Abstract
Protein folding in the endoplasmic reticulum is a complex process whose malfunction is implicated in disease and aging. Using the cell’s endogenous sensor (the unfolded protein response), we identified several hundred yeast genes with roles in endoplasmic reticulum folding and systematically characterized their functional interdependencies by measuring unfolded protein response levels in double mutants. This strategy revealed multiple conserved factors critical for endoplasmic reticulum folding including an intimate dependence on the later secretory pathway, a previously uncharacterized six-protein transmembrane complex, and a co-chaperone complex that delivers tail-anchored proteins to their membrane insertion machinery. The use of a quantitative reporter in a comprehensive screen followed by systematic analysis of genetic dependencies should be broadly applicable to functional dissection of complex cellular processes from yeast to man.
The endoplasmic reticulum (ER) is responsible for the folding and maturation of secreted and membrane proteins. External stress or mutations can compromise ER folding, contributing to diseases such as diabetes and neurodegeneration (1, 2). The specialized milieu of the ER is comprised of a large number of proteins that aid the structural maturation of itinerant proteins (3, 4). While many of these ER folding factors have been extensively studied, the full range of proteins contributing to this process is unknown, and how they function together is poorly understood.
Systematic identification of genes contributing to ER folding
We exploited the cell’s endogenous sensor of ER protein folding status, Ire1p, to identify genes in Saccharomyces cerevisiae that contribute to structural maturation of secretory proteins. In response to misfolded proteins in the ER, the transmembrane sensor Ire1p activates the transcription factor Hac1p (5), which in turn transcriptionally up-regulates a distinct set of genes (6, 7) in a process called the unfolded protein response (UPR). We used a reporter system in which a Hac1p-responsive promoter drives Green Fluorescent Protein (GFP) expression (8) (Fig. 1A). To correct for nonspecific expression changes, we co-expressed a red fluorescent protein (RFP) from a constitutive TEF2 promoter and used the ratio of GFP/RFP as our reporter of UPR signaling. A titration of the ER-stress inducing reducing agent dithiothreitol (DTT) demonstrated that this reporter quantitatively responds to misfolding of ER proteins (Fig. 1B).
Fig. 1.
Quantitative screen for gene deletions that perturb UPR signaling. (A) Strategy for quantifying UPR levels in deletion strains. (B) GFP/RFP reporter levels as a function of concentration of DTT, a reducing agent that causes protein misfolding in the ER. (C) UPR reporter levels of up-regulator hits by functional category.
Using Synthetic Genetic Array methodology (9), the reporter was introduced into ~4500 strains from the S. cerevisiae deletion library (10), and the median single-cell fluorescence of each strain was measured using high-throughput flow cytometry (11, 12) (Fig. S1, Table S1). The UPR showed significant basal induction, which allowed us to identify genes whose deletion caused either up-regulation (399 hits with p<0.01, explicitly modeling our experimental error) or down-regulation (334 hits with p<0.01) of the reporter. We found limited overlap between the genes whose deletion induces the UPR and the genes that were previously shown by microarray analysis to be transcriptionally upregulated by the UPR (7) (Fig. S2 and Table S2, see also (10, 13)). Thus although defining the UPR targets was fundamental to our understanding of how cells respond to ER stress it provides a limited view of the processes constitutively required for folding in the ER.
Overview of gene deletions affecting the UPR
Proteins whose deletion caused up-regulation of the reporter were highly enriched for localization throughout the secretory pathway (Fig. S3 and Table S3) (14), including the ER as well as the Golgi, vacuole and endosome. As expected, chaperones (15) and genes in the N-linked glycosylation (16) and ER-Associated Degradation (ERAD) (17) pathways featured among the top hits (Fig. 1C). However, deletion of genes involved in many other processes, including O-mannosylation, glycophosphatidylinositol anchor synthesis, lipid biosynthesis, multiple steps of vesicular trafficking and ion homeostasis, caused similarly high reporter inductions (Table S1). Moreover, the UPR up-regulators included several dozen poorly characterized genes, some whose deletion caused reporter induction rivaling the strongest hits.
