lungs, livers, and spleens over the course of 3 days of infection via qRT-PCR. Samples at later time points were not collected since the mice were moribund by the third day of infection. Organ homogenates were plated for CFU enumeration to ensure that at each time point, bacterial loads in the different infected organs were comparable (data not shown). IDO1 gene expression in the lungs was low at day 1 postinfection and was highly elevated 2 days postinfection. This high level of expression was maintained through day 3 of infection when the experiment was terminated (Fig. 6B). In contrast, the levels of IDO1 gene expression in infected livers and spleens remained at basal levels throughout the 3 days of infection, even though the organ bacterial loads were comparable to the bacterial loads in the lungs (Fig. 6B). In contrast, expression of the TDO or IDO2 genes was not induced in any of the infected organs (Fig. 6C and D). Taken together, our data demonstrate that host IDO1 gene induction in response to Francisella infection is lung specific. Similarly, IDO1 gene expression levels increased in the lungs of mice infected with a Gram-positive microbe, S. pneumoniae. Furthermore, the levels of S. pneumoniae-dependent induction of IDO1 gene expression were comparable to what we observed during F. novicida infections (Fig. 6E). In contrast, IDO1 expression was not induced in lungs infected with heat-killed Francisella. Taken together, our data demonstrate that the host induces expression of the host defense enzyme IDO1 specifically in the lungs in response to infection with live bacterial pathogens.
FIG. 6.
IDO1 expression is induced in an organ-specific manner during Francisella infection. Groups of 3 mice were infected with bacteria of wild-type strain U112 (A to D) via the intranasal, intraperitoneal, or intradermal routes of infection or the indicated bacterial strain (E) via the intranasal route of infection. Infected organs with similar bacterial loads were harvested at 2 days postinfection (A and C to E) or at each day postinfection (B) until the mice became moribund. The levels of IDO1 (A, B, and E) or TDO (C) or IDO2 (D) expression in the respective organs were determined via quantitative RT-PCR. The data are representative of two independent experiments. Statistical significance is based on the t test as compared to the corresponding uninfected organs: ***, P < 0.0005; **, P < 0.005; *, P < 0.05; ns, not statistically significant.
Attenuation of the ΔtrpA/B strain in the lungs is rescued in IDO1−/− mice.
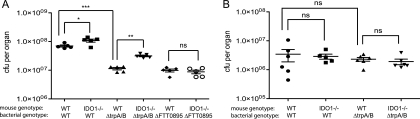
We have demonstrated that the host responds to a bacterial infection in the lung by inducing the expression of a gene that encodes for an enzyme involved in depletion of tryptophan levels, IDO1. To determine whether IDO1 induction is responsible for the attenuation of the ΔtrpA/B mutant strain in the lungs of mice, we infected mice that are deficient for IDO1 with either the wild-type or ΔtrpA/B mutant bacterial strains. Histological analysis of the infected lungs showed that there were no host background- or bacterial strain-specific differences in the composition of the cellular infiltrates. The lungs of IDO1+/+ and IDO1−/− mice infected with either the wild-type strain U112 or ΔtrpA/B mutant bacterial strains contained perivascular and paravascular inflammation made up predominantly of accumulations of polymorphonuclear neutrophils (data not shown). In addition, the levels of bacteria recovered from the lungs and skin in IDO1−/− mice were compared to the levels in IDO1+/+ mice infected with the corresponding bacterial strain via the i.n. or i.d. routes of infection, respectively, 2 days postinfection. The lungs of IDO1−/− mice infected with wild-type F. novicida via the i.n. route contained higher bacterial numbers compared to the lungs of IDO1+/+ mice infected with the same bacterial strain (∼1.5-fold) (Fig. 7A), indicating that the host enzyme IDO1 is part of the innate immune response to defend against this bacterial pathogen. Furthermore, lungs