Abstract
Human bocavirus (HBoV) was recently discovered and classified in the Bocavirus genus (family Parvoviridae, subfamily Parvovirinae) on the basis of genomic similarity to bovine parvovirus and canine minute virus. HBoV has been implicated in respiratory tract infections and gastroenteric disease in children worldwide, yet despite numerous epidemiological reports, there has been limited biochemical and molecular characterization of the virus. Reported here is the three-dimensional structure of recombinant HBoV capsids, assembled from viral protein 2 (VP2), at 7.9-Å resolution as determined by cryo-electron microscopy and image reconstruction. A pseudo-atomic model of HBoV VP2 was derived from sequence alignment analysis and knowledge of the crystal structure of human parvovirus B19 (genus Erythrovirus). Comparison of the HBoV capsid structure to that of parvoviruses from five separate genera demonstrates strong conservation of a β-barrel core domain and an α-helix, from which emanate several loops of various lengths and conformations, yielding a unique surface topology that differs from the three already described for this family. The highly conserved core is consistent with observations for other single-stranded DNA viruses, and variable surface loops have been shown to confer the host-specific tropism and the diverse antigenic properties of this family.
Human bocavirus (HBoV), a newly discovered member of the family Parvoviridae, was originally isolated in randomly selected nasopharyngeal aspirates (5). Since this initial discovery, HBoV has also been detected worldwide, predominantly in children under the age of 2 years with respiratory infections, in serum, urine, and fecal samples (40). Symptomatic children commonly exhibit acute diseases of the upper and lower respiratory tracts (7, 36, 44, 56) and, possibly, gastroenteritis (31, 56) though the link to gastroenteritis outbreaks has been questioned (12). It is still unclear if HBoV is the sole etiologic agent of respiratory disease as higher rates of coinfections with other respiratory pathogens such as human rhinovirus and Streptococcus spp. are often observed (4). However, Allander et al. recently reported (4) that HBoV was found in 19% of children with acute wheezing, thereby making it the fourth most common virus, after rhinoviruses, enteroviruses, and respiratory syncytial virus, detected in children exhibiting this symptom. These findings suggest that, at high viral load, HBoV could be an etiologic agent of respiratory tract disease (4). HBoV infection is common in the first few years of life, and clinical research suggests it may follow the primary period for acquisition of human parvovirus B19 (B19) though there is no antigenic cross-reactivity between B19 and HBoV (28, 30). By age 5, most people have circulating antibodies against HBoV, as is also true for other respiratory viruses such as respiratory syncytial virus, rhinoviruses, and human metapneumovirus (17). HBoV has also been identified in adults, with ∼63% of samples tested being seropositive, showing a positive correlation with age and a slight positive bias toward women (14).
The Parvoviridae is a family of small, nonenveloped viruses that package a single-stranded DNA (ssDNA) genome of ∼5,000 bases. These viruses are subdivided into two subfamilies: Parvovirinae and Densovirinae (Table 1). The Parvovirinae are further subdivided into five genera, all of whose members infect vertebrates. The Densovirinae (four genera) infect only invertebrates. Phylogenetic analysis places HBoV in the recently classified Bocavirus genus (Table 1). In addition to HBoV, numerous parvoviruses circulate among the human population. Among these are the following: several dependoviruses; adeno-associated virus (AAV) serotypes AAV1 to AAV3, AAV5, and AAV9; the Erythrovirus B19; and the newly discovered human parvovirus genotypes 4 (Parv4) and 5 (Parv5) (23, 27, 50). Of these, only B19 had been implicated in disease until the discovery of HBoV and Parv4, which has been isolated from patients who present symptoms of acute HIV infection (50).
TABLE 1.
Selected properties of representative members of the Parvoviridae
| Subfamily (host) and genus | Member(s)a | No. of VPsb | Groupc | Major VP(s)d |
|---|---|---|---|---|
| Parvovirinae (vertebrate) | ||||
| Parvovirus | MVM*, CPV*, FPV* | 3 | I | VP2 |
| Erythrovirus | B19*†, SPV | 2 | III | VP2 |
| Dependovirus | AAV2*†, AAV4*†, GPV | 3 | III | VP3 |
| Amdovirus | AMDV† | 2 | III | VP2 |
| Bocavirus | HBoV†, BPV, CnMV | 2 | NA | VP2 |
| Densovirinae (invertebrate) | ||||
| Densovirus | GmDNV*, JcDNV† | 4 | II | VP4 |
| Iteravirus | BmDNV† | 4-6 | II | VP1-VP4 |
| Brevidensovirus | AaeDNV, AalDNV | 2-3 | NA | VP1 or VP2/3 |
| Pefudensovirus | PfDNV | 5 | NA | VP1 |
AalDNV, Aedes albopictus densovirus; AaeDNV, Aedes aegypti densovirus; BmDNV, Bombyx mori densovirus; BPV, bovine parvovirus; CnMV, canine minute virus; CPV, canine parvovirus; FPV, feline panleukopenia virus; GPV, goose parvovirus; JcDNV, Junonia coenia densovirus; PfDNV, Periplaneta fuliginosa densovirus; SPV, simian parvovirus. *, structure determined by X-ray crystallography; †, structure determined by cryo-EM.
The number of VPs in the virion capsid.
Group refers to the surface topologies described in Results and Discussion (HBoV is currently the only bocavirus with a known structure; there is no structure available for Periplaneta fuliginosa densovirus or the brevidensoviruses). NA, structural group not assigned.
The VP(s) that comprises most of the wild-type virion.
The HBoV genome, like that of all members of the Bocavirus genus, contains three open reading frames (ORFs). The first ORF, at the 5′ end, encodes NS1, a nonstructural protein. The next ORF, unique to the bocaviruses, encodes NP1, a second nonstructural protein. The third ORF, at the 3′ end, encodes the two structural capsid viral proteins (VPs), VP1 and VP2. The HBoV VPs share 42% and 43% amino acid sequence identity with the corresponding VPs of bovine parvovirus and canine minute virus, respectively (5). More recently, two additional HBoV-like viruses, HBoV-2 and HBoV-3, were identified in stool samples from children (8, 31). The genome organization of these viruses is identical to that of HBoV, with the NS1, NP1, and VP proteins of HBoV-2 and HBoV-3 being, respectively, ∼80 and 90%, ∼70 and 80%, and ∼80 and 80% identical to the respective proteins in HBoV (8, 31).
Parvovirus genomes are packaged into a T=1 icosahedral capsid that is assembled from 60 copies of a combination of up to six types of capsid VPs (VP1 to VP6), all of which share a C terminus. VP1 is always a minor component, typically comprising about five copies per capsid, whereas the smallest VP is always the major component. The unique N-terminal region of VP1 (VP1u) contains a conserved phospholipase A2 (PLA2) motif within the first 131 amino acids that is essential for infection (49, 61). Interestingly, Aleutian mink disease virus (AMDV), the only member of the Amdovirus genus, is the only exception in that this motif is absent, which suggests that this virus employs a different mechanism to escape the endosome during infection (54).
