Abstract
A central goal of evolutionary genetics is an understanding of the forces responsible for the observed variation, both within and between species. Theoretical and empirical work have demonstrated that genetic recombination contributes to this variation by breaking down linkage between nucleotide sites, thus allowing them to behave independently and for selective forces to act efficiently on them. The Drosophila fourth chromosome, which is believed to experience no—or very low—rates of recombination has been an important model for investigating these effects. Despite previous efforts, central questions regarding the extent of recombination and the predominant modes of selection acting on it remain open. In order to more comprehensively test hypotheses regarding recombination and its potential influence on selection along the fourth chromosome, we have resequenced regions from most of its genes from Drosophila melanogaster, D. simulans, and D. yakuba. These data, along with available outgroup sequence, demonstrate that recombination is low but significantly greater than zero for the three species. Despite there being recombination, there is strong evidence that its frequency is low enough to have rendered selection relatively inefficient. The signatures of relaxed constraint can be detected at both the level of polymorphism and divergence.
Keywords: dot chromosome, recombination, gene conversion, selective constraint, relaxed constraint, purifying selection
Introduction
The comparison of genomes or genomic regions that vary in the amounts of genetic recombination that they experience has led to powerful empirical tests of several theoretical population genetic models of selection (Kliman and Hey 1993; Betancourt and Presgraves 2002; Smith and Eyre-Walker 2002; Presgraves 2005; Andolfatto 2007; Begun et al. 2007; Haddrill et al. 2007; Shapiro et al. 2007; Betancourt et al. 2009 Sella et al. 2009). The importance of testing these models stems from the insights gained into the selective forces that govern standing variation and molecular evolution, and also from furthering our understanding of the evolutionary effects of sex, and the extent to which recombination influences the efficiency of natural selection (Felsenstein 1974; Kondrashov 1988; Rice and Chippindale 2001; Bachtrog 2003; Paland and Lynch 2006; Kaiser and Charlesworth 2008).
One particularly intriguing model for the effects of recombination has been the Drosophila fourth chromosome (we will refer to it as the “fourth,” but it is also known as the “dot” or the Muller F element; Berry et al. 1991; Jensen et al. 2002; Wang et al. 2002; Wang et al. 2004; Haddrill et al. 2007; Betancourt et al. 2009). The fourth possesses unique biological features that set it apart from the other Drosophila autosomes in several ways (reviewed by Riddle and Elgin 2006). Briefly, it is usually the smallest autosome (∼5 Mb), with only a ∼1 Mb euchromatic-like region of the right arm containing ∼80 genes. Several studies have shown that the fourth may share ancestry with the X chromosome, and like the latter confers surprisingly little viability or fertility effects when segregating in more than two copies and is also associated with a chromosome-specific protein complex (“Painting of the fourth”; Larsson et al. 2001; Larsson et al. 2004). In addition, the X and fourth have been known to interact during meiosis (Mohr 1932; Sturtevant 1934; Sturtevant 1936; Sandler and Novitski 1955; Franke and Baker 1999; Ashburner et al. 2005; Gilliland et al. 2009). Despite its small size and ability to segregate with considerable variability, the fourth possesses a gene density similar to the other autosomes. The regulation of these genes is an active area of research, as replication and biochemical studies have shown that the coding region of the fourth has DNA properties that are both heterochromatic and euchromatic (Hochman 1976; Wallrath and Elgin 1995; Sun et al. 2000). This heterochromatic characterization has also historically been supported by a putative lack of recombination.
Early investigations using physical markers were unable to identify recombination events despite the inspection of tens of thousands of normal crosses. The only exceptions were lines subjected to mutagenic lab techniques such as heat shock and X-rays (Patterson and Muller 1930; Bridges 1935; Hochman 1976; Ashburner et al. 2005). Due to the putative lack of recombination, the expectation was that selection driving an allele to fixation or extinction would also drive all linked polymorphism across the chromosome with it, thus leading to an extreme overall reduction in nucleotide diversity for the fourth. Two theoretical models are capable of explaining this expectation. The first is a hitchhiking model in which mutations that are linked to a positively selected site are carried to fixation along with it (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Hudson 1990). The second model is background selection in which mutations linked to a deleterious site are purged from the population along with it (Charlesworth et al. 1993; Charlesworth et al. 1995). The extent of the effects of these two models depends on the population recombination rate ρ = 4Ner (where Ne is the effective population size and r is the per base per generation crossing over rate), the selection coefficient, s, and the rate of mutation to a beneficial or deleterious allele. The result of these linkage effects can be thought of as a local reduction in Ne, and thus, a decrease in the ability for selection to act efficiently, which can be estimated by Nes (Gordo and Charlesworth 2001).
The first population genetic support that the fourth was nonrecombining, and also that hitchhiking was driving the lack of variation, came from several early population surveys which identified no, or very little, variation within the Drosophila melanogaster subgroup (Berry et al. 1991; Hilton et al. 1994). Since these early surveys, additional polymorphism data have resulted not only in the increased likelihood that recombination has played a historical role along the fourth but also in mixed results regarding the modes of selection driving the low levels of variation (Jensen et al. 2002; Wang et al. 2002; Wang et al. 2004). Using the largest fourth chromosome population data sets available from D. melanogaster and D. simulans, Wang et al. (2002), Wang et al. (2004) observed heterogeneity in diversity levels that were consistent with recombination and estimated ρ to be 0.00016 and 0.01185 for D. melanogaster and D. simulans, respectively. Although lower than previous estimates from other chromosomes, these values were surprisingly high for the fourth. In addition, a ∼200 kb dimorphic haplotype was found near the center of D. melanogaster's chromosome, with the suggestion that it may be the target of balancing selection (Wang et al. 2002). Such a haplotype has not been observed outside D. melanogaster.
Although these latter studies cast serious doubts on claims that the fourth has been free of recombination, they were limited by the few number of loci sequenced as well as the lack of extensive outgroup sequence needed to test hypotheses regarding modes of selection. Here, we present expanded fourth chromosome population data sets for three closely related species of the D. melanogaster subgroup: D. melanogaster, D. simulans, and D. yakuba. The sister species, D. melanogaster and D. simulans, are human commensals and are found worldwide. They are estimated to have shared a common ancestor ∼2–3 million years ago (Ma). D. yakuba is limited to the African continent and is estimated to have shared a common ancestor with D. melanogaster and D. simulans ∼10 Ma (Kliman et al. 2000; Drosophila 12 Genomes Consortium 2007). We have carried out detailed analyses of recombination and have couched the data within previously published data sets, which have so far excluded the fourth, in order to test selection hypotheses for it. Our main conclusion is that recombination, while low, is a nonnegligible factor. However, despite the presence of recombination, the severity of its reduction has rendered selection relatively inefficient for all three species. Evidence for this is presented at both the polymorphism and the divergence levels.
Materials and Methods
Sequence Data
Eighty gene regions were targeted for polymerase chain reaction (PCR) and sequencing along the right arm of the fourth chromosome from D. melanogaster, D. simulans, and D. yakuba (supplementary table 1, Supplementary Material online). Primers were based on D. melanogaster's genomic Release 5 sequence. The D. melanogaster lines came from Ecuadorian and North American lines, the D. simulans lines were from worldwide samples, and the D. yakuba lines comprised Ivory Coast, Brazzaville (Congo), and Cameroon lines (supplementary table 2, Supplementary Material online). DNA alignments were generated using clustalW2 (Larkin et al. 2007) and RevTrans 1.4 (Wernersson and Pedersen 2003). All were manually inspected. Due to the number of loci targeted and difficulties in sequencing loci from the repeat-rich fourth chromosome, we were not able to resequence all putative single nucleotide polymorphisms (SNPs) in order to eliminate the possibility of sequencing errors. For this reason, we have removed singletons from some (but not all) analyses.
