Abstract
Structural studies have made significant contributions to our understanding of Sulfolobus spindle-shaped viruses (Fuselloviridae), an important model system for archaeal viruses. Continuing these efforts, we report the structure of D212 from Sulfolobus spindle-shaped virus Ragged Hills. The overall fold and conservation of active site residues place D212 in the PD-(D/E)XK nuclease superfamily. The greatest structural similarity is found to the archaeal Holliday junction cleavage enzymes, strongly suggesting a role in DNA replication, repair, or recombination. Other roles associated with nuclease activity are also considered.
Viruses are the most abundant biological units on earth (3). However, while more than 5,000 different viruses infecting bacterial and eukaryotic hosts have been described in detail, only about 50 viruses have been described from the archaeal domain of life (29, 40, 47). Interestingly, these archaeal viruses, particularly those infecting the Crenarchaea, exhibit an array of unusual morphologies (47) and unique genomic content. This has led to recognition of seven new viral families, with two additional viral families awaiting assignment (28, 48, 58, 59). However, functional annotations for most gene products in these crenarchaeal viruses have not been made, and we possess only a rudimentary understanding of their fundamental viral processes, including mechanisms of attachment, uptake, transcriptional regulation, genome replication, and virus assembly and release (29, 40, 48).
Sulfolobus spindle-shaped viruses (SSVs), or Fuselloviridae, are among the best-studied crenarchaeal viruses. Members of this family include Sulfolobus spindle-shaped virus 1 (SSV1) (32) and Sulfolobus spindle-shaped virus Ragged Hills (SSVRH) (58). SSV1 is the type virus for the family, and along with SSVRH, exhibits a characteristic 60- by 90-nm spindle- or lemon-shaped morphology with short tail fibers at one end (43, 58). These viruses contain double-stranded circular DNA genomes that encode 34 and 38 open reading frames (ORFs), respectively (58). Early analyses of the SSV1 genome revealed only two ORFs with similarity to genes of known function. ORF D335 encodes a viral integrase, although this gene appears to be nonessential (2, 11, 36), while B251 exhibits limited similarity to DnaA (23). More recently, improved bioinformatic methods have noted additional similarities (17, 48). However, 26 of 34 ORFs from SSV1 are still not reliably identified by bioinformatics.
An early study by Reiter et al. identified three proteins in the SSV1 viral particle (51). VP1 and VP3 are structural proteins (51), while VP2 is thought to function as a small, packaged, DNA binding protein (52, 58). More recently, two additional components that copurify with the viral particle have also been identified, C792 and D244 (33). C792 is a predicted membrane protein and thus likely to serve a structural role, while D244 is a soluble protein of unknown function (33). Structural studies have also contributed to our understanding of the SSV1 proteome, with crystal structures of D63, F112, and F93 revealing distant evolutionary relationships and potential functions for these proteins (26, 27, 29, 33). Unfortunately, attempts to crystallize D244 for structural annotation have been unproductive, despite significant efforts (B. J. Eilers and C. M. Lawrence, unpublished data). However, D244 orthologs are present in many Fuselloviridae (58), including SSVRH D212, which shows 80% sequence identity to SSV1 D244. Here we present the initial biochemical and structural analysis of SSVRH D212 and discuss the functional implications.
MATERIALS AND METHODS
Cloning.
ORF D212 was amplified using nested PCR, and SSVRH DNA was purified as described previously (52). Primers introduced a Shine-Dalgarno sequence, an N-terminal His6 tag, and attB sites. The internal forward and reverse primers were 5′-ATGCATCACCATCACCATCACATGACCGAAACTGATTTTAA-3′ and 5′-GTACAAGAAAGCTGGGTCCTACTTATTACTCCTAGCTAAAT-3′, respectively, while the external forward and reverse primers have been described previously (33). The amplified product was inserted into pDONR201, its sequence was verified, and then the product was transferred into pDEST14 (Invitrogen), yielding pEXP14-D212.
Expression and purification.
D212 was expressed in Escherichia coli as described for other fuselloviral proteins (26, 29, 33). Purification of D212 also followed this earlier protocol, which included cell lysis, heat denaturation, immobilized metal affinity chromatography (IMAC), and size exclusion chromatography. Lysis buffer was composed of 20 mM Tris, pH 8.0, 20 mM NaH2PO4, 1 M NaCl, 5 mM imidazole, and 0.1 mM phenylmethylsulfonyl fluoride. IMAC elution buffer was 20 mM Tris (pH 8.0), 300 mM NaCl, and 200 mM imidazole. Buffer for size exclusion chromatography was 10 mM Bis-Tris (pH 6.5) and 100 mM NaCl. Protein concentrations were determined by Bradford assay with bovine serum albumin (BSA) as a standard (33).
Crystallization and data collection.
