Abstract
The mammalian transcriptome is studded with putative noncoding RNAs, many of which are antisense to known open reading frames (ORFs). Roles in the regulation of their complementary mRNAs are often imputed to these antisense transcripts, but few have been experimentally examined, and such functions remain largely conjectural. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes two transcripts that lack obvious ORFs and are complementary to the gene (RTA) encoding the master regulator of the latent/lytic switch. Here, we show that, contrary to expectation, these RNAs do not regulate RTA expression. Rather, they are found on polysomes, and genetic analysis indicates that translational initiation occurs at several AUG codons in the RNA, leading to the presumptive synthesis of peptides of 17 to 48 amino acids. These findings underscore the need for circumspection in the computational assessment of coding potential and raise the possibility that the mammalian proteome may contain many previously unsuspected peptides generated from seemingly noncoding RNAs, some of which could have important biological functions. Irrespective of their function, such peptides could also contribute substantially to the repertoire of T cell epitopes generated in both uninfected and infected cells.
One of the most remarkable findings of recent efforts to analyze the mammalian transcriptome on a genome-wide scale has been the discovery that large regions of the genome previously thought to be noncoding are in fact represented in stable RNAs (reviewed in references 5 and 32). Use of both comprehensive cDNA identification (4) and genomic tiling arrays (2, 7) reveals that the number of transcripts is substantially greater than the number of predicted genes. In one analysis, 56% of mouse cDNAs were annotated as noncoding, based on the absence of open reading frames (ORFs) of >100 codons (4). Such studies also show that a significant fraction of the genome can produce transcripts from both strands (6, 14); in the FANTOM 3 data set, at least 25% of confirmed murine coding RNAs have well-characterized overlapping antisense transcripts; when fragmentary cDNAs are included, this percentage can rise as high as 72% (14).
The biological functions of these natural antisense transcripts (NATs) or sense-antisense pairs has been a matter of speculation for a number of years. Most discussions begin with the idea that these RNAs serve as regulators of gene expression (reviewed in reference 21), often targeting the cognate coding transcript for inhibition. Such inhibition may occur at the transcriptional level (e.g., due to impaired transcriptional elongation in the overlap region [22] or regional chromatin remodeling [23, 25, 28]) or at the posttranscriptional level (e.g., due to altered RNA processing [12, 13] or impaired translation [8, 27]). Consistent with such regulatory models, a reciprocal relationship has sometimes been seen between expression of the antisense RNA and the accumulation of its cognate RNA (14). More often, though, the two RNAs are concordantly regulated, accumulating in tandem. Very few examples of such coordinately regulated sense-antisense pairs have been rigorously studied, and proposals about their function have been largely conjectural.
The genomes of large DNA viruses (e.g., herpesviruses) may provide a useful forum in which to examine this issue. Recent genome-wide cDNA annotation in human cytomegalovirus (HCMV) (34) revealed that 45% of all viral cDNAs were predicted to be noncoding, and 55% of the cDNAs were antisense to known or predicted HCMV genes. Our own analysis of the transcriptome of Kaposi's sarcoma-associated herpesvirus (KSHV), carried out by hybridizing infected cell cDNA to a viral genomic tiling array, reveals a similar pattern of extensive hybridization to noncoding areas, including many overlapping sense-antisense RNA pairs, just as in the transcriptome of its human host (S. Chandriani, Y. Xu, and D. Ganem, unpublished results). Because the functions of many herpesviral genes are known and good assays exist for each important stage of viral replication, these viruses may provide an accessible system for probing the biological roles of antisense RNAs.
Here, we present the first detailed functional analysis of a pair of sense-antisense transcripts in KSHV. This pair was in fact discovered well before the advent of whole-genome transcription profiling, as a result of efforts in our laboratory and others to understand the control of latency in this organism. Like all herpesviruses, KSHV can produce either a latent infection (in which viral gene expression is sharply restricted and the genome persists cryptically without engendering viral progeny) or a lytic infection, in which all viral genes are expressed in a strictly regulated cascade that culminates in cell death and progeny virus release. Latency is usually the default pathway; lytic replication can be induced in latently infected cells following activation of a variety of signaling pathways. The key regulator of the latent/lytic switch is the RTA (replication and transcription activator) viral gene, whose product is a transcription factor that activates a large number of early viral promoters and sets the lytic cascade in motion. Ectopic expression of RTA in latency triggers lytic reactivation (19, 26), and null or dominant negative RTA mutants block both spontaneous and experimentally induced lytic growth (18, 31). While mapping and characterizing RTA transcripts, we discovered that RNAs antisense to RTA also accumulated during lytic KSHV replication (18). Further fine structure mapping by Saveliev and colleagues revealed that these two antisense RNAs were 5′ coterminal, unspliced, and differentially polyadenylated (24). Although these RNAs are predicted to be noncoding and have been proposed to be regulators of RTA, here we show that their expression does not impair RTA expression or function. Rather, the RNAs appear on polysomes in infected cells, and internal AUG codons in the body of the RNAs can be accessed by translating ribosomes, leading to the synthesis of small peptides of 17 to 48 amino acids (aa). The results indicate that RNAs with all the stigmata of noncoding transcripts can in fact function as mRNAs for a hitherto-neglected class of small peptides; if this proves to be true of many such RNAs, it could indicate that the mammalian proteome has even greater complexity than previously anticipated.
MATERIALS AND METHODS
Cells and virus.
293 and 293T cells were purchased from the ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and l-glutamine. BCBL1 cells were carried in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, streptomycin, glutamine, and β-mercaptoethanol. SLK(rKSHV.219) cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin, streptomycin, l-glutamine, and 10 μg/ml puromycin. For KSHV lytic expression, BCBL1 cells were induced with 0.3 mM sodium butyrate, and SLK(rKSHV.219) cells were induced with 3 mM valproic acid. Mouse RBP-Jκ−/− (OT11) and wild-type (WT) (OT13) fibroblast cell lines were kindly provided by T. Honjo and were grown in high-glucose DMEM supplemented with 10% FBS and 100 U of mouse interferon gamma (Life Technologies) per ml at 32°C.
The stable OT11-RBP-Jk cell line was generated as described previously (17).
Plasmids.
Primers used for plasmid construction are listed in Table S1 in the supplemental material.
Plasmids for RTA and 50AS expression.
