Abstract
An expanded polyglutamine (polyQ) domain in the N-terminal region of huntingtin (htt) causes misfolding and accumulation of htt in neuronal cells and the subsequent neurodegeneration of Huntington's disease (HD). Clearing the misfolded htt is critical for preventing neuropathology, and this process is mediated primarily by both the ubiquitin–proteasome system (UPS) and autophagy. Although overexpression of mutant htt can inhibit UPS activity in cultured cells, mutant htt does not inhibit global UPS activity in the brains of HD transgenic mice. These findings underscore the importance of investigating the function of the UPS and autophagy in the brain when mutant proteins are not overexpressed. When cultured PC12 cells were treated with either UPS or autophagy inhibitors, more N-terminal mutant htt fragments accumulated via inhibition of the UPS. Furthermore, in HD CAG repeat knock-in mouse brain, inhibiting the UPS also resulted in a greater accumulation of N-terminal, but not full-length, mutant htt than inhibiting autophagy did. Our findings suggest that impairment of the UPS may be more important for the accumulation of N-terminal mutant htt and might therefore make an attractive therapeutic target.
INTRODUCTION
Mounting evidence shows that N-terminal huntingtin (htt) fragments, which carry an expanded polyglutamine (polyQ) domain (>37 glutamines), are pathogenic and cause severe neurologic phenotypes in transgenic mice (1,2). Thus, the accumulation of degraded N-terminal mutant htt fragments in the human brain is probably the initial step toward the late-onset neurodegeneration in Huntington's disease (HD). Lending support to this idea, a number of proteolysis cleavage sites have been identified in the N-terminal region of htt (1,3,4), which generate small N-terminal htt fragments that can form aggregates or inclusions in the brain in an age-dependent manner. Although the role of aggregates remains controversial, their formation in the brain is correlated with the age-dependent progress of neurological symptoms (5–8). This correlation underscores the fact that clearance of toxic N-terminal htt fragments is critical for reducing or preventing HD pathology.
Misfolded proteins are primarily cleared in cells by two systems: the ubiquitin–proteasome system (UPS) and autophagy (9–11). The UPS predominantly degrades short-lived nuclear and cytosolic proteins by tagging these substrates with polyubiquitin chains; however, the narrow pore of the proteasome precludes entry of large aggregated proteins and organelles. Macroautophagy, generally referred to as autophagy, is a cellular degradative pathway for long-lived cytoplasmic proteins, protein complexes or damaged organelles. Degradation via autophagy involves a variety of proteins, including microtubule-associated protein 1 light chain-3, which has two isoforms, microtubule-associated protein 1 light chain-3 (LC3)-I and LC3-II. During autophagy activation, LC3-I is processed to produce phosphatidylethanolamine-modified LC3-II, which specially associates with the autophagosome membrane (12). A growing body of evidence indicates that autophagy can clear mutant htt and that activation of autophagy can ameliorate pathology in animal models of HD (13,14).
Since both the UPS and autophagy are found to clear mutant htt, comparing their capacities to remove misfolded proteins in the brain is important. This comparison is particularly important for understanding the roles of these two systems in clearing polyQ disease-associated proteins, as these proteins often accumulate in the nucleus that lacks autophagy. Also because the overexpression of mutant proteins can cause cellular stresses that may alter the function of cellular clearing systems in cultured cells, whether the UPS and autophagy function differently when mutant proteins are expressed at the endogenous level needs to be investigated. Here, we report that mutant htt does not significantly influence LC3 conversion in the brains of HD mice that express N-terminal mutant htt (171 amino acids with 82Q) or full-length mutant htt, suggesting that other factors that impair the function of autophagy should be considered for their roles in disease progression. We also found that inhibiting the UPS preferentially increases soluble N-terminal mutant htt fragments in the mouse brain, suggesting that improving UPS function would be more effective in reducing N-terminal mutant htt and alleviating the related neuropathology.
RESULTS
The conversion of LC3 in HEK293 cells transfected with htt
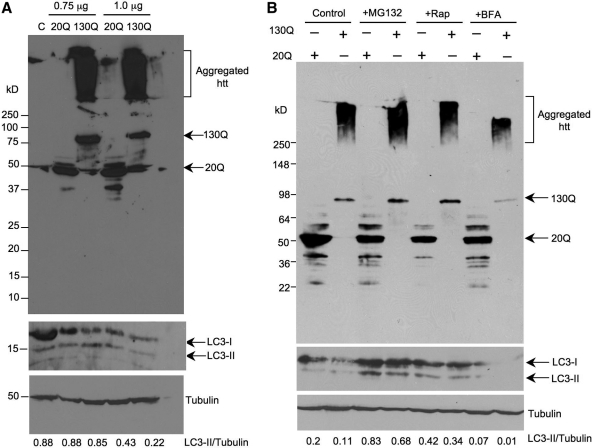
The conversion of LC3-I to LC3-II represents autophagy activation and has been used to study the relationship between autophagy and htt toxicity in a variety of cellular models. To investigate the effect of N-terminal htt fragments on LC3 conversion, we transiently transfected HEK293 cells with 0.75 or 1.0 µg pEGFP vector expressing GFP-exon 1 htt with 20Q or 130Q and examined the expression level of endogenous LC3. Mutant htt with the larger repeat (130Q) migrated slowly in sodium dodecyl sulfate (SDS) gel and also formed aggregates that stayed in the stacking gel (Fig. 1A). There are ways of measuring autophagy activity by detecting the ratio of LC3-II to LC3-I (15,16) or the ratio of LC3-II to tubulin or actin (17). Because the sensitivity for detecting LC3-II by most anti-LC3 antibodies is much higher than that for LC3-I, and because LC3-II is reliably associated with completed autophagosomes, the recommendation is that the best measure is the ratio of the relative levels of LC3-II, rather than LC3-I that is more sensitive to degradation in SDS sample buffer (18,19). Thus, we assessed the levels of LC3-II relative to tubulin by measuring the ratio of LC3-II to tubulin. There was no significant difference in this ratio between the cells that transfected with htt-20Q and htt-130Q at a dose of 0.75 µg (Fig. 1A). When a larger amount (1.0 µg) of plasmids was used for transfection, the ratio of LC3-II to tubulin seems to be reduced compared with htt-20Q, although the expression level of mutant htt is slightly lower than with the smaller dose (0.75 µg) htt-130Q (Fig. 1A). This result suggests that the expression of mutant htt did not alter autophagy activity in HEK293 cells, but a higher dose of plasmid DNA could inhibit the LC3 conversion.