The diversity of functions contributing to ER integrity presented a major obstacle in our efforts to understand how these unexpected factors function together to support protein folding in the ER. To overcome this, we explored their functional dependencies by systematically quantifying how the phenotype caused by loss of one gene was modulated by the absence of a second. Systematic analysis of genetic interactions, using growth rate as a phenotype, has been used extensively to determine gene function (9, 18-23). We sought to generalize this strategy using ER stress as a quantitative phenotype. Accordingly, we quantified UPRE-GFP reporter levels in over 60,000 strains containing pair-wise deletions among 340 of our hits (12).
Genetic interactions illuminate functional relationships
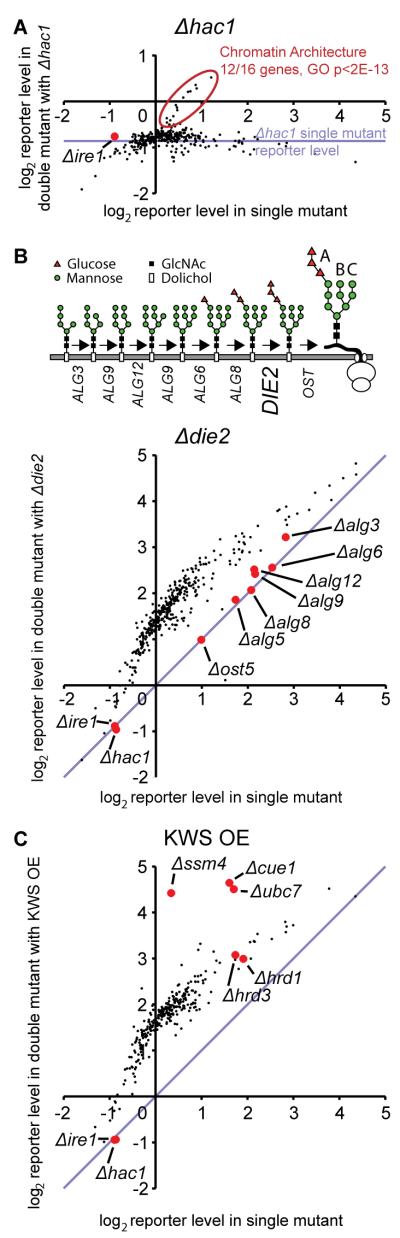
Three examples illustrate the utility of the double mutant analysis. First, comparison of the UPRE-GFP levels in the presence and absence of IRE1/HAC1 differentiated between the subset of upregulators whose deletion affected protein folding in the ER (i.e. UPRE-GFP induction was dependent on the Ire1p/Hac1p pathway) from those, like chromatin architecture genes, that were directly affecting expression of the reporter (Fig. 2A, Table S4).
Fig. 2.
Double mutant analysis provides information on functional dependencies between genes. (A) “Double mutant” (DM) plot of Δhac1. Each point represents a gene. X-coordinate represents the reporter level of a strain deleted for that gene in a WT background. Y-coordinate represents the reporter level in a double mutant lacking the same gene and additionally deleted for a second gene (in this case HAC1). The horizontal blue line indicates the reporter level in the Δhac1 single mutant. Circled in red are up-regulators whose reporter induction is HAC1-independent, which are highly enriched for chromatin architecture factors. (B) (Top) Schematic of the lumenal steps of the N-linked glycan synthesis pathway. (Bottom) DM plot for Δdie2/alg10. (C) DM plot depicting genetic interactions between deletion mutants and over-expression (OE) of the ERAD substrate KWS.
In a second example, UPR levels of pair-wise deletions with Δdie2/alg10, the enzyme that performs the last step in the synthesis pathway for N-linked glycans, illustrated our ability to define genes acting in a linear pathway (Fig. 2B). Most double mutants showed a typical increase in fluorescence which was dictated solely by the reporter levels of the single mutants. Strikingly, a specific subset of the double mutants had the same reporter level as the single mutant demonstrating “fully masking” epistatic interactions in which the function of one gene is completely dependent on the presence of a second one. Indeed, the genes that we find to be epistatic to DIE2 include the full set of factors that act immediately upstream of Die2p in the synthesis of N-linked sugars (16).
