Abstract
The human genome project has been recently complemented by whole-genome assessment sequence of 32 mammals and 24 nonmammalian vertebrate species suitable for comparative genomic analyses. Here we anticipate a precipitous drop in costs and increase in sequencing efficiency, with concomitant development of improved annotation technology and, therefore, propose to create a collection of tissue and DNA specimens for 10 000 vertebrate species specifically designated for whole-genome sequencing in the very near future. For this purpose, we, the Genome 10K Community of Scientists (G10KCOS), will assemble and allocate a biospecimen collection of some 16 203 representative vertebrate species spanning evolutionary diversity across living mammals, birds, nonavian reptiles, amphibians, and fishes (ca. 60 000 living species). In this proposal, we present precise counts for these 16 203 individual species with specimens presently tagged and stipulated for DNA sequencing by the G10KCOS. DNA sequencing has ushered in a new era of investigation in the biological sciences, allowing us to embark for the first time on a truly comprehensive study of vertebrate evolution, the results of which will touch nearly every aspect of vertebrate biological enquiry.
Keywords: ancestral state reconstruction, comparative genomics, G10K, molecular evolution, species conservation, vertebrate biology
The bold insight behind the success of the human genome project was that, although vast, the roughly 3 billion letters of digital information specifying the total genetic heritage of an individual is finite and might, with dedicated resolve, be brought within the reach of our technology (Lander et al. 2001; Venter et al. 2001; Collins et al. 2003). The number of living species is similarly vast, estimated to be between 106 and 108 for all metazoans and approximately 6 × 104 for Vertebrata, which includes our closest relatives (May 1988; Erwin 1991; Gaston 1991). With the same unity of purpose shown for the Human Genome Project, we can now contemplate reading the genetic heritage of all species, beginning today with the vertebrates. The feasibility of a “Genome 10K” (G10K) project to catalog the genomic diversity of 10 000 vertebrate genomes, approximately one for each vertebrate genus, requires only one more order of magnitude reduction in the cost of DNA sequencing, after the 4 orders of magnitude reduction we have seen in the last 10 years (Benson et al. 2008; Mardis 2008; Shendure and Ji 2008; Eid et al. 2009). The approximate number of 10 000 is a compromise between reasonable expectations for the reach of new sequencing technology over the next few years and adequate coverage of vertebrate species diversity. It is time to prepare for this undertaking.
Living vertebrate species derive from a common ancestor that lived between 500 and 600 million years ago (Ma), before the time of the Cambrian explosion of animal life. Because a core repertoire of about 10 000 genes in a genome of about a billion bases is seen in multiple, deeply branching vertebrates and close deuterostome sister groups, we may surmise that the haploid genome of the common vertebrate ancestor was already highly sophisticated. At a minimum, this genome would have consisted of 108–109 bases specifying a body plan that included, among other features: 1) segmented muscles derived from somites; 2) a notochord and dorsal hollow neural tube differentiating into primitive forebrain, midbrain, hindbrain, and spinal-chord structures; 3) basic endocrine functions encoded in distant precursors to the thyroid, pancreas, and other vertebrate organs; and 4) a highly sophisticated innate immune system (Aparicio et al. 2002; Dehal et al. 2002; Hillier et al. 2004; Sodergren et al. 2006; Holland et al. 2008; Osorio and Retaux 2008; Gregory 2009). In the descent of the living vertebrates, the roughly 108 bases in the DNA segments that specify these sophisticated features, along with more fundamental biological processes, recorded many billions of fixed changes, the outcome of innumerable natural evolutionary experiments. These and other genetic changes, including rearrangements, duplications, and losses, spawned the diversity of vertebrate forms that inhabit strikingly diverse environments of the planet today. A G10K project explicitly detailing these genetic changes will provide an essential reference resource for an emerging new synthesis of molecular, organismic, developmental, and evolutionary biology to explore the vertebrate forms of life, just as the human genome project has provided an essential reference resource for 21st century biomedicine.