To assign chromosome position, the sequenced regions were BLATed against the sequenced genomes using the UCSC BLAT server (http://genome.brc.mcw.edu/cgi-bin/hgBlat?command=start). All regions were easily assigned with exception of 11 D. simulans loci (ci, pan, Crk, CaMKI, bip2, mav, bt, unc, CG1748, CG9935, and CG1970). These regions were found to either not have hits or be out of order relative to both D. melanogaster and D. yakuba. Because no previous rearrangements have been reported in this species other than the inversion of the whole arm in D. melanogaster, which would not disrupt internal synteny, we placed these regions in their syntenic region relative to the two other species, exactly between the upstream and the downstream genes. To assign the sequence regions as either coding or noncoding, as well as each base as either silent or replacement, we used all reported D. melanogaster isoforms for these loci that were available in FlyBase's data set (http://www.flybase.org/, the FB2008_07 CDS data set was downloaded for the fourth chromosome). Additional genome information and sequence data were obtained from D. simulans' Release 1.0 and D. yakuba's Release 2.0.
Population-Level Tests and Summary Statistics
To investigate the patterns of polymorphism that exist along the fourth chromosome and to test for skews in the nucleotide site frequency spectra for these three species, we estimated several summary statistics for our sequence and haplotype data. For these analyses, we have conservatively excluded indels. The diversity estimates, θπ (Nei and Li 1979) and θw (Watterson 1975), were calculated using the “compute” program in the libsequence library (Thornton 2003). These estimates were made for the full loci (without regard to coding and noncoding regions) and for the loci partitioned into coding and noncoding regions. In addition, we estimated θπ and θw for silent and replacement sites (θπsil, θπrep, θwsil, and θwrep, respectively) within our coding regions using the “polydN/dS” program in the libsequence library (Thornton 2003). We compared D. melanogaster's fourth chromosome replacement over silent polymorphism with 225 loci from chromosome 3 which were available from the previous published data set of Shapiro et al. (2007). The same comparison was made between D. simulans's fourth and the rest of its genome using 11,445 genes from data set of Begun et al. (2007; their supplementary table 1, Supplementary Material online), after removing ten extreme outliers. The statistics Tajima's D, Fu and Li's F, and Fu and Li's D were also calculated using the compute program in the libsequence library (Thornton 2003). To test the null hypothesis that there is no variation in nucleotide diversity across the regions we sequenced, we calculated a goodness-of-fit statistic introduced by Kreitman and Hudson (1991), which is based on the observed and expected number of segregating sites within each of the sequenced regions. We partitioned D. melanogaster's haplotype data into the two dimorphic groups using Structurama (Huelsenbeck and Andolfatto 2007). For the purpose of illustrating the dimorphism in Supplementary figure 10 (see Supplementary Material online), we set the number of populations equal to 2 (model numpops = 2, mcmc ngen = 100,000, samplefreq = 100). Structurama was also used to investigate the possibility of association between haplotypes and sample locale by carrying out analyses under the models and model settings found in supplementary table 16 (see Supplementary Material online). Tests of neutrality based on haplotype configurations were carried out with haploconfig (Innan et al. 2005) (http://rosenberglab.bioinformatics.med.umich.edu/software.html).
Divergence
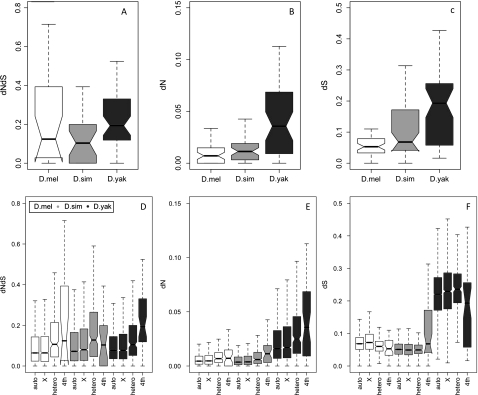
We measured the divergence of coding regions by estimating the number of nonsynonymous substitutions per nonsynonymous sites over the number of synonymous substitutions per synonymous sites (dN/dS) between orthologous sequences within our data set. Two types of alignments were available in our data set: those with two species and those with three species. Alignments overlapping less than 207 bp long were excluded. In total, we had 26 coding regions aligned between two species and 27 coding regions aligned between three species. Estimates of dN/dS were made using codeml within the PAML 4 package (Yang 2007). Pairwise estimates were generated for the two-species alignments (runmode= −2). For the three-species alignments, branch-specific (runmode =0) estimates were generated by inputting an unrooted species tree: D. melanogaster, D. simulans, and D. yakuba. Confidence intervals (CIs) were estimated using the standard error outputted by codeml (get SE = 1). To compare our divergence estimates with those from nonfourth chromosome regions, we included estimates for the same branches from Begun et al. (2007) (divergence data from their supporting data sets 1 and 6 were parsed for this purpose). Analyses of variance (ANOVAs) for the dN/ dS values were carried out on the log-transformed data, but untransformed values were plotted in figure 4.
FIG. 4.
Box plots summarizing divergence comparisons. For all comparisons the ratio dN/dS is also broken down to display dN and dS alone. Top panels are between species comparisons of the fourth chromosome. Bottom panels are within species comparisons of the fourth chromosome (4th) to other regions of the genome experiencing a range in the amount of recombination (auto = nonheterochromatic autosomal loci, X = X chromosome loci, hetero = heterochromatic autosomal loci), as defined in Begun et al. (2007).
Combining Divergence and Polymorphism
Individual MK tests (Mcdonald and Kreitman 1991) were carried out on our coding regions using the program “MKtest” (Thornton 2003) with the orthologous outgroup sequence extracted from FlyBase using D. melanogaster's gene IDs (D. melanogaster—R5.17, D. simulans—R1.3, D. yakuba—R1.3, and D. sechellia—R1.3). Estimates of α were made using the MK test v2.0 package (http://tree.bio.ed.ac.uk/software/mktest/) (Welch 2006; Betancourt et al. 2009), on groups of loci all sharing the same outgroup species. For the maximum likelihood (ML) estimation (Welch 2006; Betancourt et al. 2009), all loci took a single α value, which was estimated from the data (−a 1) and allowed θ to vary between loci (−p 2). All CIs were generated from 10,000 bootstraps (−P −10,000). For the heuristic estimators of α (−a 999), CIs based on 10,000 bootstraps were automatically outputted. We have excluded estimates of α proposed by Smith and Eyre-Walker (2002) because this estimate is known to provide overestimates if applied to data with many loci, which have low synonymous polymorphism (Smith and Eyre-Walker 2002; Welch 2006), which is true of our data set.
Codon Usage
We measured codon usage by estimating the effective number of codons (ENC; Wright 1990) using the codonw package (http://codonw.sourceforge.net/). To compare our fourth chromosome ENC data to those of normally recombining chromosomes, we parsed the ENC values previously estimated from ∼5,500 loci from chromosomes two and three from D. melanogaster's, D. simulans's, and D. yakuba's genomes (Heger and Ponting 2007). X chromosome loci were excluded because it has been shown that this chromosome possesses higher codon bias than the autosomes and lacks a positive correlation between recombination rate and codon usage bias (Singh, Arndt, et al. 2005; Singh, Davis, et al. 2005).