The X-ray crystal structures of several parvoviruses show that all VPs contain a conserved, eight-stranded β-barrel motif (βB to βI) that forms the core of the capsid (15). There is also a conserved α-helix (αA) observed in all parvovirus structures determined to date. The bulk of the VP consists of elaborate loops between the strands that form the surface of the capsid. For example, the GH loop between the βG and βH strands is ∼230 residues. The composition and topology of these loops encode several important functions, including tissue tropism, pathogenicity, and the antigenic response directed against each parvovirus during infection (2).
A number of parvoviruses have been studied by cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction in concert with and complementary to X-ray crystallographic studies (reviewed in reference 15). Reported here is the 3D structure of a recombinant HBoV capsid solved to 7.9-Å resolution using cryo-EM. The capsid of HBoV was compared to that of representative members of the Parvoviridae (Table 1) with known atomic structures (AAV2, minute virus of mice [MVM], B19, and Galleria mellonella densovirus [GmDNV]) or pseudo-atomic models built into cryo-EM reconstructed density (AMDV) to identify similarities and differences. The capsid topology of the newly emerging HBoV incorporates a combination of surface structural features seen in other members of the Parvovirinae and is closest to that of B19, the only other structurally characterized parvovirus that is pathogenic to humans. A pseudo-atomic model of the HBoV VP2, built into the reconstructed density, identified conserved core secondary structure elements, which are known to be important for parvovirus capsid assembly, and variable surface loops, which likely govern host specific interactions.
MATERIALS AND METHODS
Expression and purification.
The putative HBoV VP2 gene was constructed by de novo synthesis using the published HBoV ST1 sequence (NCBI accession number DQ000495; start codon, nucleotide [nt] 3373), and cloned into a pUC vector (Epoch Biolabs, Inc., Missouri City, TX). The vector was cut with EcoRV and HindIII to release the insert and recloned into a Fastbac1 plasmid (Invitrogen). The sequence and orientation of the resulting plasmid were confirmed, and the plasmid was used to produce a recombinant baculovirus using the standard Bac-to-Bac technology (Invitrogen).
Sf9 insect cells (ATCC) were maintained in Grace's medium (Invitrogen) with 10% fetal calf serum (FCS) and antibiotics; cells were infected with recombinant baculovirus, harvested 4 or 7 days postinfection into the medium, and collected by centrifugation. Production of baculovirus was confirmed by a combination of immunofluorescence of infected cells with mouse antibody against baculovirus (eBioscience) and the typical cytopathic effect of baculovirus-infected cells. For the immunofluorescence testing, slides were prepared by cytocentrifugation (200 × g for 8 min in a Shandon Cytospin 4) or by spotting a concentrated cell suspension onto each of eight spots of a fluorescent antibody slide (Bellco Glass, Inc.). The slides were allowed to air dry and were fixed in 1:1 acetone-methanol at −20°C for 10 min. The monoclonal antibody was diluted 1:50 in phosphate-buffered saline (PBS) plus 10% FCS, and 25 μl of the solution was incubated with each cell spot (∼200 cells) for 1 h at 37°C. After cells were washed with PBS, they were incubated with 1:100 anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody in PBS plus 10% FCS and 1:200 Evans Blue (Sigma) for 1 h at 37°C. After another washing step, the cells were examined by UV microscopy.
Sf9 cells were grown in suspension at 27°C in Sf-900 II SFM medium (Gibco/Invitrogen Corporation) and infected at a multiplicity of infection of 5 PFU per cell. Resultant HBoV virus-like particles (VLPs) were released from infected cells by three freeze-thaw cycles in lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100), with the addition of benzonase (Merck KGaA) after the second cycle. The sample was clarified by centrifugation at 12,100 × g for 15 min at 4°C. The cell lysate was pelleted through a 20% (wt/vol) sucrose cushion (in 25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.2% Triton X-100, 1 mM EDTA) by ultracentrifugation at 149,000 × g for 3 h at 4°C. The pellet from the cushion was resuspended in the same buffer overnight at 4°C. The sample was subjected to multiple low-speed spins at 10,000 × g in order to remove insoluble material. The clarified sample was purified by separation within a 10 to 40% (wt/vol) sucrose step gradient spun at 151,000 × g for 3 h at 4°C. Equilibrium dialysis was performed against 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM MgCl2 at 4°C. The final concentration of the sample was 10 mg ml−1 calculated from optical density measurements at 280 nm, with an extinction coefficient of 1.7 M−1 cm−1. The purity and integrity of the viral capsids were monitored by SDS-PAGE and negative-stain electron microscopy, respectively.
Negative-stain, transmission electron microscopy.
Small (3.5 μl) aliquots of purified VLPs (∼5 mg ml−1) were applied to a continuous carbon support film that had been glow discharged for 15 s in an Emitech K350 glow-discharge unit and subsequently stained with 1% aqueous uranyl acetate. Micrographs were recorded on a 2,048- by 2,048-pixel Gatan charge-coupled device (CCD) camera in an FEI Sphera microscope at a nominal magnification of ×40,000 at an accelerating voltage of 200 keV.
Cryo-EM and image reconstruction.
Small (3.5 μl) aliquots of purified VLPs (∼10 mg ml−1) were vitrified via standard rapid freeze-plunging procedures (1, 20). Samples were applied to Quantifoil holey grids that had been glow discharged for ∼15 s in an Emitech K350 glow-discharge unit. Grids were then blotted for ∼5 s, plunged into liquid ethane, and transferred into a precooled FEI Polara multispecimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron image film at 200 keV in an FEI Polara microscope under minimum-dose conditions (∼9 e/Å2) at a nominal magnification of ×39,000.
Sixteen micrographs exhibiting minimal astigmatism and specimen drift, recorded at underfocus settings ranging between 0.95 and 3.25 μm, were digitized at 7-μm intervals (representing 1.795-Å pixels) on a Zeiss SCAI scanner. The software package RobEM (http://cryoEM.ucsd.edu/programs.shtm) was used to extract 3,754 individual particle images (each 1872 pixels), preprocess the images, and estimate the defocus level of each micrograph (9). A random-model procedure (59) was used to generate an initial reconstruction at ∼25-Å resolution from 150 particle images. This was then used as a starting model to initiate full orientation and origin determinations and refinement of the entire set of images using AUTO3DEM (60). Corrections to compensate for the effects of phase reversals in the contrast-transfer functions of the images were performed as previously described (10, 62), but amplitude corrections were not applied. A final 3D map at an estimated resolution limit of 7.9 Å, based on the 0.5 threshold of the Fourier shell correlation (FSC0.5) criterion (55) (data not shown), was reconstructed from all particle images. To verify the new structure, we computed two, completely independent, ab initio reconstructions from separate sets of micrographs consisting of 1,770 and 1,839 particle images. The resultant reconstructions showed excellent correspondence in all coarse and fine features, as evidenced by good agreement (FSC0.5) at all spatial frequencies out to 1/9.8 Å−1. An inverse temperature factor of 1/100 Å2 was applied to the final 3D reconstruction (26) to enhance fine details and to aid in the analysis and interpretation of the capsid structure. A Gaussian function was applied to attenuate the Fourier data smoothly to zero between 7.24 and 6.75 Å. The absolute handedness of the reconstruction was set to be consistent with surface features seen in parvovirus crystal structures (see below).