Figure 1.
LC3 conversion in cultured HEK293 cells transfected with htt. (A) HEK293 cells were transfected with PRK-GFP-exon1 htt containing 20Q or 130Q at 0.75 or 1.0 µg. Total cell lysates were collected after 24 h transfection for western blotting with antibodies to htt (mEM48), LC3 (NB100-2331) and tubulin. Representative western blots are presented and the ratios of LC3-II to tubulin are shown below the blots. (B) HEK293 cells transfected with GFP-htt exon 1-20Q or GFP-exon1 htt-130Q were treated with the UPS inhibitor MG132 (10 µm) or the autophagy inhibitor brefeldin A (BFA) (100 nm) for 15 h. The cell lysates of the above drug-treated cells were analyzed by western blotting with anti-htt (mEM48) (upper panel). The same blot was then probed with anti-LC3 to reveal LC3 conversion by the above drugs (middle panel). The blot was also probed with anti-tubulin (bottom panel), and the ratios of LC3-II to tubulin are indicated under the blot. Note that MG132 increased the level of soluble mutant htt (130Q) and aggregated htt. The autophagy activator rapamycin (Rap) did not alter the extent of htt aggregation, although the treatment increases the ratio of LC3-II to tubulin. BFA appeared to reduce the expression of LC3 and mutant htt as well as its aggregation.
We also performed western blots to reveal both the soluble and aggregated forms of mutant htt in HEK293 cells after treatments with drugs that alter the activity of the UPS or autophagy. Soluble mutant htt-130Q was apparently increased by MG132. Importantly, aggregated htt in the stacking gel was also obviously increased by MG132 (Fig. 1B). Rapamycin, a drug that activates autophagy (13), did not reduce either soluble or aggregated htt compared with htt in cells without drug treatment, although it increased the levels of LC3 when compared with the controls without rapamycin. Brefeldin A (BFA) inhibited the production of LC3. However, it also inhibited the soluble mutant htt and aggregated htt, perhaps because the dose we used in the experiment inhibited the expression of transfected proteins. The same blots were probed with antibodies to LC3 and γ-tubulin to obtain the ratios of LC3-II to tubulin. The ratios confirmed that rapamycin can increase the LC3 levels, whereas BFA inhibited the LC3 levels compared with control LC3 levels. MG132 could also increase LC3 levels, perhaps because inhibiting the UPS can up-regulate autophagy activity (20,21).
Mutant htt does not alter LC3 levels in the mouse brain
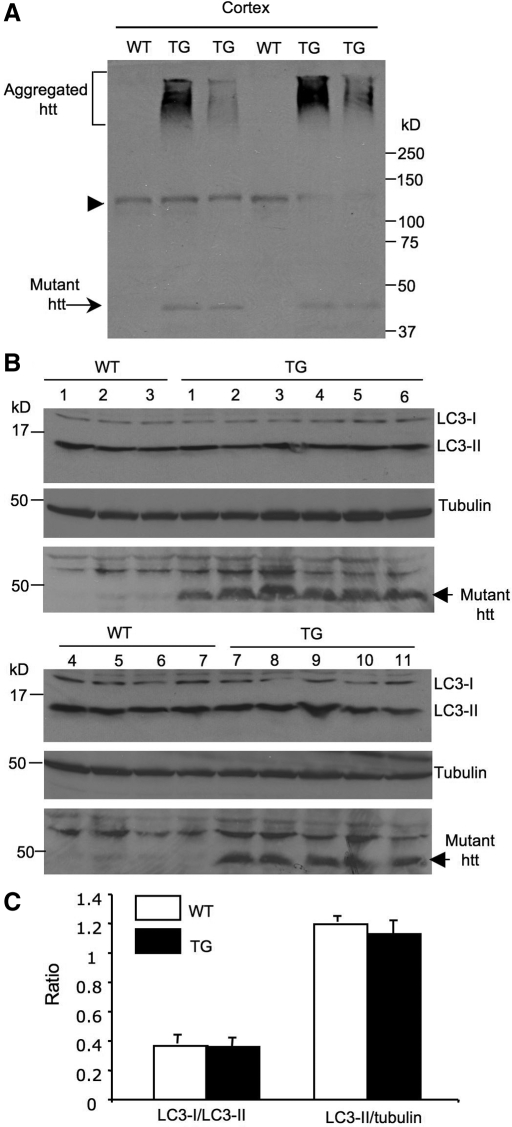
Earlier studies have shown that overexpression of mutant htt can increase autophagosome-like vacuoles in different types of cultured cells (22,23). Because overexpressed proteins can cause endoplasmic reticulum stress, which can enhance autophagy activity (24,25), cell-type-specific effects and the amounts of overexpressed proteins may account for the different results seen in our studies versus other findings. Thus, it is important to examine whether autophagy activity is altered in the mouse brain when mutant htt is not transiently overexpressed. In N171-82Q mice that express the N-terminal htt (171 amino acids) with 82Q (Fig. 2A) and show severe neurological symptoms at the age of 3–5 months, mutant htt is abundant in the cortex and displays as the soluble and aggregated forms (Fig. 2A, 26). We examined the LC3 levels (LC3-II/tubulin) and the ratio of LC3-I to LC3-II in the brain cortex tissues from multiple wild-type (WT, n = 7) and N171-82Q mice (TG, n = 11) at the age of 3–4 months. We did not see any significant difference in the conversion of LC3-I to LC3-II (LC3-I/LC3-II ratio) or the relative levels of LC3-II (LC3-II/tubulin ratio) between WT and TG mice (Fig. 2B and C).
Figure 2.
LC3 conversion is not altered in the brain of N171-82Q mice. (A) Western blot analysis of the brain cortex of wild-type (WT) and N171-82Q (TG) mice at the age of 3–4 months. The samples were probed with mEM48 to verify the expression of mutant htt in HD transgenic mice. Arrow indicates soluble mutant htt and bracket indicates aggregated htt. Arrowhead indicates nonspecific bands. (B) Western blot analysis of LC3-I and LC3-II levels in the brain cortex of 7 wild-type (WT) and 11 N171-82Q (TG) mice at 3–4 months of age (upper panel). The blots were also probed with the antibody to tubulin (middle panel). The same samples were also probed with the 1C2 antibody to reveal the expression of transgenic htt (low panel). (C) The ratios of LC3-I to LC3-II and LC3-II to tubulin are shown. There is no statistical significance (P > 0.19) between WT and TG mice.