Beyond elaborations of ancient biochemical and developmental pathways, vertebrate evolution is characterized by stunning innovations, including adaptive immunity, multichambered hearts, cartilage, bones, and teeth, an internal skeleton that has given rise to the largest aquatic and terrestrial animals on the planet, a variety of sensory modalities that detect and process external stimuli, and specialized endocrine organs such as the pancreas, thyroid, thymus, pituitary, adrenal, and pineal glands (Shimeld and Holland 2000). At the cellular level, the neural crest, sometimes referred to as a fourth germ layer, is unique to vertebrates and gives rise to a great variety of structures, including some skeletal elements, tendons and smooth muscle, neurons and glia of the autonomic nervous system, melanocytes in the skin, dentin in the teeth, parts of endocrine-system organs, and connective tissue in the heart (Meulemans and Bronner-Fraser 2002; Baker 2008). Integration of sophisticated vertebrate sensory, neuroanatomical and behavioral elaborations coupled with often dramatic anatomical and physiological changes allowed exploitation of oceanic, terrestrial, and aerial ecological niches. Anticipated details of expansions and losses of specific gene families revealed by the G10K project will provide new insights into the molecular mechanisms behind these extraordinary innovations.
Adaptive changes in noncoding regulatory DNA also play a fundamental role in vertebrate evolution and understanding these changes represents an even greater challenge for comparative genomics (Hoekstra and Coyne 2007; Stranger et al. 2007). Almost no part of the known noncoding vertebrate gene regulatory apparatus bears any discernable resemblance at the DNA level to analogous systems in our deuterostome distant cousins. Yet, noncoding DNA segments represents the majority of the bases found to be under selection for the removal of deleterious alleles, and are likely to form the majority of the functional units in vertebrate genomes (Waterston et al. 2002; Siepel et al. 2005). Noncoding DNA segments are also hypothesized to be the major source of evolutionary innovation within vertebrate subclades (King and Wilson 1975; Holland et al. 2008). The origins and evolutionary trajectory of the subset of noncoding functional elements under the strongest selection to remove deleterious alleles can be traced deep into the vertebrate tree (Bejerano et al. 2004), in many cases to its very root, whereas other noncoding functional elements have uniquely arisen at the base of a particular class, order or family of vertebrate species. Within vertebrate lineages that evolved from a common ancestor in the last 100 My, such as placental mammals (∼5000 species), modern birds (∼10 000 species), and acanthomorphan fishes (∼16 000 species), evolutionary coalescence to a common ancestral DNA segment can be reliably determined even for segments of noncoding DNA. This enables detailed studies of base-by-base evolutionary changes throughout the genome, in both coding and noncoding DNA. Thus, the G10K project will provide power to address critical hypotheses concerning the origin and evolution of functional noncoding DNA segments and their role in molding physiological and developmental definitions of living animal species.
Through comprehensive investigation of vertebrate evolution, the G10K project will also lay the foundation needed to understand the genetic basis of recent and rapid adaptive changes within species and between closely related species. Coupled with evolutionary studies of recently diversifying clades, it will help address an increasingly urgent need to predict species’ responses to climate change, pollution, emerging diseases, and invasive competitors (Stockwell et al. 2003; Bell et al. 2004; Kohn et al. 2006; Thomas et al. 2009). It will enable studies of genomic phylogeography and population genetics that are crucial to assessment, monitoring, and management of biological diversity, especially of threatened and endangered species (Brito and Edwards 2009). Recent studies validate some of the potential contributions that the availability of genome sequences can provide to endangered species conservation efforts (Hillier et al. 2004; Romanov et al. 2009). Whole-genome sequence assemblies will be essential to facilitate genome-wide single nucleotide polymorphism discovery and to enable studies of historical demography, population structure, disease risk factors, and a variety of other conservation-related biological attributes. Species for which assembled whole-genome sequences are available will immediately be more amenable to a variety of biological studies that can contribute to assessments and science-based management. Such understanding could help curb the accelerating extinction crisis and slow the loss of biodiversity worldwide. Thus, as many threatened or endangered species should be included in the G10K project as is feasible.