Generation of density maps for representative members of the Parvoviridae genera.
Density maps were generated from the crystal structure Cα coordinates of AAV2 (Protein Data Bank [PDB] code 1LP3) (58), B19 (PDB code 1S58) (33), GmDNV (PDB code 1DNV) (51), and MVM (PDB code 1Z1C) (38) and from the Cα pseudo-atomic model of AMDV (42). The coordinates for each VP2 subunit were used to generate a complete icosahedral capsid using the oligomer generator in VIPERdb (13). The CCP4 software suite was used to calculate structure factors (7.9-Å resolution cutoff; B-factor of 100 Å2) and electron density maps, using the SFALL and FFT (fast Fourier transform) routines, respectively (18). The calculated maps were rendered with RobEM (http://cryoEM.ucsd.edu/programs.shtm) and Chimera (48).
Sequence analysis and generation of unrooted phylogenic tree.
VP2 sequences of representative members of the six Parvoviridae genera (taken from the PDB accession files listed above, AMDV [NCBI accession number ABG20974], and HBoV [NCBI accession number ABN46897]) were uploaded to the Biology Workbench (52). A multiple sequence alignment was conducted using ClustalW (53) with default values and with B19 as the reference sequence. An unrooted phylogenetic tree was then created using the PHYLIP algorithm (22).
Generation and docking of a homology model of HBoV VP2 into cryo-reconstructed density.
A homology model was built for the HBoV VP2 (amino acids 37 to 542) using the Swiss-PdbViewer Deep View, version 3.7, software program (25) with the VP2 sequence (NCBI accession number ABN46897) and the B19 VP2 coordinates (PDB code 1S58) supplied as a template. B19 VP2 was chosen as a template for building the HBoV VP2 pseudo-atomic model because the comparison of density maps generated for the members of the different Parvoviridae genera (see above) identified B19 as being the most structurally similar to HBoV even though the AAV2 VP2 sequence had a slightly higher sequence identity.
The HBoV VP2 homology model was used to generate the coordinates for a complete, 60-subunit capsid through icosahedral matrix multiplication in VIPERdb (13). The 60-mer was then visualized within the HBoV cryo-EM density map in COOT (21). The core β-barrel (β-strands B to I) and helix αA elements fitted into density contoured at a 3.3-σ threshold without further adjustment. The loops between β-strands D and E (DE loop) and between β-strands H and I (HI loop) of a reference VP2 monomer were manually adjusted to fit into density contoured at a threshold of 1.0 σ guided by their distinct structural features. The remaining surface loops were adjusted to be within the reconstructed envelope at a density threshold of 0.5 σ. Further model building included addition of amino acids at the N terminus of the VP2 monomer (residues 28 to 36) to satisfy density (0.5-σ threshold) inside the capsid under the icosahedral 5-fold axes. Following these adjustments, the geometry of the model was regularized under idealized geometric constraints in COOT (21). The noncrystallographic symmetry (NCS) function in COOT was used to generate the remaining 59 icosahedral symmetry-related VP2 monomers to verify their fit within the cryo-EM density map. The pseudo-atomic capsid model coordinates were used to generate a density map for comparison with the HBoV cryo-EM density in MAPMAN (37) using the program's similarity function (http://xray.bmc.uu.se/usf/mapman_man.html).
Comparison of parvovirus VP2 structures.
The structures of VP2 from representative members of six Parvoviridae genera (Table 1) were aligned by superposition of the Cα positions of their atomic coordinates (AAV2, B19, GmDNV, and MVM) or pseudo-atomic coordinates built into cryo-reconstructed densities (AMDV and HBoV) using the MatcherMaker tool (43) in Chimera (48).
RESULTS AND DISCUSSION
HBoV capsid structure.
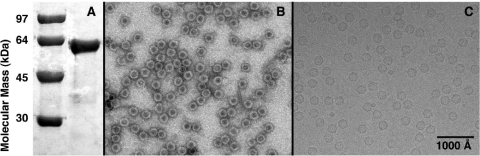
Self-assembled VP2 VLPs expressed and purified from insect cells were checked for composition and purity by SDS-PAGE (Fig. 1A) and for integrity by negative-stain electron microscopy (Fig. 1B) prior to sample vitrification and cryo-EM (Fig. 1C). A 3D reconstruction of the HBoV capsid, at an estimated resolution limit of 7.9 Å, was computed from 3,754 individual particle images (Fig. 2) and compared to the capsid structures of representative members of the Parvovirinae (B19, AAV2, AMDV, and MVM) and Densovirinae (GmDNV) subfamilies (Fig. 3).
FIG. 1.
Characterization of recombinant HBoV VLPs. (A) A 10% SDS-PAGE gel of HBoV VLPs (right lane) and molecular mass markers (left lane), showing that VLPs consist solely of VP2 (∼61 kDa). (B) Micrograph of negatively stained VLPs shows that most capsids exhibit a circular profile and are filled with stain, consistent with their being empty, spherical shells. (C) Micrograph of unstained, vitrified VLP sample highlights that these HBoV particles are intact, empty, and spherical.
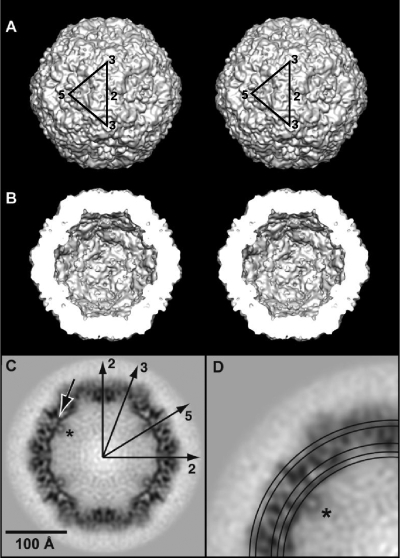
FIG. 2.