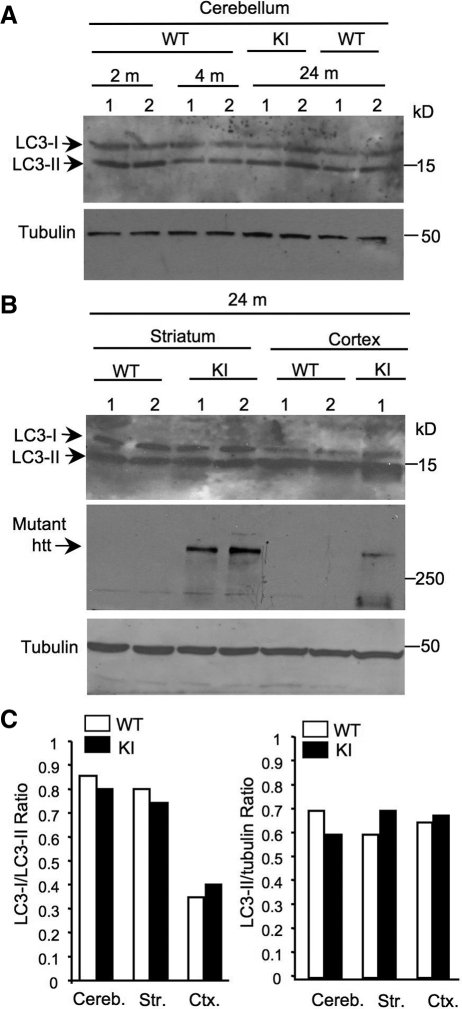
A more important question is whether there is any altered autophagy function in HD mice that express full-length mutant htt at the endogenous level. Thus, we further examined the LC3 levels in HD CAG150 knock-in (KI) mice that express full-length mutant htt with 150Q under the control of the endogenous mouse htt gene promoter and display mild neurological phenotypes (27). We did not find any noticeable changes in LC3 levels in the cerebellum between 2, 4 and 24 months or between WT and KI mice at 24 months (Fig. 3A). Similarly, in the striatum and cortex of mice at the age of 3–4 months (data not shown) and 24 months, we could not find any significant difference in LC3 levels between WT and KI mice (Fig. 3B). Since these mice express an expanded polyQ domain in endogenous mouse htt, which cannot be detected by our monoclonal anti-htt antibody mEM48 (8), we used 1C2, which reacts with an expanded polyQ domain, to confirm the expression of mutant htt in KI mouse brain (middle panel in Fig. 3B). Quantitation of the ratios of LC3-I/LC3-II and LC3-II/tubulin did not reveal any significant difference in the conversion of LC3-I to LC3-II between WT and KI mice (Fig. 3C).
Figure 3.
LC3 conversion is not altered in the brain of HD knock-in mice. (A–B) Representative western blots showing the expression of LC3 in the cerebellum (A), cortex and striatum (B) of wild-type (WT) and HD CAG150 knock-in (KI) mice at the age of 2, 4 and 24 months. 1C2 was also used to verify the expression of mutant htt (arrow) in KI mice (middle panel in B). (C) The ratios of LC3-I to LC3-II and LC3-II to tubulin are shown. The data were obtained from three to four mice per group. Cereb., cerebellum; Str., striatum; Ctx, cortex.
UPS inhibition causes a greater accumulation of mutant htt than autophagy inhibition does
Several groups found that there is no inhibition of the global UPS activity in the HD and polyQ disease mouse brains (28–31). As other factors, such as aging, may impair the activities of the UPS and autophagy (30,32), we compared the changes in mutant htt levels after cells were treated with UPS and autophagy inhibitors. There were two important issues we wanted to address. One is whether UPS and autophagy inhibitions have different effects on the clearance of normal and mutant htt fragments; the second is whether full-length and N-terminal mutant htt fragments are differentially affected by these inhibitors.
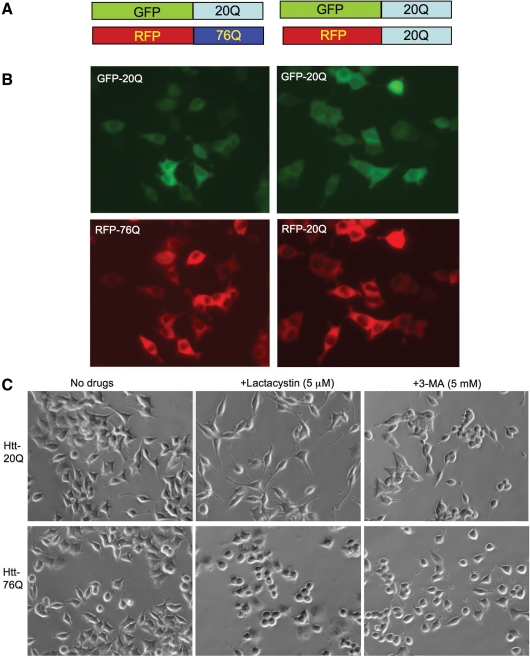
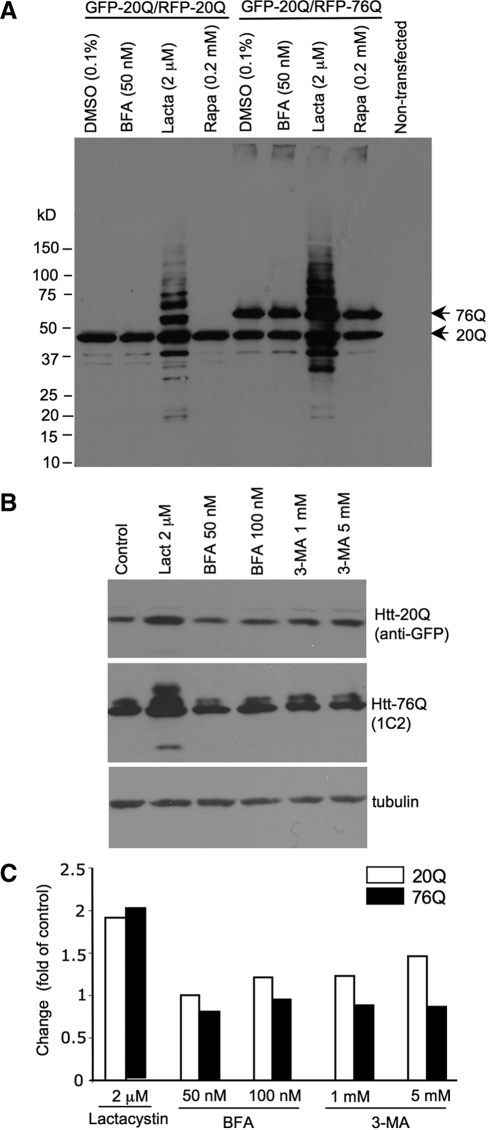
To address the first issue, we established stably transfected PC12 cells that co-express GFP-exon1 htt-20Q and RFP-exon1 htt-76Q. Tagging fluorescent proteins to htt would allow us to examine the expression of normal and mutant htt in living cells and to ensure that both normal and mutant htt fragments are expressed in the same cells. For the control, we also established PC12 cells that coexpress GFP-exon1 htt-20Q and RFP-exon1 htt-20Q (Fig. 4A and B). Because these cells express both normal and mutant htt fragments, the changes in these two proteins after drug treatments can rigorously be compared in the same cells under the same conditions. Transfection of htt into PC12 cells and selection of stably transfected cells did not cause significant differences in LC3 levels between these cell lines (Supplementary Material, Fig. S1). Treating these cells with lactacystin (5 µm) for 15 h revealed that lactacystin stimulated neurites of htt-20Q cells (Fig. 4C), a phenomenon that has been reported in normal PC12 cells (33). However, lactacystin failed to elongate the neurites of cells (GFP-20Q/RFP-76Q or htt-76Q-E12) expressing htt-76Q (Fig. 4C). Two other independent cell lines expressing GFP-20Q/RFP-76Q (htt-76Q-A9 and htt-76Q-E10) also show defective neurite extension following the lactacystin treatment (Supplementary Material, Fig. S2). These results support the previous findings that mutant htt can cause a neurite outgrowth defect (34,35). Cells treated with 3-methyladenine (3-MA, 5 mm) for 15 h showed no obvious morphological changes or degeneration. Thus, these drug treatments allowed us to examine the changes in the expression levels of transfected htt that were not due to cell degeneration.
Figure 4.
Coexpression of exon1 htt with a normal (20Q) or expanded (76Q) polyQ domain in stably transfected PC12 cells. (A) DNA structures of GFP- or RFP-fused exon1 htt that contains either 20Q or 76Q. (B) Fluorescent microscopy showing the expression of both GFP- and RFP-htt corresponding to htt constructs in (A) in the same PC12 cells. (C) Cell morphology of stably transfected PC12 cells showing that cells expressing RFP-htt-76Q (E12 line) failed to respond to lactacystin treatment to extend long neurites, compared with the control cells expressing GFP-htt-20Q.
Next, we compared the levels of normal and mutant htt via western blotting after inhibiting either the UPS or autophagy to assess the changes in htt levels relative to its levels before drug treatment or to the loading control proteins. mEM48 western blots clearly show that UPS inhibition by lactacystin dramatically increased the levels of htt fragments in the cells expressing htt-76Q when compared with other drug treatments (Fig. 5A). However, htt-20Q also appeared to be increased by lactacystin. To quantify the changes in transfected htt, we probed the blots separately with antibodies to GFP, which reacts with GFP-htt-20Q, and 1C2 antibody, which specifically reacts with mutant htt (RFP-htt-76Q). As shown in Figure 5B, the htt-76Q level was much higher after lactacystin treatment than after treatment with the autophagy inhibitors BFA or 3-MA. The blots were also probed with the antibody to tubulin to normalize the loading protein amount in each lane. The changes in transfected htt after drug treatments were measured as the fold change of htt in control cells without drug treatments. This quantification revealed that inhibiting the UPS could increase the levels of htt-20Q and htt-76Q with a slightly greater increase of htt-76Q (Fig. 5C), which is consistent with an earlier finding that the UPS can cleave expanded polyQ peptides completely and efficiently as for normal polyQ peptides (36). Inhibiting autophagy by BFA and 3-MA also increased the levels of htt fragments to some extent. However, the UPS inhibitor lactacystin caused a greater increase in htt fragments than BFA and 3-MA did.
Figure 5.
UPS inhibition leads to a greater increase of transfected htt than autophagy inhibition does. (A) Western blot analysis of the levels of transfected htt after various drug treatments for 15 h. Mutant htt (76Q) and normal htt (20Q) are indicated. Note that lactacystin caused more transfected htt to accumulate than other drugs. (B) The cell lysates of stably transfected cells were probed by anti-GFP to reveal the level of normal htt (20Q) or probed by 1C2 to reveal mutant htt (76Q). The same samples were also probed with antibody to tubulin. (C) The changes in the levels of transfected htt in PC12 cells after drug treatments. The control is cells treated with the drug vehicle DMSO. Densitometry analysis of the htt levels was used to obtain the fold of control for htt levels in cells that were treated with drugs, as indicated.