Recombination
We carried out several analyses to investigate the extent to which recombination has shaped the fourth chromosome. Calculations for simple two loci estimates of linkage disequilibrium (LD) (r2, Hill and Robertson 1966 and D', Lewontin 1964) were carried out using the “genotype” function within R's genetics package version 1.3.2 and visualized with R's LDheatmap package version 0.2–6. Regression analyses of LD over distance were carried out using the lm function in R 2.9.0 (http://www.R-project.org). The permutation tests for the significance of the r2 values were carried out by collecting the estimates for each of 10,000 site-permuted haplotypes. The minimum number of recombination events along the chromosome arm was estimated by the method of Hudson and Kaplan (Rm; Hudson and Kaplan 1985) and the method of Myers and Griffiths (Rh; Myers and Griffiths 2003), using the RecMin software (http://www.stats.ox.ac.uk/∼myers/RecMin.html).
In addition to the above estimates, we calculated the population estimate of LD, ρ = 4Ner, where Ne is the effective population size and r is the per generation, per base, rate of crossing over, and the relative rate of gene conversion, f = C/ρ, where C = 4Ne c and c is the per generation per base conversion rate. These calculations were carried out using the composite likelihood approach (Hudson 2001) with the maxhap software (http://home.uchicago.edu/rhudson1/source/maxhap.html). The points on the grids that we searched over for D. melanogaster and D. simulans were ρ = 0–0.0001, incrementing by 0.000001 and f = 0–600, incrementing by 12. The values for D. yakuba were ρ =0–0.0001, incrementing by 0.000001 and f =0–1,500, incrementing by 30. Maxhap was also used for jackknife resampling, where we set the tract length equal to the value that produced the ML from above.
We also implemented a second method for the estimation of ρ and f, which is based on a rejection sampling scheme introduced by Padhukasahasram et al. (2004), in C using the GNU Scientific Library (http://www.gnu.org/software/gsl/). Because conversion is expected to affect LD only over short distances, this method is most sensitive to estimating conversion if the distance between the outer SNPs for the two patterns is kept short. The further apart these SNPs, the more the method becomes an estimate of crossing over (Padhukasahasram et al. 2004; Padhukasahasram et al. 2006). We calculated the number of triplets and quadruplets and the frequencies of pattern a and pattern b (see Padhukasahasram et al. 2004) from all three species with the outer SNP distances equal to 5, 10, 12, 15, 25, and 50 kb. Because D. simulans is the only species that had nonzero values for these summary statistics at relatively close outer SNP distances (10 kb), we carried out this rejection method on D. simulans alone. Initially, we simulated data sets sparsely over a grid of ρ values from 0 to 70 and f values from 0 to 900. Based on these results, we then narrowed our grid to ρ = 4–50, incremented by 2, and f =50–650, incrementing by 50. Seven thousand replicates were simulated for each point on the grid. We accept simulated data sets if the frequency of patterns a and b were within 20% of the empirical values (pattern a = 0.019–0.029, pattern b = 0.020–0.030).
To facilitate simulating the structure of the data set we have (small fragments spaced over a reasonably large chromosomal region), we used a modified version of ms (Hudson 2002), which we call msREG (Tomoyuki Kado, unpublished). Essentially, the modifications involve inputing the coordinates of the regions sequenced and then moving any recombination between those regions that arises over the genealogy to the border of the closest region end. It also ignores any conversion event that occurs between regions. This has the effect of maintaining the linkage generated in the simulation but saves computation time by ignoring “unobserved” events between regions.
Based on the low ρ values that we estimated for these three species, two additional questions arise: 1) Are these values significantly greater than 0; are the data compatible with no recombination? and 2) Could conversion alone account for our low values of ρ? To address the first question, we simulated data sets without gene conversion (C = 0) over a range of 4Ner values and asked at what values of ρ could we significantly reject ρ = 0. We simulated 10,000 data sets for each 4Ner value using msREG and estimated ρ using maxhap as described above. Because we excluded singletons for our recombination estimates (above), we also removed singletons from our simulated data. The removal of singletons, both in our empirical data and simulations, violates the standard neutral model that the composite likelihood method assumes (Hudson 2002); however, previous simulation studies have shown it to be robust to violations where SNP ascertainment shifts the SNP frequency spectrum toward intermediate frequency (Smith and Fearnhead 2005). To address the second question, we simulated data sets with no crossing over (ρ = 0) over a range of values for C. The conversion tract was set to values compatible with those estimated from the true data (D. simulans = 400 bp, D. melanogaster = 300 bp, and D. yakuba = 300 bp). The grid varied with each species but was reasonably fine (∼61 points for ρ and ∼45 points for f), and the upper limits were set so that only very rarely did the ML estimate involve them. We simulated 5,000 data sets for each 4Ner value (the computation time was considerably longer for these high C values than for the high r values) using msREG and removed singletons and estimated ρ using maxhap as described above.
Results and Discussion
Sequencing Results
Eighty orthologous gene regions were targeted for PCR and sequencing from D. melanogaster, D. simulans, and D. yakuba (supplementary table 1, Supplementary Material online). In total, 20 lines were used from each species (supplementary table 2, Supplementary Material online). The total number of regions that were successfully sequenced was 58 for D. melanogaster, 64 for D. simulans, and 55 for D. yakuba. The average length of these reads for all three species was ∼700 bp, amounting to ∼40 kb of total DNA sequence from each line. Recurrent mutations were not a major issue as only a single triallelic site was found in D. simulans' Crk locus, and a single triallelic site in D. yakuba's yellow-h locus. In addition, there was no shared polymorphism between species. If all 20 lines are included, and singletons are included, the total number of SNPs equals 87 for D. melanogaster, 181 for D. simulans, and 96 for D. yakuba. If singletons are excluded, the counts drop to 55, 98, and 38, respectively. However, after manual inspection of all sequenced regions, it was clear that four lines from D. melanogaster and five lines from D. yakuba were not completely inbred and thus had residual heterozygosity. Not much data are lost by eliminating the heterozygous lines. If only homozygous lines are considered, and if singletons are retained, the total number of SNPs equals 84 for D. melanogaster, 181 for D. simulans, and 81 for D. yakuba. If singletons are removed from the homozygous lines, the remaining number of SNPs is 54, 96, and 35, respectively. For the analyses presented below, unless otherwise stated, only the homozygous lines excluding singletons have been used (see Materials and Methods; supplementary tables 3–5, Supplementary Material online). DNA sequences have been submitted to GenBank.
Recombination Has Played a Historical Role along the Fourth
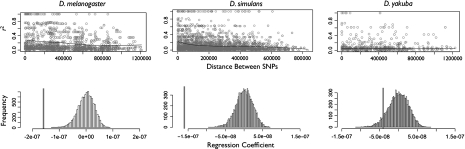
One of the central aims of this analysis was to more thoroughly examine the role that recombination has had in shaping patterns of polymorphism along the fourth chromosome. In this study, we significantly expanded on the number of loci as well as added D. yakuba samples in order to provide additional tests of the previous claims that recombination occurs along the fourth at an appreciable frequency. We first examined the amount of LD present over the loci using two statistics, r2 (Hill and Robertson 1966) and D' (Lewontin 1964) (Supplementary fig. 1, Supplementary Material online). Visual inspection of heatplots of these statistics qualitatively indicates blocks of intermediate and high LD interspersed with lower LD, possibly focused near the center of sequenced region. To provide a better measure of this, we plotted r2 against SNP distance and computed the regression coefficients (fig. 1). If recombination is present, LD should decrease with SNP distance, and a negative slope would be observed. All three species exhibited a negative slope over increasing SNP distance (D. melanogaster's regression coefficient =−1.58 × 10−7, D. simulan's regression coefficient = − 1.61 × 10−7, and D. yakuba's regression coefficient = − 4.06 × 10−8). To determine the significance of the slope, we permutated the sites and recalculated the regression. This should remove the effects of any true linkage over distance and provide a null distribution against which to compare our observed values. Comparisons of our true estimates to the empirical distributions suggested that the decay with distance is very significant for both D. melanogaster (P = 0) and D. simulans (P = 0) but marginally significant for D. yakuba's (P = 0.071) (fig. 1).