of IDO1−/− mice infected with the ΔtrpA/B mutant strain contained significantly higher bacterial numbers than those of IDO1+/+ mice infected with the same bacterial strain (∼3.0-fold). Thus, the attenuation of the F. novicida tryptophan auxotroph is less severe in IDO1−/− mice (∼2-fold fewer CFU) than the attenuation of the auxotroph in IDO1+/+ mice (∼6-fold fewer CFU) (Fig. 7A), indicating that the tryptophan auxotroph was partially rescued in the lungs of IDO1−/− mice. In contrast, the numbers of wild-type and ΔtrpA/B mutant bacteria were not significantly different in the skin (Fig. 7B) or spleens (data not shown) of IDO1+/+ and IDO1−/− mice that had been inoculated via the i.d. route. In addition, we infected IDO1+/+ or IDO1−/− mice with an FTT0895 mutant bacterial strain (the ΔFTT0895 mutant). FTT0895 is a gene essential for purine biosynthesis and the ΔFTT0895 mutant is attenuated in the lungs to a similar degree as the ΔtrpA/B mutant strain. However, the attenuation of the ΔFTT0895 mutant bacterial strain was not rescued in the lungs of IDO1−/− mice (Fig. 7A). Taken together, these results suggest that lung-specific induction of IDO1 gene expression during F. novicida infection plays a role in controlling bacterial replication and specifically accounts for a portion of the attenuation of the ΔtrpA/B strain in the lungs.
FIG. 7.
Attenuation of trpA/B mutants in the lungs is partially complemented in IDO1−/− mice. Groups of 5 mice of the indicated genotype were infected intranasally (A) or intradermally (B) with the indicated bacterial strain. WT, wild type. Infected lungs (A) or skins (B) were harvested at 2 days postinfection and plated to enumerate CFU. The data are representative of two independent experiments (A) or one independent experiment (B). Statistical significance is shown based on the t test as compared as indicated: ***, P < 0.0001; **, P < 0.005; *, P < 0.05; ns, not statistically significant.
DISCUSSION
Francisella is a highly infectious pathogen that infects hosts via multiple routes of inoculation. Once the bacterium enters the body, it travels to the draining lymph nodes and then spreads to the liver, lungs, and spleen of infected humans or animals, where it replicates to high numbers (9, 37). In this study, we report the use of a microarray-based, negative-selection screen in an intranasal model of infection to identify genes that contribute to F. novicida growth and/or survival in the lungs (Fig. 1; see Table S1 in the supplemental material). We demonstrated that F. novicida tryptophan biosynthetic genes are required for bacterial growth and/or survival specifically in the lungs (Fig. 4A to D). In addition, we have shown that indoleamine 2,3-dioxygenase 1 (IDO1) gene expression is specifically induced in the lungs of mice that have been infected with live bacteria (Fig. 6A and B). Heat-killed F. novicida failed to induce pulmonary IDO1 gene expression, while a Gram-positive microbe, S. pneumoniae, induced high levels of IDO1 gene expression that are comparable to those observed during F. novicida infection (Fig. 6E), indicating that the induction of IDO1 expression is an innate immune response to live bacteria and is not restricted to Gram-negative bacteria.
In addition, we demonstrate that the attenuation of a tryptophan biosynthesis mutant, and not a purine biosynthesis mutant, was partially rescued in the lungs of IDO1−/− mice, demonstrating that IDO1 specifically inhibits growth of the F. novicida tryptophan auxotrophs in the lungs of infected mice (Fig. 7A and B). As IDO1 has previously been shown to exert an antimicrobial effect against bacteria, viruses, and parasites in vitro by depleting intracellular levels of tryptophan (21, 22, 29), we believe that IDO1 is exerting its antimicrobial effect against F. novicida in the lungs via tryptophan deprivation. This is further supported by previous observations that IDO1 gene expression is induced in mice infected with Toxoplasma gondii, which is associated with tryptophan depletion in the lungs (11, 36).