Full-length 50AS was reverse transcription-PCR (RT-PCR)-amplified from BCBL1 induced total RNA. RT-PCR was carried out using a one-step RT-PCR kit (Stratagene) according to the manufacturer's protocol. The final, 3-kb PCR product was TA cloned into the pCR4 vector and recloned into pCDNA3.1(+) by EcoRI digestion and religation. The resulting construct was named pCMV-50AS. The RTA 3-kb genomic sequence (nucleotides [nt] 71498 to 74629) was PCR amplified using KSHV genomic DNA as a template. The 3-kb PCR product was TA cloned into the pTarget vector first and then recloned into the pCS2+ vector by EcoRI digestion and ligation. The final construct was named pCMV-RTA-3k. The RTA 5-kb genomic sequence (nucleotides 71498 to 76479) was PCR amplified using KSHV genomic DNA as a template, and the 5-kb PCR product was TA cloned into the pTarget vector first and then recloned into the pCS2+ vector by EcoRI digestion and ligation. The final construct was named pCMV-RTA-5k. The following steps were carried out to construct the pCMV-RTA-4.5k plasmid, which contains the full length of RTA3k and part of its downstream sequence, with the 500-bp 50AS promoter deleted. A 1.5-kb RTA downstream sequence was PCR amplified using pCMV-RTA-5k as a template. The PCR product was cut with NheI and XbaI and ligated into pCMV-RTA-3k treated with NheI and XbaI. The final construct was named pCMV-RTA-4.5k. The structures of all constructs were confirmed by sequencing.
Plasmids for 50AS promoter study.
All plasmids constructed for 50AS promoter study used pGL3-basic as a vector. Each 50AS potential promoter sequence was PCR amplified using KSHV genomic DNA as a PCR template and the same reverse primer as that listed in Table S1 in the supplemental material but different forward primers. All constructs were confirmed by sequencing.
Plasmids for 50AS-S ORF study.
The enhanced green fluorescent protein (EGFP) gene (without ATG) was PCR amplified using the EGFP-N1 plasmid as a template. EGFP PCR product was cleaved with EcoRI and XbaI and ligated into EcoRI/XbaI-treated pCDNA3.1(+), resulting in plasmid pCDNA-GFP(−)ATG. Each potential 50AS-S ORF gene was PCR amplified using pCMV-50AS as a template and the same forward primer as that listed in Table S1 in the supplemental material but different reverse primers. Each ORF PCR product was cleaved with HindIII and EcoRI and ligated into HindIII/EcoRI-treated pCDNA-GFP(−)ATG to fuse in-frame with the EGFP gene.
RACE.
A FirstChoice RLM-RACE kit (Ambion) was used for a rapid amplification of cDNA ends (RACE) assay to determinate the 5′ and 3′ ends of 50AS transcripts. BCBL1 cells were induced with sodium butyrate, and total RNA was harvested at 48 h postinduction (hpi). 5′ and 3′ RACE reactions were carried out according to the manufacturer's protocol. DNA fragments obtained in the reactions were cloned into the pCR4-topo vector and sequenced. The primers used for the RACS were as follows: the 5′ RACE gene-specific inner primer (5′-AGGTGTGCCGTGTAGAGATTCAAC-3′), the 5′ RACE gene-specific outer primer (5′-AGTTCACAGACGGTGTCAGTCAA-3′), the 3′ RACE gene-specific inner primer for 50AS-L (5′-ACGAGGACTTTCAGGATACAGA-3′), the 3′ RACE gene-specific outer primer for 50AS-L (5′-GAGAGAGTGGCGTGTCATAGT-3′), the 3′ RACE gene-specific inner primer for 50AS-S (5′-GGAGTGGACGCTGACATTAC-3′), and the 3′ RACE gene-specific outer primer for 50AS-S (5′-TGAATCTCTACACGGCACAC-3′).
Luciferase assay.
293 cells were seeded into 24-well tissue culture plates 24 h prior to transfection. Cotransfections were performed using Fugene (Roche) according to the manufacturer's instructions. 50AS promoter luciferase (LUC) reporter constructs were transfected with and without the RTA expression plasmid pCDNA3-FLc50 (18). The total amount of DNA and the amount of each plasmid used in the cotransfection mixture were kept constant by using pcDNA3.1(+) as control filler plasmid DNA. Total cell lysates were harvested at 48 h posttransfection (hpt), and luciferase assays were carried out with a luciferase assay system (E4530; Promega) as recommended by the manufacturer. Data are reported as fold induction over the level for the basic control and represent the means ± standard deviations (SD) of results from three separate experiments performed in triplicate.
Cotransfection assay.
293 cells were plated into 6-well plates 24 h prior to transfection. pCMV-RTA-3k was cotransfected with an increased amount of pCMV-50AS into 293 cells using Fugene (Roche) according to the manufacturer's instructions. The total amount of DNA used in the cotransfection mixture was kept constant by using pCDNA3.1(+) as control filler plasmid DNA. RNA or protein was harvested at 48 hpt for Northern blot analysis or Western blot analysis.
Northern blots.
Total RNA was harvested from cells using RNA-BEE (Tel-Test, Inc.); poly(A) mRNA was further purified using an Oligotex mRNA midi kit (Qiagen), separated by 1% agarose-formaldehyde gel electrophoresis, and transferred onto a Nytran membrane. Nonisotopic riboprobes were generated using biotin-16-UTP and a MAXIscript in vitro transcription kit (Ambion). Hybridization was carried out in ULTRAhyb solution (Ambion) overnight at 65°C. The probe signals were detected by using a BrightStar BioDetect kit (Ambion). For detecting 50AS transcripts, plasmid RTA-DE (16) was cleaved with XbaI and transcribed in vitro using T7 polymerase to generate riboprobes which hybridized with 50AS transcripts at nt 485 to 1258. For detecting RTA RNA, plasmid pCR4ORF50 (1) was cleaved with PmeI and transcribed in vitro using T7 polymerase to generate riboprobes which hybridized with RTA RNA at nt 478 to 1234.
Western blots.
293T cells were transfected with various expression constructs and cell lysates were harvested at 48 h posttransfection (hpt). Cell lysates were prepared by rinsing cells twice with ice-cold phosphate-buffered saline (PBS), scraping adherent cells from the plate, spinning cells at 1,000 rpm for 5 min, and resuspending pellets in 2 to 3 times the pellet volume of radioimmunoprecipitation assay (RIPA) lysis buffer. The protein content of the lysates was quantified using the Bio-Rad protein assay. Twenty micrograms of protein was run on 4 to 15% polyacrylamide gels, and protein was transferred to polyvinylidene difluoride (PVDF) membranes. Blots were blocked for 1 h at room temperature in 5% nonfat dry milk in Tris-buffered saline (TBS) plus 0.1% Tween. Primary antibodies were incubated overnight at 4°C at 1:2,000 anti-RTA, 1:1,000 anti-β-actin (sc-1615; Santa Cruz Biotechnology); secondary antibodies (anti-rabbit-horseradish peroxidase [HRP] and anti-goat-HRP; Santa Cruz Biotechnology) were incubated at 1:5,000 for 1 h at room temperature. For green fluorescent protein (GFP) fusion protein detection, HRP-conjugated GFP-FL antibody (where “FL” is full-length) was incubated overnight at 4°C at 1:5,000 (sc-8334HRP; Santa Cruz Biotechnology); signal was detected using ECL detection reagent (Amersham).
siRNA.