UPS inhibition preferentially increased the accumulation of N-terminal mutant htt fragments compared with full-length mutant htt
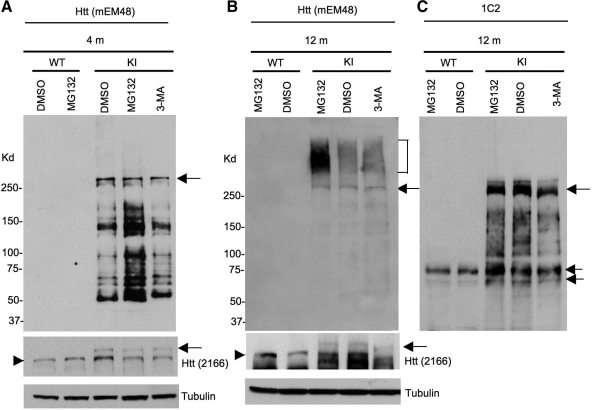
Although mutant htt may not affect the UPS and autophagy in the brain, the function of these two systems may decline with age or is affected by other cellular factors. To compare the effects of inhibiting the UPS or autophagy on the endogenous levels of full-length and truncated fragments of mutant htt in the mouse brain, we analyzed the expression of mutant htt in another HD KI mouse model that expresses human exon1 htt with 140Q in endogenous mouse htt (7,37). We chose 4- and 12-month-old HD KI mice for examination, as HD KI mice show more htt aggregation and obvious behavioral phenotypes at 12 months of age than 4-month-old mice. The expression of human exon1 htt sequences allows for the detection of both soluble and aggregated forms of transgenic htt via the monoclonal antibody mEM48. Microinjection of MG132 and 3-MA into mouse brains is known to inhibit UPS and autophagy function, respectively (38,39). Thus, we delivered the UPS inhibitor MG132 and the autophagy inhibitor 3-MA via stereotaxic injection into the striatum of HD KI mice. After 24 h, we isolated the striatum and performed western blotting with mEM48, which selectively reacts with polyQ-expanded human htt but cannot detect normal mouse htt (8). In the striatum of HD KI mice at 4 months of age, we saw a drastic increase in multiple soluble htt fragments after inhibiting the UPS by MG132, but not after inhibiting autophagy by 3-MA. However, full-length mutant htt levels were not different in the control (DMSO, dimethyl sulfoxide)-, MG132- and 3-MA-injected striatum (Fig. 6A). In old HD mice at the age of 12 months, mEM48 detected more aggregated htt, which was evident in the stacking gel, than soluble htt fragments (Fig. 6B), suggesting that the majority of mEM48 immunoreactive products are the aggregated form of htt in aged mouse brain or the oligomerized htt that may be less sensitive to mEM48. To verify this, we re-probed the same blot with 1C2 that reacts much stronger with soluble and oligomerized mutant htt than with aggregated htt (8). The majority of degraded htt products that reacted with 1C2 appeared as smear on the blot, reflecting the oligomerization of htt fragments (Fig. 6C). The levels of oligomerized htt, except for small htt bands (double arrows in Fig. 6C), were not significantly increased by MG132, suggesting that the oligomerized htt fragments are less stable or can form aggregates more quickly in the older mouse brain. In support of this possibility, aggregated htt is significantly increased by MG132, but not DMSO or 3-MA (Fig. 6B). Importantly, mEM48 and 1C2 western blots show that the full-length mutant htt did not significantly increase in the striatum of old mice after the treatment with MG132 or 3-MA (Fig. 6B and C). We also performed immunohistochemical analysis of the injected brain regions with the antibody to htt (mEM48). Because nuclear inclusions are easily identified, we quantified the density of nuclear inclusions and found that MG132 injection significantly (P < 0.01) increased the relative numbers (134.7 ± 19.5) of nuclear inclusions per image field (by 20 times) when compared with those of DMSO (67.4 ± 16.3) or BFA (59.8 ± 12.9) injection.
Figure 6.
The UPS inhibitor MG132 selectively increases the levels of degraded htt products in the striatum of HD CAG140 knock-in mice. One microliter of vehicle DMSO, MG132 (100 µm) or 3-MA (100 mm) was microinjected into the striatum of wild-type (WT) or HD CAG140 knock-in (KI) mice at 4 or 12 months of age. After 24 h, the striatum was isolated for western blotting with mEM48 (A–B), which reacts with polyQ-expanded human htt and its aggregates but is unable to detect normal mouse htt. The blot of 12-month mouse brain samples was also probed by 1C2 (C), which selectively reacts with expanded polyQ and detected soluble (double arrows) and oligomerized, but not aggregated, mutant htt on the blot. Full-length mutant htt was also detected by the mouse antibody 2166 or 1C2 and is indicated by arrows. Endogenous mouse htt is indicated by arrowheads. Aggregated htt is indicated by the bracket. An increased amount of degraded htt fragments was seen in the striatum of 4-month-old HD KI mice after MG132 injection (A). Note that in old HD KI mice at 12 months of age (B), aggregated htt is predominantly seen and also increased by MG132. Two male mice of each group were examined.
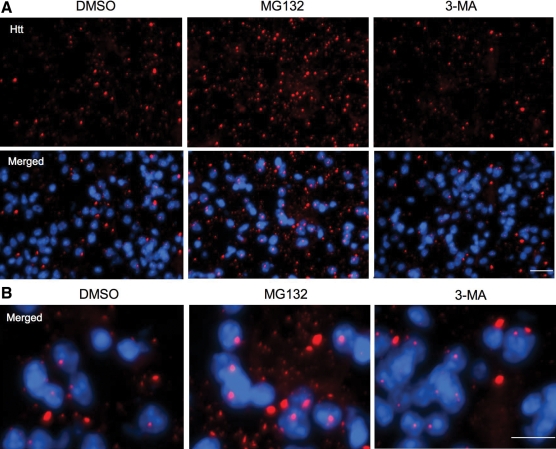
These results also show that MG132, but not 3-MA, was able to increase the density of htt aggregates in the striatum of HD KI mouse brains (Fig. 7). Although 3-MA injection could reduce LC3 puncta in the injected site when compared with the control, htt aggregates were not increased by the 3-MA injection (Supplementary Material, Fig. S3). Taken together, impaired UPS activity can preferentially increase the accumulation of toxic N-terminal htt fragments and aggregated htt, but not full-length mutant htt.
Figure 7.
Immunofluorescent staining of the striatal sections of the 12-month-old HD KI mouse brain. The striatum of HD KI mice was injected with DMSO, the UPS inhibitor MG132, or the autophagy inhibitor 3-MA. After 24 h, the striatal sections were isolated and fixed for immunofluorescent staining with the antibody to htt (mEM48) and Hoechst dye to label nucleus (blue). Note that mutant htt forms nuclear inclusions and neuropil aggregates that are small and outside the nucleus. MG132, but not 3-MA, increased the density of htt aggregates seen in the low (A) and high (B) magnification images. Scale bars: 20 µm in (A) and 10 µm in (B). Two female mice each group were examined.
DISCUSSION
Although both the UPS and autophagy clear mutant htt, it remains unknown whether the activity of autophagy in the brain can be affected by mutant htt. Here we did not find that transgenic mutant htt can alter the conversion of LC3-I to LC3-II, suggesting that the brain autophagy function may not be particularly affected by the expression of mutant htt. In addition, our findings suggest that the UPS is more important than autophagy for removing toxic N-terminal mutant htt fragments, which could have implications for the development of a more effective treatment.