Induction of IDO1 gene expression has previously been observed in the lungs of mice after the administration of immunostimulatory DNA sequences (14), pokeweed mitogen (48), or lipopolysaccharide (47) or during influenza virus (49) and Toxoplasma gondii infection (11, 36). However, we are the first study to report that IDO1 gene expression is induced specifically in the lungs in response to bacterial infection. As IDO1 gene expression appears to be induced in response to a wide range of immune stimuli and pathogens, this suggests that IDO1 functions as a general innate immune defense mechanism of the lungs to deal with potential infection. The host's use of such a nutriprive (from the latin privare, to deprive of nutrients) antimicrobial mechanism as a general innate immune defense mechanism to infection is perhaps not surprising given that such nutriprive antimicrobial mechanisms are usually effective against both auxotrophic and prototrophic microbes, as evidenced by the higher numbers of wild-type bacteria in the lungs of IDO1−/− mice compared to the IDO1+/+ mice (Fig. 7A). This is because amino acid synthesis is costly to microbes in terms of energy expenditure, a consideration especially pertinent for tryptophan, an amino acid that requires approximately seven times the amount of energy needed to produce compared to other amino acids like glycine or alanine (50). Furthermore, microbes usually prefer to use environmental amino acids if available rather than synthesize them de novo. Thus, in the presence of such nutriprive antimicrobial mechanisms, even the prototrophic microbes have to expend more energy upregulating expression of the appropriate metabolism genes (46) and thus are still slightly restricted for replication.
Our work has also demonstrated that bacterial metabolic pathways can have differential importance in various organs where we observed that tryptophan metabolism in F. novicida is only important for bacterial colonization of the lungs and not in other organs. Similar observations in other bacterial species have been observed. For instance, glutathione and/or glutamine metabolism is needed for Campylobacter jejuni colonization of the intestinal tract but not the liver (15). Such data indicate that specific metabolic pathways can enhance the ability of a pathogen to colonize specific tissues. Given that many primary pulmonary pathogens like S. pneumoniae, Mycobacterium tuberculosis, and Legionella pneumoniae all contain genes coding for a functional tryptophan biosynthetic pathway, it is possible that tryptophan metabolism contributes to making these microbes highly virulent pulmonary pathogens.
Organ-specific immunity is a concept that arose on the basis that host immune responses have to be closely and differentially regulated, such that an effective and adequate immune response is launched during infection, yet any overt immune responses are dampened so as not to cause uncontrolled inflammation and immunopathology. However, what is considered an appropriate or overt immune response highly depends on the organ in question. Different organs have vastly different physiological functions and face various bacterial challenges. Those factors in turn shape each organ's unique and customized immune response to infection (32). As an example, inflammation levels in the skin can be much higher than that of the lungs before it becomes detrimental to the survival of the host. Our demonstration of IDO1 as a host innate immune response that is induced specifically in the lungs during infection with Francisella tularensis further supports this concept of organ-specific immunity. Apart from the antimicrobial role via tryptophan starvation discussed thus far, IDO1 also has an immunoregulatory role involving negative feedback mechanisms that dampen any excess adaptive inflammatory responses that could be potentially pathological (23). Although IDO1-dependent immunosuppressive functions could be important during Francisella infections, we believe that since these functions of IDO1 are important during the adaptive immune phase of infections (20, 23, 24), IDO1 is likely acting to starve the bacteria of tryptophan at the 48-h time point we looked at in our studies. In addition, we identified IDO1 as an organ-specific innate immune factor. Polymorphisms in the human IDO1 gene that result in significantly reduced protein expression and nearly complete loss of IDO1 activity in vitro have been reported (1). It would be interesting to determine if these polymorphisms would in turn lead to increased susceptibility of the affected individuals to pulmonary infections.
The ΔtrpA/B mutant bacterial strain was partially complemented in the lungs of IDO1−/− mice (Fig. 7A), suggesting that another IDO1-independent innate immune mechanism may be important during an acute bacterial infection in the lungs. Francisella can infect and replicate within a range of different cell types in the lungs (13), as well as extracellularly (10). The lung contains more than 40 cell types (4, 7); it is possible that there are IDO1-independent killing mechanisms in the lungs that are acting on the attenuated F. novicida tryptophan auxotroph and are not present in other tissues.
