Abstract
The α- and β-globin gene families of jawed vertebrates have diversified with respect to both gene function and the developmental timing of gene expression. Phylogenetic reconstructions of globin gene family evolution have provided suggestive evidence that the developmental regulation of hemoglobin synthesis has evolved independently in multiple vertebrate lineages. For example, the embryonic β-like globin genes of birds and placental mammals are not 1:1 orthologs. Despite the similarity in developmental expression profiles, the genes are independently derived from lineage-specific duplications of a β-globin pro-ortholog. This suggests the possibility that other vertebrate taxa may also possess distinct repertoires of globin genes that were produced by repeated rounds of lineage-specific gene duplication and divergence. Until recently, investigations into this possibility have been hindered by the dearth of genomic sequence data from nonmammalian vertebrates. Here, we report new insights into globin gene family evolution that were provided by a phylogenetic analysis of vertebrate globins combined with a comparative genomic analysis of three key sauropsid taxa: a squamate reptile (anole lizard, Anolis carolinensis), a passeriform bird (zebra finch, Taeniopygia guttata), and a galliform bird (chicken, Gallus gallus). The main objectives of this study were 1) to characterize evolutionary changes in the size and membership composition of the α- and β-globin gene families of tetrapod vertebrates and 2) to test whether functional diversification of the globin gene clusters occurred independently in different tetrapod lineages. Results of our comparative genomic analysis revealed several intriguing patterns of gene turnover in the globin gene clusters of different taxa. Lineage-specific differences in gene content were especially pronounced in the β-globin gene family, as phylogenetic reconstructions revealed that amphibians, lepidosaurs (as represented by anole lizard), archosaurs (as represented by zebra finch and chicken), and mammals each possess a distinct independently derived repertoire of β-like globin genes. In contrast to the ancient functional diversification of the α-globin gene cluster in the stem lineage of tetrapods, the physiological division of labor between early- and late-expressed genes in the β-globin gene cluster appears to have evolved independently in several tetrapod lineages.
Keywords: Anolis, gene duplication, gene family evolution, genome evolution, hemoglobin, zebra finch
Introduction
The functional and regulatory divergence of duplicated genes is known to play an extremely important role in evolutionary innovation (Ohno 1970; Zhang 2003; Taylor and Raes 2004; Lynch 2007). Repeated rounds of gene duplication and divergence can lead to the functional diversification of multigene families, whereby different gene family members acquire distinct biochemical functions and/or patterns of expression. The α- and β-globin gene families of jawed vertebrates (gnathostomes) provide an excellent example of the kind of physiological versatility that can be attained through functional and regulatory divergence of duplicated genes that encode different subunit polypeptides of the same heteromeric protein. The hemoglobin of gnathostome vertebrates is a heterotetramer composed of two α-chain subunits and two β-chain subunits that are encoded by members of two paralogous gene families. The progenitors of the α- and β-globin gene families were produced by tandem duplication of an ancestral single-copy hemoglobin gene ∼450–500 Ma in the stem lineage of gnathostomes (Goodman et al. 1975, 1987; Czelusniak et al. 1982). The ancestral linkage arrangement of the proto α- and β-globin genes has been retained in amphibians (Hentschel et al. 1979; Jeffreys et al. 1980; Kay et al. 1980; Hosbach et al. 1983; Fuchs et al. 2006) and teleost fish (Chan et al. 1997; Gillemans et al. 2003; Pisano et al. 2003). In birds and placental mammals, by contrast, the α- and β-globin gene clusters are located on different chromosomes due to transposition of the proto β-globin gene to a new genomic location sometime after the stem lineage of amphibians split from the stem lineage of amniotes (Hardison 2008; Patel et al. 2008). In monotremes and marsupials, a vestige of the ancestral linkage arrangement is present in the form of an orphaned β-like globin gene (ω-globin) that is located at the 3′ end of the α-globin gene cluster (Wheeler et al. 2001, 2004; De Leo et al. 2005; Hoffmann et al. 2008b; Patel et al. 2008).
Over the course of vertebrate evolution, subsequent rounds of gene duplication and divergence gave rise to families of α- and β-like globin genes that are ontogenetically regulated, such that functionally distinct hemoglobin isoforms are expressed in embryonic and adult erythroid cells (Collins and Weissman 1984; Hardison 1998, 2001). The α- and β-like globin genes of Xenopus are differentially expressed in the larval and adult stages (Banville and Williams 1985a, 1985b; Fuchs et al. 2006), and the α- and β-like globin genes of birds and mammals are also differentially expressed during prenatal development and postnatal life (Hardison 2001; Alev et al. 2008, 2009).
In the α-globin gene family, the physiological division of labor between early- and late-expressed genes was established in the common ancestor of tetrapod vertebrates, and it appears to have been retained in all descendant lineages. The ancestral arrangement of the tetrapod α-globin gene cluster consists of three linked genes, 5′-αE, αD, and αA-3′ (Hoffmann and Storz 2007). The αE- and αA-globin genes originated via tandem duplication of an ancestral proto α-globin gene after the stem lineage of fishes split from the stem lineage of tetrapods, and the αD-globin gene originated subsequently via tandem duplication of the αE-globin gene in the common ancestor of tetrapods (Hoffmann and Storz 2007). All tetrapods examined to date possess an ortholog of the early-expressed αE-globin gene at the 5′ end of the α-globin gene cluster and an ortholog of the late-expressed αA-globin gene at the 3′ end of the cluster (Flint et al. 2001; Hughes et al. 2005; Cooper et al. 2006; Fuchs et al. 2006; Hoffmann and Storz 2007; Hoffmann et al. 2008b). In mammals, the αE- and αA-globin genes are often present in multiple copies, but the αD-globin gene has been deleted independently in multiple lineages (Hughes et al. 2005; Cooper et al. 2006; Hoffmann et al. 2008b). The αD-globin gene has also been secondarily lost from Xenopus (Fuchs et al. 2006; Hoffmann and Storz 2007), and it remains to be seen whether this gene is absent from the genomes of all amphibians. The αE-globin gene (known as αL-globin in amphibians, π-globin in birds, and ζ-globin in mammals) is exclusively expressed in primitive erythroid cells derived from the yolk sac, and the adult αA-globin gene is expressed in definitive erythroid cells during later stages of prenatal development and postnatal life (Proudfoot et al. 1982; Higgs et al. 1989; Hardison 2001). The αA- and αD-globin genes are also present in squamate reptiles, and both genes exhibit elevated rates of amino acid substitution relative to their orthologous counterparts in turtles, tuatara, and birds (Gorr et al. 1998). In birds, the αD-globin gene is expressed in both embryonic and adult erythrocytes (Alev et al. 2008, 2009). By contrast, even in mammals that possess an intact, transcriptionally active copy of αD-globin, the product of this gene is not known to be assembled into functional hemoglobin tetramers (Goh et al. 2005).