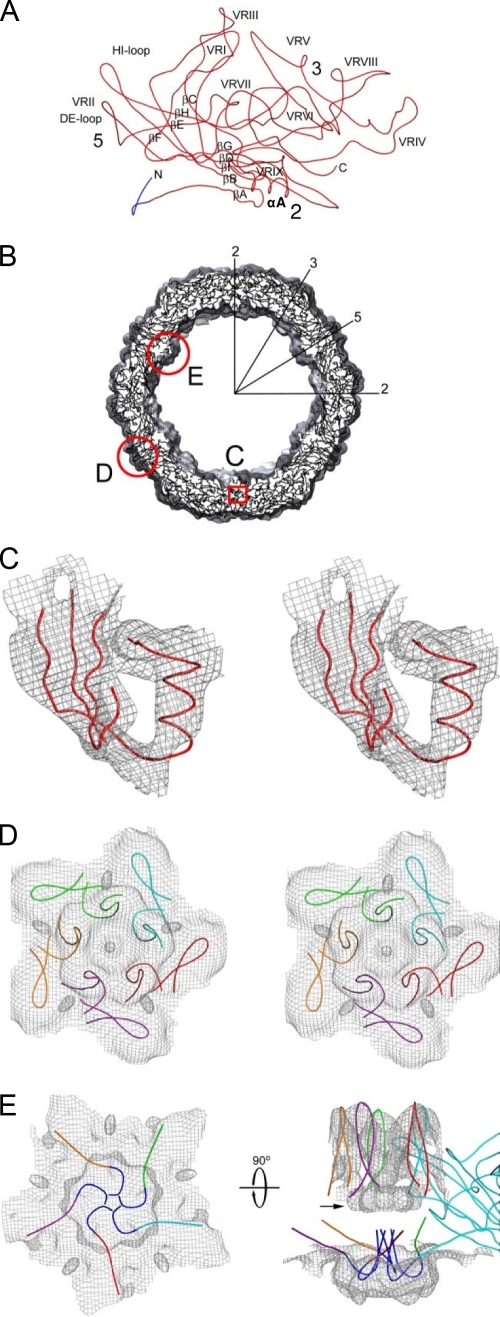
HBoV cryo-reconstruction. (A) Stereo, shaded-surface representation of the entire capsid viewed along a 2-fold direction. The black equilateral triangle bounded by two 3-fold (labeled 3) axes, separated by a 2-fold (labeled 2) axis, and by a 5-fold (labeled 5) axis identifies one asymmetric unit of the icosahedral capsid. (B) Same structure as in panel A but with the front half removed and generated as a perspective view. (C) Central cross-section of the reconstructed map, with highest- and lowest-density features depicted in black and white, respectively. Long arrows highlight the two- (2), three- (3), and 5-fold (5) axes that lie in the plane of the cross-section in one quadrant. Density “plugging” the 5-fold channel at low radii is indicated by the short, black arrow for one channel. (D) Enlarged view of upper left quadrant from panel C with circles drawn at radii corresponding to the radial density projections shown in Fig. 4. Density at the base of the channel modeled as residues 28 to 36 is indicated by an asterisk in panels C and D.
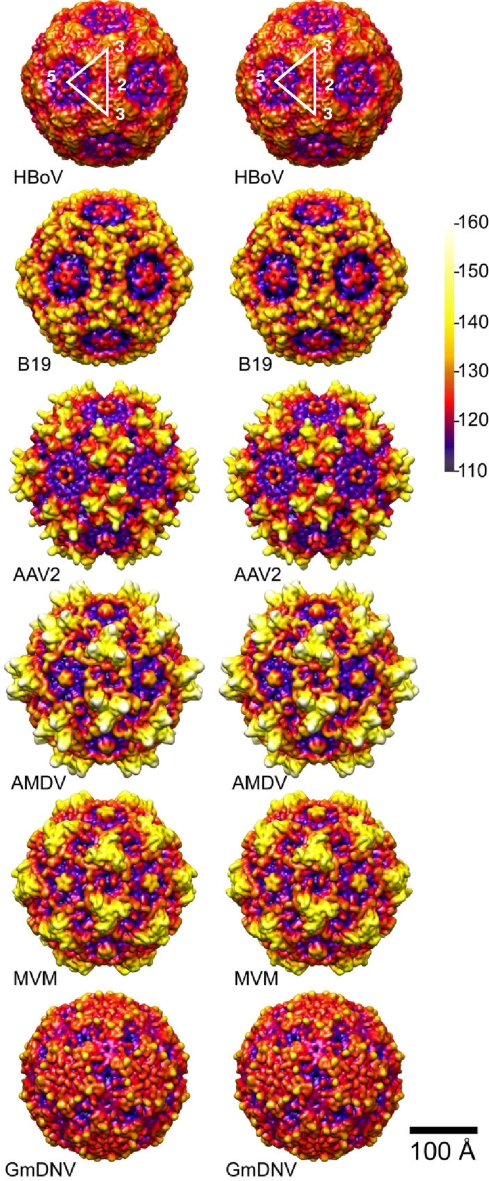
FIG. 3.
Comparison of six representative parvovirus capsid structures. Stereo, radially color-cued, surface representation of HBoV cryo-reconstruction shown with corresponding 2-fold views of B19, AAV2, AMDV, MVM, and GmDNV highlights similarities and differences in the outer surfaces of the capsids. Density maps of the latter five virus capsids were computed from atomic (B19, AAV2, MVM, and GmDNV) or pseudo-atomic (AMDV) coordinates, and all are rendered at the same resolution (7.9 Å) and magnification. Vertical bar depicts color cueing as a function of particle radius in Å.
The HBoV capsid exhibits three characteristic features common to other vertebrate parvoviruses. The first of these is a dimple-like depression at each icosahedral 2-fold axis (Fig. 2A and 3). The second is a large, trimeric protrusion that surrounds each 3-fold axis or is located at the 3-fold axis (Fig. 2A and 3). The third is a channel at each 5-fold axis, whose outermost opening is formed by a small, pentameric structure encircled by a wide, canyon-like region (Fig. 2A and 3). While the dimple is also observed among the invertebrate parvoviruses, these members lack the 3-fold protrusions and canyon around the 5-fold channel (e.g., GmDNV in Fig. 3).
The external diameter of the HBoV capsid ranges from ∼215 Å at the lowest points of the dimple and canyon to ∼280 Å at the top of the protrusion. For comparison, the maximum diameters of AMDV and GmDNV (the largest and smallest capsids of the representative parvoviruses being compared) are 310 and 270 Å, respectively, and the minimum diameter calculated from the capsid density maps generated from the atomic or pseudo-atomic coordinates of all the viruses being compared is ∼220 Å (Fig. 3). The spherical, relatively smooth topology of the invertebrate GmDNV clearly distinguishes it from the others, especially those with more prominent surface features such as AMDV, MVM, and AAV2. HBoV appears to be morphologically most similar to B19 since both viruses display features intermediate to the above extremes.