All small interfering RNA (siRNA) oligonucleotides were designed and produced by Applied Biosystems as silencer select siRNAs. The control siRNAs were negative control 1 (catalog no. 4390843), negative control 2 (catalog no. 4390846), and GAPDH siRNA (catalog no. 4390849). The following silencer select siRNAs targeting 50ASs were designed and produced: si50AS-L1 (sense, 5′-GAUCGUAGAUUGUUUCGUA-3′ [nt 1939 to 1957 of 50AS]), si50AS-L2 (sense, 5′-GGAAGAGGUUAAUAUCAUA-3′ [nt 1550 to 1568 of 50AS]), si50AS-B1 (sense, 5′-GGUAACCUGCAACAACGUA-3′ [nt 746 to 764 of 50AS]), and si50AS-B2 (sense, 5′-CGGUCAAAGCCUUACGCUU-3′ [nt 475 to 493 of 50AS]). We transfected cells with 5 nM siRNA using DharmaFECT-1 (Dharmacon) according to the manufacturer's instructions.
Lytic reactivation and viral production.
SLK(rKSHV.219) cells were transfected with control siRNAs or siRNAs targeting 50ASs and induced with 3 mM valproic acid. The level of lytic reactivation of recombinant KSHV (rKSHV) was assayed by flow cytometry analysis of the cells for GFP and red fluorescent protein (RFP) expression at 72 h postinduction (hpi). Viruses were harvested from filtered supernatant of each sample before the flow analysis at 72 h postinduction (hpi). Viral titer was assayed by infection of 293 cells and flow cytometry analysis for GFP expression at 48 h postinfection. RNA and protein samples for each siRNA transfection were taken at 48 hpi from duplicated wells and subjected to Northern blot analysis or Western blot analysis for 50AS and RTA expression level.
Polyribosomal fractionation assay.
293T cell extracts used for polysome gradient centrifugation were prepared as described previously (3, 15). In brief, 293T cells cultured in 10-cm culture dishes were harvested 48 h after transfection by replacing the culture medium with fresh medium containing cycloheximide (Sigma) at a final concentration of 50 μg/ml for 30 min. Cytoplasmic extracts for polysome analysis were prepared as described previously (3), loaded onto a 10% to 50% sucrose gradient, and centrifuged in a SW41Ti rotor at 36,000 rpm for 2 h at 4°C. To disrupt polyribosomes in a separate control experiment to distinguish between free mRNP and polysomal mRNA, cytoplasmic extracts were incubated with 0.02 M EDTA (pH 7.4) at 4°C for 10 min and loaded on a separate gradient. For viral experiments, BCBL1 cells were induced with 0.3 mM sodium butyrate for 24 h. Cells were treated with 50 μg/ml cycloheximide (Sigma) for 30 min. Cytoplasmic extraction and sucrose gradient fractionation were carried out as described above. Gradient fractions were collected in 0.5-ml aliquots from the top of the gradients, and RNA profiles were determined by monitoring absorbance at 260 nm. Each fraction was treated with 200 μg/ml proteinase K and 1% SDS for 30 min at 42°C. Total RNA from each fraction was isolated using TriLS reagent (sigma) and subjected to Northern blot analysis for 50AS transcripts. Northern blots of actin and ethidium bromide (EtBr) staining of rRNA were served as controls.
Site-directed mutagenesis.
All mutations were generated using a QuikChange multisite-directed mutagenesis kit (Stratagene). Mutagenesis was performed according to the manufacturer's protocol.
p271m and p364m were generated using the ORF-417-EGFP plasmid as a template and the primers 5′-GACCCGACATCCCATATTTCAGGGCCCGC-3′ (for 271m) and 5′-GTTGTGCCTCCGATAGTCCCTCAGACGTGG-3′ (for 364m), respectively. For p599m, the ORF-715-EGFP plasmid was used as a template with the primer 5′-CCTTGGTCTCCGTCATAACCGCCCGGAAGC-3′. In all cases, a single G nucleotide was replaced by an A nucleotide (underlined).
RESULTS
Expression of RTA antisense transcripts in BCBL1 cells.
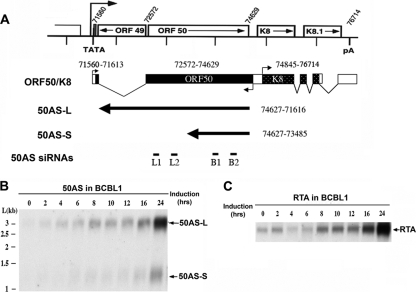
Figure 1A shows the genomic organization of the RTA locus. The RTA gene is encoded by KSHV ORF 50; its mRNA is spliced at its 5′end and is polyadenylated at a poly(A) site 3′ to the adjacent ORFs K8 and K8.1. To examine the kinetics of antisense RNA production following lytic reactivation, we exposed latently infected BCBL1 cells to a known inducer, sodium butyrate, and harvested polyadenylated RNA at various times postinduction. This RNA was examined by Northern blotting with a riboprobe specific for the noncoding strand of the ORF 50 region. As shown in Fig. 1B, two antisense (AS) transcripts of 3.0 and 1.2 kb were expressed in BCBL1 cells, which we designate 50AS-L (large) and 50AS-S (small), respectively. Both RNAs accumulate steadily during lytic replication in a fashion that parallels the accumulation of RTA mRNA itself (Fig. 1C). This indicates that RTA and the 50AS transcripts are coordinately (rather than reciprocally) expressed, like most host sense-antisense pairs. cDNAs of both 50AS transcripts were reverse transcribed from induced BCBL1 RNA, cloned into the pCR4 vector, and sequenced. Sequencing of these cDNAs affirmed earlier findings that both transcripts are unspliced, unedited, and differentially polyadenylated, as summarized in Fig. 1A. cDNA cloning and 5′ RACE (not shown) confirmed that the two transcripts initiate at a common start site, in our hands at nt 74627. (This start site is located just a few nucleotides upstream of the RTA stop codon on the opposite strand.) Although our results are very similar to earlier results (24), the 5′ end that we mapped on 50AS RNA was 109 nt upstream of the site indicated by Saveliev et al. This discrepancy is most likely due to the fact that their analysis was carried out using BC1 cells, which harbor a KSHV isolate different from that in BCBL1 cells.
FIG. 1.
50AS RNA structures. (A) Mapping of 50AS transcripts on the ORF 50 locus. Open boxes depict genomic coding regions. Black rectangles depict the cDNA structure of RTA mRNA, with nucleotide positions of exons indicated above. Black arrows depict the cDNA structure of 50AS RNAs, with the nucleotide positions that they span indicated at right. (Bottom line) Positions of 50AS siRNAs B1, B2, L1, and L2 (cf. Fig 4). (B, C) Northern blot analysis of 50AS and RTA RNAs expressed in BCBL1 cells. BCBL1 cells were induced with sodium butyrate, and mRNAs were harvested at the indicated time points postinduction. RNAs were separated on 1% agarose-formaldehyde gel and transferred to nylon membranes; these were then probed with specific riboprobes to detect the 50AS transcripts (B) or RTA (C).