In contrast to the ancient diversification of the α-globin gene family, available evidence suggests that the physiological division of labor between early- and late-expressed β-like globin genes may have been repeatedly reinvented in different vertebrate lineages. For example, the embryonic “ε-globin” gene of chicken (Gallus gallus) is not a 1:1 ortholog of the embryonic ε-globin gene of placental mammals as they were independently derived from lineage-specific duplications of a β-globin pro-ortholog (Goodman et al. 1987; Reitman et al. 1993; Aguileta et al. 2004; Patel et al. 2008; Opazo et al. 2008b). Indeed, even within mammals, early- and late-expressed β-like globin genes appear to have evolved independently in egg-laying monotremes and therians (placental mammals + marsupials; Opazo et al. 2008b). These patterns suggest that the developmental regulation of β-chain hemoglobin synthesis may have evolved multiple times independently. Until recently, investigations into this possibility were hindered by the fact that chicken was the only nonmammalian tetrapod for which a complete genome sequence was available. Fortunately, the availability of complete genome sequences from an amphibian (western clawed frog, Xenopus tropicalis), a squamate reptile (anole lizard, Anolis carolinensis), and a passeriform bird (zebra finch, Taeniopygia guttata) now permits a more comprehensive and detailed reconstruction of globin gene family evolution in tetrapod vertebrates. Here, we report new insights into globin gene family evolution that were provided by a phylogenetic analysis of vertebrate globins combined with a comparative analysis of genome sequences from three sauropsid taxa: anole lizard, zebra finch, and chicken. Comparative genomic analyses of the three sauropsid taxa can be expected to yield important insights into globin gene family evolution because lepidosaur reptiles (represented by anole lizard) diverged from the stem lineage of archosaurs (represented by birds) 275 Ma (Hedges 2009) and passeriform and galliform birds (represented by zebra finch and chicken, respectively) diverged ∼100 Ma (Hedges et al. 2006).
The main objectives of this study were 1) to characterize evolutionary changes in the size and membership composition of the α- and β-globin gene families of tetrapod vertebrates and 2) to test whether functional diversification of the globin gene clusters occurred independently in different tetrapod lineages. Results of our comparative genomic analysis revealed intriguing patterns of gene turnover in the orthologous globin gene clusters of different taxa and also revealed pronounced differences between the α- and β-globin gene families in the pattern and timing of diversification. Specifically, our results revealed that the β-like globin genes diversified independently in several major lineages of tetrapod vertebrates: amphibians, lepidosaurs, archosaurs, and mammals.
Materials and Methods
Genomic Sequence Data and Annotation
We obtained genomic DNA sequences spanning the α- and β-globin gene clusters of two bird species: zebra finch (unpublished genome, the Genome Center at Washington University) (α-globin gene cluster, chromosome 14: 2987641–2979228; β-globin gene cluster, chromosome 1B: 784737–915857) and chicken (α-globin gene cluster, AY016020; β-globin gene cluster, L17432). We also searched the current assembly of the green anole lizard genome (unpublished genome, Broad Institute, release 54 of the ensembl database) for the presence of α- and β-like globin genes. In all cases, α- and β-like globin genes were annotated by comparison with other globin genes that were already present in the database. We also used the program Genscan (Burge and Karlin 1997) to identify and annotate additional genes lying upstream and downstream of the globin gene clusters. In most cases, coding sequences derived from the genome assembly were validated by comparison with the corresponding EST database.
In addition to the genome sequences from anole lizard, zebra finch, and chicken, we also obtained sequences from a phylogenetically diverse collection of other vertebrate species in order to reconstruct pathways of globin gene family evolution during the radiation of tetrapods. In the case of the α-like globin genes, we annotated sequences of α-like globin genes from human (Homo sapiens [NG_000006]) and platypus (Ornithorhynchus anatinus [AC203513]), and we obtained sequences of α-like globin genes from frog (X. tropicalis [NM_001005092 and NM_203529]), salamander (Pleurodeles waltl [M13365 and X14226], yellow-foot tortoise (Geochelone denticulata [AF499739]), and zebrafish (Danio rerio [NM_131257]). In the case of the β-like globin genes, we obtained genomic sequences spanning the β-globin gene cluster of two placental mammals (human, H. sapiens [AC104389] and cat, Felis catus [AC129072]), one amphibian (western clawed frog, X. tropicalis [scaffold 733: 145910–445992]), and four fishes (zebrafish, D. rerio [chromosome 3: 52848044–52988682]; medaka, Oryzias latipes [chromosome 19: 1414512–1555137]; fugu, Takifugu rubripes [chromosome Un: 14351875–14492550]; and pufferfish, Tetraodon nigroviridis [chromosome 2: 3478461–3619109]). For each species, we identified globin genes in the unannotated genomic sequence by using the program Genscan (Burge and Karlin 1997) and by comparing known exon sequences to genomic contigs using the program BLAST2, version 2.2 (Tatusova and Madden 1999).
In order to obtain a more detailed view of the evolutionary history of the β-like globin genes in sauropsid vertebrates, we also assembled a data set of amino acid sequences that included β-globin sequences from a diverse array of reptiles. For comparative purposes, this data set included conceptual translations of all functional β-like globin genes from anole lizard, chicken, zebra finch, human, and cat, as well as amino acid sequences from an additional squamate reptile (common iguana, Iguana iguana, [P18987]), a sphenodont reptile (tuatara, Sphenodon punctatus [P10060 and P10061]), crocodilians (Nile crocodile, Crocodylus niloticus [P02129], and American alligator, Alligator mississippiensis [P02130]), and testudines (loggerhead turtle, Caretta caretta [Q10733], and Galapagos tortoise, G. nigra [P83123]). We also used new amino acid sequences from paralogous βI- and βII-globins of the Indian python, Python molurus (Squamata: Boidae; Stoeckelhuber 1992; Gorr 1993) and the βII-globin paralog of the common iguana (see below). Two β-globin paralogs from the western clawed frog were included as outgroup sequences.
Amino Acid Sequencing
We determined the complete primary sequence of the βII-globin paralog of the common iguana (I. iguana) and the Galapagos marine iguana (Ambylrhynchus cristatus) by means of automated Edman degradation in conjunction with amino acid analyses of peptides separated by high-performance liquid chromatography (HPLC). Specifically, we conducted N-terminal sequencing of the isolated and lyophilized native chains (Rücknagel et al. 1988) together with gas-phase sequencing of HPLC-purified tryptic peptides from reduced pyridine-ethylated chains (Gorr 1993; Stoeckelhuber et al. 2002). This latter reaction uses vinyl pyridine to alkylate reactive cyteinyl SH-groups in order to irreversibly block disulfide-based aggregates of β-chain subunits. The βII-globin amino acid sequences from the common iguana and the marine iguana were deposited in the UniProt Knowledgebase under accession nos. P86390 and P86391.