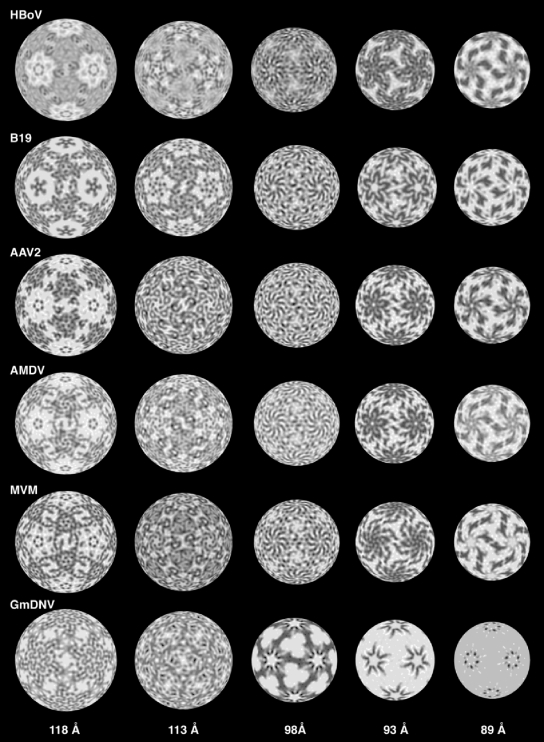
Radial density projections of the six representative Parvoviridae capsids reveal that structural similarity among them is greatest at low radii and most divergent at the capsid surface (Fig. 4). Of note, all the vertebrate parvoviruses have strikingly similar structures at radii of <98 Å, suggesting that they share a core. Again, GmDNV stands out among the parvoviruses as having a structure that is most distinct from the others at all radii due to minor positional differences in conserved core secondary structural elements and significant differences in the surface topology of its major capsid VP (see below).
FIG. 4.
Radial density projections of six representative parvoviruses. The virus labels are as defined in the text and in Table 1. Density distributions at different radii (Fig. 2D) illustrate similarities (inside) and differences (toward the outside) among the virus capsids. Contrast is the same as that depicted in Fig. 2C.
Pseudo-atomic model of HBoV VP2 built based on the B19 VP2 crystal structure.
The HBoV capsid surface topology was observed to be most similar to that of B19 (Fig. 3) even though the AAV2 VP2 protein sequence is closer to that of HBoV in percentage identity (Table 2). Preliminary docking of the crystal structure of the VP2 subunit for both AAV2 and B19 into the HBoV cryo-reconstruction showed that the β-barrel and αA helix in both viruses fitted into the HBoV density (with a MAPMAN [37] correlation coefficient of 0.6), but for AAV2, the loop regions that form each protrusion extended outside the HBoV envelope (data not shown), consistent with HBoV's smoother surface (Fig. 3). Thus, the B19 crystal structure coordinates (VP2 residues 19 to 554) (33) were used to construct the initial homology model of HBoV VP2 (residues 37 to 542) for docking into the cryo-EM density map (Fig. 5A and B).
TABLE 2.
Pairwise sequence alignment of HBoV with representative members of Parvoviridae for which structures are available
| Virus | Comparison with HBoV sequencea |
|||
|---|---|---|---|---|
| % Identity | % Strong similarity | % Weak similarity | % Different | |
| AAV2 | 23 | 17 | 14 | 46 |
| B19 | 19 | 17 | 14 | 50 |
| MVM | 16 | 17 | 16 | 51 |
| AMDV | 14 | 17 | 13 | 56 |
| GmDNV | 12 | 13 | 11 | 64 |
Percentages of identity, similarity, and difference are as defined in reference 53.
FIG. 5.
Pseudo-atomic model of HBoV VP2. (A) A Cα trace of the VP2 pseudo-atomic model. Labels highlight the β-strand regions (BIDG-CHEF) that form the core β-barrel, the α-helix A (αA), the DE and HI loops, and the nine predicted variable surface regions (VR-I to VR-IX). The approximate 2-, 3-, and 5-fold axes are shown by the numbers 2, 3, and 5, respectively. The dark-blue, N-terminal portion of the model represents nine residues that were not included in the initial HBoV homology model but that can be fit into the HBoV cryo-reconstruction. (B) An equatorial slab of pseudo-atomic HBoV VP2 monomers docked into the reconstructed density map. Several icosahedral symmetry axes are labeled as described in the legend of Fig. 2B. Regions outlined in red are enlarged in subsequent panels to further emphasize the fit of the model. (C) Stereo view of a small portion of the VP2 model (in red) showing the structurally conserved, β-barrel core (β-strands A, B, I, D, and G) and αA helix modeled into the reconstructed density (gray mesh, contoured at a threshold of 3.3 σ). (D) Same as panel C for a view of the capsid region surrounding the 5-fold axis, showing the fit of five, symmetry-related, DE loop β-ribbon models that form the cylindrical channel and the fit of five HI loops that form the petal-like structures on the canyon floor. The five VP2 monomers are distinguished by color. (E) The left model shows the same region as panel D but viewed from inside the capsid and highlighting the channel plug. Residues 28 to 36 in the HBoV VP2 model are shown in blue. The model at right is the same but viewed from the side to show the model of one monomer and the DE loops of four symmetry-related monomers at the pore of the 5-fold channel. N-terminal residues 28 to 36 (in blue) were modeled as extending up into the 5-fold channel. The color scheme is the same as in panel D.
A map of HBoV at subnanometer resolution enabled precise, rigid-body fitting of the conserved secondary structural elements (βA, β-barrel core, and αA helix) as well as the DE and HI loops, which required only small manual adjustments, followed by bond geometry refinement (Fig. 5B to D). The remaining interstrand loops and the C terminus of the VP2, including variable regions (VRs) (as defined in reference 24), were adjusted to conform to the HBoV density envelope (Fig. 5A and B). The initial homology model of HBoV started at residue 37, which corresponds to B19 residue 19, the first N-terminal residue reported in the crystal structure (33). However, a clearly defined region of density at the base of the channel adjacent to residue 37 of the initial HBoV model remained uninterpreted following the fitting procedure. This density (Fig. 2C and D, asterisk) was modeled as residues 28 to 36 contributed by five VP2 monomers (Fig. 5E). These nine residues were modeled as a small loop that projects into the additional density and extends toward the channel, as seen in B19 and MVM (3, 32) (Fig. 5A and E, dark blue loop). This interpretation positions residue 28 of the HBoV model toward the remaining uninterpreted density that lies close to the entrance of the channel and appears to form a “plug-like” structure inside the capsid (Fig. 2C and 5E, black arrow). The correlation coefficient between the map of the 60-subunit capsid (generated from the final HBoV VP2 pseudo-atomic model; residues 28 to 542) and the cryo-EM density map is 0.71. This represents an improvement over 0.6 for the initial B19 model (see above) as a result of model adjustment guided by the reconstructed map. A correlation coefficient of <1.0 is consistent with the inability to precisely place the variable surface loops at the current resolution.