The utility of aggravating genetic interactions, in which pairs of deletions lead to exaggerated folding defects, was illustrated in a third example in which we over-expressed the constitutively misfolded and rapidly degraded membrane protein KWS (24) in deletion strains that were hits in our screen (Fig. 2C). KWS degradation is mediated by a well-defined subset of the ERAD machinery, including the E3 ubiquitin ligase Ssm4p/Doa10p, and the E2 ubiquitin-conjugating complex Ubc7p and Cue1p (24). The role of these factors in mitigating the stress caused by over-expression of KWS is revealed in our data by the strongly aggravating interaction between their deletions and KWS over-expression. In contrast, other ERAD components, which do not act on KWS, including the Hrd1p/Hrd3p E3 ligase complex (24), show typical reporter levels.
Phenotypic interaction score (π-score) quantifies functional relationships
Strongly aggravating and fully masking interactions as described above are only a subset of the broader range of possible genetic interactions in which pairs of perturbations lead to a continuum of exacerbated (aggravating) or attenuated folding defects. We sought to quantify these systematically by developing a phenotypic interaction score or “π-score” which describes the degree to which a double mutant UPR reporter level differs from that expected from the two single mutant levels. A simple empirical multiplicative model accurately predicted the typical double mutant reporter levels when we accounted for saturation of the reporter (Fig. 3A, see (12)). The π-score for each double mutant was given by the difference between the typical levels expected from the reporter levels of the two single mutants and the measured UPR reporter levels in the double mutant. Thus negative π-scores (exaggerated inductions) represent aggravating interactions and positive π-scores (unusually low inductions) represent alleviating interactions with the fully masking/epistatic interactions being a subset of the positive π-scores.
Fig. 3.
Systematic identification of genetic interactions. (A) Generalized DM plot illustrating the distribution of reporter levels in double mutants Δxxx Δyfg plotted against reporter levels in single mutants Δxxx. A red curve traces the typical double mutant reporter level as a function of the single mutant reporter level. The interaction value (π-score) is determined by the difference between the expected and measured UPR levels in a double mutant. Double mutants with unusually high fluorescence (blue dots), typical fluorescence (black dots), or unusually low fluorescence (yellow dots) represent aggravating, no, or alleviating genetic interactions, respectively. Fully masking interactions are found either on the horizontal blue line (Δyfg fully masks Δxxx) or on the diagonal blue line (Δxxx fully masks Δyfg). (B) Hierarchical clustering of a genetic interaction map based on systematic π-score analysis. To the right of the map, functional clusters are labeled (Table S5). Clusters referred to in the text are highlighted in red; those containing novel components are marked in italics.
Systematic identification of functional groups through phenotypic interaction maps
In growth-based studies, the pattern of genetic interactions of a mutation provides a signature that can be used to group genes by function (20, 21, 25). Analogous hierarchical clustering on the double mutant π-scores yielded a map with a high density of precise functional clusters (Fig. 3B, S4). This analysis accurately grouped over 100 of the previously well-characterized genes into 22 functions spanning a wide range of processes (Table S5). Among genes whose deletions directly affected the ER folding environment (i.e. caused Ire1p/Hac1p-dependent reporter induction) our map grouped not only the ERAD and glycosylation machinery discussed above, but also many other processes including those in the distal secretory pathway (Fig. 3B). Our map also accurately clustered multiple functions that act downstream of HAC1 including the Chromatin Assembly Complex, core histones and histone chaperones.
Genetic interactions identify functional hierarchies
Within the functional groups defined above, the specific double mutant phenotypes revealed the extent to which the activities of individual components depended on each other. For example, all of the known components of the “ERAD-L” machinery needed for disposal of misfolded lumenal proteins (17) formed a tight cluster. The double mutant phenotypes of Δhrd3 revealed the expected full dependence of YOS9, DER1, and USA1 on HRD3 (Fig. 4A) (17). In contrast, only partial epistasis was seen with the E2 Ubc7p and its membrane anchor Cue1p, consistent with their known roles in other branches of ERAD and the ability of another E2, Ubc1p, to partially substitute in their absence (26). In addition, the clustering analysis suggested that YLR104W is a novel component of ERAD that acts upstream of HRD3 and USA1 (perhaps by delivering a subset of ERAD targets to the Hrd1p ligase) (Table S6). Our complete list of genetic interactions, which includes over 500 full masking relationships among 213 genes, should provide a resource of functional predictions for the community (Table S6). We also provide a MATLAB script to display double mutant plots for any gene in our dataset (12).