Phylogeny of α- and β-Like Globin Genes
Sequences of vertebrate globin genes were aligned using the program MUSCLE (Edgar 2004), as implemented in the European Bioinformatics Institute Web server (http://www.ebi.ac.uk). Phylogenetic relationships were inferred using Bayesian and maximum likelihood approaches. Bayesian analyses were conducted in Mr. Bayes v.3.1.2 (Ronquist and Huelsenbeck 2003) running six simultaneous chains for 5,000,000 generations, sampling every 1,000 generations, and using default priors. Support for the nodes and parameter estimates were derived from a majority rule consensus of the last 2,500 trees. The average standard deviation of split frequencies remained less than 0.01 after the burn-in threshold. Maximum likelihood analyses were conducted in Treefinder, version October 2008 (Jobb et al. 2004), and support for the nodes was evaluated with 1,000 bootstrap pseudoreplicates. In both Bayesian and maximum likelihood analyses, different models of nucleotide substitution were permitted for each codon position. We used the “propose model” tool of Treefinder to select the best fitting model of nucleotide substitution for each codon position in the maximum likelihood analysis. Model selection was based on the Akaike information criterion with correction for small sample size. In the case of the amino acid data set, the phylogeny reconstruction was conducted with Mr. Bayes v.3.1.2 (Ronquist and Huelsenbeck 2003) using a JTT model of amino acid substitution, running eight simultaneous chains for 5,000,000 generations, sampling every 1,000 generations, and using default priors. Support for the nodes and parameter estimates were derived from a majority rule consensus of the last 2,500 trees. The average standard deviation of split frequencies remained less than 0.01 after the burn-in threshold. For the phylogenetic analysis of β-globin amino acid sequences, we only included sequences from reptile taxa for which we successfully recovered all expressed adult β-globins. For this reason, we excluded marine iguana from the analysis because we successfully sequenced only one of the two adult β-globin paralogs from this species.
Inferring Orthologous Relationships
To examine the genomic structure of the α- and β-globin gene clusters of zebra finch, we conducted pairwise comparisons of sequence similarity with the orthologous chicken gene cluster using the Pipmaker program (Schwartz et al. 2000). Because interparalog gene conversion is typically restricted to the coding regions of globin genes, we inferred orthologous relationships among β-like globin genes by reconstructing phylogenetic relationships of flanking sequence and intron 2 sequence (Hardison and Gelinas 1986; Hardison and Miller 1993; Storz et al. 2007; Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b; Opazo et al. 2009; Runck et al. 2009). We conducted phylogeny reconstructions on three nonoverlapping sequence alignments: 1 kb of flanking sequence immediately upstream of the initiation codon, 2385 bp of intron 2 sequence, and 1 kb of flanking sequence immediately downstream of the termination codon. Because previous studies have demonstrated that ectopic recombinational exchanges are restricted to the 5′ ends of chicken β-like globin genes (including exons 1 and 2) and upstream flanking regions (Dodgson et al. 1983; Reitman et al. 1993), sequence variation in intron 2 and the downstream flanking region should be especially useful for the purpose of assigning orthologous relationships. Phylogenetic analyses of each data partition were conducted in a maximum likelihood framework as described above.
Results
Genomic Structure and Evolution of the Sauropsid α-Globin Gene Cluster
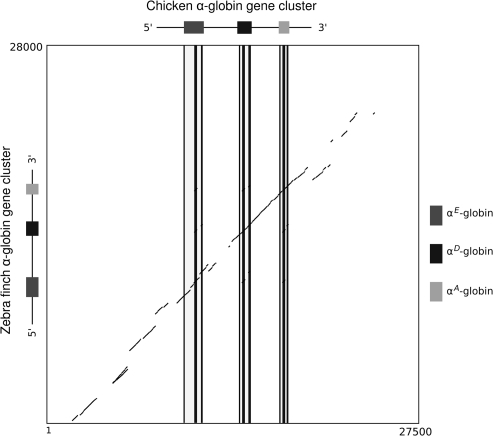
The two bird species included in our study, zebra finch (Passeriformes: Estrildidae) and chicken (Galliformes: Phasianidae), have both retained the ancestral complement of developmentally regulated α-like globin genes: 5′-αE, αD, and αA-3′. Pairwise dot plots between the α-globin gene clusters of these two species revealed high levels of sequence conservation across both coding and noncoding regions (fig. 1). Phylogenies based on coding sequence indicate that the three α-like globin genes in the zebra finch cluster are 1:1 orthologs of the three α-like genes in the chicken cluster (fig. 2). The α-globin gene clusters of both species also retained very similar patterns of intergenic spacing: From the initiation codon of αE-globin to the termination codon of αA-globin, the gene cluster spanned 8,414 bp in zebra finch and 7,379 bp in chicken. In both species, the α-globin gene cluster was flanked by an ortholog of chromosome 16 open reading frame 35 (C16Orf35) on the 5′ side and by an ortholog of the transmembrane protein 6 on the 3′ side.
FIG. 1.
Dot plot of sequence similarity between the α-globin gene clusters of chicken (Gallus gallus) and zebra finch (Taeniopygia guttata).
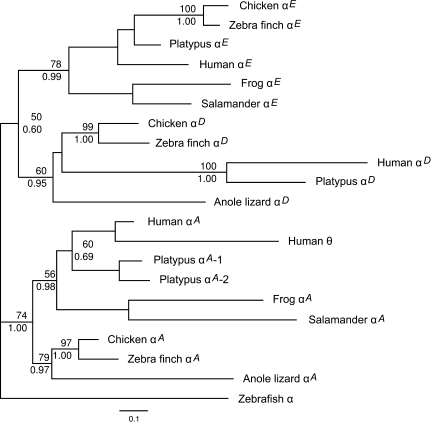
FIG. 2.
Maximum likelihood phylogram depicting relationships among the α-globin sequences of vertebrates based on nucleotide sequence data. The sequence of the zebrafish (Danio rerio) was used as outgroup. Values on the nodes denote bootstrap support values (above) and Bayesian posterior probabilities (below).
In the anole lizard genome, we identified two putatively functional α-like globin genes that were located in different scaffolds (2790: 1–15038 and 1188: 1–163036). Phylogenies based on coding sequence indicate that one of the genes is orthologous to the αA-globin gene of other tetrapods, and the other gene is orthologous to the αD-globin gene of other amniotes (fig. 2). We did not find an ortholog of the embryonic αE-globin gene in the anole lizard genome. The αD-globin gene of this species is located on scaffold 2790, and its position is syntenic with the α-globin gene cluster of other vertebrates: On the 5′ end, the gene is flanked by an ortholog of C16Orf35, and on the 3′ end, it is flanked by a highly distinct globin gene that exhibited a strong match to the recently discovered “globin Y” (GbY) gene. Bioinformatic analyses reported by Fuchs et al. (2006) indicated that the GbY gene is most closely related to cytoglobin, a respiratory globin protein that is expressed in fibroblast-related cells and distinct neuronal cells (Burmester et al. 2004; Hankeln et al. 2004; Nakatani et al. 2004; Schmidt et al. 2004; Hankeln et al. 2005; Schmidt et al. 2005). To date, the GbY gene has only been characterized in Xenopus (Fuchs et al. 2006) and platypus (Patel et al. 2008). The apparently orthologous GbY genes of Xenopus, anole lizard, and platypus are located in syntenic positions at the 3′ end of the α-globin gene cluster. The anole lizard GbY gene exhibits the typical 3-exon/2-intron globin gene structure, and a conceptual translation of the coding sequence yields a polypeptide of 151 amino acids that shows 35% identity/60% similarity to the GbY sequences from Xenopus and platypus and 28% identity/50% similarity to tetrapod cytoglobins. Despite its close linkage to the α-like globin genes, the GbY gene does not encode a subunit polypeptide of hemoglobin and its function remains a mystery (Fuchs et al. 2006).