50AS expression is controlled by an RTA-activated, bidirectional promoter.
To better understand the regulation of 50AS expression, 1-kb and 2-kb fragments of DNA upstream of the 50AS transcription start site were cloned in front of a luciferase (LUC) reporter gene in the construct pGL3-basic. Cells were transfected with these constructs in the absence or presence of plasmid vectors encoding RTA, and luciferase expression was assayed. As shown in Fig. 2A, the 1-kb and 2-kb 50AS DNA fragments display clear basal promoter activity in the absence of RTA and were strongly upregulated (215- to 330-fold) in the presence of RTA. Interestingly, this same genomic region directly precedes ORF K8 (which is transcribed from the opposite strand) (Fig. 1A) and was previously characterized as the K8 promoter, which is also known to be RTA responsive (30). To confirm that this region indeed contains a bidirectional promoter element, the 1-kb 50AS promoter sequence was cloned into the pGL3-basic vector in the reverse orientation. As expected, this construct also displayed basal promoter activity and was activated by RTA over 107-fold (Fig. 2A, 50AS-P-1kb-rev).
FIG. 2.
The 50AS promoter is RTA responsive and bidirectional. (A) Luciferase assays showing that the 50AS promoter and its inverted version are activated by RTA. 293 cells were transfected with luciferase reporter constructs containing the indicated 50AS promoter segments, with or without the RTA expression plasmid. The luciferase assay was performed as described in Materials and Methods. Data are presented as the fold induction over the level for the basic vector control. (B) Schematic representation of the luciferase reporter constructs with serial deletions of the 50AS promoter. The arrow indicates the transcription start site of 50ASs. The number shows the position of each deletion relative to the 50AS transcription start site. The hatched box shows the RBP-Jκ binding sites on K8 promoter identified by Wang and Yuan (30). (C) The essential region for RTA activation on 50AS promoter is positions −194 to −94. 293 cells were cotransfected with the indicated luciferase constructs, containing different 50AS promoter deletions with or without the RTA expression plasmid. The luciferase assay was performed, and luciferase activities were compared. Data are presented as the fold induction over the level for the basic vector control. (D) RBP-Jκ function is required for the RTA activation of the 50AS promoter. WT (OT13), RBP-Jκ-null (OT11), or RBP-Jκ rescued (OT11-RBP-Jκ) cells were cotransfected with the 50AS-D7 luciferase reporter construct, a β-galactosidase (β-Gal) expression construct with or without the RTA expression plasmid. Luciferase and β-galactosidase assays were performed. Transfection efficiency was normalized by β-Gal activity. Data are presented as the fold induction by RTA. All data represent the means ± SD of results from three separate experiments performed in triplicate.
To determine the essential region of the 50AS promoter required for RTA activation, serial 5′ deletion mutants were made (Fig. 2B) in the 1-kb promoter fragment-luciferase reporter construct. Each promoter deletion construct was then cotransfected into 293 cells with or without RTA. As shown in Fig. 2C, a construct (50AS-D7) bearing only 194 bp upstream of the RNA start site was sufficient for full inducibility by RTA. Like many RTA-responsive promoters, this region contains no sites homologous to the RTA recognition element (RRE) of the PAN promoter for RTA but does contain two binding sites for the transcription factor RBP-Jk (or CSL-1). We have earlier shown that RTA can be directed to RBP-Jk/CSL-1 sites on DNA by protein-protein interactions with RBP-Jk and that this interaction leads to activation of many target genes in the KSHV genome (16). (Interestingly, Wang and Yuan [30] have previously found that RBP-Jk/CSL-1 binding to this region is also required for RTA activation of K8 transcription in the opposite direction).
To affirm that RBP-Jκ binding to this region is also important for RTA activation of the 50AS promoter, the 50AS-D7 luciferase reporter construct was also tested for RTA inducibility in a mouse embryo fibroblast cell line (OT11) derived from an RBP-Jκ−/− mouse (20) and the corresponding wild type cell line (OT13). As shown in Fig. 2D, 67-fold activation by RTA was observed on 50AS-D7 in WT (OT13) MEF cells, and this activation was nearly ablated in the RBP-Jκ/CSL-1 knockout cells (OT11); substantial activation was restored by stable reexpression of recombinant RBP-Jκ/CSL-1 in the OT11 line (OT11-RBP-Jκ).
50AS RNAs do not inhibit RTA expression.
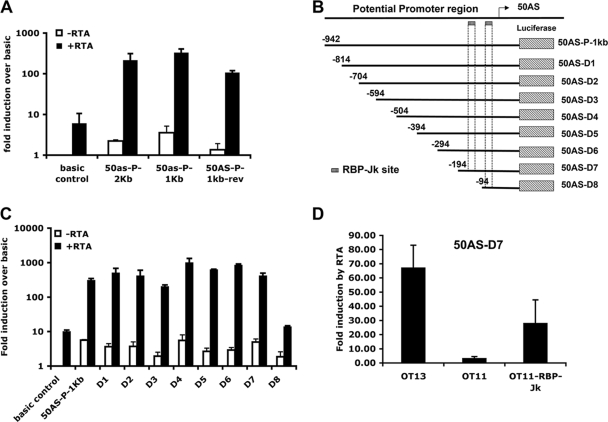
To investigate whether 50AS transcripts have any regulatory function in RTA gene expression, expression constructs for RTA and 50 AS RNAs were made (Fig. 3A, top panel). pCMV-50AS expresses both 50AS-L and 50AS-S from an adjacent cytomegalovirus (CMV) promoter. pCMV-RTA-3k expresses RTA from a 3-kb genomic fragment in which a heterologous poly(A) site is placed 3′ to the RTA coding region. With these constructs, we asked if expression of 50AS RNAs impaired accumulation of RTA mRNA or protein. Cotransfection of increasing amounts of pCMV-50AS with a fixed amount of pCMV-RTA-3k led to accumulation of increasing amounts of 50AS-L and 50AS-S (Fig. 3A, bottom left panel) but did not reduce the accumulation of RTA mRNA (Fig. 3A, bottom left panel) or RTA protein (Fig. 3A, bottom right panel).
FIG. 3.