FIG. 1.
Upper panel: Regression of the recombination estimate r2 over SNP distance. Lower panel: Empirical distribution of r2 values for permuted r2 versus distance samples. Dark vertical line indicates the true estimate.
To calculate a lower bound on the number of recombination events that have occurred in the genealogy of our samples, we computed the statistics Rm (Hudson and Kaplan 1985) and Rh (Myers and Griffiths 2003). These two statistics were both nonzero (the minimum number of incompatabilities for the three species being 12 in D. melanogaster's and the maximum being 51 in D. simulans), providing another line of evidence that recombination has occurred within D. melanogaster and D. simulans as well as D. yakuba (table 1). These events were not limited to intergenic regions: 17 of 51 were inferred to be intragenic within D. simulans, 4 of 22 within D. yakuba, and 6 of 19 within D. melanogaster (Supplementary fig. 2, Supplementary Material online).
Table 1.
Minimum Estimates for Crossing over for Drosophila melanogaster’s, D. simulans’s, and D. yakuba’s Fourth Chromosome. The Lower Bound Estimates Were Estimated Assuming the Length of the Euchromatic Region of the Chromosomes Right Arm is 1,156 kb for the Three Species
| D. melanogaster | D. simulans | D. yakuba | |
| Rm | 12 | 28 | 14 |
| Rh | 19 | 51 | 22 |
| Lower bound on Rm density | 0.010/kb/chromosome | 0.024/kb/chromosome | 0.012/kb/chromosome |
| Lower bound on Rh density | 0.016/kb/chromosome | 0.044/kb/chromosome | 0.019kb/chromosome |
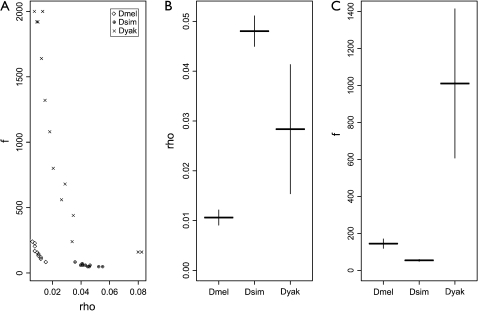
Recombination events can be resolved either by crossing over (reciprocal) or by conversion (nonreciprocal) (Szostak et al. 1983), and the relative usage of the two pathways can be estimated with polymorphism data. We estimated the population recombination parameters ρ(4Ner) and f, the ratio of conversion to crossing over (f = C/ ρ, where C = 4Nec and c is the per generation per base conversion rate), using two different methods. We first obtained estimates using the composite likelihood approach (Hudson 2001). Consistent with the inferred minimum number of recombination events, ρ was estimated to be greatest for D. simulans (0.000085/bp/generation or ∼80/chromosome/generation), followed by D. yakuba (0.000024/bp/generation or ∼33/chromosome/generation), and then by D. melanogaster (0.000012/bp/generation or ∼16/chromosome/generation) (Supplementary fig. 3, Supplementary Material online). For each of the models we ran, those with gene conversion always provided higher likelihoods. The conversion tract lengths that generated the MLs were 400 bp for D. melanogaster and D. yakuba and 300 bp for D. simulans. At these tract lengths f = 144 for D. melanogaster, f = 60 for D. simulans, and f = 960 for D. yakuba.
There is evidence suggesting that differences between D. melanogaster and D. simulans, and possibly D. yakuba, for both ρ and f are detectable. Figure 2 displays the pseudovalues from jackknife resampling and CIs surrounding the jackknife means for the two estimates. The ρ estimates have been scaled by the respective species' silent diversity (θwsil) to control for differing Ne. As can be seen by the large CIs for D. yakuba's estimates in figure 2b and c, and the spread of points in figure 2a, there are large variances associated with this species. This is not surprising as there are few SNPs for it. However, D. melanogaster and D. simulans both have tighter CIs, and although we remain cautious due to the small sample size, they imply that the rate of crossing over and conversion may be different between at least these two species, with D. simulans experiencing a higher ρ but lower f. This is also consistent with D. simulans having significantly higher nucleotide diversity. We also note that for each species, the CIs from the jackknife resampling do not include zero, lending support for recombination.
FIG. 2.
Jackknife resampling of the composite likelihood estimates for the population recombination rate, ρ, and the ratio of conversion to crossing over, f. To facilitate comparisons across species with different effective population sizes, ρ has been scaled by the respective species silent diversity, θs. (A) Pseudovalues for the joint distributions of ρ and f. (B) Jackknife means and 95% CIs for ρ. (C) Jackknife means and 95% CIs for f.
To investigate the power that the composite likelihood method has in discriminating very low estimates of ρ from zero, we simulated coalescent events to match our data structure under both a pure recombination model (C =0) and a pure conversion model (ρ = 0) (see Materials and Methods). We note that this approach is not completely satisfactory as these are coestimated variables, and it is currently an active area of research to independently estimate each. Nonetheless, we can still ask whether conversion alone could account for our ρ estimates, and at what values either ρ or C can produce ρ estimates significantly different than zero. The results of our pure recombination simulations suggest that the probability of observing ρ = 0 is significantly unlikely around the simulated ρ value of ∼0.000004/bp/generation (∼4 crossovers/chromosome/generation) for all species (Supplementary fig. 4, Supplementary Material online). This is three times lower than the smallest estimate that we observed from D. melanogaster of 0.000012/bp/generation (12 crossovers/chromosome/generation). Similarly, simulations under the pure conversion model only rarely produced ρ estimates approaching those that we obtained with the true data set when C is set unbelievably high (4Nec > 0.03) (Supplementary fig. 5, Supplementary Material online). These two simulation results lend additional support that our ρ estimates are significantly different than zero.
Due to the conversion bias for the fourth, we implemented a second approach to estimate f that used a rejection sampling scheme based on incompatibilities between triplet and quadruplet sets of SNPs (Padhukasahasram et al. 2004). Because only the D. simulans data provide reasonably spaced SNPs for this method, we limited the approach to this species (supplementary table 6, Supplementary Material online). We simulated genealogies over a grid of ρ and f values and accepted data sets if the frequencies of matches to the triplet and the quadruplet patterns fell within 20% of our empirical values. The resulting posterior distribution had a maximum at f = 250, and ρ = 0.000019/bp (∼18/chromosome), though the distribution was fairly flat on a ridge with ρ = 10–18 and f > 200 (Supplementary fig. 6, Supplementary Material online). Because this method is tailored more for estimating conversion, it may not be surprising that its estimate of ρ is less than the composite likelihood estimate above; however, it suggests that f may be higher than 60.
In summary, our recombination analyses motivate a view of the fourth in which rare but appreciable recombination is shared by the three species, as is the predominance of conversion relative to crossing over (f > 1). Limited data within Drosophila for both the tract length and the relative rate of gene conversion to crossing over limit our ability to make a strong comparative statement regarding our estimates for the fourth. That said, estimates from multiple organisms (including Drosophila) suggest that the fourth's tract lengths fall within the previously reported range (∼50–2 kb; Hilliker et al. 1994; Frisse et al. 2001; Jeffreys and May 2004; Yin et al. 2009). And compared with f estimates from the su(s) and su(wa) loci from D. melanogaster's X chromosome, which range from 7.1 to 48 (Gay et al. 2007; Yin et al. 2009), a somewhat higher ratio for the fourth might be suggested, even when excluding D. yakuba.