Two other groups have previously reported the use of negative-selection screens to identify Francisella genes that play a role in survival in the lungs of mice (17, 39). Although the genes that we identified in this study overlap with the previous studies to a certain extent, any difference could be due to differences in the techniques used (e.g., signature-tagged mutagenesis [STM] [39] and microarray tracking of transposon mutants [MATT] [17]), route of inoculation, and strains utilized. We did not perform TraSH analysis on systemic organs like the liver and spleen following pulmonary infection, as it has previously been reported by Su et al. that a significant bottleneck exists for bacterial spread systemically from the lungs (39).
In conclusion, we have demonstrated how a microarray-based, negative-selection approach can be applied to different tissues in animal models of infection and that this can lead to the identification of organ-specific bacterial factors that are important for bacterial growth and/or survival in a specific host tissue. In addition, we have shown that the further characterization of mutants identified by this approach can lead to the identification of organ-specific host innate immune defenses. Furthermore, our results highlight some relatively unexplored areas of host-pathogen interactions. For example, although bacterial metabolism has been the subject of intense study for decades, we still know very little about how metabolic differences may affect the ability of pathogenic bacteria to colonize different tissues or how the different nutritional requirements contribute to evasion of host defenses. A better understanding of the metabolic requirements of bacterial pathogens in various tissues could potentially lead to the development of novel antimicrobial strategies.
Supplementary Material
Acknowledgments
We thank Donna Bouley for assistance in the interpretation of the lung histology sections and thank Elizabeth Joyce, David Weiss, and Anna Brotcke for experimental assistance; Stanley Falkow, Manuel Amieva, and Thomas Henry for helpful discussions; and Sara Fisher for administrative assistance.
Kaitian Peng was supported by a predoctoral fellowship from the Agency for Science, Technology and Research, Singapore. This work was supported by grants AI063302 and AI065359 from the NIH-NIAID to Denise Monack.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 12 April 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Arefayene, M., S. Philips, D. Cao, S. Mamidipalli, Z. Desta, D. A. Flockhart, D. S. Wilkes, and T. C. Skaar. 2009. Identification of genetic variants in the human indoleamine 2,3-dioxygenase (IDO1) gene, which have altered enzyme activity. Pharmacogenet. Genomics 19:464-476. [DOI] [PubMed] [Google Scholar]
- 2.Ball, H. J., H. J. Yuasa, C. J. Austin, S. Weiser, and N. H. Hunt. 2009. Indoleamine 2,3-dioxygenase-2: a new enzyme in the kynurenine pathway. Int. J. Biochem. Cell Biol. 41:467-471. [DOI] [PubMed] [Google Scholar]
- 3.Barker, J. R., and K. E. Klose. 2007. Molecular and genetic basis of pathogenesis in Francisella tularensis. Ann. N. Y. Acad. Sci. 1105:138-159. [DOI] [PubMed] [Google Scholar]
- 4.Bergen, H. T., T. C. Cherlet, P. Manuel, and J. E. Scott. 2002. Identification of leptin receptors in lung and isolated fetal type II cells. Am. J. Respir. Cell Mol. Biol. 27:71-77. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crapo, J. D., B. E. Barry, P. Gehr, M. Bachofen, and E. R. Weibel. 1982. Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 126:332-337. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, M. F., D. Niks, H. Ngo, T. R. Barends, and I. Schlichting. 2008. Tryptophan synthase: the workings of a channeling nanomachine. Trends Biochem. Sci. 33:254-264. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forestal, C. A., M. Malik, S. V. Catlett, A. G. Savitt, J. L. Benach, T. J. Sellati, and M. B. Furie. 2007. Francisella tularensis has a significant extracellular phase in infected mice. J. Infect. Dis. 196:134-137. [DOI] [PubMed] [Google Scholar]