50AS RNAs have no effect on RTA expression level in transient transfection assays. (A) (Top row) diagrammatic structures of the 50AS and RTA expression vectors. (Bottom) The pCMV-RTA-3k plasmid was cotransfected with the indicated (increasing) amounts of pCMV-50AS plasmid into 293 cells. RNA and protein were harvested at 48 h posttransfection and subjected to Northern blot analysis for 50AS or RTA RNAs (left panel) and Western blot analysis for RTA protein expression (right panel). (B) (Top) Schematic representation of constructs used for analysis of RTA and 50AS expression in cis. Arrows show the direction of the RTA transcripts. (Bottom left) Northern blot analysis showing 50AS and RTA transcripts following transfection of indicated plasmids into 293 cells. (Bottom right) Western blot analysis showing RTA expression levels by immunoblotting in cells transfected with the indicated plasmids. Actin expression is shown as a loading control.
It is important to note that in these experiments, 50AS RNAs were being supplied in trans from a separate plasmid. In vivo, however, 50AS RNAs may be transcribed from the same DNA template as RTA mRNA, offering the possibility of interactions in cis. To examine this possibility, we constructed additional plasmids in which the sense and antisense RNAs could be produced from the same DNA template. pCMV-RTA-5k contains 5 kb of KSHV genomic DNA spanning both RTA exons and the intervening intron and includes 1,793 bp of 3′ KSHV DNA. This plasmid produces RTA from a CMV-promoted transcript that closely mimics the authentic RTA mRNA; it also can produce both 50AS-L and 50AS-S, since it retains the 50AS promoter. pCMV-RTA-4.5k is derived from pCMV-RTA-5k via deletion of a 500-bp region spanning the 50AS promoter. As such, it should express the RTA message but not the 50AS transcripts. To verify that the two genomic RTA vectors express the expected RNAs, each construct was transfected into 293 cells and total RNA was harvested at 48 h posttransfection (hpt). Poly(A)+ RNA was purified and subjected to Northern blot analysis using specific riboprobes for ORF 50 and 50ASs, respectively. Figure 3B, bottom left panel, shows that pCMV-RTA-5k produces both 50AS transcripts in addition to RTA mRNA, while pCMV-RTA-4.5k produced little or no 50AS RNA but did efficiently express RTA mRNA. (The RTA mRNA produced by pCMV-RTA-4.5k is smaller than its WT counterpart due to the 500-bp deletion in the former.) In keeping with the RTA transcript levels, immunoblotting studies showed that the accumulation of RTA protein from the 4.5k vector was identical to that produced by the 5k vector (Fig. 3B, bottom right panel). We conclude that the 50AS transcripts do not strongly regulate RTA production. However, we acknowledge that this experiment does not perfectly recreate the cis regulation as it occurs in infected cells, as the CMV promoter driving RTA expression in our plasmids is stronger than the native RTA promoter.
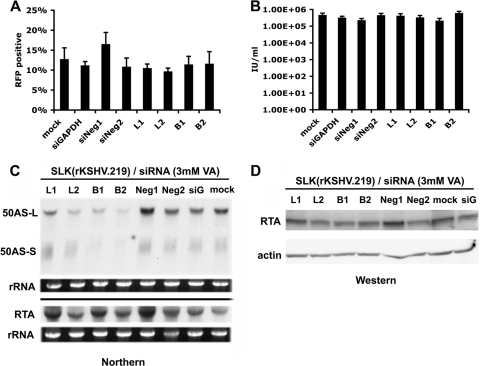
siRNA knockdown of 50AS RNAs does not affect RTA accumulation in infected cells or strongly influence the lytic cycle.
The aforementioned analyses all took place with cells transfected with RTA or 50AS expression vectors. To examine the effects of the 50AS RNAs in their natural context, we treated latently infected cells with siRNAs designed to knock down either 50AS-L or both 50AS-L and 50AS-S and examined the course of lytic KSHV infection in such cells. If 50AS transcripts negatively regulate RTA mRNAs, then selective ablation of 50AS RNAs should upregulate RTA and lead to enhanced sensitivity to lytic induction. To assess this with rigorous quantitation, we employed a genetically marked KSHV genome (rKSHV.219) (29) in which GFP is expressed constitutively in latency while an RFP gene is under the control of a lytic promoter responsive to RTA. We infected SLK endothelial cells with this virus and established a latently infected mass culture [termed SLK(rKSHV.219)] (35); as expected, 100% of these cells were GFP positive, but the frequency of spontaneous lytic reactivation, as judged by RFP expression, was low (<0.5%).
We designed several siRNA constructs to target the 50AS RNAs. siRNAs B1 and B2 target the region common to both 50AS-L and 50AS-S, while siRNAs L1 and L2 target the region unique to 50AS-L and should spare 50AS-S (Fig. 1A, bottom panel). When SLK(rKSHV.219) cells were transfected with these siRNAs, no impact was seen on the percentage of cells undergoing spontaneous lytic reactivation (not shown), not surprisingly, perhaps, since most transfected cells are not detectably expressing 50AS transcripts. When SLK(rKSHV.219) cells transfected with these siRNAs were induced with a submaximal concentration of the inducer valproic acid (3 mM), conditions sufficient to induce 10 to 15% of the mock-transfected culture to RFP positivity, no consistent effect on the frequency of lytic induction was observed (Fig. 4A). In support of this conclusion, when virus in the supernatants of these induced cultures was assayed for infectivity, similar yields of infectious virus were produced, irrespective of the siRNA used for the transfection (Fig. 4B). Figure 4C confirms that in these induced, KSHV-infected cells, transfection with siRNA B1 or B2 strongly reduced accumulation of both 50AS-L and 50AS-S (relative to cells transfected with each of 3 control siRNAs) but that transfection of siRNAs L1 and L2 selectively reduced accumulation of 50AS-L. Finally, neither siRNAs B1 and B2 nor siRNAs L1 and L2 significantly or consistently influenced the accumulation of RTA mRNA (Fig. 4C) or protein (Fig. 4D) in these induced cultures. We conclude that 50AS transcripts have little effect on RTA expression or virus yield in the context of viral infection in culture.
FIG. 4.
Analysis of siRNA-mediated knockdown of 50AS RNAs in SLK(rKSHV.219) cells. SLK endothelial cells expressing a recombinant KSHV virus with RFP under the control of a lytic promoter [SLK(rKSHV.219] were transfected with no siRNA (mock), negative-control siRNAs (Neg1 and Neg2), GAPDH siRNA (siG), or each of the 50AS siRNAs (L1, L2, B1, and B2) depicted in Fig. 1A. Cells were induced with 3 mM valproic acid for 72 h. (A) Cells were analyzed by flow cytometry for expression of RFP as a reporter of lytic induction. Data represent the means ± SD of results from four separate experiments. (B) Viruses were harvested from filtered supernatant of each sample at 72 h postinduction. Viral titer was assayed by infection of 293 cells and flow cytometry for GFP expression. Viral production is shown as numbers of infectious units (IU) per ml. All data represent the means ± SD of results from four separate experiments. (C) RNA was harvested from each sample at 48 h postinduction and probed for 50AS and RTA expression, respectively. rRNA level was shown by ethidium bromide (EtBr) staining as an input control. (D) Proteins were harvested and assayed for RTA protein expression. Actin expression is shown as an input control.