D212 was concentrated to 10 mg/ml using Amicon spin concentrators and crystallized using hanging drop vapor diffusion. Drops were set up at 24°C using 2 μl of D212 and 2 μl of well solution consisting of 0.1 M HEPES (pH 7.5), 0.05 M K2HPO4, and 10% (wt/vol) polyethylene glycol (PEG) 8000. Crystals up to 0.1 × 0.05 × 0.02 mm in size were obtained in 2 weeks. The crystals were transferred to well solution containing 25% glycerol as a cryoprotectant for 10 min and then flash-frozen in liquid nitrogen. Heavy atom soaks were carried out for 1 h in 10 mM K2PtCl4 or 30 s in 0.5 M NaBr in phosphate-free well solution containing 25% glycerol. A single-wavelength data set centered on the Br K edge and a three-wavelength data set at the Pt L edge were collected at the Stanford Synchrotron Radiation Laboratory. A native data set was also collected. Data were integrated, scaled, and reduced in space group P21 to a resolution of 2.4 Å using HKL-2000 (41) (Table 1 ). For the Mn2+ metal soaks, native D212 crystals were transferred to phosphate-free well solution augmented with 25% glycerol and 10 mM MnCl2. The crystals were soaked overnight at 24°C and flash-frozen. Data were collected at 100 K using our Cu-Kα home source and a MAR345 image plate detector.
TABLE 1.
| Parameter | Value for data set: |
|||||
|---|---|---|---|---|---|---|
| D212 | D212-K2PtCl4 |
D212-NaBr peak | D212-MnCl2 | |||
| Inflection | Peak | Remote | ||||
| Wavelength (Å) | 0.95369 | 1.07270 | 1.07203 | 0.82653 | 0.91163 | 1.54 |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell constants | ||||||
| a, b, c (Å) | 56.68, 82.11, 60.29 | 56.55, 82.02, 60.18 | 56.31, 81.76, 59.93 | 56.49, 81.88, 60.10 | 56.57, 81.59, 60.53 | 56.40, 80.48, 59.78 |
| α, β, γ (degrees) | 90.0, 115.48, 90.0 | 90.0, 115.41, 90.0 | 90.0, 115.44, 90.0 | 90.0, 115.40, 90.0 | 90.0, 115.03, 90.0 | 90.0, 114.84, 90.0 |
| Resolution range (Å) | 50-2.4 (2.49-2.40) | 50-3.05 (3.16-3.05) | 50-2.8 (2.9-2.8) | 50-3.35 (3.47-3.35) | 50-2.8 (2.9-2.8) | 50-3.7 (3.83-3.70) |
| No. of unique reflections | 18,121 (1,868) | 9,530 (958) | 12,089 (1,207) | 7,233 (708) | 12,107 (1,186) | 4,952 (501) |
| Avg redundancy | 3.1 (3.0) | 6.3 (6.4) | 6.3 (6.3) | 6.4 (6.4) | 7.3 (5.9) | 3.1 (2.9) |
| I/σ | 32.6 (4.3) | 41.4 (7.5) | 45.2 (7.1) | 29.06 (6.9) | 28.6 (3.7) | 9.2 (2.7) |
| Completeness (%) | 92.9 (95.7) | 99.9 (100) | 99.2 (100) | 98.8 (99.4) | 99.5 (96.9) | 93.9 (96.5) |
| Rsymc (%) | 3.4 (32.6) | 6.2 (29.3) | 6.5 (30.8) | 7.9 (31.7) | 7.2 (33.2) | 11.5 (31.0) |
Data were integrated, scaled, and reduced using the HKL-2000 software package (41).
Numbers in parentheses refer to the highest-resolution shell.
Rsym = 100·ΣhΣi|I(h) − <I(h)>|/ΣhI(h) where Ii(h) is the ith measurement of reflection h and <I(h)> is the average value of the reflection intensity.
Structure determination and refinement.
The structure was solved using multiple isomorphous replacement with anomalous scattering (MIRAS). SOLVE (55) was used to determine the positions of two Pt atoms and one Br atom per asymmetric unit and to calculate initial phases. RESOLVE (54) was used for density modification and initial model building. Cycles of iterative model building with COOT (15) and refinement with REFMAC5 (35) led to the final model. Translation/libration/screw (TLS) parameters (42) were included in the refinement with each D212 subunit divided into 15 TLS groups. The final model, which includes 75 water molecules, results in crystallographic (Rcryst) and free (Rfree) R factors of 20.9% and 24.5%, respectively (Table 2 ). Model quality was assessed with MolProbity (31), with 98.1% of the residues in the most favored regions and the remaining 1.9% in additionally allowed regions of the Ramachandran plot. Figures were generated with PYMOL (14) and TOPDRAW (7).
TABLE 2.
Model refinement
| Parameter | Value |
|---|---|
| Rcrysta (%) | 20.9 |
| Rfreea (%) | 24.5 |
| Real space CCb (%) | 94.5 |
| Mean B value (overall; Å2) | 32.2 |
| Coordinate error (based on maximum likelihood; Å) | 0.169 |
| RMSD from ideality | |
| Bonds (Å) | 0.005 |
| Angles (degrees) | 0.698 |
| Ramachandran plotc | |
| Most favored (%) | 98.1 |
| Additional allowed (%) | 1.9 |
| PDB accession code | 2W8M |
Rcryst = Σ||Fo| − Fc||/ΣFo|, where Fo and Fc are the observed and calculated structure factor amplitudes used in refinement, respectively. Rfree is calculated like Rcryst but using the “test” set of structure factor amplitudes (5%) that were withheld from refinement (35).
Correlation coefficient (CC) is agreement between the model and the 2mFo-DFc electron density map (35).
Calculated using MolProbity (12).
Holliday junction cleavage assay.