Interestingly, the Anolis ortholog of the adult αA-globin gene is located on scaffold 1188, and it is flanked by an ortholog of the chicken adenylate cyclase 9 gene (ADCY9; located on chromosome 14: 13294809–13353192) on the 5′ end, and an ortholog of the chicken GSG1L gene (located on chromosome 14: 12411725–1274104) on the 3′ end. The chicken ADCY9 and GSG1L genes are located on opposite sides of the chicken α-globin cluster, which is located on chromosome 14: 12722493–12730496. This seemingly anomalous arrangement of α-like globins in anole lizard is probably attributable to an error in the current genome assembly. If the position of the lizard αA-globin gene is not an artifact of the current assembly, it would represent a major departure from the canonical gene arrangement of all other tetrapod vertebrates studied to date, where the α-like globin genes are always flanked by the genes N-methylpurine-DNA glycosylase and C16Orf35 at the 5′ end of the gene cluster (Flint et al. 2001; Hughes et al. 2005; Hardison 2008; Patel et al. 2008; Hoffmann et al. 2008b). An alignment of α-globin amino acid sequences from Anolis, chicken, and zebra finch is provided in supplementary figure S1A (Supplementary Material online).
Genomic Structure and Evolution of the Sauropsid β-Globin Gene Cluster
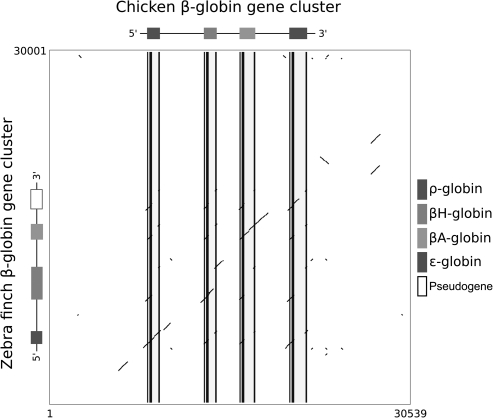
In the zebra finch, the β-globin gene cluster contains four genes (from 5′ to 3′): ρ-globin, βH-globin, βA-globin, and an ε-globin pseudogene-3′ (fig. 3). As with the α-globin gene cluster, the β-globin gene clusters of zebra finch and chicken have remained very similar in terms of intergenic spacing: From the initiation codon of ρ-globin to the termination codon of ε-globin, the zebra finch cluster spans 13,231 bp and the chicken cluster spans 13,691 bp. The zebra finch β-globin gene cluster is flanked on both sides by multiple odorant receptors genes, as is the case with all other amniotes studied to date (Bulger et al. 1999; Hardison 2008; Patel et al. 2008). Pairwise sequence comparison between zebra finch and chicken revealed that high levels of sequence conservation were largely restricted to coding regions and proximal flanking regions (fig. 3).
FIG. 3.
Dot plot of sequence similarity between the β-globin gene clusters of chicken (Gallus gallus) and zebra finch (Taeniopygia guttata).
The annotation of β-like globin genes in zebra finch revealed that two of the four predicted β-like globin genes, βA- and ε-globin, contained insertions and deletions in the coding sequence that would render them nonfunctional. The βA-globin sequence contained two single nucleotide insertions, and the ε-globin sequence contained multiple frame-shifting indels. In comparison with the other early-expressed gene, ρ-globin, the putative ε-globin pseudogene contained four indels in exon 2, ranging 6–50 bp in length, and three indels in exon 3, ranging 1–17 bp in length. To verify the authenticity of these inactivating indels in the zebra finch βA- and ε-globin genes, we searched the corresponding EST database and found a sequence corresponding to the βA-globin gene that had a perfectly intact open reading frame. We therefore used the sequence from the EST record for all phylogenetic analyses. We did not find an EST record that matched the ε-globin sequence, which suggests that this gene is not expressed in zebra finch.
In the anole lizard genome, we found two putatively functional β-like globin genes that were located in different scaffolds (7008: 1–4809 and 3777: 1–10310), and Genscan searches did not reveal the presence of any additional genes upstream or downstream of the β-like globin genes in either scaffold. Thus, the current state of the anole lizard genome assembly does not permit inferences regarding patterns of conserved synteny in the β-globin gene cluster. The two β-globin paralogs of Anolis were distinguished by a total of 36 amino acid substitutions (24.49% sequence divergence). An alignment of β-globin amino acid sequences from Anolis, chicken, and zebra finch is provided in supplementary figure S1B (Supplementary Material online).
Inferring Orthologous Relationships among β-Like Globin Genes
If the functional diversification of the β-globin gene family predated the radiation of tetrapod vertebrates, then we should recover a phylogenetic tree with the following topological features: 1) The early-expressed genes of all taxa group together in one clade of orthologous sequences, 2) the late-expressed genes from these same taxa group together in a sister clade of orthologous sequences, and 3) sequences within each of the two paralogous sister clades independently recover the known organismal phylogeny: (amphibians (mammals (lepidosaurs, archosaurs))). Contrary to these expectations, maximum likelihood and Bayesian phylogeny reconstructions revealed that β-like globin genes of amphibians, reptiles, birds, and mammals were all reciprocally monophyletic relative to one another (fig. 4). This phylogenetic pattern indicates that the β-globin gene clusters diversified independently in each of these different tetrapod lineages. Each of the main groups of tetrapods inherited an ortholog of the same proto β-globin gene, which then underwent one or more rounds of duplication and divergence to produce distinct repertoires of β-like globin genes in each descendant lineage. These lineage-specific patterns had been previously documented in the case of birds (as represented by chicken alone) and mammals (Patel et al. 2008; Opazo et al. 2008a, 2008b). The addition of genomic sequences from Xenopus and anole lizard revealed that amphibians and squamate reptiles have also invented their own distinct repertoires of β-like globin genes. The addition of genomic sequence from zebra finch revealed that passeriform and galliform birds have retained the same ancestral complement of β-like globin genes. This was not a foregone conclusion because the estimated date of the duplication event that gave rise to the ρ-/ε-globin gene pair in chicken was reported to be 49.4–55.1 Ma (Aguileta et al. 2006), whereas the estimated date of divergence between passeriformes and galliformes is thought to be ∼100 Ma (Hedges et al. 2006).
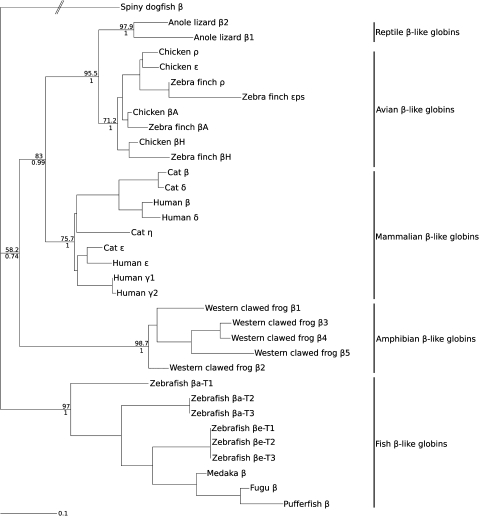
FIG. 4.
Maximum likelihood phylogram depicting relationships among the β-globin sequences of vertebrates based on nucleotide sequence data. The sequence of the spiny dogfish (Squalus acanthias) was used as outgroup. Values on the nodes denote bootstrap support values (above) and Bayesian posterior probabilities (below).