Polymorphism Data Provide Evidence that the Fourth Has Experienced Relaxed Purifying Selection
As we have shown, recombination is low but present along the fourth chromosome, and linkage is fairly strong but not complete across the loci we sampled. Because the linkage effects amount to a reduced Ne, the extent of the effects can be measured by estimates of nucleotide diversity. Two estimates of nucleotide diversity were calculated, θπ (Nei and Li 1979) and θw (Watterson 1975). These estimates were made for three different data divisions: 1) for the unpartitioned loci (without regard to coding potential), 2) for the loci divided into coding and noncoding regions, and 3) for the silent and replacement sites of the coding regions. Overall, we observed low levels of diversity, with many sequenced regions having no segregating polymorphism (supplementary tables 7–9, Supplementary Material online). The mean diversity for D. melanogaster was θw = 0.00062, θπ = 0.000614, for D. simulans θw = 0.00114, θπ = 0.00092, and for D. yakuba θw = 0.00065, θπ = 0.00049. Considering the unpartitioned loci “within” species, there was no evidence of heterogeneity in diversity levels between loci (D. melanogaster χ2 [57, N =58] = 42.68, P =0.93; D. simulans χ2 [62, N = 63] =70.39, P =0.22; and D. yakuba χ2 [54, N = 55] = 49.85, P = 0.64; supplementary tables 10–12, Supplementary Material online).
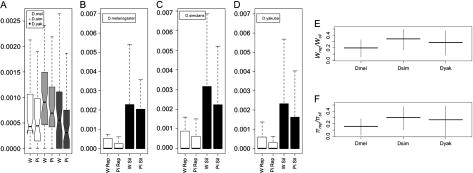
Despite these low estimates, when comparing the unpartitioned loci “between” species, ANOVA showed a significant species effect on θw, F(2,174) = 8.60, P = 0.0003, and on θπ, F(2,174) = 5.91, P = 0.0033; figure 3a. Posthoc Tukey's Honestly Significant Difference Tests for these two statistics showed that D. simulans's values were significantly higher (adjusted P < 0.05 for all comparisons involving D. simulans; supplementary table 13, Supplementary Material online). Because our population sampling for D. simulans was the most diverse of the three species (supplementary table 2, Supplementary Material online), it might be questioned whether these elevated estimates are representative of any one of the populations. However, these results are consistent with previous findings for the fourth (both local and worldwide samples) as well as the rest of the genome and likely reflects a larger Ne (which could also be influenced by its potentially higher ρ; Akashi 1995; Wang et al. 2004).
FIG. 3.
Comparisons of nucleotide diversity estimates. (A) Boxplot of Wattersons θ (W) and π (Pi) for the unpartitioned loci between the three species. (B–D) Boxplots of replacement (rep) and silent (sil) diversity. For each species, there is significantly greater silent diversity. (E–F) Comparisons of the replacement/silent diversity between the three species. Vertical bars represent the 95% bootstrap CI.
These average estimates of nucleotide diversity for D. melanogaster and D. simulans were lower than previous estimates from a smaller number of loci, where the mean θπ for D. melanogaster was 0.0021 and for D. simulans was 0.0024 (Wang et al. 2002; Wang et al. 2004). Much of this difference can be accounted for by the larger number of loci for which no segregating sites were obtained in our current data set (supplementary tables 7–9, Supplementary Material online). However, in D. melanogaster, there are some striking differences between our current samples that were also sequenced in an earlier study, for example, unc-13 and toy, which were both found to be approximately five times higher than our current estimates. These differences likely result from the fact that the previous study came from a worldwide population instead of lines mostly derived from local North American ones (North Carolina) as we have here.
Comparison between coding and noncoding regions indicated that there is no significant difference for θπ or θw between these partitions for any of the species and regardless of whether singletons were included or not (all Wilcoxon P > 0.1; fig. 3). This likely can be explained by the fact that most of our data are coming from the coding regions as well as the fact that the noncoding regions are either intronic or directly 5′ or 3′ of the genes where purifying selection for regulatory elements may exist. However, when the data are further broken down into the replacement and silent sites of the coding regions, there was significantly more variation at the silent sites for all three species, suggesting that purifying selection is operating more strongly on amino acid changing substitutions (all Wilcoxon tests for θπ and θw;P < 0.005) (fig. 3b–d). The comparisons of replacement diversity over silent diversity indicated that there are no statistically significant differences between species (fig. 3e and f; mean ratios for D. melanogaster: θwrep/θwsil = 0.199, θπrep / θπsil = 0.160; D. simulans: θwrep / θwsil = 0.343, θπrep / θπsil = 0.300; and D. yakuba: θwrep / θwsil =0.285, θπrep / θπsil = 0.264; ANOVA for θwrep / θwsil F(2,68) =2.59, P = 0.082 and ANOVA for θπrep / θπsil F(2,68) =2.76, P = 0.071).
Though these values are indicative of selective constraint, they are higher than the values from data sets containing loci from normally recombining chromosomes. For example, data from D. melanogaster's chromosome 3 (Shapiro et al. 2007) provided a significantly lower mean for θwrep / θwsil (0.133, 95% bootstrapped CI =0.092–0.173) and a marginally lower mean for θπrep / θπsil (0.119, 95% bootstrapped CI = 0.077 –0.161). Comparisons with D. simulans' genomic data (Begun et al. 2007) also indicate a significantly lower mean θπrep / θπsil for the X (0.113, 95% bootstrapped CI = 0.103 –0.1233), chromosome 2 (0.087, 95% bootstrapped CI = 0.083 –0.091), and chromosome 3 (0.090, 95% bootstrapped CI = 0.083 –0.091). Genomic population data are not currently available for similar D. yakuba comparisons.
This elevation in θrep / θsil seen on the fourth is consistent with a reduction in the efficiency of selection resulting in an increased number of mildly deleterious mutations segregating. However, in principle—though unlikely given the results above—it is also possible that positive selection could be driving the excessive number of nonsynonymous polymorphism. If positive selection is responsible, and if the selective events were recent, we would expect to see this reflected in tests of the nucleotide site frequency spectrum and possibly with the use of divergence data (below). To test our population samples for departures from neutrality, we calculated Tajima's D (Tajima 1989), Fay and Wu's H (Fay and Wu 2000), Fu and Li's F (Fu and Li 1993), and Fu and Li's D (Fu and Li 1993). No individual loci exhibited a significant skew for any of these statistics, regardless of whether singletons were excluded or included (supplementary tables 7–9; Supplementary figs. 7–9, Supplementary Material online). We note that if singletons are included, and all regions are considered together, there is a significant excess of negative Tajima's D values for D. simulans and D. yakuba (Supplementary figs. 8–9; supplementary table 14, Supplementary Material online). For D. yakuba, this excess is contributed to by the coding regions (binomial test, P = 0.002) and, to a marginal extent, the noncoding regions (binomial test, P = 0.057). For D. simulans, an excess is only observed when the two categories are combined (binomial test, P = 0.01349). Though the mean Tajima's D was negative for D. melanogaster (−0.011), consistent with its well-recognized genome-wide trend (Haddrill et al. 2005; Glinka et al. 2003; Andolfatto 2007), there was not an excess of either negative or positive values (all binomial tests P > 0.6; Supplementary fig. 7, supplementary table 14, Supplementary Material online).
The observation that there is an excess of negative Tajima's D values for D. simulans and D. yakuba suggests an overall excess of low-frequency mutations. Though this might be interpreted as weak evidence for positive selection, it could also result from demographic factors, background selection, or a combination of these factors. Negative Tajima's D values have been reported for a number of Drosophila species, including those studied here (Kliman et al. 2000; Bachtrog and Andolfatto 2006), and simulation studies have shown that background selection in areas of high linkage can likewise lead to negative Tajima's D values (Kaiser and Charlesworth 2008). Given the results from the previous analyses and those below, we argue that our summary statistics of the frequency spectrum do not provide support for positive selection to have acted recently at any of the individual loci and do not support a sweep model for the chromosome of these species as a whole.