Fixed and mobile four-way junctions were assembled and purified as described in the work of Birkenbihl et al. (6). Holliday junction cleavage activity was assayed as described for the Sulfolobus islandicus rod-shaped virus 1 (SIRV1) and SIRV2 Holliday junction cleavage enzymes with only minor modifications (6). Specifically, the 5′-32P-end-labeled fixed and mobile Holliday junctions (10 nM) were incubated at 25°C for 2 h in a 10-μl reaction mixture that also contained 1 μM D212, 10 mM Bis-Tris (pH 6.5), 100 mM NaCl, 1 mM dithiothreitol (DTT), and 10 mM divalent cation, either MgCl2 or MnCl2. T4 endonuclease VII was used as a positive control.
Protein structure accession number.
Coordinates and structure factors were deposited in the Protein Data Bank (PDB ID 2W8M).
RESULTS
The D212 construct consists of 212 residues of the full-length protein plus an N-terminal His6 tag, giving a calculated mass of 25,895 Da. It elutes from the size exclusion column with an apparent molecular mass of 50 kDa, suggesting that D212 is present in solution as a homodimer. D212 crystallizes in space group P21 with two copies of the D212 protomer in the asymmetric unit. Lacking methionine residues, the structure was determined at 2.4-Å resolution by multiple isomorphous replacement with anomalous scattering (MIRAS). Consistent with size exclusion chromatography, the structure reveals a homodimer.
Structure of the D212 protomer.
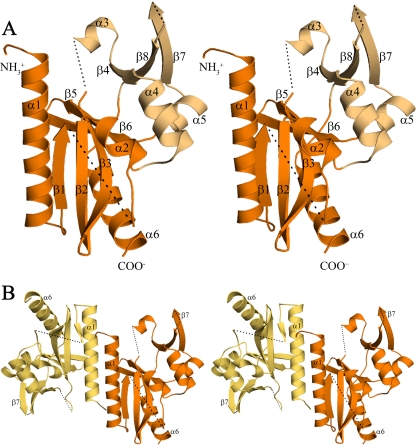
The D212 polypeptide folds to form a mixed α/β structure composed of five α-helices and eight β-strands (Fig. 1) that can be divided into two subdomains which we refer to as the “catalytic domain” and the “DNA binding domain,” respectively (see below and reference 45). The catalytic domain is the central domain of the dimeric protein and is a doubly wound α/β structure. It is composed of a predominantly antiparallel five-stranded β-sheet (β1, β2, β3, β5, and β6) that is flanked on one side by helices α1 and α6 and on the other by helix α2. The second, smaller domain, the DNA binding domain (45), is composed of a 3-stranded antiparallel β-sheet (β4, β7, and β8) with helix α3 lying along one side and helices α4 and α5 running across one face. In addition, a number of the connecting loops are disordered (shown as dotted lines in Fig. 1). In chain B, disordered regions include residues 1 to 18 at the N terminus, residues 58 to 67 in the β1-α2 loop, residues 100 to 106 connecting β3 to α3, residues 172 to 174 in the β7-β8 loop, and residues 204 to 212 at the C terminus (Fig. 1). Although exact residue numbers differ, similar disorder is seen for chain A. The two subunits show similar topology over most of the structure and superimpose with a root mean square deviation (RMSD) of 0.62 Å. The only notable difference between the two subunits is the absence or presence of the short α3 helix, which is disordered in chain A. The dimer interface is formed by several N-terminal residues, helix α1, and strand β1. The α1 helix lies at the heart of the interface, where it interacts with the symmetry-related helix in an antiparallel side-by-side fashion. Together, these elements form a hydrophobic interface that buries ∼1,090 Å2 of the subunit surface.
FIG. 1.
The structure of D212 (stereo images). (A) The D212 protomer (chain B) adopts the restriction endonuclease fold. The central “catalytic” domain (orange) is composed of a mixed, five-stranded β-sheet (β1, β2, β3, β5, and β6) packed between α-helices (α1/α6 and α2). The smaller “DNA recognition” domain (light orange) is composed of an antiparallel three-stranded β sheet (β4, β7, and β8) along with three α-helices, α3, α4, and α5. Dashed lines represent chain breaks. (B) The D212 dimer oriented as in panel A, with chain A in yellow-orange and chain B in orange. The dimer interface is formed by residues at the N terminus, the α1 helix, and strand β1. It is primarily hydrophobic and buries ∼12% of the subunit surface area.
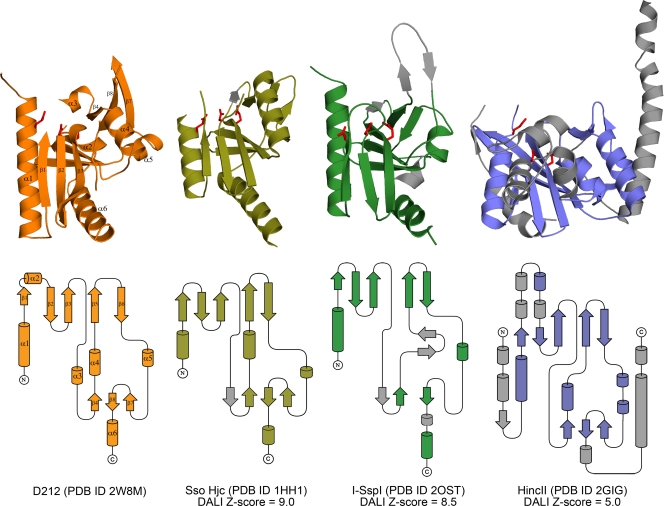
Structural homology.