50AS RNAs are associated with polyribosomes.
Examination of the nucleotide sequence of 50AS-S RNA showed that it harbors no large open reading frames (ORFs). As suggested by the genomic map of Fig. 1, sequences spanning KSHV ORF 49, which are located antisense to the intron of RTA mRNA, are represented at the 3′ end of 50AS-L. However, in infected cells ORF 49 is expressed from a separate, 1.3-kb mRNA that initiates 15 nt upstream of the ORF 49 initiation codon (11). Consistent with this, in vitro translation of 50AS-L mRNA in a rabbit reticulocyte lysate did not engender ORF 49 protein (or any other conventional polypetide) (see Fig. S1 in the supplemental material). Nonetheless, our finding that 50AS RNAs do not regulate RTA expression led us to consider the possibility that they might be unconventional mRNAs, perhaps producing proteins by unusual translational strategies (e.g., the use of non-AUG initiation codons, ribosomal frameshifting, or both).
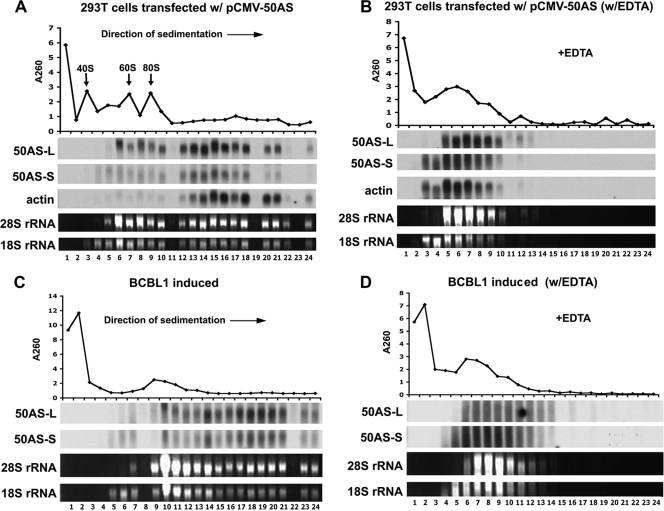
To investigate this, we first asked if 50AS transcripts can be found on polyribosomes in vivo. 293T cells were transfected with pCMV-50AS, and cell extracts were harvested at 48 h posttransfection and separated on 10 to 50% linear sucrose density gradients. After fractionation, RNA extracted from each fraction was examined by Northern blotting using 50AS-specific riboprobes. Figure 5A shows that substantial amounts of the 50AS-L and 50AS-S transcripts were associated with fractions expected to contain polyribosomes; analysis of each fraction by blotting for actin mRNA demonstrated that actin messages display a similar profile. To confirm that this sedimentation profile is indeed reflective of polysome association, extracts were pretreated before centrifugation with EDTA to dissociate the ribosomes and then analyzed by sucrose velocity sedimentation as before. The A260 profile of the gradient, as well as the analysis of the rRNA and actin mRNA content of each fraction, confirmed polysome dissociation (Fig. 5B). With EDTA treatment, both 50AS transcripts, like actin mRNA, were redistributed to the top of the gradient. To confirm that 50ASs associated with polyribosomes in the context of authentic viral replication, latently infected BCBL1 cells were induced to lytic KSHV growth with sodium butyrate; after 24 h, cell lysates were prepared and subjected to a polyribosome fractional assay with or without EDTA pretreated. Figure 5C and D show that in the viral context, 50AS transcripts are again associated with polyribosomes. These data provide strong circumstantial evidence that these transcripts are competent for translation.
FIG. 5.
50AS transcripts are associated with polyribosomes. (A, B) 293T cells transfected with pCMV-50AS were subjected to polyribosomal fractionation assays without (A) or with (B) EDTA treatment. (C, D) BCBL1 cells induced with 0.3 mM sodium butyrate for 24 h were subjected to polyribosomal fractionation assays without (C) or with (D) EDTA treatment. Total RNA was purified from each fraction and subjected to Northern blotting for 50AS-L, 50AS-S, and actin, using a specific riboprobe for each transcript. The 28s rRNA and 18s rRNA levels are also shown by ethidium bromide (EtBr) staining. The A260 profile for each gradient is shown at the top of each panel.
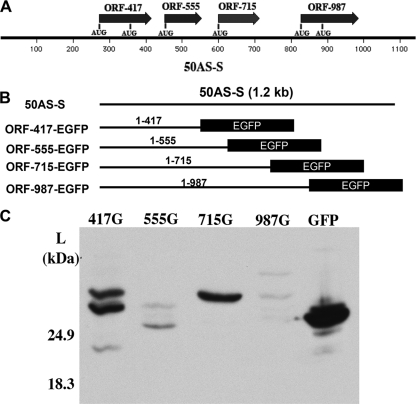
50AS-S encodes at least three small peptides.
To study the possible protein products from 50AS translation, we focused on the smaller 50AS-S transcript. Sequence inspection showed that there are at least four small potential ORFs with AUG initiators located on 50AS-S (Fig. 6A) (for reasons that will become clear below, each ORF is designated with a number corresponding to the last nucleotide of the ORF). To investigate whether these small ORFs can be translated, each potential ORF was fused (in-frame) to a GFP coding sequence (with the authentic GFP ATG start codon deleted); each fusion protein is named according to the nucleotide at the 50AS/GFP fusion junction. To maintain the original context of the 50AS-S transcript, each construct began at the 50AS-S transcriptional start site (Fig. 6B). 293T cells were transfected with each construct, and total cell lysates from each transfection were subjected to Western blot analysis using EGFP antibody. As shown in Fig. 6C, fusions to ORFs 555 and 987 generated only trace levels of GFP-related proteins, most of which were of insufficient size to carry the predicted fusion protein; the significance of these bands is doubtful. In contrast, two major GFP fusion proteins were efficiently generated from the ORF 417-GFP construct (Fig. 6C, lane 417G), and a single polypeptide was generated from the ORF 715-GFP construct (Fig. 6C, lane, 715G). Each of these GFP fusion proteins was larger than GFP protein itself (Fig. 6C, lane GFP), indicating that these are fusion proteins likely translated from the upstream 50AS-S ORFs.
FIG. 6.
GFP fusion proteins are expressed from two potential open reading frames (ORFs) of 50AS-S RNA. (A) Potential ORFs on 50AS-S. Each ORF was named by its stop codon nucleotide position on the 50AS-S RNA. The potential translation start codons (AUGs) for each ORF are also indicated. (B) Schematic representation of the EGFP fusion constructs. Each potential 50AS ORF with its own stop codon deleted was fused in-frame with EGFP and expression driven by the CMV promoter. All constructs begin at the transcription start site of 50AS-S. (C) A Western blot with GFP antibody shows the expression of the GFP fusion proteins. 293T cells were transfected with individual GFP fusion constructs. Total cell lysates were harvested at 48 hpt and subjected to Western blotting for GFP fusion protein expression.