- 11.Fujigaki, S., K. Saito, M. Takemura, N. Maekawa, Y. Yamada, H. Wada, and M. Seishima. 2002. l-Tryptophan-l-kynurenine pathway metabolism accelerated by Toxoplasma gondii infection is abolished in gamma interferon-gene-deficient mice: cross regulation between inducible nitric oxide synthase and indoleamine-2,3-dioxygenase Infect. Immun. 70:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, V., and B. A. Cunha. 1997. Tularemia pneumonia. Semin. Respir. Infect. 12:61-67. [PubMed] [Google Scholar]
- 13.Hall, J. D., R. R. Craven, J. R. Fuller, R. J. Pickles, and T. H. Kawula. 2007. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect. Immun. 75:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, T., S. P. Rao, K. Takabayashi, J. H. Van Uden, R. S. Kornbluth, S. M. Baird, M. W. Taylor, D. A. Carson, A. Catanzaro, and E. Raz. 2001. Enhancement of innate immunity against Mycobacterium avium infection by immunostimulatory DNA is mediated by indoleamine 2,3-dioxygenase Infect. Immun. 69:6156-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofreuter, D., V. Novik, and J. E. Galan. 2008. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4:425-433. [DOI] [PubMed] [Google Scholar]
- 16.Keim, P., A. Johansson, and D. M. Wagner. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105:30-66. [DOI] [PubMed] [Google Scholar]
- 17.Kraemer, P. S., A. Mitchell, M. R. Pelletier, L. A. Gallagher, M. Wasnick, L. Rohmer, M. J. Brittnacher, C. Manoil, S. J. Skerett, and N. R. Salama. 2009. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect. Immun. 77:232-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Hälltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjöstedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 19.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luft, T., E. Maraskovsky, and M. Schnurr. 2005. IDO production, adaptive immunity, and CTL killing. Blood 106:2228-2229. [Google Scholar]
- 21.Mackenzie, C. R., C. B. Willberg, and W. Daubener. 1998. Inhibition of group B streptococcal growth by IFN gamma-activated human glioblastoma cells. J. Neuroimmunol. 89:191-197. [DOI] [PubMed] [Google Scholar]
- 22.Mehta, S. J., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-gamma: role of tryptophan catabolism. J. Infect. Dis. 177:1326-1331. [DOI] [PubMed] [Google Scholar]
- 23.Mellor, A. 2005. Indoleamine 2,3 dioxygenase and regulation of T cell immunity. Biochem. Biophys. Res. Commun. 338:20-24. [DOI] [PubMed] [Google Scholar]
- 24.Mellor, A. L., D. Munn, P. Chandler, D. Keskin, T. Johnson, B. Marshall, K. Jhaver, and B. Baban. 2003. Tryptophan catabolism and T cell responses. Adv. Exp. Med. Biol. 527:27-35. [DOI] [PubMed] [Google Scholar]
- 25.Miles, E. W. 1979. Tryptophan synthase: structure, function and subunit interaction. Adv. Enzymol. Relat. Areas Mol. Biol. 49:127-186. [DOI] [PubMed] [Google Scholar]
- 26.Nano, F. E., and C. Schmerk. 2007. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105:122-137. [DOI] [PubMed] [Google Scholar]
- 27.Owen, C. R., E. O. Buker, W. L. Jellison, D. B. Lackman, and J. F. Bell. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 12:967-978. [DOI] [PubMed] [Google Scholar]
- 29.Pfefferkorn, E. R., M. Eckel, and S. Rebhun. 1986. Interferon-gamma suppresses the growth of Toxoplasma gondii in human fibroblasts through starvation for tryptophan. Mol. Biochem. Parasitol. 20:215-224. [DOI] [PubMed] [Google Scholar]
- 30.Raboni, S., S. Bettati, and A. Mozzarelli. 2009. Tryptophan synthase: a mine for enzymologists. Cell. Mol. Life Sci. 66:2391-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rafice, S. A., N. Chauhan, I. Efimov, J. Basran, and E. L. Raven. 2009. Oxidation of L-tryptophan in biology: a comparison between tryptophan 2,3-dioxygenase and indoleamine 2,3-dioxygenase. Biochem. Soc. Trans. 37:408-412. [DOI] [PubMed] [Google Scholar]