Consistent with its calculated pI of 9.0, a search for structurally similar proteins using the DALI (20) server identified members of the PD-(D/E)XK nuclease superfamily as the closest structural homologues. This diverse endonuclease family was initially identified in structurally characterized type II restriction enzymes but has subsequently been recognized in many other enzymes involved in the “3 Rs” of DNA replication, repair, and recombination (38). Consistent with the lack of functional annotation for D212 and its orthologs, members of the PD-(D/E)XK nuclease superfamily can exhibit extreme variability in their amino acid sequences, making it difficult to recognize highly diverged members (38). Among this family, D212 was found to be most similar to the Holliday junction cleavage enzyme from Sulfolobus solfataricus (Sso Hjc, PDB ID 1HH1), giving a DALI Z-score of 9.0, with an RMSD of 3.0 Å for 115 equivalent residues showing 12% identity (8). Other proteins with significant similarity include additional archaeal Holliday junction cleavage enzymes, an atypical homing endonuclease (I-SspI), and several type II restriction endonucleases. I-SspI, the atypical bacterial homing endonuclease (PDB ID 2OST), gives a DALI Z-score of 8.5, with an RMSD of 3.2 Å for 117 equivalent residues showing 12% identity (61). Among the restriction endonucleases, HincII (PDB 2GIG) gives a DALI Z-score of 5.0 and an RMSD of 3.4 Å for 116 equivalent residues with 9% identity (21). Importantly, the structures of I-SspI and HincII have been determined in complex with DNA.
Comparison of the S. solfataricus Holliday junction cleavage enzyme (Sso Hjc) with D212 highlights the similarity between the two folds (Fig. 2). Despite the additional length, 212 residues compared to ∼145 in the archaeal Holliday junction cleavage enzymes, D212 largely lacks additional secondary structural elements. A minor difference is replacement of helix α3 in D212 with strand β4 in Sso Hjc (Fig. 2). Other than this, the increased size of D212 is largely a result of increased loop lengths in regions that connect secondary structural elements and an additional 10 residues at the N terminus. Interestingly, the N-terminal residues are disordered in both D212 (18 disordered residues) and Sso Hjc (8 disordered residues), where these residues have been implicated in DNA binding and are thought to be ordered only upon interaction with DNA (8). Similarly, I-SspI (atypical homing endonuclease) with 151 amino acids is also largely similar, although, consistent with the lower Z score, it exhibits additional differences (Fig. 2). These are particularly noticeable in the DNA recognition domain (61), where I-SspI inserts two additional β-strands and loses helix α4.
FIG. 2.
Topology diagrams. Cartoon and topology representations for SSVRH D212 (orange), Sso Hjc (olive), I-SspI (green), and HincII (blue). Secondary structural elements that are not equivalent to D212 are colored in gray. Side chains corresponding to acidic residues in the E-PD-(D/E)XK motif are shown in red, demonstrating the conserved active site location. Overall, the relative similarities to D212 are well reflected by the DALI Z-scores.
In contrast to Sso Hjc and I-SspI, HincII (type 2 restriction endonuclease) with 257 amino acids is significantly larger than D212. Nevertheless, the secondary structural elements in both domains of D212 match closely to the core nuclease structure in this restriction enzyme (21), and the major differences between the structures are the presence of several embellishments in the HincII structure. These include the addition of two α-helices and a β-strand at the N-terminal end, a small saposin-like domain that is inserted following the first β-strand, and 2 α-helices at the C-terminal end (21). In addition, the last helix in D212 adopts a different orientation than that seen in HincII.
Active site architecture.
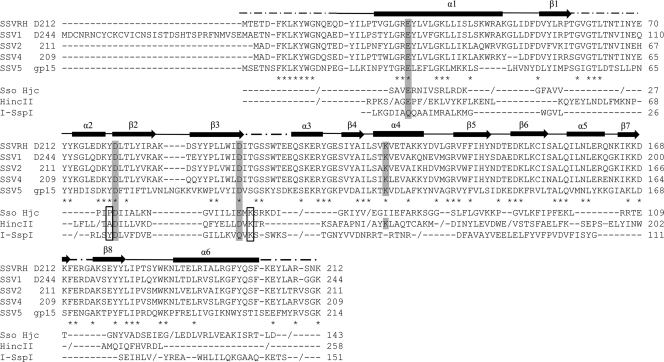
The archaeal Holliday junction cleavage enzymes, homing endonuclease (I-SspI), and type II restriction enzymes that show structural similarity to D212 are all members of a subclass of the PD-(E/D)XK nuclease family that contain an extended motif with an extra N-terminal acidic residue (E), yielding an E-PD-(D/E)XK active site motif that contains a trio of acidic residues. However, many of the individual residues in this motif are not strictly conserved, particularly the proline and lysine residues. Superposition of Sso Hjc and HincII on D212 identifies an equivalent active site motif in D212 (Fig. 3 and 4). Specifically, Glu28, Asp79, and Asp97 in D212 correspond to residues Glu12, Asp42, and Glu55 in Sso Hjc, respectively, while in HincII, these residues correspond to Glu35, Asp114, and Asp127. Importantly, these residues are also strictly conserved in all of the D212 fuselloviral orthologs, including SSV1 D244. In contrast, I-SspI makes several substitutions, utilizing Asp11, Asp36, and Gln49. These clusters of acidic active site residues generally function to coordinate a metal ion, either Mg2+ or Mn2+, that is thought to play multiple roles in catalysis. These include facilitating the interaction with substrate DNA, activating water for nucleophilic attack, and stabilizing the transition state (1, 10, 25, 39, 61).