A similar density plug at the base of the 5-fold channel was also observed in the cryo-reconstruction of B19 VLPs comprised solely of VP2 (32). This plug appeared as negative difference density when the B19 VLP cryo-reconstruction was subtracted from reconstructions of wild-type full (VP1, VP2, and DNA) and empty (VP1 and VP2) B19 capsid structures (32). The location of the first ordered N-terminal residue in the B19 VP2 VLP crystal structure (starting with residue 18) overlapped this negative density, suggesting that the remaining, unordered residues lie inside the VLP capsid (32). In the B19 report, no effort was made to model the missing residues into the negative difference density. However, positive difference density was observed in the B19 full and empty capsid minus VLP density map subtractions, which projected into the channel toward the capsid surface, emerging between the DE loop β-ribbons onto the canyon at the capsid surface. This density was modeled as B19 VP2 residues 1 to 17 (32). The differences in locations of the N-terminal residues in B19 VLPs versus capsids with or without DNA and VP1 are likely dictated by the presence of the DNA and/or VP1. The VP1u motif was not observed in B19 capsids (32), most likely because only a few copies are present per particle, and their contributions to the density map would be averaged out by the icosahedral symmetry imposed during structure determination. The B19 channel appears to be closed or tightly constricted at the capsid surface (Fig. 3) (32, 33). Thus, disposition of the N-terminal residues of the B19 capsid proteins on the outer surface is consistent with previous reports that placed VP1u on the B19 surface (6, 29, 34), and this differs from other parvoviruses in which externalization is proposed to occur via the channel. In HBoV the channel is constricted at its base but open at the top (Fig. 2C and D, and 3). Thus, it is difficult to assess whether the HBoV VP2 and/or VP1 N termini are also already located on the capsid surface. Our current model of HBoV suggests that the N-terminal residues (Fig. 5E) may protrude through the channel in virions. Structural studies of capsids that contain VP1 or both VP1 and DNA are needed to verify this prediction.
HBoV VP2 shares low sequence identity but high structural similarity to other Parvovirinae VP2s.
Sequence alignments and an unrooted phylogenetic tree were generated to quantitatively compare the HBoV VP2 sequence with the corresponding VP sequences in five other parvovirus genera (Table 1) and to help guide homology modeling based on the HBoV cryo-EM density map. The pairwise sequence identities ranged between 12 and 23%, with GmDNV, the sole invertebrate member in the comparison, being the most divergent from HBoV (Table 2). Phylogenetic analysis based on a multiple sequence alignment of the six viruses being compared placed them almost equidistant from each other in an unrooted tree (data not shown), suggesting high divergence.
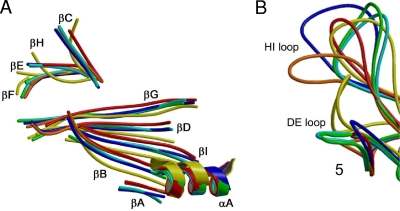
Structural superimposition of the Cα positions of the atomic (B19, AAV2, and MVM) and pseudo-atomic (AMDV) coordinates of the Parvovirinae VP2s with the pseudo-atomic model generated for HBoV (residues 28 to 542) showed that, despite the high (∼50%) sequence variability, the eight-stranded β-barrel (BIDG-CHEF) core, βA, and αA helix are conserved in all the viruses (Fig. 6A). Most VRs in the VP2 structures of the Parvovirinae being compared to HBoV are located within the loops between the β-strands and occur at and around the 3-fold axis (15). In the HBoV pseudo-atomic model, the VRs are also predicted to occur around this axis (Fig. 5A). However, at the current resolution of the HBoV density map, the exact positions of the loop structures are unknown and thus preclude a comparison with those of the other viruses. As with the Parvovirinae members, the core β-strands are structurally very similar between HBoV and GmDNV though the exact positions of αA differ slightly (Fig. 6A).
FIG. 6.
Superposition of structurally conserved VP2 regions from six representative members of the Parvoviridae. (A) Superimposition of the eight-stranded β-barrel motif (BIDG and CHEF sheets), strand βA, and helix αA of AAV2 (blue), MVM (green), B19 (orange), and GmDNV (yellow) and the pseudo-atomic models for AMDV (cyan) and HBoV (red). Structural elements are labeled as described in the legend of Fig. 5A. (B) Superimposition of the DE and HI loops of the six viruses colored as in panel A.
HBoV capsid surface topology is unique but shares features common to other vertebrate parvoviruses.
Based on the overall surface morphologies of their capsids, parvoviruses have been structurally assigned to three groups (47). Groups I and III comprise members of the Parvovirinae subfamily, all of which share three features as described earlier: dimples at 2-fold axes, protrusions at or surrounding 3-fold axes, and channels surrounded by canyons at the 5-fold axes (Fig. 3). The primary difference between these two groups occurs in the vicinity of each 3-fold axis. Group I capsids have a single, relatively flat, pinwheel-shaped protrusion, whereas group III capsids have three distinct protrusions. Current structural data place members of the Parvovirus genus in group I and members of the Erythrovirus, Dependovirus, and Amdovirus genera in group III (Table 1) (47). A third topology (group II), seen in GmDNV and other members of the Densovirinae subfamily (11, 51), is characterized by capsids which are relatively spherical and featureless compared to the vertebrate parvoviruses (Table 1) (47). This morphology, exemplified by GmDNV, results from the reduced length of the surface loops compared to those of members of the Parvovirinae (51).
HBoV possesses features common to groups I and III and also others that are unique. One distinguishing feature in HBoV arises from a difference in surface topology at the 2-fold axis, where the HBoV dimple is narrower and shallower than that in the other parvoviruses (Fig. 3). However, like B19, the HBoV dimple is flanked by wider “walls” between the 2- and 5-fold depressions than are present in the other viruses, and this may reflect a conformational loop difference in the modeled VR-IX located close to the 2-fold axis (Fig. 5A). Residues that form the walls of the parvovirus dimple have been implicated in receptor attachment for MVM (41) and as determinants of tissue tropism and pathogenicity for other members of the Parvovirus genus (reviewed in reference 38). There is no evidence, however, that this region plays an analogous role in B19 or HBoV.
Other characteristic parvovirus features at the 2-fold region are conserved in HBoV. Helix αA, observed in all parvovirus structures determined to date (reviewed in reference 15), is also present in HBoV (Fig. 5C and 6A). This helix forms stabilizing interactions at the icosahedral 2-fold interface (24). Even though there are fewer interactions at the 2-fold interface than at the 3-fold interface, they are proposed to play a crucial role in parvovirus capsid assembly (24, 57). Conservation of this structural feature in the HBoV VP2 thus suggests that it plays a similar, central role in capsid assembly.