Fig. 4.
Genetic interactions identify functional dependencies of uncharacterized proteins. (A) DM plot of Δhrd3. (Inset) Enlargement of a region of Fig. 3B, showing genetic interactions of the ERAD cluster. (B) Selected genetic interactions of the ER Membrane Complex (EMC). (C) SDS PAGE analysis of immunoprecipitation of Emc3p -FLAG and associated proteins; protein identities were determined by mass spectrometry. *The specificity of the Por1p interaction has not been evaluated.
Analysis of phenotypic interactions reveals novel pathways important for ER protein folding
Using this systematic approach we have discovered a pathway involving a conserved (Table S9) multi-protein transmembrane complex. The poorly characterized genes YCL045C, YJR088C, YKL207W, YGL231C, KRE27 and YLL014W all clustered together and showed strongly alleviating interactions among themselves (Fig. 4B), a signature of factors that cooperate to carry out a single function (21). Immunoprecipitation of FLAG-tagged Ykl207wp revealed that proteins encoded by these genes form an apparently stoichiometric complex (Fig. 4C). Accordingly we termed this the ER Membrane protein Complex (EMC) and named the genes from this cluster EMC1 through EMC6. While the precise biochemical roles of the EMC will have to wait for future studies, our data suggests that loss of the EMC leads to accumulation of misfolded membrane proteins: the pattern of genetic interactions of strains deleted for EMC members most closely resembled that seen in a strain over-expressing the misfolded transmembrane protein Sec61-2p (a mutated form of the Sec61 translocon) (27) and is similar to the pattern of a strain over-expressing the misfolded transmembrane protein KWS (24). This shared pattern includes strong aggravating interactions with Δubc7 and Δcue1, whose gene products are known to be involved in elimination of misfolded membrane proteins (17), but minimal interactions with other ERAD components.
A second cluster containing two conserved yet uncharacterized proteins (Yer140wp and Slp1p), show robust alleviating interactions with EMC components (Fig. 4B) as well as with each other. In support of a functional link between Yer140wp and Slp1p, these two proteins are suggested to be in a physical complex (28). The finding of two conserved protein complexes that are functionally dependent on each other underscores the value of this genetic data in identifying uncharacterized pathways required for ER folding.
Genetic interactions identify components of the tail-anchored protein biogenesis machinery
As a final example, we focused on Yor164cp/Get4p and Mdy2p/Tma24p/Get5p, as our analysis implicated them in tail-anchored (TA) protein biogenesis. TA proteins are an important class of transmembrane proteins, which includes SNARE trafficking factors (29, 30). TA proteins have a single C terminal transmembrane domain, which is inserted into the ER membrane through the action of the recently discovered GET pathway: the Get3p/Arr4p ATPase (and its mammalian homolog Asna1/TRC40) binds newly synthesized TA proteins and brings them to the ER via the ER membrane receptor complex formed by Get1p/Mdm39p and Get2p/Hur2p/Rmd7p (31-33). Our double mutant analysis pointed to a role of Yor164c/Get4p and its physical interaction partner Mdy2p/Get5p (34) in the GET pathway as Δget4 and Δmdy2/get5 tightly clustered with Δget3 (Fig. S5). Additionally, loss of GET3 fully masked the effect of Δget4 and Δmdy2/get5 (Fig. 5A). Moreover, these deletions partially suppressed the UPR induction of Δget1 and Δget2, a phenomenon previously seen with other phenotypes for Δget3 (32).
Fig. 5.