FIG. 3.
Sequence alignments for SSVRH D212 (58) and its fuselloviral orthologs, SSV1 D244 (43), SSV2 ORF 211 (52), SSV4 ORF 209 (44), and SSV5 gp15 (50). The asterisks indicate strictly conserved residues in the 5 fuselloviral orthologs. Structure-based sequence alignments (DALI equivalences [20]) are also shown for D212 and representative structural homologs: Sso Hjc, I-SspI, and HincII. D212 secondary structural elements are shown above the alignments where slashes (/) specify chain breaks. The conserved acidic residues of the E-PD-(D/E)XK motif and the repositioned active site lysine in the fuselloviral proteins are shaded. Boxed residues indicate the predicted positions of the proline and lysine residues. For SSV1 D244, the extended N terminus and the presence of a conserved Met residue equivalent to the start codon in the remaining orthologs suggest that D244 may be misannotated, perhaps explaining our inability to crystallize D244.
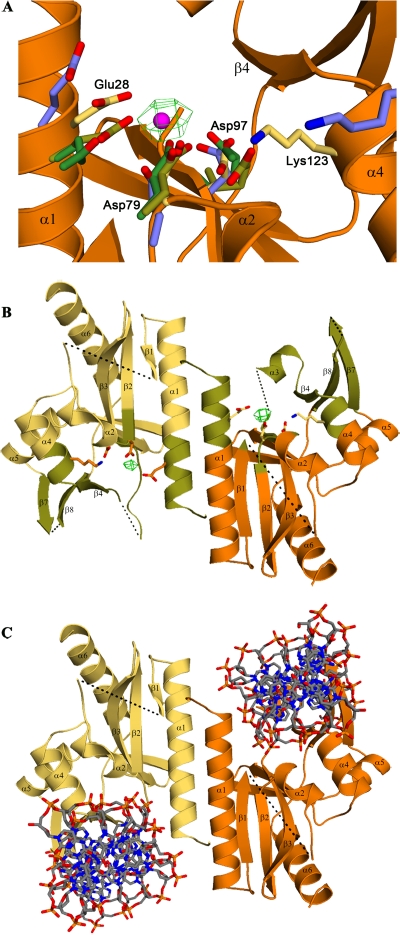
FIG. 4.
Active site residues. (A) Active site residues from Sso Hjc (olive), I-SspI (green), and HincII (blue) are superimposed on D212 (yellow side chains, orange backbone) using the conserved acidic residues highlighted in Fig. 3 and their immediate neighbors, i.e., D212 B-chain residues 27 to 29, 78 to 80, and 96 to 98, and the corresponding residues in Sso Hjc, I-SspI, and HincII. The superposition shows that the E-PD-(D/E)XK nuclease active site motif is structurally conserved in D212. The predicted Mn2+ position is shown in magenta, centered within the green mesh of the 5σ difference electron density seen in the Mn2+ soaked crystals. (B) The D212 dimer is shown in orange and yellow, with regions predicted to be involved in DNA recognition colored in olive. Active site residues and Mn2+ are also depicted as described above. The two Mn2+ ions are ∼25 Å apart, similar to the distances seen in Holliday junction cleavage enzymes. However, this distance is significantly greater than that seen in restriction and homing endonucleases which bring two active sites to bear on a single piece of double-stranded DNA. (C) DNA from the structure of HincII/DNA complex (21) is docked by superposition on the D212 dimer, highlighting the location of putative DNA binding channels which might interact with the arms of a junctional complex.
Similarly, the lysine residue in the E-PD-(D/E)XK motif can also serve in substrate recognition, to orient and stabilize the attacking hydroxyl nucleophile or to play a role in transition state stabilization (25, 61). While structure-based sequence alignments do not show a lysine residue in the equivalent position in D212, Lys123 is in close proximity to the three acidic active site residues and might serve a similar role. This would be consistent with reports on the migration of active site lysine residues in type II restriction endonucleases (9, 16, 46, 53). Interestingly, HincII also shows a lysine at this position, which is also strictly conserved in the fuselloviral orthologs (Fig. 3). Finally, the proline residue of the motif is present in Sso Hjc but is absent from I-SspI, HincII, D212, and all D212 orthologs.
The structural similarity to Sso Hjc and HincII suggested that Glu28 and Asp79 of D212 would also serve to coordinate Mg2+ or Mn2+. This was tested by soaking D212 crystals in 10 mM MnCl2 and collecting a 3.7-Å data set on our home source diffractometer. Indeed, the difference density map for the Mn2+ soaked crystals (Fig. 4) revealed a single manganese ion per subunit at the predicted metal binding site, further confirming the role of these residues in D212. Importantly, the two active sites in the D212 dimer are separated by about 25 Å (Fig. 4B). While this separation is similar to that seen in Holliday junction cleavage enzymes, this is significantly greater than that seen in type II restriction endonucleases and the homing endonucleases (I-SspI), where two active sites come together to cleave opposing strands in a single piece of double-stranded DNA.