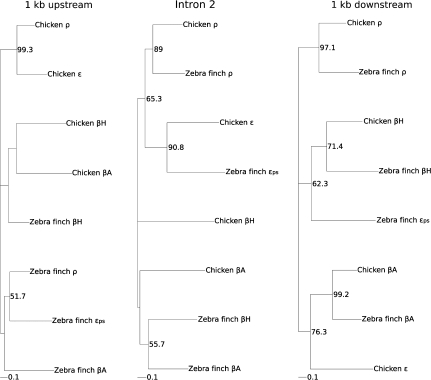
Within birds, phylogenies based on coding sequence were able to resolve orthologous relationships for the adult β-globin genes but not the early-expressed genes (fig. 5). Phylogeny reconstructions based on coding sequence and upstream flanking sequence depict a clear history of concerted evolution between the two early-expressed genes, as paralogous ρ- and ε-globin sequences from the same species group together in the tree to the exclusion of orthologous sequences in other species (fig. 5). Pairwise alignment of the zebra finch ρ- and ε-globin sequences shows an increased sequence similarity in the upstream sequence that is attributable to ρ→ε gene conversion. The conversion tract was 433 bp long, starting 90 bp upstream of the start codon and spanning exon 2 of the ε-globin gene (Dodgson et al. 1983). Because intron 2 and downstream flanking sequence have not experienced a history of gene conversion, phylogeny reconstructions of these noncoding regions can be expected to recover the true history of gene duplication and species divergence. Accordingly, phylogeny reconstructions based on intron 2 and downstream flanking sequence conclusively demonstrate that the ρ-, βH-, βA-, and ε-globin genes of zebra finch and chicken are 1:1 orthologs (fig. 5).
FIG. 5.
Maximum likelihood phylogram depicting relationships among β-like globin genes in chicken and zebra finch based on 1 kb of the 5′ flanking sequence (left), intron 2 (center), and 1 kb of 3′ flanking sequence (right). Measures for support for the relevant nodes are presented as bootstrap values.
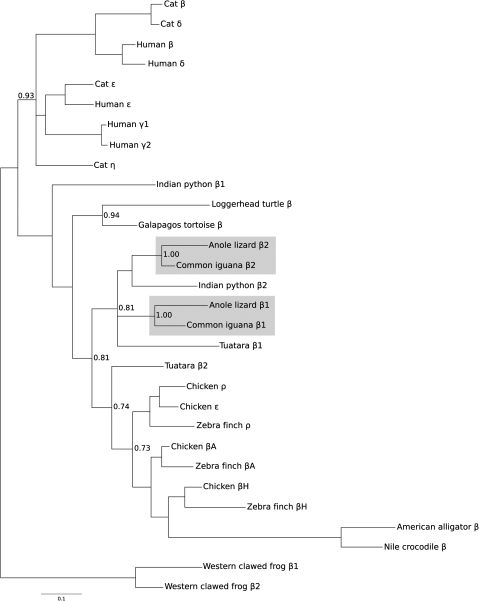
The phylogeny reconstruction based on amino acid sequences revealed that reptilian β-globins form a single monophyletic group, although support for the corresponding node was relatively weak (fig. 6). In the tree based on amino acid sequences, archosaur β-globins grouped together in a clade that was sister to the βD-globin paralog of the tuatara, and lepidosaur β-globins grouped together in a clade that also included the β2-globin paralog of the Indian python and the βA-globin paralog of the tuatara. In addition, the two testudine β-globins form a monophyletic group and the β1-globin paralog of the Indian python represents the most basal lineage. This phylogeny reconstruction indicates that the β1 globin genes of anole lizard and common iguana are 1:1 orthologs, as are the β2 globin genes from the same species pair (fig. 6). The β1 and β2 globin genes of anole lizard and iguana appear to be products of a lizard-specific duplication event (and the two iguana β-globin sequences correspond to the “βIIa” and “βIIb” sequences in Gorr et al. 1998). The phylogenetic positions of β-globin paralogs from tuatara and Indian python suggest that the common ancestor of sauropsids may have possessed a fairly diverse repertoire of β-like globin genes and that the differential loss and retention of these genes may account for much of the observed variation in gene content among contemporary reptiles. Due to the limited resolution afforded by amino acid sequences, we will ultimately need genomic sequence data from turtles, tuatara, and snakes to reconstruct pathways of β-globin gene family evolution during the basal radiation of nonavian reptiles. We also reconstructed phylogenetic relationships using a mixed model approach. The estimated topology showed two minor discrepancies relative to the original tree. The first discrepancy concerns the outgroup sequences, where relationships among the mammalian ε-, γ-, and η-globin genes were not resolved. The second discrepancy is that the chicken and zebra finch βA-globin genes were not grouped together, and instead the zebra finch βA-globin gene was placed sister to the crocodilian β-globin genes. This latter result is clearly incorrect because trees based on DNA sequence variation in intron 2 and the 3′ flanking region also demonstrate that the chicken and zebra finch βA-globin genes are 1:1 orthologs (fig. 5).
FIG. 6.
Bayesian phylogram depicting relationships among the β-like globin genes of amniote vertebrates based on amino acid sequence data. Two β-globin sequences from western clawed frog (Xenopus tropicalis) were used as outgroups. Values on the nodes correspond to Bayesian posterior probabilities.
Discussion
In recent years, comparative analyses of completely sequenced genomes have revealed extensive variation in the size and membership composition of multigene families among species (Demuth and Hahn 2009). This variation results from differential gene gains via duplication and gene losses via deletion or inactivation. In many cases, the resultant changes in gene family composition may account for important phenotypic differences between species. Our comparative genomic analysis of the α- and β-globin gene clusters revealed a number of significant differences in gene content among vertebrate taxa. These differences were especially pronounced in the case of the β-globin gene cluster, as our phylogenetic reconstructions revealed that amphibians, lepidosaurs (as represented by anole lizard), archosaurs (as represented by zebra finch and chicken), and mammals each possess a distinct independently derived repertoire of β-like globin genes.
Variation in Gene Family Size and Membership Composition among Taxa
In the case of the α-globin gene family, the anole lizard emerged as the only gnathostome vertebrate species yet examined that does not possess an ortholog of the embryonic αE-globin gene. Given that the αD-globin gene originated via duplication of a proto αE-globin gene that had an ancestral larval/embryonic function (Hoffmann and Storz 2007), it may be that αD-containing hemoglobin (HbD) performs the necessary oxygen transport functions during early stages of embryonic development in lizards and other reptiles that do not possess an ortholog of αE-globin. Developmental studies of hemoglobin synthesis will be necessary to test this hypothesis about the interchangeability of αE- and αD-globin in reptiles.
Although zebra finch and chicken possess an identical complement of αE-, αD-, and αA-globin genes, the αD-globin gene exhibits very different patterns of prenatal expression in these two species (Alev et al. 2009). In primitive erythroid cells of zebra finch embryos, the rank order of transcript abundance is αD > αE > αA, and in erythroid cells of chicken at the same stage of development, the rank order of transcript abundance is αE > αA > αD (Alev et al. 2008, 2009). In the definitive erythroid cells of adult birds, the HbD isoform typically constitutes the minor fraction of adult hemoglobin and αA-containing hemoglobin typically constitutes the major fraction, although the relative abundance of the two isoforms is quite variable among species (Borgese and Bertles 1965; Brown and Ingram 1974; Hiebl et al. 1987; Ikehara et al. 1997). It will be important to achieve a more complete understanding of the physiological division of labor among the αE-, αD-, and αA-containing hemoglobin isoforms during different stages of development in birds and nonavian reptiles.