HD is one among a family of age-dependent neurological disorders that includes eight other polyQ diseases (1), Alzheimer's disease, Parkinson's disease and amyotrophic lateral sclerosis. All these diseases share the phenomena of late-onset accumulation of toxic proteins and selective neurodegeneration. Because these disease proteins are cleared largely by the UPS and autophagy, considerable effort has been put forth to investigate the relationship between the misfolded disease proteins and the function of the UPS or autophagy. Early studies using cellular models of polyQ diseases or in vitro systems showed that expanded polyQ proteins can impair UPS function (40–42); however, in vivo studies of polyQ disease mouse models revealed no global impairment of the UPS in the mouse brains (28–31). Instead, an age-dependent decline in UPS activity is found to correlate with the age-related accumulation and aggregation of mutant htt in HD mouse brains (30,32). Thus, overexpressed mutant protein in the in vitro system may undermine the capacities of the UPS, whereas transgenic mutant proteins in the brain are unlikely to have a significant impact on the global UPS activity because of their slow accumulation over time. These studies underscore the importance of investigating the cellular autophagy function when mutant proteins are not overexpressed. By examining LC3 conversion, which results from autophagy activation, we saw no obvious difference in LC3 conversion in the brains of WT versus HD mice. Thus, as with the UPS, autophagy may not be affected significantly by the expression of mutant htt in the brain. This possibility also stresses the need to compare the relative abilities of the UPS and autophagy to clear misfolded proteins in the brain, especially for mutant polyQ proteins, which often accumulate in the nucleus, where autophagy is absent.
Autophagy inhibitors are known to increase the accumulation of transfected htt in different types of cultured cells. For example, inhibition of autophagy in COS-7 and PC12 cells by 3-MA can raise the level of mutant htt (43). By comparing the effects of commonly used inhibitors of the UPS and autophagy, we found that inhibiting UPS function leads to more accumulation of mutant htt in transfected HEK293 cells than autophagy inhibition does. Since the intracellular autophagy degradative capacity varies with cell type, age, transformation and/or disease (44), we must compare the consequences of inhibiting the UPS versus autophagy in mouse brains expressing mutant htt at the endogenous level. Our finding indicates that inhibiting brain UPS also causes more mutant htt to accumulate in the brain than inhibiting autophagy.
Although the drugs we used are well-established tools in various systems to alter UPS and autophagy function, these drugs may also have off-target effects or affect multiple pathways. For example, rapamycin is found to inhibit protein synthesis to alleviate the accumulation of misfolded proteins (45). However, many groups have provided strong evidence that autophagy plays an important role in removing aggregate-prone forms of mutant htt or other mutant proteins in the cytoplasm. There is compelling evidence that loss of autophagy can increase the accumulation of misfolded proteins in the cytoplasm and lead to neurodegeneration (46,47). Further, we also know that the UPS and autophagy interact with each other to regulate the cellular clearance function (48,49). Thus, loss of one of these two systems would lead to severe cellular dysfunction and alter the bulk degradation of proteins or degenerated organelles. Since most polyQ-expanded proteins, unlike other disease proteins, can accumulate in the nucleus in which autophagy is absent, the UPS and autophagy are likely to have different effects on polyQ disease proteins. Also, because mutant htt does not affect the global activity of the UPS (29–31) and LC3 conversion in HD mouse brains, we need to distinguish which system plays a more important role in removing soluble toxic htt fragments that can initiate early neuropathological events. Interestingly, our examination of HD mouse brains revealed that inhibiting the UPS increased the aggregation of mutant htt to a much greater extent than autophagy inhibition did. This is perhaps because, unlike transfected proteins that are largely expressed in the cytoplasm, transgenic htt in the mouse brain can accumulate in the nucleus in which the UPS is the major degradation pathway to remove misfolded proteins. It is possible that aggregation of mutant htt is dynamically mediated by soluble mutant htt, which is more efficiently removed by the UPS whereas autophagy is more important for clearing aggregate-prone forms of mutant htt in the cytoplasm.
Our findings suggest that impaired UPS activity can preferentially increase soluble N-terminal fragments of mutant htt. This conclusion is supported by the fact that there is selective accumulation of degraded htt fragments, but not full-length htt, after the brain UPS is inhibited by MG132. Since truncated N-terminal mutant htt fragments are prone to misfolding, the UPS may target these misfolded proteins more efficiently for degradation. Although mutant htt does not seem to affect the function of the UPS and the conversion of LC3-I to LC3-II, the age-dependent decline of UPS function in the brain could contribute to the late-onset accumulation of proteolytic mutant htt fragments and the formation of htt aggregates. It remains to be investigated whether age-related alteration of autophagy function also contributes to the accumulation of mutant proteins or their aggregates in the brain. Since the aggregation of disease proteins may confer protection against their toxicity by reducing their interactions with other cellular proteins or subcellular compartments, it would be more important to prevent the accumulation of the soluble form of disease proteins, such that early pathological events can be inhibited.
MATERIALS AND METHODS
Mice
Hdh (CAG)150 (27) or 140 (7), KI mice and N171-82Q mice (26) were bred and maintained in the animal facility at Emory University in accordance with institutional guidelines.
Plasmids, antibodies and reagents
GFP-LC3 was provided by Dr. Zhenyu Yue at Rockefeller University (50). PRK-exon1 htt with 20Q and 150Q plasmid, PRK-RFP-exon1 htt with 20Q, 76Q or 150Q plasmid, and pEGFP-exon1 htt-20Q or 130Q were produced in our earlier studies (30,32). The rabbit polyclonal anti-LC3 antibodies (NB100-2331, NB600-1384, Novus Biologicals, Littleton, CO, USA) and L7543 (Sigma-Aldrich, St. Louis, MO, USA) were used at 1:1000 for western blotting. The mouse anti-htt antibody (mEM48) was previously produced in our laboratory (8,32). The mouse anti-htt 1C2 and 2166 antibodies were purchased from Millipore (Temecula, CA, USA). The mouse anti-gamma-tubulin antibody was purchased from Sigma-Aldrich and used at 1:50 000 dilution. Secondary antibodies were peroxidase-conjugated donkey anti-mouse or donkey anti-rabbit IgG (H + L) from Jackson ImmunoResearch (West Grove, PA, USA). Drugs [MG132, lactacystin, bafilomycin A1 (BFA), rapamycin and 3-methyladenine (3-MA)] were obtained from Sigma-Aldrich.