DNA binding sites.
The structure of an archaeal Holliday junction cleavage enzyme in complex with DNA has not been reported. Structures for two bacteriophage enzyme/DNA complexes, T7 endonuclease I (18) and T4 endonuclease VII (5), have been reported. Unfortunately, their lack of structural similarity to D212 makes it difficult to model a complex with D212. However, structures have been reported for I-SspI and HincII in complex with a single piece of double-stranded DNA, and these complexes can be docked on D212 by structural superposition. The fit of the HincII DNA appears to be a superior model, although it does clash with elements of the β3/β4 connection that are distal to the active site, which in the B chain includes helix α3. Nevertheless, the docking suggests that DNA is bound within the rather prominent clefts on the “sides” of the D212 dimer (Fig. 4) and is mediated by residues at the N terminus of helix α1, the α2/β2 loop, residues lying within the extended connection between β3 and β4, the N terminus of α4, β4, and the β7-β8 loop. Many of these elements lie in disordered or poorly ordered areas and are likely to become better ordered upon interaction with DNA. With regard to the clash of the β3-β4 connection, this mobile loop might rearrange to avoid clashes with the DNA. Alternatively, it might serve to discriminate between double-stranded DNA and a junctional DNA complex. This model predicts that the disordered β7-β8 loop will lie within the DNA major groove. A similar loop in HincII, known as the recognition or R loop, participates in base-specific interactions, suggesting a similar role for the β7-β8 loop in D212 (30). The surface electrostatics in the presence of Mn2+ are consistent with this binding model (not shown). In addition, we also note a very basic surface feature running across the “back side” of the dimer, between the two active sites, which might further facilitate substrate recognition.
The locations of the subunit interfaces in Hjc, HincII, and I-SspI differ substantially from those in D212. HincII recognizes a degenerate 8-bp sequence to make a blunt-ended cut. Thus, the dimer interface is positioned such that two active sites are brought to bear on opposite sides of a single piece of double-stranded DNA. Similarly, I-SspI utilizes two symmetrically arranged active sites to cleave a 23-bp target sequence, yielding 3-base, 3′ overhangs. In contrast, the active sites in D212 and Hjc are much further apart, ∼25 Å. In the case of D212, this allows double-stranded DNA to be docked into each of the active sites (Fig. 4), perhaps representing two arms of a junctional complex. These differences suggest that D212 is more likely to catalyze junctional cleavage during DNA replication, repair, or recombination than as a restriction enzyme or homing endonuclease.
Due to the similarity with Sso Hjc, we tried to demonstrate D212 Holliday junction cleavage (Hjc) activity with both fixed and mobile Holliday junctions (6). However, using freshly purified D212 and an array of divalent cations, Hjc activity was not observed. In contrast, Hjc activity was observed for a positive control, T4 endonuclease VII. However, the potential for nuclease activity is clearly present, as older protein preparations that were proteolytically clipped did show metal-dependent nuclease activity (data not shown). But because this activity required excessively high protein concentrations, the activity of the clipped protein was not characterized further. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of the crystals used to collect the native data set indicated that the crystal structure represents the intact protein rather than this degraded form.
DISCUSSION
The structure of D212 clearly reveals a type II restriction endonuclease fold and identifies the hallmark PD-(D/E)XK catalytic motif that is spatially conserved in this nuclease superfamily. This fold and its associated catalytic residues are also found in other classes of nucleases, where they play roles in DNA replication, repair, recombination, and other processes requiring nuclease activity (10). Structural examples include MutH (4); Vsr (56); RecB and λ-exonuclease (24), which are involved in DNA repair; archaeal Holliday junction cleavage enzymes like Hjc and Hje (8, 34, 37); TnsA, which is a component of TN7 transposase (19); and the homing endonuclease I-SspI (61). In addition, bioinformatics, biochemical, and/or genetic approaches have identified the PD-(D/E)XK motif in non-long terminal repeat (LTR) retrotransposases (60) and in proteins associated with excisionase activity (10, 49, 57).
The similarity of D212 to the archaeal Holliday junction cleavage enzymes suggests that it may function as a junctional resolvase. However, the characterized archaeal enzymes position their two active site centers on a relatively flat surface (8, 34, 37) and are predicted to recognize a planar, stacked-X Holliday junction (22, 34) like that seen in the structure of T4 endonuclease VII (5). In contrast, the predicted DNA binding channels in D212 run at oblique angles down opposite sides of the dimer. This is reminiscent of the structure of the T7 endonuclease I and suggests that D212 recognizes a junctional complex with substantially different geometry than that recognized by previously characterized archaeal enzymes, perhaps explaining the lack of activity in the Holliday junction cleavage assay. Like T7 endonuclease I, D212 may show strong selectivity for a DNA branch point with a specific geometric structure (18) and might exhibit significant discrimination for particular sequences within the junctional arms (13).
This putative activity might be employed to resolve intermediates in replication of the fuselloviral genome. Alternatively, D212 may function in a DNA repair pathway, and expression of D212 might play a role in correcting replication defects or other damage to the viral genome. In this light, it is interesting that SSV1 D244, a D212 ortholog, is thought to be packaged within the SSV1 viral particle (33). The presence of D212 in the viral particle could ensure availability upon infection of a new host cell, helping to protect the integrity of the viral genome from the acidic, hyperthermal environments in which these viruses reside.