YOR164C/GET4 and MDY2/GET5 function in the pathway of tail-anchored protein insertion. (A) DM plot depicting the functional dependencies of MDY2/GET5. (B) In-vitro translocation assay. Sec22p was translated in cytosol from wild-type (WT) or Δmdy2/get5 strains. Error bars represent +/− SEM, N=3 (C) GFP-Sed5p localization defect in Δget3, Δget4 and Δmdy2/get5 strains. The images of at least 20 cells per strain with similar average fluorescence were quantified to determine the distribution of each strain’s total fluorescence across pixels of different intensities. (D) Silver stain of immunoprecipitation of Get3-FLAGp from ER microsomes and cytosol; protein identities were determined by mass spectrometry.
Several observations support a role for GET4 and MDY2/GET5 in TA protein biogenesis. First, cytosolic extracts from strains lacking Mdy2p/Get5p had a defect in insertion of the model TA substrate Sec22p into ER microsomes (Fig. 5B). Second, several of the in vivo phenotypes characteristic of loss of GET members are also observed in Δget4 and Δmdy2/get5 strains. These include a highly significant (p<10−30, Mann-Whitney U) re-localization of the TA protein GFP-Sed5p from punctate Golgi structures to a more diffuse pattern (Fig. 5C, S7) (12) as well as mis-localization of the peroxisomal TA protein GFP-Pex15p to mitochondria (Fig. S8) (32). Consistent with a defect in Sed5 biogenesis, loss of Get4p or Mdy2p/Get5p led to secretion of HDEL proteins, a phenotype that is seen in other GET deletion strains (Fig. S9). Third, immunoprecipitation revealed that Get4p and Mdy2p/Get5p bind Get3p in the cytosol (Fig. 5D). Mdy2p/Get5p also co-localized with Get3p and TA proteins to punctate protein aggregates that form in Δget1 strains (32) (Fig. S10, S11). Localization of Get3 to these puncta is dependent on Get4p and Mdy2p/Get5p but not vice versa (Fig. S10-12), suggesting that Get4p and Mdy2p/Get5p help deliver TA proteins to Get3p in the cytosol for trafficking to the ER membrane. Interestingly, Get4p and Mdy2p/Get5p have been suggested to be peripherally associated with ribosomes (34) where they could potentially capture nascent TA proteins. Thus, while Get4p and Mdy2p/Get5p are localized outside of the secretory pathway and initially may have appeared to be false positives, our double mutant analysis revealed how they impact ER protein folding.
Perspective
Our work reveals the range of processes that make the ER a robust folding compartment yielding both a list of components and a blueprint for their functional interdependence. These factors include a wide range of activities such as chaperones, glycosylation enzymes, and ERAD components as well as trafficking pathways, transcriptional regulatory networks, modulators of lipid and ion composition, and vacuolar function. The diversity of activities found supports and extends the recent view in which ER protein folding homeostasis (proteostasis) emerges from the dynamic interplay between folding, degradation and export processes (35, 36). From a practical perspective, our studies provide a rational starting point for efforts to modulate the ER folding capacity to intervene in disease (36).
More broadly, dissecting complex cellular processes represents a major challenge in cell biology. Deletion libraries and RNAi approaches now make it possible to identify important factors rapidly (37). But this in turn creates a bottleneck in their functional characterization, which classically required specialized gene-by-gene follow-up studies. Our approach in effect allows hundreds of different secondary screens to be carried out in parallel to explore systematically the functional interdependencies of hits, thus providing a foundation for focused mechanistic investigations. Given the large number of potential ways of creating proximal reporters for different aspects of biology, our strategy for generalizing systematic quantitative genetic analysis should be broadly applicable to other processes and organisms including mammals through the use of double RNAi treatments.
Summary.
Genetic interactions reveal the functional relationships of genes required for protein folding in the endoplasmic reticulum.