PC12 cell lines stably transfected with htt
PC12 cells were stably transfected with EGFP-exon-1 htt-20Q and RFP-exon1 htt-130Q constructs. Due to the instability of the CAG repeat, the cloned PC12 cells expressed mutant htt with a reduced number (76 CAGs or RFP-exon1 htt-76Q) of the repeat. The control cells expressed EGFP-exon-1 htt-20Q and RFP-exon1 htt-20Q constructs. Positive cells were selected under fluorescent microscopy to ensure that the cells expressed both GFP and RFP signals. Western blotting was performed to verify the expression of transfected htt in the selected cell lines. Of three cell lines (htt-76Q-A9, htt-76Q-E12 and htt-76Q-E10) that express mutant htt at similar levels, htt-76Q-E12 cells were focused for further analysis. The positive PC12 cell lines were maintained in DMEM/F12 medium with 10% fetal bovine serum and 5% horse serum, 100 U/ml penicillin and 100 µg/ml streptomycin sulfate at 37°C in an atmosphere of 5% CO2 and 95% humidity. All the drug treatments were performed in 6-well plates with a density of 1 × 106 cells/well.
Cell culture, transfection and drug treatment
Human embryonic kidney (HEK) 293 cells were maintained in DMEM/F12 (Invitrogen, Carlsbad, CA, USA) medium with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin sulfate at 37°C in an atmosphere of 5% CO2 and 95% humidity. Cells seeded at an 80% confluent onto 6-well plates were transiently transfected with htt or GFP-LC3 plasmids using lipofectamine reagent (Invitrogen). After 48 h of culture, cells were treated with 200 ng/ml of rapamycin, 5–10 µm MG132, 2–5 µm lactacystin, 50–100 nm BFA or 1–10 mm 3-MA in the fresh medium for 15 h before fluorescence microscopy and western blot analyses.
Western blotting and immunofluorescent microcopy
Cultured cells were rinsed with phosphate-buffered saline (PBS) three times at room temperature and 250 µl 1× SDS loading buffer were added immediately to the cell pellet, sonicated for 5–8 s, and heated at 100°C for 5 min before western blot analysis. The striatum, cortex and cerebellum of WT and HD mice were dissected and immediately homogenized in the homogenization buffer (50 mm Tris, pH 8.8,100 mm NaCl, 5 mm MgCl2, 1 mm ethylene-diamine-tetraacetic acid, pH 8, 0.5% NP40, 2 mm Na3VO4, 5 mm NaF) with 1 × protease inhibitor cocktails, Pierce 78430 (Rockford, IL, USA) and 0.1 mm phenylmethanesulfonyl fluoride (Sigma-Aldrich). Samples were diluted to 5 mg/ml of protein concentrations. Protein samples were boiled for 5 min in 1× gel-loading buffer containing 2% SDS for western blot analysis. The samples (75–100 µg of protein) were separated on a 4–12% (for htt and other proteins) or 18% (for LC3) Tris–glycine SDS–polyacrylamide gel (Invitrogen). Proteins were transferred to a nitrocellulose membrane (Amersham, Piscataway, NJ, USA). The nitrocellulose membranes were blocked with 5% non-fat dry milk/phosphate buffered saline with 0.1% Tween-20 (PBST) for 30 min, and the blots were rinsed with 1× PBST and incubated with primary antibodies 24–72 h at 4°C. Secondary HRP-conjugated anti-rabbit or -mouse IgGs were incubated with the blot in 5% milk/PBS for 1 h at room temperature. ECL-plus was then used to reveal immunoreactive bands on the blots. To verify protein loading amounts, blots were also re-probed with mouse anti-gamma-tubulin antibody.
For immunofluorescent microscopy, Htt-transfected HEK293 cells were fixed for 7 min with 4% paraformaldehyde/PBS and then subjected to immunofluorescent microscopy examination. Immunofluorescent staining of mouse brain sections was performed using the method described previously (8). After mice were anesthetized, brains were removed and sectioned at 20 µm using a freezing microtome. Free-floating sections were fixed in 4% paraformaldehyde in 0.1 m phosphate buffer for 10 min and preblocked in 4% normal goat serum in PBS, 0.1% Triton-X, then incubated with mEM48 antibody for immunohistochemical analysis. Light micrographs were taken using a microscope (Axiovert 200 MOT; Carl Zeiss, Inc.) equipped with a digital camera (Orca-100; Hamamatsu) and the image acquisition software OpenLAB (Improvision).
Stereotaxic injection of the UPS or autophagy inhibitors into mouse brain
We anesthetized mice with intraperitoneal injections of 2.5% Avertin (0.012 ml/g body weight) and performed lateral stereotaxic injections of the UPS inhibitor MG132 and the autophagy inhibitor 3-MA into the striatum. Using a 5 µl Hamilton syringe, we injected 1.0 µl of DMSO, MG132 at 100 µm in DMSO or 3-MA dissolved at 100 mm in saline into the striatum at the following coordinates: 0.8 mm rostral to bregma, 2.0 mm lateral to the midline and 3.5 mm ventral to the dural surface. The injection rate was 0.5 µl/2 min, and the needle was left in place for an additional 5 min before being removed. After 24 h, the injected animals were sacrificed, and the striatum was isolated for western blotting analysis. We used the same gender of mice for each experiment and at least two mice of each group were examined.
Statistical analysis
All data were expressed as mean ± SEM. Statistical results were analyzed by GraphPad Prism (Version 5) software, and statistical significance (P < 0.05) was assessed using Student's t-test or one-way analysis of variance, followed when appropriate by a post-hoc analysis using Dunnett's test.