Another possible function is suggested by the presence of the PD-(D/E)XK motif in ExiS from the Mycoplasma MAV1 phage and XhisH from Anabaena. ExisS is thought to participate in excision of the MAV1 genome, while XhisH is implicated in excision of the fdxN element from the Anabaena chromosome (10, 49, 57). SSVRH, SSV1, and other Fuselloviridae utilize a virally encoded integrase to integrate their viral DNA. However, the attP sites utilized for this reside within the integrase gene, with integration resulting in disruption of the integrase gene. As long as episomal copies of the viral genome remain, they can provide the necessary integrase needed to excise the viral genome. However, in their absence, it has not been clear if, or how, the viral genome might be rescued. This work raises the possibility that D212 might play a role in excising the viral genome. Finally, D212 might facilitate interviral recombination events between fuselloviruses, which are thought to be relatively frequent, or even participate in the excision of a covalent closed circular DNA virus from a concatemer of two integrated fuselloviruses, as suggested by Redder et al. (50).
Acknowledgments
This work was supported by the National Science Foundation (MCB-0628732 and MCB-0920312). Portions were carried out at the Stanford Synchrotron Radiation Laboratory with support from the Department of Energy and the National Institutes of Health.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. Narayana, L. S. Pierson III, et al. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 4.Ban, C., and W. Yang. 1998. Structural basis for MutH activation in E. coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 17:1526-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biertumpfel, C., W. Yang, and D. Suck. 2007. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449:616-620. [DOI] [PubMed] [Google Scholar]
- 6.Birkenbihl, R. P., K. Neef, D. Prangishvili, and B. Kemper. 2001. Holliday junction resolving enzymes of archaeal viruses SIRV1 and SIRV2. J. Mol. Biol. 309:1067-1076. [DOI] [PubMed] [Google Scholar]
- 7.Bond, C. S. 2003. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19:311-312. [DOI] [PubMed] [Google Scholar]
- 8.Bond, C. S., M. Kvaratskhelia, D. Richard, M. F. White, and W. N. Hunter. 2001. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. U. S. A. 98:5509-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozic, D., S. Grazulis, V. Siksnys, and R. Huber. 1996. Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 A resolution. J. Mol. Biol. 255:176-186. [DOI] [PubMed] [Google Scholar]
- 10.Bujnicki, J. M., and L. Rychlewski. 2001. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 3:69-72. [PubMed] [Google Scholar]
- 11.Clore, A. J., and K. M. Stedman. 2007. The SSV1 viral integrase is not essential. Virology 361:103-111. [DOI] [PubMed] [Google Scholar]
- 12.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Declais, A. C., J. Liu, A. D. Freeman, and D. M. Lilley. 2006. Structural recognition between a four-way DNA junction and a resolving enzyme. J. Mol. Biol. 359:1261-1276. [DOI] [PubMed] [Google Scholar]
- 14.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 15.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 16.Feder, M., and J. M. Bujnicki. 2005. Identification of a new family of putative PD-(D/E)XK nucleases with unusual phylogenomic distribution and a new type of the active site. BMC Genomics 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frols, S., P. M. Gordon, M. A. Panlilio, C. Schleper, and C. W. Sensen. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48-59. [DOI] [PubMed] [Google Scholar]
- 18.Hadden, J. M., A. C. Declais, S. B. Carr, D. M. Lilley, and S. E. Phillips. 2007. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature 449:621-624. [DOI] [PubMed] [Google Scholar]
- 19.Hickman, A. B., Y. Li, S. V. Mathew, E. W. May, N. L. Craig, and F. Dyda. 2000. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell 5:1025-1034. [DOI] [PubMed] [Google Scholar]
- 20.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 21.Joshi, H. K., C. Etzkorn, L. Chatwell, J. Bitinaite, and N. C. Horton. 2006. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J. Biol. Chem. 281:23852-23869. [DOI] [PubMed] [Google Scholar]
- 22.Khuu, P. A., A. R. Voth, F. A. Hays, and P. S. Ho. 2006. The stacked-X DNA Holliday junction and protein recognition. J. Mol. Recognit. 19:234-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. 1992. Archaebacterial virus SSV1 encodes a putative DnaA-like protein. Nucleic Acids Res. 20:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovall, R., and B. W. Matthews. 1997. Toroidal structure of lambda-exonuclease. Science 277:1824-1827. [DOI] [PubMed] [Google Scholar]
- 25.Kovall, R. A., and B. W. Matthews. 1999. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 3:578-583. [DOI] [PubMed] [Google Scholar]
- 26.Kraft, P., D. Kummel, A. Oeckinghaus, G. H. Gauss, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Structure of D-63 from Sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function. J. Virol. 78:7438-7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft, P., A. Oeckinghaus, D. Kummel, G. H. Gauss, J. Gilmore, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J. Virol. 78:11544-11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson, E. T., B. Eilers, S. Menon, D. Reiter, A. Ortmann, M. J. Young, and C. M. Lawrence. 2007. A winged-helix protein from Sulfolobus turreted icosahedral virus points toward stabilizing disulfide bonds in the intracellular proteins of a hyperthermophilic virus. Virology 368:249-261. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, C. M., S. Menon, B. J. Eilers, B. Bothner, R. Khayat, T. Douglas, and M. J. Young. 2009. Structural and functional studies of archaeal viruses. J. Biol. Chem. 284:12599-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little, E. J., and N. C. Horton. 2005. DNA-induced conformational changes in type II restriction endonucleases: the structure of unliganded HincII. J. Mol. Biol. 351:76-88. [DOI] [PubMed] [Google Scholar]