The dimorphic haplotypes that were previously observed in D. melanogaster near the CG1793-toy loci (Wang et al. 2002) were also identified in this data set (Supplementary fig. 10, Supplementary Material online). The previous identification of these haplotypes was surprising not only because of the nonrecombining status of the chromosome but also because it was unclear how they have been maintained. Tests of neutrality based on the number of haplotypes present in the sample of Wang et al. (2002) suggested nonneutral forces such as balancing selection might be responsible, but there were no significant skews of the nucleotide site frequency spectrum supporting the existence of balancing selection in the recent past (Wang et al. 2002). Interestingly, our data demonstrate that the haplotype extends well beyond the CG1793-toy region and appears to be present, though with decreasing strength, across most of the chromosome (Supplementary fig. 10, Supplementary Material online). However, unlike previous results (Wang et al. 2002), when we examined the statistical properties of the haplotypes within subregions of our data set using the absolute frequency of the most common haplotype (M; Depaulis and Veuille 1998), the total number of unique haplotypes (K; Depaulis and Veuille 1998), and the haplotype diversity (H; Hudson et al. 1994), we did not observe evidence for selection (all test P > 0.1; supplementary table 15, Supplementary Material online). It should be recognized, though, that the previous claims of balancing selection were restricted to a Israelian population where local selective or demographic forces could differ from our current sample (Wang et al. 2002). Overall, it remains unclear why these haplotypes remain but because our samples all come from North American (and most from North Carolina), they are retained in close geographic proximity. Although evidence of admixture has not been reported for the rest of the D. melanogaster genome, it is possible that admixture combined with low levels of recombination may be responsible. Because admixture would produce a genome-wide effect, if such events have occurred and their signatures still exist off of the fourth, they should be readily detectable for these same lines using the whole genome data soon to be released for the Drosophila Genetic Reference Panel (http://service004.hpc.ncsu.edu/mackay/Good_Mackay_site/DBRP.html).
Similar to what was seen for D. melanogaster, variable haplotype clusters could be identified within the D. simulans and D. yakuba data sets, depending on the model and parameters used (see Materials and Methods, supplementary table 16, Supplementary Material online). However, as in D. melanogaster, haplotypes were frequently assigned across sample locales and no clear associations were discerned (data not shown).
Divergence Data Provide Evidence that the Fourth Has Experienced Relaxed Purifying Selection
If the fourth has experienced a reduction in the efficiency of selection, as was suggested by our polymorphism data, we would expect there to be an accelerated rate of protein evolution due to the fixation of mildly deleterious substitutions between species when compared with loci from regions of higher recombination. On the other hand, if positive selection has operated, we might observe a significant excess of nonsynonymous substitutions per nonsynonymous site divided by the number of synonymous substitutions per synonymous site (dN / dS > 1). Divergence data, therefore, can provide another test of the relaxed constraint hypothesis.
In order to investigate the evolutionary dynamics across the coding portion of the fourth chromosome, we generated alignments for all overlapping coding regions we sequenced for which a reliable alignment could be made. To maximize the number of three-species alignments, we extracted the missing regions within our data set from the corresponding genome database (see Materials and Methods). In total, our data set comprises 46 three-species alignments, along with five pairwise alignments. From these alignments, we estimated branch-specific dN/dS values.
In general, we observed purifying selection (dN /dS < 1) along all branches of the topology for all three species (fig. 4a, Supplementary fig. 11, Supplementary Material online). Focusing on the three-species alignments, as expected given the greater divergence, D. yakuba had significantly higher dN and dS values than either D. melanogaster or D. simulans (fig. 4b and c). However, the mean dN/ dS was significantly less than 1 for all branches, and not significantly different from each other, with mean dN/dS equal to 0.213 for D. melanogaster, 0.257 for D. simulans, and 0.318 for D. yakuba (ANOVA F(2,130) = 0.490, P = 0.613) (fig. 4a, Supplementary fig. 11, Supplementary Material online). The mean dN/dS for the pairwise alignments was 0.16 (n = 5, supplementary table 17, Supplementary Material online). Though we observed four coding regions that had dN/ dS values >1 (pho, CG1674, lgs, and Sox102F), each had very high standard errors. In addition, for three of these loci for which we could carry out MK tests, none were significant (data not shown).
To ask if these divergence values were different than those found in regions of increased recombination, we compared them with divergence estimates from nonheterochromatic autosomal loci, X chromosome loci, and heterochromatic loci (see Methods and Materials). We observed highly significant heterogeneity in the divergences between genomic regions for all species (fig. 4d–f, supplementary table 18, Supplementary Material online). The most striking differences came from comparisons of dN and dN/dS from loci on the fourth or within heterochromatic regions to those on the X or (nonheterochromatic) autosomal loci. A consistent trend for dS was not observed, however: For D. melanogaster and D. yakuba, dS for the fourth and heterochromatic loci were reduced when compared with most of the other regions, whereas in D. simulans, there was only evidence for an elevated dS between the fourth and all other regions. Depressed dS values for D. yakuba's and D. melanogaster's fourth are similar to previous reports (Haddrill et al. 2007) and likely have to do with the specific estimator employed in PAML (Bierne and Eyre-Walker 2004; Haddrill et al. 2007).
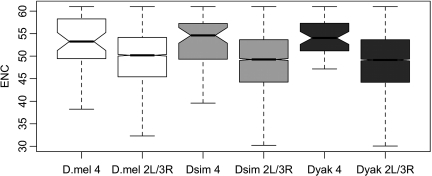
A related expectation of loci within regions of reduced recombination is that levels of codon bias should decrease (Akashi 1995). This can result from the reduced efficiency of selection for preferred codon usage, and it has been shown that genomic heterogeneity in measures of codon bias is positively correlated with cross over rates. Not surprisingly (Powell and Moriyama 1997), codon bias (using the ENC; Wright 1990) is significantly decreased within our fourth chromosome data set when compared with loci from chromosome 3 and 2 (D. melanogaster, Wilcoxon P = 3.3 × 10−5; D. simulans, Wilcoxon P = 2.5 × 10−6; and D. yakuba, Wilcoxon P = 3.4 × 10−9; fig. 5).
FIG. 5.
Box plots comparing estimates of codon bias (effective number of codons, ENC) from chromosomes 2 and 3 to estimates from the fourth chromosome.
The general conclusion from our divergence results provides another line of evidence in support of the hypothesis that the fourth has experienced relaxed constraint as a result of its reduced levels of recombination and argue against positive selection acting during the divergences of these three species. This latter point is bolstered by a recent study between D. melanogaster and D. yakuba, which also found evidence for an increased rate of deleterious fixation within regions of low or no recombination (Haddrill et al. 2007).
Combining Polymorphism and Divergence Data Indicates a Reduced Role for Positive Selection
The combined use of polymorphism and divergence data has significantly contributed to increasing evidence that a surprisingly large proportion of fixed differences between Drosophila species has been driven by positive selection (Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Sella et al. 2009). MK tests, and its derivatives used to estimate the proportion of amino acid substitutions driven to fixation by positive selection (α), have provided important methodological advancement toward quantifying these amounts. To date, studies have provided estimates of α that are roughly ∼0.45 for D. simulans and D. melanogaster, thus suggesting that ∼45% of amino acid substitutions between these species have been positively selected (Mcdonald and Kreitman 1991; Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Welch 2006; Andolfatto 2007; Begun et al. 2007; Haddrill et al. 2008). Variation in α estimates results from the particular method and outgroup used (Bierne and Eyre-Walker 2004). Given our previous analyses providing a lack of evidence for positive selection and a reduced efficiency of purifying selection, α for the fourth chromosome should concordantly be lower than previous estimates as the samples have come from regions of higher recombination.