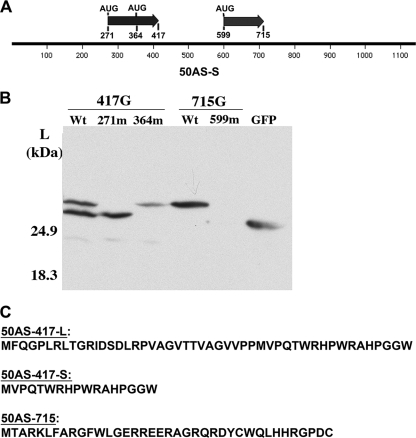
To confirm this hypothesis, each of the potential AUG initiation codons on the ORF 417-GFP and ORF 715-GFP constructs was mutated to AUA to disrupt the translation of the corresponding GFP fusion protein (Fig. 7A). When the first AUG of ORF 417 (located at nt 271) was mutated, the larger of the two GFP fusion proteins was no longer expressed (Fig. 7B, lane 271m). When the second AUG of ORF 417 (located at nt 364) was inactivated, the lower band of the doublet disappeared (Fig. 7B, lane 364m). Similarly, mutation of the sole AUG of ORF 715 (located at nt 599) led to ablation of the cognate GFP fusion protein (Fig. 7B). These results confirm that the AUGs on ORF 417 and ORF 715 of 50AS-S are translationally competent in the context of 50AS transcripts and are responsible for the expression of the corresponding GFP fusion protein.
FIG. 7.
50AS-S RNA encodes at least three small peptides. (A) Diagram showing ORF 417, ORF 715, and their potential translation initiation codons on 50AS-S RNA. Also shown are the relative nucleotide positions of each starting codon on 50AS-S. (B) Point mutations in the AUG codons disrupt the expression of the corresponding GFP fusion proteins. Each AUG codon on ORF 417 and ORF 715 was mutated to AUA and transfected into 293T cells. Total cell lysates were harvested at 48 hpt and subjected to Western blotting for GFP fusion protein expression. (C) Amino acid sequence of the three 50AS-S-encoded peptides.
The predicted peptide sequences encoded by these three ORFs are shown in Fig. 7C. All are tiny; the predicted products of ORF 417 are only 48 and 17 amino acids, and that of ORF 715 is only 38 aa long. One common feature of these putative peptides is that they are all extremely basic, with pI values of 11.82 for the 48-aa peptide of ORF 417, 12.48 for the 17-aa peptide of ORF 417-S, and 10.06 for the product of ORF 715. A scan of their predicted amino acid sequences yielded no predicted motifs that might help assign potential functions, and efforts to detect the peptides in infected or transfected cells have been frustrated by their poor immunogenicity, small size, and likely low abundance. However, all 3 peptides are completely conserved, at both the amino acid level (Fig. 8) and the nucleotide level (not shown), in the four independent isolates of KSHV for which DNA sequences are available.
FIG. 8.
Conservation of 50AS peptides in different strains of KSHV. Amino acid sequences of peptides 417 (A) and 715 (B) from different KSHV strains are aligned. The GenBank reference numbers for sequence data are as follows: for BC1, U75698; for strain GK18, AF148805; and for 219BAC, GQ994935.1.
DISCUSSION
The RTA and 50AS RNAs of KSHV provide an instructive example of a sense-antisense transcript pair of the type that is now being widely recognized in the mammalian transcriptome. Absent any direct experimental tests, one would have been very likely to ascribe to 50AS RNAs a role in regulation of RTA: RTA is, after all, the master regulator of the lytic switch and an obvious node in any regulatory network governing viral replication. However, every experimental test that we could design proved inconsistent with the notion that the primary function of 50AS RNAs is to regulate RTA expression. These RNAs did not regulate RTA RNA accumulation in cis or in trans in transfected cells, and knockdown of the 50AS RNAs had no significant effects on virus reactivation, the function controlled by RTA; viral infectivity was likewise unaffected by siRNAs directed against 50AS transcripts. These results are consistent with the facts that RTA and 50AS RNAs accumulate in tandem during the viral replicative cycle (Fig. 1B and C) and that RTA protein levels rise throughout the cycle despite the accumulation of 50AS RNAs (18). While it is difficult to decisively exclude a contributory regulatory role for 50AS transcripts under at least some conditions in vivo, our data strongly point in a different direction, namely, that 50AS transcripts are unconventional mRNAs that direct the production of small peptides of as yet unknown function.
Despite extensive efforts, we have not yet been able to detect these peptides biochemically in infected cells. The peptides have proven to be poorly immunogenic following conjugation to carrier proteins, and the antisera that we have raised have not been able to identify the free peptides by immunoblotting or immune precipitation. However, the presence of 50AS RNA on polysomes (Fig. 5) and the expression data shown in Fig. 6 and 7 leave little doubt that their AUG codons are accessed by the translational machinery. As to the function of these peptides, the preservation of viral induction and infectivity following siRNA knockdown of 50AS transcripts (Fig. 4) argues against an essential role in viral replication, assembly, or release (though this conclusion must be qualified by the fact that siRNA experiments do not ablate expression of their target completely). Nonetheless, the conservation of their coding sequences in all isolates of KSHV speaks of a likely role in vivo, perhaps involving an aspect of infection not assayed in culture, e.g., virion stability, mucosal persistence, person-to-person spread, or immune evasion.
Our findings have several additional implications that do not revolve around any putative functions of the peptides. First, these results underscore the importance of direct experimental examination of sense-antisense pairs on a case-by-case basis and emphasize that computational assignments of RNAs as noncoding should always be considered provisional. RNAs annotated as noncoding usually have many AUG-directed ORFs smaller than the window set computationally; moreover, ribosomal frameshifting (9) has the potential to generate additional products not predicted by simple enumeration of predicted ORFs. (In fact, we carried out extensive searches for such frameshifted products from 50AS mRNAs [using epitope-tagging strategies] but failed to identify any such species [Y. Xu, unpublished data].) Second, given the large number of putative noncoding RNAs in the mammalian transcriptome, there may well exist in many cells a substantial collection of small peptides derived not by proteolysis or abortive translation (33) but by directed translation of small ORFs. In keeping with this idea, a recent paper has suggested that the tal RNA of Drosophila, previously annotated as a noncoding RNA (though it is not an antisense transcript), may encode peptides as short as 11 amino acids (10). On the basis of these studies, it is likely that many more examples of this phenomenon will surface as individual “antisense” or “noncoding” transcripts undergo closer experimental scrutiny.