In the case of the β-globin gene family, the one notable difference between the two birds included in our analysis is that the ortholog of the chicken ε-globin gene is a pseudogene in zebra finch. This may seem surprising because ε-globin is the most highly expressed β-like globin gene during early stages of chicken embryogenesis (Alev et al. 2008), and early-expressed globin genes are typically subject to especially stringent levels of functional constraint (Aguileta et al. 2004). However, the ρ- and ε-globin genes are very similar in sequence due to a history of interparalog gene conversion (supplementary fig. S1, Supplementary Material online). Concerted evolution between the two early-expressed paralogs may have produced a degree of functional redundancy that would have helped mitigate the effects of inactivating the ε-globin gene. Similarly, differential loss and retention of embryonic β-like globin genes during the basal radiation of placental mammals have produced extensive variation in the complement of early-expressed genes in contemporary species (Opazo et al. 2008a).
Results of our comparisons between zebra finch and chicken indicate that the size and membership composition of the α- and β-globin gene families are far more stable in birds than in mammals. Zebra finch (Passeriformes) and chicken (Galliformes) represent two highly distinct avian lineages that are thought to have diverged ∼100 Ma (Hedges et al. 2006). Despite this ancient divergence, zebra finch and chicken have each retained an identical complement of three orthologous α-like globin genes. By contrast, over a similar timescale of divergence, gene turnover in the mammalian α-globin gene family has produced extensive variation in gene copy number among species (Hoffmann et al. 2008b). For example, guinea pig and rat, which diverged ∼60 Ma (Steppan et al. 2004), possess three and seven functional α-like globin genes, respectively (Storz et al. 2008). Zebra finch and chicken have also retained an identical complement of orthologous β-like globin genes, with the exception that ε-globin has been inactivated in zebra finch. Again, the stability of the avian β-globin gene cluster stands in marked contrast to the pattern of gene turnover that has been documented in mammals (Opazo et al. 2008a, 2008b, 2009). Within rodents, for example, guinea pig and rat possess two and eight functional β-like globin genes, respectively (Hoffmann et al. 2008a).
In addition to the pronounced differences in genomic stability, the globin gene clusters of birds and mammals also differ with respect to the association between linear gene order and the temporal order of expression during development. Genes in the mammalian α- and β-globin gene clusters are almost always colinear with their temporal order of expression: The early-expressed genes are located at the 5′ end of the cluster and the later-expressed genes are located at the 3′ end. The only known exceptions to this pattern are attributable to en bloc duplications of multiple genes, where early-expressed genes in the 3′ duplication block are located downstream of late-expressed genes in the 5′ duplication block (Townes et al. 1984; Cheng et al. 1987; Hoffmann et al. 2008a). In the chicken β-globin gene cluster, by contrast, the embryonic ρ- and ε-globin genes demarcate the 5′ and 3′ ends of the gene cluster, respectively, and the late-expressed βH- and βA-globin genes are sandwiched in between (Bruns and Ingram 1973a, 1973b; Reitman et al. 1993).
Different Timescales of Functional Diversification in the α- and β-Globin Gene Families
Results of our phylogenetic analysis confirm that the α- and β-like globin gene families of tetrapods have diversified over very different timescales. In the α-globin gene cluster, a division of labor between early- and late-expressed genes was established in the common ancestor of tetrapods, and all descendant lineages appear to have retained the same basic mechanism for the developmental regulation of α-chain hemoglobin synthesis: The α-chain subunits of embryonic hemoglobin are encoded by αE-globin and the α-chain products of adult hemoglobin are encoded by αA-globin. The anole lizard represents the one notable exception to this pattern due to the apparent absence of αE-globin in this species.
In contrast to the ancient functional diversification of the α-globin gene cluster, the physiological division of labor between early- and late-expressed genes in the β-globin gene cluster appears to have evolved independently in several different tetrapod lineages. In addition to the wealth of data regarding the developmental regulation of hemoglobin synthesis in humans, mice, and other mammals (reviewed by Hardison 2001; Nagel and Steinberg 2001; Brittain 2002), developmental patterns of hemoglobin synthesis have also been characterized in Xenopus (Banville and Williams 1985a, 1985b; Fuchs et al. 2006), chicken (Nakazawa et al. 2006; Alev et al. 2008; McIntyre et al. 2008; Nagai and Sheng 2008), and zebra finch (Alev et al. 2009). These data on developmental expression patterns take on special significance in light of our discovery regarding the phylogenetic history of the β-globin gene clusters in each of these taxa. Becasue the early-expressed β-like globin genes in amphibians, birds, and mammals are not orthologous to one another, our results indicate that stage-specific expression in primitive erythroid cells must have evolved independently in each of these lineages. Likewise, well-characterized functional differences between the products of early- and late-expressed β-like globin genes also must have evolved independently. Although the two β-like globin genes of anole lizard appear to be products of a lizard-specific duplication event (figs. 4 and 6), no data are yet available regarding the developmental expression profiles of these genes. To the best of our knowledge, there are no published reports on developmental patterns of hemoglobin synthesis in any nonavian reptiles. Further study is required to determine whether the two β-like globin genes of lizards have evolved developmentally regulated expression differences.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank M. Stoeckelhuber for sharing the β1- and β2-globin sequences of Indian python, the Broad Institute Genome Sequencing Platform and Genome Sequencing and Analysis Program for permission to use unpublished genome sequence data for Anolis carolinensis, and F. Di Palma, K. Lindblad-Toh, and W. Warren from The Genome Center at Washington University for permission to use unpublished genome sequence data for Taeniopygia guttata. We are also grateful to S. V. Edwards and two anonymous reviewers for helpful comments on the manuscript. This work was funded by grants to F.G.H. from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 476739/2008-0), to J.F.S. from the National Institutes of Health, the National Science Foundation, and the Nebraska Research Council, fellowships to T.A.G. by the Studienstiftung des Deutschen Volkes and the Max-Planck-Gesellschaft and partnership in a collaborative grant (“Euroxy”) of the EU’s 6th Framework Programme, and to J.C.O. by the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 11080181), Programa Bicentenario en Ciencia y Tecnología (PSD89), and the American Society of Mammalogists.