SUPPLEMENTARY MATERIAL
FUNDING
X.L. is supported in part by the China Scholarship Council. This work was supported by grants of National Institutes of Health AG019206, NS041669 (X.J.L.), NS045016 (S.H.L.), and grant from the National Natural Science Foundation of China 30430260 to H.L.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Michael Levine for providing breeding pairs of HD CAG140 knock-in mice, Zhenyu Yue for providing GFP-LC3 plasmid and Moe H. Aung for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Li X., Li H., Li X.J. Intracellular degradation of misfolded proteins in polyglutamine neurodegenerative diseases. Brain Res. Rev. 2008;59:245–252. doi: 10.1016/j.brainresrev.2008.08.003. doi:10.1016/j.brainresrev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heng M.Y., Detloff P.J., Albin R.L. Rodent genetic models of Huntington disease. Neurobiol. Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. doi:10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Ratovitski T., Nakamura M., D'Ambola J., Chighladze E., Liang Y., Wang W., Graham R., Hayden M.R., Borchelt D.R., Hirschhorn R.R., et al. N-terminal proteolysis of full-length mutant huntingtin in an inducible PC12 cell model of Huntington's disease. Cell Cycle. 2007;6:2970–2981. doi: 10.4161/cc.6.23.4992. [DOI] [PubMed] [Google Scholar]
- 4.Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L.M., Trottier Y., et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington's disease. J. Biol. Chem. 285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. doi:10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Li H., Li S.H., Cheng A.L., Mangiarini L., Bates G.P., Li X.J. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum. Mol. Genet. 1999;8:1227–1236. doi: 10.1093/hmg/8.7.1227. doi:10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- 7.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. doi:10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 8.Wang C.E., Tydlacka S., Orr A.L., Yang S.H., Graham R.K., Hayden M.R., Li S., Chan A.W., Li X.J. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum. Mol. Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. doi:10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. doi:10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 10.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. doi:10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 11.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. doi:10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. doi:10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar S., Rubinsztein D.C. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. doi:10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A.D., Xie X., Ma D., et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl Acad. Sci. USA. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. doi:10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl Acad. Sci. USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. doi:10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D., de Pablo R., Tacchetti C., Rubinsztein D.C., Ballabio A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. doi:10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwata A., Riley B.E., Johnston J.A., Kopito R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. doi:10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 21.Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl Acad. Sci. USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. doi:10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kegel K.B., Kim M., Sapp E., McIntyre C., Castano J.G., Aronin N., DiFiglia M. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen A., Larsen K.E., Behr G.G., Romero N., Przedborski S., Brundin P., Sulzer D. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum. Mol. Genet. 2001;10:1243–1254. doi: 10.1093/hmg/10.12.1243. doi:10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 24.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. doi:10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin L., Wang Z., Tao L., Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 6 doi: 10.4161/auto.6.2.11062. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. doi:10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 27.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. doi:10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 28.Bowman A.B., Yoo S.Y., Dantuma N.P., Zoghbi H.Y. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin–proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 2005;14:679–691. doi: 10.1093/hmg/ddi064. doi:10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- 29.Bett J.S., Goellner G.M., Woodman B., Pratt G., Rechsteiner M., Bates G.P. Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington's disease mice: exclusion of proteasome activator REGgamma as a therapeutic target. Hum. Mol. Genet. 2006;15:33–44. doi: 10.1093/hmg/ddi423. doi:10.1093/hmg/ddi423. [DOI] [PubMed] [Google Scholar]
- 30.Tydlacka S., Wang C.E., Wang X., Li S., Li X.J. Differential activities of the ubiquitin–proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J. Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. doi:10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maynard C.J., Bottcher C., Ortega Z., Smith R., Florea B.I., Diaz-Hernandez M., Brundin P., Overkleeft H.S., Li J.Y., Lucas J.J., et al. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl Acad. Sci. USA. 2009;106:13986–13991. doi: 10.1073/pnas.0906463106. doi:10.1073/pnas.0906463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H., Cao F., Wang Z., Yu Z.X., Nguyen H.P., Evans J., Li S.H., Li X.J. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J. Cell Biol. 2003;163:109–118. doi: 10.1083/jcb.200306038. doi:10.1083/jcb.200306038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obin M., Mesco E., Gong X., Haas A.L., Joseph J., Taylor A. Neurite outgrowth in PC12 cells. Distinguishing the roles of ubiquitylation and ubiquitin-dependent proteolysis. J. Biol. Chem. 1999;274:11789–11795. doi: 10.1074/jbc.274.17.11789. doi:10.1074/jbc.274.17.11789. [DOI] [PubMed] [Google Scholar]
- 34.Wyttenbach A., Swartz J., Kita H., Thykjaer T., Carmichael J., Bradley J., Brown R., Maxwell M., Schapira A., Orntoft T.F., et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum. Mol. Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. doi:10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 35.Rong J., McGuire J.R., Fang Z.H., Sheng G., Shin J.Y., Li S.H., Li X.J. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J. Neurosci. 2006;26:6019–6030. doi: 10.1523/JNEUROSCI.1251-06.2006. doi:10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michalik A., Van Broeckhoven C. Proteasome degrades soluble expanded polyglutamine completely and efficiently. Neurobiol. Dis. 2004;16:202–211. doi: 10.1016/j.nbd.2003.12.020. doi:10.1016/j.nbd.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Levine M.S., Klapstein G.J., Koppel A., Gruen E., Cepeda C., Vargas M.E., Jokel E.S., Carpenter E.M., Zanjani H., Hurst R.S., et al. Enhanced sensitivity to N-methyl-d-aspartate receptor activation in transgenic and knockin mouse models of Huntington's disease. J. Neurosci. Res. 1999;58:515–532. doi:10.1002/(SICI)1097-4547(19991115)58:4<515::AID-JNR5>3.0.CO;2-F. [PubMed] [Google Scholar]
- 38.Zhang J.Y., Peng C., Shi H., Wang S., Wang Q., Wang J.Z. Inhibition of autophagy causes tau proteolysis by activating calpain in rat brain. J. Alzheimers Dis. 2009;16:39–47. doi: 10.3233/JAD-2009-0908. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X.D., Wang Y., Wu J.C., Lin F., Han R., Han F., Fukunaga K., Qin Z.H. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy. 2009;5 doi: 10.4161/auto.5.3.8174. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin–proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. doi:10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 41.Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E.E. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A.L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. doi:10.1016/S1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 43.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. doi:10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 44.Chu C.T. Autophagic stress in neuronal injury and disease. J. Neuropathol. Exp. Neurol. 2006;65:423–432. doi: 10.1097/01.jnen.0000229233.75253.be. doi:10.1097/01.jnen.0000229233.75253.be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King M.A., Hands S., Hafiz F., Mizushima N., Tolkovsky A.M., Wyttenbach A. Rapamycin inhibits polyglutamine aggregation independently of autophagy by reducing protein synthesis. Mol. Pharmacol. 2008;73:1052–1063. doi: 10.1124/mol.107.043398. doi:10.1124/mol.107.043398. [DOI] [PubMed] [Google Scholar]
- 46.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 48.Nedelsky N.B., Todd P.K., Taylor J.P. Autophagy and the ubiquitin–proteasome system: collaborators in neuroprotection. Biochim. Biophys. Acta. 2008;1782:691–699. doi: 10.1016/j.bbadis.2008.10.002. doi:10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. doi:10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q.J., Ding Y., Kohtz D.S., Mizushima N., Cristea I.M., Rout M.P., Chait B.T., Zhong Y., Heintz N., Yue Z. Induction of autophagy in axonal dystrophy and degeneration. J. Neurosci. 2006;26:8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. doi:10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.