It is conceivable that many of these peptides may not be synthesized for any express biological purpose; rather, their association with polysomes might represent the “background noise” of the translational initiation machinery, reflecting the access of occasional scanning ribosomes to internal initiation codons. Even if so, this would not be a trivial finding when considered on a genome-wide scale. For example, such host-derived peptides would be expected to enter the major histocompatibility complex (MHC) class I antigen presentation pathway, resulting in still more autoantigens than have previously been appreciated, in a fashion analogous to that described by Yewdell and Nicchitta for peptides resulting from aborted elongation (or other aberrant events) during translation of conventional ORFs (33). For genes from exogenous pathogens, like viruses, this could mean that many more cytotoxic T lymphocyte (CTL) responses could be generated during an infection than are currently measured. Clearly, much more attention will need to be paid to the possibility that “noncoding” RNAs may engender noncanonical translation products. Our approach of examining their association with polysomes provides a useful way of beginning such an investigation, one which, if married to appropriately crafted microarrays or large-scale cDNA cloning, can potentially be expanded to a genome-wide scale.
Supplementary Material
Acknowledgments
We thank J. Vieira (University of Washington) for Vero cells producing rKSHV.219 and J. Myoung (UCSF) for generating the SLK cells infected with this virus.
Footnotes
Published ahead of print on 31 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bechtel, J., A. Grundhoff, and D. Ganem. 2005. RNAs in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:10138-10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertone, P., V. Stolc, T. E. Royce, J. S. Rozowsky, A. E. Urban, X. Zhu, J. L. Rinn, W. Tongprasit, M. Samanta, S. Weissman, M. Gerstein, and M. Snyder. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306:2242-2246. [DOI] [PubMed] [Google Scholar]
- 3.Bor, Y. C., J. Swartz, Y. Li, J. Coyle, D. Rekosh, and M.-L. Hammarskjöld. 2006. Northern blot analysis of mRNA from mammalian polyribosomes. Nat. Protoc. doi: 10.1038/nprot.2006.216. [DOI]
- 4.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V. B. Bajic, S. E. Brenner, S. Batalov, A. R. Forrest, M. Zavolan, M. J. Davis, L. G. Wilming, V. Aidinis, J. E. Allen, A. Ambesi-Impiombato, R. Apweiler, R. N. Aturaliya, T. L. Bailey, M. Bansal, L. Baxter, K. W. Beisel, T. Bersano, H. Bono, A. M. Chalk, K. P. Chiu, V. Choudhary, A. Christoffels, D. R. Clutterbuck, M. L. Crowe, E. Dalla, B. P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C. F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T. R. Gingeras, T. Gojobori, R. E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T. K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S. P. Krishnan, A. Kruger, S. K. Kummerfeld, I. V. Kurochkin, L. F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- 5.Carninci, P., J. Yasuda, and Y. Hayashizaki. 2008. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 20:274-280. [DOI] [PubMed] [Google Scholar]
- 6.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, J., P. Kapranov, J. Drenkow, S. Dike, S. Brubaker, S. Patel, J. Long, D. Stern, H. Tammana, G. Helt, V. Sementchenko, A. Piccolboni, S. Bekiranov, D. K. Bailey, M. Ganesh, S. Ghosh, I. Bell, D. S. Gerhard, and T. R. Gingeras. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149-1154. [DOI] [PubMed] [Google Scholar]
- 8.Ebralidze, A. K., F. C. Guibal, U. Steidl, P. Zhang, S. Lee, B. Bartholdy, M. A. Jorda, V. Petkova, F. Rosenbauer, G. Huang, T. Dayaram, J. Klupp, K. B. O'Brien, B. Will, M. Hoogenkamp, K. L. Borden, C. Bonifer, and D. G. Tenen. 2008. PU. 1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 22:2085-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh, P. J. 2000. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 64:131-170. [DOI] [PubMed] [Google Scholar]
- 10.Galindo, M. I., J. I. Pueyo, S. Fouix, S. A. Bishop, and J. P. Couso. 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González, C. M., E. L. Wong, B. S. Bowser, G. K. Hong, S. Kenney, and B. Damania. 2006. Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 80:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings, M. L., H. A. Ingle, M. A. Lazar, and S. H. Munroe. 2000. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J. Biol. Chem. 275:11507-11513. [DOI] [PubMed] [Google Scholar]
- 13.Hastings, M. L., C. Milcarek, K. Martincic, M. L. Peterson, and S. H. Munroe. 1997. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 25:4296-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y., Y. C. Bor, Y. Misawa, Y. Xue, D. Rekosh, and M. L. Hammarskjold. 2006. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 443:234-237. [DOI] [PubMed] [Google Scholar]
- 16.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 20.Oka, C., T. Nakano, A. Wakeham, J. L. de la Pompa, C. Mori, T. Sakai, S. Okazaki, M. Kawaichi, K. Shiota, T. W. Mak, and T. Honjo. 1995. Disruption of the mouse RBP-J. kappa gene results in early embryonic death. Development 121:3291-3301. [DOI] [PubMed] [Google Scholar]
- 21.Prasanth, K. V., and D. L. Spector. 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 21:11-42. [DOI] [PubMed] [Google Scholar]
- 22.Prescott, E. M., and N. J. Proudfoot. 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 99:8796-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sado, T., Y. Hoki, and H. Sasaki. 2005. Tsix silences Xist through modification of chromatin structure. Dev. Cell 9:159-165. [DOI] [PubMed] [Google Scholar]
- 24.Saveliev, A., F. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-314. [DOI] [PubMed] [Google Scholar]
- 25.Sun, B. K., A. M. Deaton, and J. T. Lee. 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21:617-628. [DOI] [PubMed] [Google Scholar]
- 26.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thenie, A. C., I. M. Gicquel, S. Hardy, H. Ferran, P. Fergelot, J. Y. Le Gall, and J. Mosser. 2001. Identification of an endogenous RNA transcribed from the antisense strand of the HFE gene. Hum. Mol. Genet. 10:1859-1866. [DOI] [PubMed] [Google Scholar]
- 28.Tufarelli, C., J. A. Stanley, D. Garrick, J. A. Sharpe, H. Ayyub, W. G. Wood, and D. R. Higgs. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34:157-165. [DOI] [PubMed] [Google Scholar]
- 29.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225-240. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jkappa in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 359:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79:3479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasuda, J., and Y. Hayashizaki. 2008. The RNA continent. Adv. Cancer Res. 99:77-112. [DOI] [PubMed] [Google Scholar]
- 33.Yewdell, J. W., and C. V. Nicchitta. 2006. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 27:368-373. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, G., B. Raghavan, M. Kotur, J. Cheatham, D. Sedmak, C. Cook, J. Waldman, and J. Trgovcich. 2007. Antisense transcription in the human cytomegalovirus transcriptome. J. Virol. 81:11267-11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegelbauer, J. M., C. S. Sullivan, and D. Ganem. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41:130-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.