References
- Aguileta G, Bielawski JP, Yang Z. Gene conversion and functional divergence in the β-globin gene family. J Mol Evol. 2004;59:177–189. doi: 10.1007/s00239-004-2612-0. [DOI] [PubMed] [Google Scholar]
- Aguileta G, Bielawski JP, Yang Z. Evolutionary rate variation among vertebrate β-globin genes: implications for dating gene family duplication events. Gene. 2006;380:21–29. doi: 10.1016/j.gene.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Alev C, McIntyre BAS, Nagai H, Shin M, Shinmyozu K, Jakt LM, Sheng G. BetaA, the major β-globin in definitive red blood cells, is present from the onset of primitive erythropoiesis in chicken. Dev Dyn. 2008;237:1193–1197. doi: 10.1002/dvdy.21510. [DOI] [PubMed] [Google Scholar]
- Alev C, Shinmyozu K, McIntyre BAS, Sheng G. Genomic organization of zebra finch α- and β-globin genes and their expression in primitive and definitive blood in comparison with globins in chicken. Dev Genes Evol. 2009;219:353–360. doi: 10.1007/s00427-009-0294-8. [DOI] [PubMed] [Google Scholar]
- Banville D, Williams JG. Developmental changes in the pattern of larval β-globin gene expression in Xenopus laevis. Identification of two early larval beta-globin mRNA sequences. J Mol Biol. 1985a;184:611–620. doi: 10.1016/0022-2836(85)90307-9. [DOI] [PubMed] [Google Scholar]
- Banville D, Williams JG. The pattern of expression of the Xenopus laevis tadpole α- globin genes and the amino acid sequence of the three major tadpole α-globin polypeptides. Nucleic Acids Res. 1985b;13:5407–5421. doi: 10.1093/nar/13.15.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese TA, Bertles JF. Hemoglobin heterogeneity: embryonic hemoglobin in the duckling and its disappearance in the adult. Science. 1965;148:509–511. doi: 10.1126/science.148.3669.509. [DOI] [PubMed] [Google Scholar]
- Brittain T. Molecular aspects of embryonic hemoglobin function. Mol Aspects Med. 2002;23:293–342. doi: 10.1016/s0098-2997(02)00004-3. [DOI] [PubMed] [Google Scholar]
- Brown JL, Ingram VM. Structural studies on chick embryonic hemoglobins. J Biol Chem. 1974;249:3960–3972. [PubMed] [Google Scholar]
- Bruns GA, Ingram VM. Erythropoiesis in the developing chick embryo. Dev Biol. 1973a;30:455–459. doi: 10.1016/0012-1606(73)90102-4. [DOI] [PubMed] [Google Scholar]
- Bruns GA, Ingram VM. The erythroid cells and haemoglobins of the chick embryo. Philos Trans R Soc Lond B Biol Sci. 1973b;266:225–305. doi: 10.1098/rstb.1973.0050. [DOI] [PubMed] [Google Scholar]
- Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human β-globin loci: the β-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burmester T, Haberkamp M, Mitz S, Roesner A, Schmidt M, Ebner B, Gerlach F, Fuchs C, Hankeln T. Neuroglobin and cytoglobin: genes, proteins, and evolution. IUBMB Life. 2004;56:703–707. doi: 10.1080/15216540500037257. [DOI] [PubMed] [Google Scholar]
- Chan FY, Robinson J, Brownlie A, Shivdasani RA, Donovan A, Brugnara C, Kim J, Lau BC, Witkowska HE, Zon LI. Characterization of adult α- and β-globin genes in the zebrafish. Blood. 1997;89:688–700. [PubMed] [Google Scholar]
- Cheng JF, Raid L, Hardison RC. Block duplications of a zeta-zeta-alpha-theta gene set in the rabbit α-like globin gene cluster. J Biol Chem. 1987;262:5414–5421. [PubMed] [Google Scholar]
- Collins FS, Weissman SM. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- Cooper SJB, Wheeler D, De Leo A, Cheng J, Holland RAB, Marshall Graves JA, Hope RM. The mammalian αD-globin gene lineage and a new model for the molecular evolution of α-globin gene clusters at the stem of the mammalian radiation. Mol Phylogenet Evol. 2006;38:439–448. doi: 10.1016/j.ympev.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Czelusniak J, Goodman M, Hewett-Emmett D, Weiss ML, Venta PJ, Tashian RE. Phylogenetic origins and adaptive evolution of avian and mammalian haemoglobin genes. Nature. 1982;298:297–300. doi: 10.1038/298297a0. [DOI] [PubMed] [Google Scholar]
- De Leo AA, Wheeler D, Lefevre C, Cheng J, Hope R, Kuliwaba J, Nicholas KR, Westerman M, Graves JAM. Sequencing and mapping hemoglobin gene clusters in the Australian model dasyurid marsupial Sminthopsis macroura. Cytogenet Genome Res. 2005;108:333–341. doi: 10.1159/000081528. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. The life and death of gene families. Bioessays. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- Dodgson JB, Stadt SJ, Choi OR, Dolan M, Fischer HD, Engel JD. The nucleotide sequence of the embryonic chicken β-type globin genes. J Biol Chem. 1983;258:12685–12692. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, et al (14 co-authors) Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α-globin cluster. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 2006;112:296–306. doi: 10.1159/000089884. [DOI] [PubMed] [Google Scholar]
- Gillemans N, McMorrow T, Tewari R, et al. (12 co-authors) Functional and comparative analysis of globin loci in pufferfish and humans. Blood. 2003;101:2842–2849. doi: 10.1182/blood-2002-09-2850. [DOI] [PubMed] [Google Scholar]
- Goh S, Lee YT, Bhanu NV, Cam MC, Desper R, Martin BM, Moharram R, Gherman RB, Miller JL. A newly discovered human α-globin gene. Blood. 2005;106:1466–1472. doi: 10.1182/blood-2005-03-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Czelusniak J, Koop BF, Tagle DA, Slightom JL. Globins: a case study in molecular phylogeny. Cold Spring Harb Symp Quant Biol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- Goodman M, Moore GW, Matsuda G. Darwinian evolution in the genealogy of haemoglobin. Nature. 1975;253:603–608. doi: 10.1038/253603a0. [DOI] [PubMed] [Google Scholar]
- Gorr TA. Hämoglobin: Sequenz und Phylogenie. Die Primärstruktur von Globinketten des Quastenflossers (Latimeria chalumnae) sowie folgender Reptilien: Galapagos -Meerechse (Amblyrhynchus cristatus), Grüner Leguan (Iguana iguana), Indigonatter (Drymarchon corais), Glattstirnkaiman (Paleosuchus palpebrosus) 1993 [Ph.D thesis]. [Munich (Germany)]: University of Munich. [Google Scholar]
- Gorr TA, Mable BK, Kleinschmidt T. Phylogenetic analysis of reptilian hemoglobins: trees, rates, and divergences. J Mol Evol. 1998;47:471–485. doi: 10.1007/pl00006404. [DOI] [PubMed] [Google Scholar]
- Hankeln T, Ebner B, Fuchs C, et al. (23 co-authors) Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J Inorg Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Hankeln T, Wystub S, Laufs T, Schmidt M, Gerlach F, Saaler-Reinhardt S, Reuss S, Burmester T. The cellular and subcellular localization of neuroglobin and cytoglobin - a clue to their function? IUBMB Life. 2004;56:671–679. doi: 10.1080/15216540500037794. [DOI] [PubMed] [Google Scholar]
- Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- Hardison R. Organization, evolution, and regulation of the globin genes. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge: Cambridge University Press; 2001. pp. 95–116. [Google Scholar]
- Hardison R, Miller W. Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters. Mol Biol Evol. 1993;10:73–102. doi: 10.1093/oxfordjournals.molbev.a039991. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Globin genes on the move. J Biol. 2008;7:35. doi: 10.1186/jbiol92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Gelinas RE. Assignment of orthologous relationships among mammalian α-globin genes by examining flanking regions reveals a rapid rate of evolution. Mol Biol Evol. 1986;3:243–261. doi: 10.1093/oxfordjournals.molbev.a040392. [DOI] [PubMed] [Google Scholar]