HBoV topology at the 3-fold axis is intermediate between that of groups I and III. The group III viruses, AAV2 and AMDV, have large protrusions that arise from the EF and GH loops. The protrusions in HBoV are less pronounced than in these two parvoviruses, which may result from shorter sequences and/or conformational differences in the HBoV EF and GH loops. This results in a less pronounced depression in this region (Fig. 3). The group I viruses, e.g., MVM, also lack depressions at their 3-fold axes (Fig. 3). Residues near the 3-fold axes in AAV2 and B19 have been implicated as receptor attachment sites (16, 35, 39, 45, 46). The protrusions surrounding these axes are responsible for the antigenic reactivity of members of the Parvovirinae subfamily (reviewed in reference 2). Given this antigenic role of the group I and III surface protrusions, the lack of similar structures in group II viruses is consistent with the fact that invertebrate viruses are not subject to adaptive pressures of the host immune response (51).
As mentioned above, the capsid structures of members of the Parvovirinae have a conserved channel at the icosahedral 5-fold axis, surrounded by a canyon (Fig. 3). The channel is a β-cylinder formed by five symmetry-related β-ribbons inserted between β-strands D and E with a loop at the top (referred to as the DE loop). In HBoV the DE loops form finger-like protrusions that surround the channel (Fig. 5A and D and 6B) and project away from the 5-fold axis in a manner similar to the conformation observed in AAV2, resulting in an open channel (Fig. 3). In contrast, for the other Parvovirinae structures, the DE loops cluster closer to the 5-fold axis and constrict the channel (Fig. 3 and 6B). This channel in parvoviruses is postulated to serve several functions, for example, as the site of (i) VP2 externalization in members of the Parvovirus genus (e.g., MVM, in which VP3 is generated by a maturation cleavage event), (ii) VP1u externalization (except in B19 and GmDNV) for its PLA2 function, and (iii) genomic DNA packaging. However, because the diameter of the channel varies between ∼5 and 25 Å at its narrowest and widest points, respectively, it is likely too small to accommodate extrusion of the ∼130 amino acids in the PLA2 domain. It has thus been proposed that flexibility and structural rearrangement of the DE loop (and possibly the HI loop; see below) are required to facilitate these functions. Conservation of this channel in HBoV suggests that it performs analogous functions. Notably, HBoV appears to have the largest surface opening of the parvovirus structures determined to date (Fig. 3).
The canyon depression in HBoV is less pronounced than the canyons seen in all other parvovirus structures except for GmDNV, which lacks that feature (Fig. 3). In HBoV the HI loop lies in the canyon and is structurally most similar to the HI loop of AAV2 and, although shorter in HBoV, generates a similar surface topology for the two viruses (Fig. 3 [red loop features on the canyon floor], 5D, and 6B). The HI loop conformation differs significantly in B19 because it projects radially from the capsid surface and lies very close to the DE loop to form a continuous, raised surface in the canyon (Fig. 3). This B19 HI loop conformation is similar to that proposed to occur in AAV2 following receptor attachment (39). The HI loops in MVM and AMDV, which also lie on the canyon floor, are structurally similar to each other but differ from the HI loop of HBoV (Fig. 6B). In GmDNV the HI loop lies very close to the DE loop, and together they “fill in” the canyon floor (Fig. 3 and 6B). The HI loop, which has been proposed to undergo structural rearrangements in AAV2 to facilitate VP1 externalization upon receptor binding, has also been reported to play a role in capsid assembly (19, 39).
Summary.
The structure of the HBoV capsid presented here represents the first for a member of the newly established Bocavirus genus of the Parvoviridae. The subnanometer resolution obtained in the cryo-reconstruction clearly reveals that the newly identified HBoV assembles a capsid that retains the highly conserved structural core adopted by all other parvoviruses and does so despite low sequence similarity between its major capsid protein and the capsid proteins of the other viruses that have existed for many years. Conservation of core structural features in the major coat protein, including the eight-stranded β-barrel, the αA helix, the DE loop, and the HI loop, is consistent with their contributing essential functions in capsid assembly and stability. Conservation of the β-cylinder channel formed by the DE loop strongly suggests that this channel is essential for productive HBoV infection as it likely serves as a portal for VP1u externalization and DNA packaging, as reported for other parvoviruses.
Overall, HBoV shares most of its capsid surface features with the two other human parvoviruses (B19 and AAV2) in this comparative study, and these features result in a unique capsid morphology whose functional domains are yet to be verified. The resemblance of B19 and AAV2 capsids at the 3-fold axes suggests that this region confers similar, receptor attachment capability. On the other hand, the structural difference of the HBoV 5-fold axis from that of B19 (reported to have its VP1/VP2 localized on the capsid surface) and its similarity to that of AAV2 (proposed to externalize its VP1u for its PLA2 function via the channel) suggest that the HBoV capsid is required to undergo conformational transitions similar to the AAV2 capsid during cellular trafficking. Variable HBoV VP2 surface loops, as modeled based on known parvovirus structures (15) and constrained by the reconstructed density envelope, likely reflect adaptations related to host cell recognition and evasion of the adaptive immune response. The structure of HBoV provides an additional framework for the molecular annotation of parvovirus VP structures, which have evolved to facilitate infectivity through the utilization of structural elements and capsid surface features that fulfill myriad functions.
Acknowledgments
We thank Jinghua Tang for helpful discussions and for verifying the absolute scale of the HBoV density map by quantitative comparison to the B19 crystal structure.
This work was supported in part by NSF grant MCB-0718948 (M.A.-M. and R.M.), by NIH grants R21 AI81072341 (M.A.-M. and R.M.), R37 GM-33050, and 1S10 RR020016 (T.S.B.), by NIH NRSA fellowship F32A1078624 (K.N.P.), and by support from the University of California, San Diego (UCSD), and the Agouron Foundation (T.S.B.) to establish cryo-EM facilities at UCSD.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., and M. S. Chapman. 2006. Corrleating structure with function in the viral capsid, pp. 125-139. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, NY.