- 32.Raz, E. 2007. Organ-specific regulation of innate immunity. Nat. Immunol. 8:3-4. [DOI] [PubMed] [Google Scholar]
- 33.Saslaw, S., H. T. Eigelsbach, H. E. Wilson, J. A. Prior, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107:689-701. [DOI] [PubMed] [Google Scholar]
- 34.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702-714. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu, T., S. Nomiyama, F. Hirata, and O. Hayaishi. 1978. Indoleamine 2,3-dioxygenase. Purification and some properties J. Biol. Chem. 253:4700-4706. [PubMed] [Google Scholar]
- 36.Silva, N. M., C. V. Rodrigues, M. M. Santoro, L. F. Reis, J. I. Alvarez-Leite, and R. T. Gazzinelli. 2002. Expression of indoleamine 2,3-dioxygenase, tryptophan degradation, and kynurenine formation during in vivo infection with Toxoplasma gondii: induction by endogenous gamma interferon and requirement of interferon regulatory factor 1. Infect. Immun. 70:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sjostedt, A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1-29. [DOI] [PubMed] [Google Scholar]
- 38.Steinemann, T. L., M. R. Sheikholeslami, H. H. Brown, and R. W. Bradsher. 1999. Oculoglandular tularemia. Arch. Opthalmol. 117:132-133. [DOI] [PubMed] [Google Scholar]
- 39.Su, J., J. Yang, D. Zhao, T. H. Kawula, J. A. Banas, and J. R. Zhang. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75:3089-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan, J. T., E. F. Jeffery, J. D. Shannon, and G. Ramakrishnan. 2006. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188:3785-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takikawa, O., R. Yoshida, R. Kido, and O. Hayaishi. 1986. Tryptophan degradation in mice initiated by indoleamine 2,3-dioxygenase J. Biol. Chem. 261:3648-3653. [PubMed] [Google Scholar]
- 42.Taylor, M. W., and G. S. Feng. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5:2516-2522. [PubMed] [Google Scholar]
- 43.Thackray, S. J., C. G. Mowat, and S. K. Chapman. 2008. Exploring the mechanism of tryptophan 2,3-dioxygenase. Biochem. Soc. Trans. 36:1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner, E. R., G. Bitterlich, D. Fuchs, A. Hausen, G. Reibnegger, G. Szabo, M. P. Dierich, and H. Wachter. 1987. Human macrophages degrade tryptophan upon induction by interferon-gamma. Life Sci. 41:273-280. [DOI] [PubMed] [Google Scholar]
- 46.Wood, H., C. Fehlner-Gardner, J. Berry, E. Fischer, B. Graham, T. Hackstadt, C. Roshick, and G. McClarty. 2003. Regulation of tryptophan synthase gene expression in Chlamydia trachomatis. Mol. Microbiol. 49:1347-1359. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, R., and O. Hayaishi. 1978. Induction of pulmonary indoleamine 2,3-dioxygenase by intraperitoneal injection of bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A. 75:3998-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida, R., T. Oku, J. Imanishi, T. Kishida, and O. Hayaishi. 1986. Interferon: a mediator of indoleamine 2,3-dioxygenase induction by lipopolysaccharide, p(I) X poly (C), and pokeweed mitogen in mouse lung. Arch. Biochem. Biophys. 249:594-604. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, R., Y. Urade, M. Tokuda, and O. Hayaishi. 1979. Induction of indoleamine 2,3-dioxygenase in mouse lung during virus infection. Proc. Natl. Acad. Sci. U. S. A. 76:4084-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zegarra-Moran, O., C. Folli, B. Manzari, R. Ravazzolo, L. Varesio, and L. J. Galietta. 2004. Double mechanism for apical tryptophan depletion in polarized human bronchial epithelium. J. Immunol. 173:542-549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.