We have combined divergence and polymorphism data by carrying out MK tests on individual loci and also estimated α for the full chromosome region. The number of loci for the individual MK tests was 28 for D. melanogaster, 39 for D. simulans, and 22 for D. yakuba. Two of D. simulans' loci (CG1901 and CG1922) and 2 of D. yakuba's loci (CG2052 and CG2177) were significant (P <0.05); however, this number is expected given a false discovery rate of 5% (4.45).
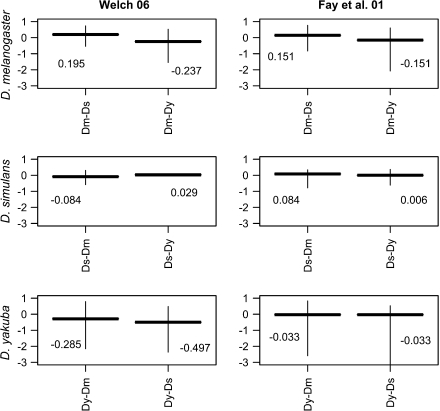
Next, two different α estimates were computed for the subset of fourth chromosome loci that share the same outgroup species (Fay et al. 2001; Smith and Eyre-Walker 2002; Welch 2006; fig. 6). Our mean estimates of α were considerably lower than the previous estimates of ∼0.45 from D. melanogaster and D. simulans when using the methods of Fay et al. (2001) and Welch (2006) and often negative (though not significantly different than zero due to the large CIs). The negative mean α values could result from both sampling errors and violations of the assumptions on which the underlying models are based—that most mutations lie in the extreme tails (either strongly deleterious or strongly beneficial)—or that most mutations are neutral (Mcdonald and Kreitman 1991; Bierne and Eyre-Walker 2004). Given the evidence in the above sections for ineffective selection acting along the fourth, these low estimates of alpha are consistent with a relaxation of selective constraint and likely reflect a higher frequency of deleterious substitutions.
FIG. 6.
Estimates of α, the proportion of amino acid substitutions driven by positive selection. The method for the two estimates is provided above each column (see Materials and Methods). Two values for each estimate are provided, each one using a different outgroup species (Dy, D. yakuba; Dm, D. melanogaster; and Ds, D. simulans). Vertical bars are 95% boostrap CIs.
The higher α estimates from autosomal and X chromosome data sets compared with our fourth chromosome data, although strongly suggestive could be further motivated by autosomal samples from the same populations from which we obtained our fourth chromosome loci, similar to what was recently done by Betancourt et al. (2009). Arguing for a stable population size for D. americana, Betancourt et al. (2009) justified pooling samples and were thus able to employ a likelihood-based test for differences in α between fourth and nonfourth loci. In doing so, they also found a reduction in α for the fourth chromosome, thus providing evidence that the reduction in the efficacy of positive selection as a result of low levels of recombination exists outside of the D. melanogaster subgroup.
Conclusions
The Drosophila fourth chromosome's peculiar biology has made it an important model for several fundamental genetic phenomena (Riddle and Elgin 2006). Here, we have focused on its status as a putatively nonrecombining chromosome and the potential effects that this has on its standing nucleotide variation and divergence. Regarding the presence of recombination, our results support it as being a historically important process for the fourth chromosomes of D. melanogaster, D. simulans, and D. yakuba. Although significantly reduced when compared with other regions of the genome that contain similar gene density, signatures of recombination are still detectable within our population genetic data. In particular, our data, and several other accounts, suggest that gene conversion is the predominant resolution of its recombination events (Jensen et al. 2002; Wang et al. 2002; Wang et al. 2004). These low levels of recombination combined with a significant conversion bias could explain the inability to identify recombination events using only physical markers. Additional cytological support for some form of recombination comes from recent work analyzing fixed and live oocytes in which heterochromatic DNA threads were found to form between homologous fourth chromosomes (and occasionally between the fourth and the X) during meiosis (Hughes et al. 2009). How the threads are resolved remains an open question; however, considering the frequency with which the threads were observed to form, considered together with infrequent crossing over, it is reasonable to suspect nonreciprocal exchanges.
By contrasting our sequence data from the fourth with data from regions of the genomes experiencing increased recombination rates, we have been able to test evolutionary predictions regarding recombination's role in either helping to efficiently fix beneficial mutations or efficiently remove deleterious ones. Our results provide striking evidence that the frequency of recombination is low enough to have rendered selection relatively inefficient. The signatures of relaxed constraint can be detected at the level of polymorphism (where there is an increased frequency of nonsynonymous mutations segregating), at the level of divergence (where coding regions have diverged more quickly), and when the two data sets are combined (where estimates of α, the proportion of nonsynonymous fixations driven by positive selection, is considerably lower). Although positive selection may be capable of driving some of these patterns, there is very little evidence from our data to evoke a nonneutral explanation; tests of the frequency spectrum, dN/ dS, and MK tests do not lead to rejections of neutrality. Demographic effects could potentially influence these analyses, especially because regions with a low Ne will be dominated by drift. However, the fact that there were no strong signatures of a bottleneck in our polymorphism data (no significantly positive Tajima's D estimates) and that the general conclusions held across all three species indicate that demographic influences are an unlikely explanation. Furthermore, similar results were recently reported for D. americana's fourth, a distantly related species, suggesting a more general trend for inefficient selection along this chromosome (Betancourt et al. 2009). The overall lack of evidence for positive selection acting on the fourth and instead strong evidence for a reduction in the efficiency of selection for four Drosophila species points toward background selection as the primary force driving the lack of nucleotide diversity. Additional support for this claim comes from a recent theoretical adjustment to the background selection model, which accounts for multiple sites that are under relatively strong selection within regions of low recombination (Kaiser and Charlesworth 2008). This modification results in levels of variability consistent with those observed for the fourth from the three species studied here as well as the fourth of D. americana (Kaiser and Charlesworth 2008; Betancourt et al. 2009).
Supplementary Data
Supplementary tables 1–18 and figures 1–11 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We would like to thank Dick Hudson and Marty Kreitman for helpful discussions and advice over several of the analyses. David Turissini kindly provided the population sequences for D. melanogaster's third chromosome. Members of the M. Long lab provided helpful comments throughout the project's completion; in particular, Margarida Cardoso Moreira who also read and provided important suggestions on earlier drafts. We are also thankful to John Welch for suggestions and correspondence over the estimates of α. This work was supported by National Institute of Health grants (R01GM065429-01A1 and R01GM078070-01A1) awarded to M.L., an NSFC key grant (30430400) and funding from the Chinese Academy of Sciences OOCS fund awarded to W.W., and grants awarded to H.I. from JSPS and the Graduate School of Advanced Studies. J.R.A. was supported by a University of Chicago Harpers Fellowship and a GHANN grant awarded to the Committee on Evolutionary Biology. JSPS's EAPSI program provided a fellowship to J.R.A. to carry out a portion of the work in the lab of H.I.