- 3.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 4.Allander, T., T. Jartti, S. Gupta, H. G. M. Niesters, P. Lehtinen, R. Osterback, T. Vuorinen, M. Waris, A. Bjerkner, A. Tiveljung-Lindell, B. G. van den Hoogen, T. Hyypiä, and O. Ruuskanen. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44:904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, S., M. Momoeda, M. Kawase, S. Kajigaya, and N. S. Young. 1995. Peptides derived from the unique region of B19 parvovirus minor capsid protein elicit neutralizing antibodies in rabbits. Virology 206:626-632. [DOI] [PubMed] [Google Scholar]
- 7.Arnold, J. C., K. K. Singh, S. A. Spector, and M. H. Sawyer. 2006. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin. Infect. Dis. 43:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur, J. L., G. D. Higgins, G. P. Davidson, R. C. Givney, and R. M. Ratcliff. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman, V. D., E. S. Chase, A. W. E. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruemmer, A., F. Scholari, M. Lopez-Ferber, J. F. Conway, and E. A. Hewat. 2005. Structure of an insect parvovirus (Junonia coenia Densovirus) determined by cryo-electron microscopy. J. Mol. Biol. 347:791-801. [DOI] [PubMed] [Google Scholar]
- 12.Campe, H., C. Hartberger, and A. Sing. 2008. Role of human bocavirus infections in outbreaks of gastroenteritis. J. Clin. Virol. 43:340-342. [DOI] [PubMed] [Google Scholar]
- 13.Carrillo-Tripp, M., C. M. Shepherd, I. A. Borelli, S. Venkataraman, G. Lander, P. Natarajan, J. E. Johnson, C. L. Brooks, and V. S. Reddy. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 37:D436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cecchini, S., A. Negrete, T. Virag, B. S. Graham, J. I. Cohen, and R. M. Kotin. 2009. Evidence of prior exposure to human bocavirus as determined by a retrospective serological study of 404 serum samples from adults in the United States. Clin. Vaccine Immunol. 16:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman, M. S., and M. Agbandje-McKenna. 2006. Atomic structures of viral particles, pp. 107-123. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. L. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, NY.
- 16.Chipman, P. R., M. Agbandje-McKenna, S. Kajigaya, K. E. Brown, N. S. Young, T. S. Baker, and M. G. Rossmann. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. U. S. A. 93:7502-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow, B. D. W., and F. P. Esper. 2009. The human bocaviruses: a review and discussion of their role in infection. Clin. Lab. Med. 29:695-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 19.DiPrimio, N., A. Asokan, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 82:5178-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 21.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 23.Fryer, J. F., A. Kapoor, P. D. Minor, E. Delwart, and S. A. Baylis. 2006. Novel parvovirus and related variant in human plasma. Emerg. Infect. Dis. 12:151-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govindasamy, L., E. Padron, R. McKenna, N. Muzyczka, N. Kaludov, J. A. Chiorini, and M. Agbandje-McKenna. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80:11556-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 26.Havelka, W. A., R. Henderson, and D. Oesterhelt. 1995. Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 247:726-738. [DOI] [PubMed] [Google Scholar]
- 27.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn, J. S., D. Kesebir, S. F. Cotmore, A. D'Abramo, C. Cosby, C. Weibel, and P. Tattersall. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J. Infect. Dis. 198:41-50. [DOI] [PubMed] [Google Scholar]
- 29.Kajigaya, S., H. Fujii, A. Field, S. Anderson, S. Rosenfeld, L. J. Anderson, T. Shimada, and N. S. Young. 1991. Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad. Sci. U. S. A. 88:4646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kantola, K., L. Hedman, T. Allander, T. Jartti, P. Lehtinen, O. Ruuskanen, K. Hedman, and M. Söderlund-Venermo. 2008. Serodiagnosis of human bocavirus infection. Clin. Infect. Dis. 46:540-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapoor, A., E. Slikas, P. Simmonds, T. Chieochansin, A. Naeem, S. Shaukat, M. M. Alam, S. Sharif, M. Angez, S. Zaidi, and E. Delwart. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann, B., P. R. Chipman, V. A. Kostyuchenko, S. Modrow, and M. G. Rossmann. 2008. Visualization of the externalized VP2 N termini of infectious human parvovirus B19. J. Virol. 82:7306-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. U. S. A. 101:11628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawase, M., M. Momoeda, N. S. Young, and S. Kajigaya. 1995. Most of the VP1 unique region of B19 parvovirus is on the capsid surface. Virology 211:359-366. [DOI] [PubMed] [Google Scholar]
- 35.Kern, A., K. Schmidt, C. Leder, O. J. Müller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kesebir, D., M. Vazquez, C. Weibel, E. D. Shapiro, D. Ferguson, M. L. Landry, and J. S. Kahn. 2006. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J. Infect. Dis. 194:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleywegt, G. J., and T. A. Jones. 1996. xdlMAPMAN and xdlDATAMAN—programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D Biol. Crystallogr. 52:826-828. [DOI] [PubMed] [Google Scholar]
- 38.Kontou, M., L. Govindasamy, H. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy, H. C., V. D. Bowman, L. Govindasamy, R. McKenna, K. Nash, K. Warrington, W. Chen, N. Muzyczka, X. Yan, T. S. Baker, and M. Agbandje-McKenna. 2009. Heparin binding induces conformational changes in adeno-associated virus serotype 2. J. Struct. Biol. 165:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindner, J., and S. Modrow. 2008. Human bocavirus—a novel parvovirus to infect humans. Intervirology 51:116-122. [DOI] [PubMed] [Google Scholar]
- 41.López-Bueno A., M. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng, E. C., E. F. Pettersen, G. S. Couch, C. C. Huang, and T. E. Ferrin. 2006. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteny, M., H. G. M. Niesters, H. A. Moll, and M. Y. Berger. 2007. Human bocavirus in febrile children, The Netherlands. Emerg. Infect. Dis. 13:180-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Donnell, J., K. A. Taylor, and M. S. Chapman. 2009. Adeno-associated virus-2 and its primary cellular receptor-Cryo-EM structure of a heparin complex. Virology 385:434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opie, S. R., K. H. Warrington, M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 49.Qu, X., W. Liu, Z. Qi, Z. Duan, L. Zheng, Z. Kuang, W. Zhang, and Y. Hou. 2008. Phospholipase A2-like activity of human bocavirus VP1 unique region. Biochem. Biophys. Res. Commun. 365:158-163. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, B., J. F. Fryer, J. Oldenburg, H. Brackmann, S. A. Baylis, and A. M. Eis-Hübinger. 2008. Frequency of contamination of coagulation factor concentrates with novel human parvovirus PARV4. Haemophilia 14:978-986. [DOI] [PubMed] [Google Scholar]
- 51.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramaniam, S. 1998. The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins 32:1-2. [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tijssen, P., J. Szelei, and Z. Zádori. 2006. Phospholipase A2 domains in structural proteins of parvoviruses, p. 95-105. In J. Kerr, S. Cotmore, M. E. Bloom, R. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, NY.
- 55.van Heel, M., and M. Schatz. 2005. Fourier shell correlation threshold criteria. J. Struct. Biol. 151:250-262. [DOI] [PubMed] [Google Scholar]
- 56.Vicente, D., G. Cilla, M. Montes, E. G. Pérez-Yarza, and E. Pérez-Trallero. 2007. Human bocavirus, a respiratory and enteric virus. Emerg. Infect. Dis. 13:636-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan, X., K. A. Dryden, J. Tang, and T. S. Baker. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan, X., R. S. Sinkovits, and T. S. Baker. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zádori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariépy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 10:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]