- Hedges SB. The timetree of life. 2009. Oxford: Oxford University Press. [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hentschel CC, Kay RM, Williams JG. Analysis of Xenopus laevis globins during development and erythroid cell maturation and the construction of recombinant plasmids containing sequences derived from adult globin mRNA. Dev Biol. 1979;72:350–363. doi: 10.1016/0012-1606(79)90124-6. [DOI] [PubMed] [Google Scholar]
- Hiebl I, Braunitzer G, Schneeganss D. The primary structures of the major and minor hemoglobin-components of adult Andean goose (Chloephaga melanoptera, Anatidae): the mutation Leu→Ser in position 55 of the β-chains. Biol Chem Hoppe Seyler. 1987;368:1559–1569. doi: 10.1515/bchm3.1987.368.2.1559. [DOI] [PubMed] [Google Scholar]
- Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ. A review of the molecular genetics of the human α-globin gene cluster. Blood. 1989;73:1081–1104. [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 2008a;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008b;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Storz JF. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol Biol Evol. 2007;24:1982–1990. doi: 10.1093/molbev/msm127. [DOI] [PubMed] [Google Scholar]
- Hosbach HA, Wyler T, Weber R. The Xenopus laevis globin gene family: chromosomal arrangement and gene structure. Cell. 1983;32:45–53. doi: 10.1016/0092-8674(83)90495-6. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Cheng J, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci USA. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehara T, Eguchi Y, Kayo S, Takei H. Isolation and sequencing of two α-globin genes α(A) and α(D) in pigeon and evidence for embryo-specific expression of the α(D)-globin gene. Biochem Biophys Res Commun. 1997;234:450–453. doi: 10.1006/bbrc.1997.6667. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Wood D, Simons JP, Kay RM, Williams JG. Linkage of adult α- and β-globin genes in X. laevis and gene duplication by tetraploidization. Cell. 1980;21:555–564. doi: 10.1016/0092-8674(80)90493-6. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kay RM, Harris R, Patient RK, Williams JG. Molecular cloning of cDNA sequences coding for the major α- and β-globin polypeptides of adult Xenopus laevis. Nucleic Acids Res. 1980;8:2691–2707. doi: 10.1093/nar/8.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- McIntyre BAS, Alev C, Tarui H, Jakt LM, Sheng G. Expression profiling of circulating non-red blood cells in embryonic blood. BMC Dev Biol. 2008;8:21. doi: 10.1186/1471-213X-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Sheng G. Definitive erythropoiesis in chicken yolk sac. Dev Dyn. 2008;237:3332–3341. doi: 10.1002/dvdy.21746. [DOI] [PubMed] [Google Scholar]
- Nagel RL, Steinberg MH. Role of epistatic (modifier) genes in the modulation of the phenotypic diversity of sickle cell anemia. Pediatr Pathol Mol Med. 2001;20:123–136. [PubMed] [Google Scholar]
- Nakatani K, Okuyama H, Shimahara Y, Saeki S, Kim D, Nakajima Y, Seki S, Kawada N, Yoshizato K. Cytoglobin/STAP, its unique localization in splanchnic fibroblast -like cells and function in organ fibrogenesis. Lab Invest. 2004;84:91–101. doi: 10.1038/labinvest.3700013. [DOI] [PubMed] [Google Scholar]
- Nakazawa F, Nagai H, Shin M, Sheng G. Negative regulation of primitive hematopoiesis by the FGF signaling pathway. Blood. 2006;108:3335–3343. doi: 10.1182/blood-2006-05-021386. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin (Germany): Springer-Verlag; 1970. [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci USA. 2008a;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci USA. 2008b;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF. Origin and ascendancy of a chimeric fusion gene: the β/δ-globin gene of paenungulate mammals. Mol Biol Evol. 2009;26:1469–1478. doi: 10.1093/molbev/msp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VS, Cooper SJB, Deakin JE, Fulton B, Graves T, Warren WC, Wilson RK, Graves JAM. Platypus globin genes and flanking loci suggest a new insertional model for β-globin evolution in birds and mammals. BMC Biol. 2008;6:34. doi: 10.1186/1741-7007-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano E, Cocca E, Mazzei F, Ghigliotti L, di Prisco G, Detrich HW, 3rd, Ozouf-Costaz C. Mapping of α- and β-globin genes on Antarctic fish chromosomes by fluorescence in-situ hybridization. Chromosome Res. 2003;11:633–640. doi: 10.1023/a:1024961103663. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Gil A, Maniatis T. The structure of the human ζ-globin gene and a closely linked, nearly identical pseudogene. Cell. 1982;31:553–563. doi: 10.1016/0092-8674(82)90311-7. [DOI] [PubMed] [Google Scholar]
- Reitman M, Grasso JA, Blumenthal R, Lewit P. Primary sequence, evolution, and repetitive elements of the Gallus gallus (chicken) β-globin cluster. Genomics. 1993;18:616–626. doi: 10.1016/s0888-7543(05)80364-7. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rücknagel KP, Braunitzer G, Wiesner H. Hemoglobins of reptiles. The primary structures of the α I- and β I-chains of common iguana (Iguana iguana) hemoglobin. Biol Chem Hoppe Seyler. 1988;369:1143–1150. doi: 10.1515/bchm3.1988.369.2.1143. [DOI] [PubMed] [Google Scholar]
- Runck AM, Moriyama H, Storz JF. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol Biol Evol. 2009;26:2521–2532. doi: 10.1093/molbev/msp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Gerlach F, Avivi A, et al. (11 co-authors) Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279:8063–8069. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Laufs T, Reuss S, Hankeln T, Burmester T. Divergent distribution of cytoglobin and neuroglobin in the murine eye. Neurosci Lett. 2005;374:207–211. doi: 10.1016/j.neulet.2004.10.071. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Stoeckelhuber M. [diploma thesis] [Munich (Germany)]: University of Munich; 1992. Sequenzermittlung von Schlangenhaemoglobinen. Primaerstrucktur von Globinketten der Indigonatter (Drymarchon corais erebenus) und des Tigerpython (Python molurus bivittatus) [Google Scholar]
- Stoeckelhuber M, Gorr T, Kleinschmidt T. The primary structure of three hemoglobin chains from the indigo snake (Drymarchon corais erebennus, Serpentes): first evidence for αD chains and two β chain types in snakes. Biol Chem. 2002;383:1907–1916. doi: 10.1515/BC.2002.214. [DOI] [PubMed] [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Hoffmann FG, Opazo JC, Moriyama H. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics. 2008;178:1623–1638. doi: 10.1534/genetics.107.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Townes TM, Shapiro SG, Wernke SM, Lingrel JB. Duplication of a four-gene set during the evolution of the goat β-globin locus produced genes now expressed differentially in development. J Biol Chem. 1984;259:1896–1900. [PubMed] [Google Scholar]
- Wheeler D, Hope R, Cooper SB, Dolman G, Webb GC, Bottema CD, Gooley AA, Goodman M, Holland RA. An orphaned mammalian β-globin gene of ancient evolutionary origin. Proc Natl Acad Sci USA. 2001;98:1101–1106. doi: 10.1073/pnas.98.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D, Hope RM, Cooper SJB, Gooley AA, Holland RAB. Linkage of the β-like ω-globin gene to α-like globin genes in an Australian marsupial supports the chromosome duplication model for separation of globin gene clusters. J Mol Evol. 2004;58:642–652. doi: 10.1007/s00239-004-2584-0. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.