References
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at "silent" sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic K, Hawley SR. Cold Spring Harbor (NY) 2nd ed. Laboratory Press; 2005. Drosophila: a laboratory handbook. [Google Scholar]
- Bachtrog D. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in drosophila. Nat Genet. 2003;34:215–219. doi: 10.1038/ng1164. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Andolfatto P. Selection, recombination and demographic history in Drosophila miranda. Genetics. 2006;174:2045–2059. doi: 10.1534/genetics.106.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (10 co-authors. Population genomics: whole-genome analysis of polymorphism and divergence in drosophila simulans. PLoS Biol. 2007 doi: 10.1371/journal.pbio.0050310. 5:e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A, Ajioka J, Kreitman M. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics. 1991;129:1111–1117. doi: 10.1093/genetics/129.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Betancourt AJ, Presgraves CD. Linkage limits the power of natural selection in drosophila. Proc Natl Acad Sci. 2002;99:13616–13620. doi: 10.1073/pnas.212277199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Bridges C. The mutants and linkage data of chromosome four of Drosophila melanogaster. Biol Zh. 1935;4:401–420. [Google Scholar]
- Charlesworth B, Morgan M, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Morgan M. The pattern of neutral molecular variation under the background selection model. Genetics. 1995;141:1619–1632. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaulis F, Veuille M. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol Biol Evol. 1998;15:1788. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. Hitchhiking under positive darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wycoff G, Wu C-I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Baker BS. The rox1 and rox2 rnas are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Rienzo AD. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay JC, Myers S, McVean G. Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007;177:881–894. doi: 10.1534/genetics.107.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland WD, Hughes SF, Viettia DR, Hawley RS. Congression of achiasmate chromosomes to the metaphase plate in Drosophila melanogaster oocytes. Dev. Biol. 2009;325:122–128. doi: 10.1016/j.ydbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Glinka S, Ometto L, Mousset S, Stephan W, De Lorenzo D. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics. 2003;165:1269–1278. doi: 10.1093/genetics/165.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I, Charlesworth B. Genetic linkage and molecular evolution. Curr Biol. 2001;11:R684–R686. doi: 10.1016/s0960-9822(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Haddrill P, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding dna in Drosophila simulans. Mol Biol Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P, Halligan D, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Variable strength of translational selection among 12 Drosophila species. Genetics. 2007;177:1337–1348. doi: 10.1534/genetics.107.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hilliker AJ, Harauz G, Reaume AG, Gray M, Clark SH, Chovnick A. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics. 1994;137:1019–1026. doi: 10.1093/genetics/137.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton H, Kliman R, Hey J. Using hitchhiking genes to study adaptation and divergence during speciation within the Drosophila melanogaster species complex. Evolution. 1994;48:1900–1913. doi: 10.1111/j.1558-5646.1994.tb02222.x. [DOI] [PubMed] [Google Scholar]
- Hochman B. The fourth chromosome of Drosophila melanogaster. Genet Biol Drosophila. 1976;1:903–928. [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. Evidence for positive selection in the superoxide dismutase (sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. Gene genealogies and the coalescent process. In: Futuyma DJ, Antonovics J, editors. Oxford Surveys in Evolutionary Biology. Vol. 7. Oxford (UK): Oxford University Press; 1990. pp. 1–44. [Google Scholar]
- Hudson R. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson R, Kaplan N. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process prior. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Gilliland WD, Cotitta JL, Takeo S, Collins KA, Hawley RS. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000348. 5:e1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Zhang K, Marjoram P, Tavare S, Rosenberg NA. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics. 2005;169:1763–1777. doi: 10.1534/genetics.104.032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Jensen M, Charlesworth B, Kreitman M. Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster and D. simulans. Genetics. 2002;160:493–507. doi: 10.1093/genetics/160.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Charlesworth B. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 2008;25:9–12. doi: 10.1016/j.tig.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Hudson R, Langley C. The "Hitchhiking Effect" revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ, McCarter J, Wakeley J, Hey J. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics. 2000;156:1913–1931. doi: 10.1093/genetics/156.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman R, Hey J. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Hudson R. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics. 1991;127:565–582. doi: 10.1093/genetics/127.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, et al. (10 co-authors. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. Clustal W and Clustal X version 2.0. [DOI] [PubMed] [Google Scholar]
- Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V. Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci USA. 2001;98:6273–6278. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Svensson M, Stenberg P, Makitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci USA. 2004;101:9728. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Mcdonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mohr O. Genetical and cytological proof of somatic elimination of the fourth chromosome in Drosophila melanogaster. Genetics. 1932;17:60–80. doi: 10.1093/genetics/17.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Griffiths R. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B, Marjoram P, Nordborg M. Estimating the rate of gene conversion on human chromosome 21. Am J Hum Genet. 2004;75:386–397. doi: 10.1086/423451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B, Wall J, Marjoram P, Nordborg M. Estimating recombination rates from single-nucleotide polymorphisms using summary statistics. Genetics. 2006;174:1517. doi: 10.1534/genetics.106.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paland S, Lynch M. Transitions to asexuality result in excess amino-acid substitutions. Science. 2006;311:990–992. doi: 10.1126/science.1118152. [DOI] [PubMed] [Google Scholar]
- Patterson J, Muller H. Are “Progressive” mutations produced by X-rays? Genetics. 1930;15:495–577. doi: 10.1093/genetics/15.6.495f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Moriyama EN. Evolution of codon usage bias in Drosophila. Proc Natl Acad Sci USA. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Recombination enhances protein adaptation in drosophila melanogaster. Curr Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- Rice WR, Chippindale AK. Sexual recombination and the power of natural selection. Science. 2001;294:555–559. doi: 10.1126/science.1061380. [DOI] [PubMed] [Google Scholar]
- Riddle N, Elgin S. The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosome Res. 2006;14:405–416. doi: 10.1007/s10577-006-1061-6. [DOI] [PubMed] [Google Scholar]
- Sandler L, Novitski E. Evidence for genetic homology between chromosomes I and IV in Drosophila melanogaster, with a proposed explanation for the crowding effect in triploid. Genetics. 1955;41:189–193. doi: 10.1093/genetics/41.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the drosophila genome? PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000495. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J, Huang W, Zhang C, et al. (12 co-authors. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci USA. 2007;104:2271. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Genomic heterogeneity of background substitutional patterns in Drosophila melanogaster. Genetics. 2005;169:709–722. doi: 10.1534/genetics.104.032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Davis D, Jerel C, Petrov DA. X-linked genes evolve higher codon bias in drosophila and caenorhabditis. Genetics. 2005;171:145–155. doi: 10.1534/genetics.105.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Eyre-Walker A. Adaptive protein evolution in drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Fearnhead P. A comparison of three estimators of the population-scaled recombination rate: accuracy and robustness. Genetics. 2005;171:2051–2062. doi: 10.1534/genetics.104.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. Preferential segregation of the fourth chromosomes in Drosophila melanogaster. Proc Natl Acad Sci USA. 1934;20:515–518. doi: 10.1073/pnas.20.9.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. Preferential segregation in Triplo-IV Females of Drosophila melanogaster. Genetics. 1936;21:444–466. doi: 10.1093/genetics/21.4.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Cuaycong M, Craig C, Wallrath L, Locke J, Elgin S. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci USA. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;17:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Wallrath L, Elgin S. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Wang W, Thornton K, Berry A, Long M. Nucleotide variation along the Drosophila melanogaster fourth chromosome. Science. 2002;295:134–137. doi: 10.1126/science.1064521. [DOI] [PubMed] [Google Scholar]
- Wang W, Thornton K, Emerson J, Long M. Nucleotide variation and recombination along the fourth chromosome in drosophila simulans. Genetics. 2004;166:1783–1794. doi: 10.1534/genetics.166.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Welch JJ. Estimating the genome-wide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. Revtrans: multiple alignment of coding dna from aligned amino acid sequences. Nucl Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yin J, Jordan MI, Song YS. Joint estimation of gene conversion rates and mean conversion tract lengths from population SNP data. Bioinformatics. 2009:i231–239. doi: 10.1093/bioinformatics/btp229. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.