Supplementary Material
Acknowledgments
We thank C.-S. Chin for flow cytometry software development; C. Boone, D. Breslow, N. Krogan, D. Ng, C. Wang for reagents; K. Thorn for microscopy assistance; S. Zhou, D. King, G. Cagney, N. Krogan for mass spectrometry analyses; J. Metz; P. Nittler, A. Carroll, J. Hill, J. Ihmels, S. Wang, C. Chu for technical help; R. Andino, T. Aragon, A. Battle, N. Bradshaw, J. DeRisi, N. Friedman, J. Hollien, J. Newman, D. Koller, R. Kupferman, H. Madhani, L. Osherovich, F. Papa, and members of the Weissman, Walter and Schwappach Labs for stimulating discussion; B. Toyama for graphics; M. Bassik, D. Breslow, O. Brandman, S. Churchman, S. Neher, for comments on the manuscript. This work was supported by the HHMI (J.S.W., and P.W.), the NSF (M.C.J., E.Q.), the International Human Frontier Science Program (M.S., J.W.), an NIH K99/R00 award (M.S.), and an EMBO long-term fellowship (J.W.), the Deutsche Forschungsgemeinschaft (V.S.), and the Wellcome Trust (B.S.).
References
- 1.Lin JH, Walter P, Yen TS. Annu Rev Pathol. 2008;3:399. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheuner D, Kaufman RJ. Endocr Rev. 2008 May;29:317. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebert DN, Molinari M. Physiol Rev. 2007 Oct;87:1377. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 4.Trombetta ES, Parodi AJ. Annu Rev Cell Dev Biol. 2003;19:649. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 5.Ron D, Walter P. Nat Rev Mol Cell Biol. 2007 Jul;8:519. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 6.Casagrande R, et al. Mol Cell. 2000 Apr;5:729. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- 7.Travers KJ, et al. Cell. 2000 Apr 28;101:249. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 8.Pollard MG, Travers KJ, Weissman JS. Mol Cell. 1998 Jan;1:171. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 9.Tong AH, et al. Science. 2001 Dec 14;294:2364. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 10.Giaever G, et al. Nature. 2002 Jul 25;418:387. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 11.Newman JR, et al. Nature. 2006 Jun 15;441:840. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 12.Materials and methods are available as supporting material on Science Online
- 13.Winzeler EA, et al. Science. 1999 Aug 6;285:901. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 14.Huh WK, et al. Nature. 2003 Oct 16;425:686. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 15.Bukau B, Weissman J, Horwich A. Cell. 2006 May 5;125:443. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Helenius A, Aebi M. Annu Rev Biochem. 2004;73:1019. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 17.Nakatsukasa K, Brodsky JL. Traffic. 2008 Jun;9:861. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decourty L, et al. Proc Natl Acad Sci U S A. 2008 Apr 15;105:5821. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girgis HS, Liu Y, Ryu WS, Tavazoie S. PLoS Genet. 2007 Sep;3:1644. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, et al. Cell. 2006 Mar 10;124:1069. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 21.Schuldiner M, et al. Cell. 2005 Nov 4;123:507. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Segre D, Deluna A, Church GM, Kishony R. Nat Genet. 2005 Jan;37:77. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 23.St Onge RP, et al. Nat Genet. 2007 Feb;39:199. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vashist S, Ng DT. J Cell Biol. 2004 Apr;165:41. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong AH, et al. Science. 2004 Feb 6;303:808. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 26.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. Nat Cell Biol. 2000 Jul;2:379. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 27.Sommer T, Jentsch S. Nature. 1993 Sep 9;365:176. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 28.Collins SR, et al. Mol Cell Proteomics. 2007 Mar;6:439. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Burri L, Lithgow T. Traffic. 2004 Jan;5:45. doi: 10.1046/j.1600-0854.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 30.Borgese N, Brambillasca S, Soffientini P, Yabal M, Makarow M. Biochem Soc Trans. 2003 Dec;31:1238. doi: 10.1042/bst0311238. [DOI] [PubMed] [Google Scholar]
- 31.Favaloro V, Spasic M, Schwappach B, Dobberstein B. J Cell Sci. 2008 Jun 1;121:1832. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuldiner M, et al. Cell. 2008 Aug 22;134:634. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanovic S, Hegde RS. Cell. 2007 Mar 23;128:1147. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 34.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Genes Dev. 2006 May 15;20:1294. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2008 Feb 15;319:916. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 36.Mu TW, et al. Cell. 2008 Sep 5;134:769. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutros M, Ahringer J. Nat Rev Genet. 2008 Jul;9:554. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.