- 31.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 32.Martin, A., S. Yeats, D. Janekovic, W. D. Reiter, W. Aicher, and W. Zillig. 1984. SAV 1, a temperate UV-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 3:2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menon, S. K., W. S. Maaty, G. J. Corn, S. C. Kwok, B. J. Eilers, P. Kraft, E. Gillitzer, M. J. Young, B. Bothner, and C. M. Lawrence. 2008. Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376:270-278. [DOI] [PubMed] [Google Scholar]
- 34.Middleton, C. L., J. L. Parker, D. J. Richard, M. F. White, and C. S. Bond. 2004. Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 32:5442-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 36.Muskhelishvili, G., P. Palm, and W. Zillig. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol. Gen. Genet. 237:334-342. [DOI] [PubMed] [Google Scholar]
- 37.Nishino, T., K. Komori, D. Tsuchiya, Y. Ishino, and K. Morikawa. 2001. Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition. Structure 9:197-204. [DOI] [PubMed] [Google Scholar]
- 38.Niv, M. Y., D. R. Ripoll, J. A. Vila, A. Liwo, E. S. Vanamee, A. K. Aggarwal, H. Weinstein, and H. A. Scheraga. 2007. Topology of type II REases revisited; structural classes and the common conserved core. Nucleic Acids Res. 35:2227-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlowski, J., and J. M. Bujnicki. 2008. Structural and evolutionary classification of type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 36:3552-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortmann, A. C., B. Wiedenheft, T. Douglas, and M. Young. 2006. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat. Rev. Microbiol. 4:520-528. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. Carter and R. Sweet (ed.), Macromolecular crystallography, part A, vol. 276. Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 42.Painter, J., and E. A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439-450. [DOI] [PubMed] [Google Scholar]
- 43.Palm, P., C. Schleper, B. Grampp, S. Yeats, P. McWilliam, W. D. Reiter, and W. Zillig. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242-250. [DOI] [PubMed] [Google Scholar]
- 44.Peng, X. 2008. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology 154:383-391. [DOI] [PubMed] [Google Scholar]
- 45.Pingoud, A., and A. Jeltsch. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29:3705-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pingoud, V., E. Kubareva, G. Stengel, P. Friedhoff, J. M. Bujnicki, C. Urbanke, A. Sudina, and A. Pingoud. 2002. Evolutionary relationship between different subgroups of restriction endonucleases. J. Biol. Chem. 277:14306-14314. [DOI] [PubMed] [Google Scholar]
- 47.Prangishvili, D., P. Forterre, and R. A. Garrett. 2006. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4:837-848. [DOI] [PubMed] [Google Scholar]
- 48.Prangishvili, D., R. A. Garrett, and E. V. Koonin. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52-67. [DOI] [PubMed] [Google Scholar]
- 49.Ramaswamy, K. S., C. D. Carrasco, T. Fatma, and J. W. Golden. 1997. Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol. Microbiol. 23:1241-1249. [DOI] [PubMed] [Google Scholar]
- 50.Redder, P., X. Peng, K. Brugger, S. A. Shah, F. Roesch, B. Greve, Q. She, C. Schleper, P. Forterre, R. A. Garrett, and D. Prangishvili. 2009. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ. Microbiol. 11:2849-2862. [DOI] [PubMed] [Google Scholar]
- 51.Reiter, W. D., P. Palm, A. Henschen, F. Lottspeich, W. Zillig, and B. Grampp. 1987. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol. Genet. Genomics 206:144-153. [Google Scholar]
- 52.Stedman, K. M., Q. She, H. Phan, H. P. Arnold, I. Holz, R. A. Garrett, and W. Zillig. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154:295-302. [DOI] [PubMed] [Google Scholar]
- 53.Tamulaitiene, G., A. Jakubauskas, C. Urbanke, R. Huber, S. Grazulis, and V. Siksnys. 2006. The crystal structure of the rare-cutting restriction enzyme SdaI reveals unexpected domain architecture. Structure 14:1389-1400. [DOI] [PubMed] [Google Scholar]
- 54.Terwilliger, T. C. 2000. Maximum likelihood density modification. Acta Crystallogr. D 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsutakawa, S. E., T. Muto, T. Kawate, H. Jingami, N. Kunishima, M. Ariyoshi, D. Kohda, M. Nakagawa, and K. Morikawa. 1999. Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell 3:621-628. [DOI] [PubMed] [Google Scholar]
- 57.Voelker, L. L., and K. Dybvig. 1998. Characterization of the lysogenic bacteriophage MAV1 from Mycoplasma arthritidis. J. Bacteriol. 180:5928-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiedenheft, B., K. Stedman, F. Roberto, D. Willits, A. K. Gleske, L. Zoeller, J. Snyder, T. Douglas, and M. Young. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J. Virol. 78:1954-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiang, X., L. Chen, X. Huang, Y. Luo, Q. She, and L. Huang. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J. Virol. 79:8677-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. U. S. A. 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, L., R. P. Bonocora, D. A. Shub, and B. L. Stoddard. 2007. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 26:2432-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]