Proposal
To this end, we propose to assemble a “virtual collection” of frozen or otherwise suitably preserved tissues or DNA samples representing on the order of 10 000 extant vertebrate species, including some recently extinct species that are amenable to genomic sequencing (Table 1). This collection represents combined specimen materials from at least 43 participating institutions (Table 2). In many cases, we have collected both male and female samples and for certain species several samples that reflect geographic diversity and/or diversity within localized populations.
Table 1.
Counts of vertebrate species stipulated for Genome 10K collection from G10KCOS
| Groups | Orders |
Families |
Genera |
Species | ||||||||
| With G10K samples | Total | % of total | With G10K samples | Total | % of total | With G10K samples | Total | % of total | With G10K samples | Total | % of total | |
| Mammals | 27 | 27 | 100 | 145 | 150 | 97 | 763 | 1230 | 62 | 1826 | 5416 | 34 |
| Birds | 32 | 34 | 94 | 182 | 199 | 91 | 1587 | 2172 | 73 | 5074 | 9723 | 52 |
| Amphibians | 3 | 3 | 100 | 50 | 56 | 89 | 301 | 510 | 59 | 1760 | 6570 | 27 |
| Reptiles | 4 | 4 | 100 | 63 | 65 | 97 | 751 | 1087 | 69 | 3297 | 9002 | 37 |
| Fishes | 62 | 62 | 100 | 424 | 532 | 80 | 1777 | 4956 | 36 | 4246 | 31 564 | 13 |
| Totals | 128 | 130 | 98 | 864 | 1002 | 86 | 5179 | 9955 | 52 | 16 203 | 62 275 | 26 |
Species and other taxa numbers are initially from NCBI taxonomy (www.ncbi.nlm.nih.gov) (Wheeler et al. 2008), as specified by Wilson and Reeder (2005) for the mammals, Hackett et al. (2008); Howard et al. (2003) for birds, Marjanovic and Laurin (2007); AmphibiaWeb (2009) for Amphibia, Catalog of Fishes (Eschmeyer 1998) for fishes, The TIGR Reptile Database (Uetz et al. 2007) for reptiles.
Table 2.
List of collections and participating institutions
| Institutions | Steward(s) | Web address |
| Academy of Natural Sciences, Philadelphia | Nate Rice | http://www.ansp.org/ |
| American Museum of Natural History | Joel Cracraft | http://research.amnh.org/ornithology/personnel/jlc.htm |
| Australian National University | Jennifer A. Marshall Graves | http://www.rsbs.anu.edu.au/ResearchGroups/CGG/index.php |
| Australian National Wildlife Collection, Canberra | Leo Joseph | http://www.csiro.au/places/ANWC.html |
| Bell Museum of Natural History, University of Minnesota | F. Keith Barker | http://www.bellmuseum.org/ |
| Biodiversity Institute of Ontario | Alexei Borisenko | http://www.biodiversity.ca |
| Biodiversity Research Institute, University of Kansas | Edward O. Wiley | http://www.nhm.ku.edu/fishes/ |
| Burke Museum, University of Washington | To be determined | http://www.washington.edu/burkemuseum/collections/genetic/index.php |
| California Academy of Sciences | Jens V. Vindum | NA |
| CIBIO, University of Porto, Portugal | Albano Beja-Pereira | http://cibio.up.pt/ |
| Círculo Herpetológico de Panamá | Roberto Ibáñez | http://ara.inbio.ac.cr/SSTN-IABIN/welcome.htm |
| CSIRO Marine and Atmospheric Research | Alastair Graham | http://www.cmar.csiro.au/anfc/ |
| Departamento de Zoologia, I.B., UNESP, Sao Paulo | Célio F. B. Haddad | NA |
| Field Museum of Natural History, Chicago | Harold K. Voris | http://www.fieldmuseum.org/research_collections/zoology/ |
| George Washington University | Guillermo Orti | http://www.gwu.edu/~biology/faculty/orti.cfm |
| Inst of Chemical Biology and Fundamental Medicine, SB RAS | Alexander S. Graphodatsky | http://www.niboch.nsc.ru/eng_index.html |
| Institut de Recherche pour le Développement, Paris, France | Philippe Gaubert | http://pgaubert.perso.neuf.fr/ |
| Institute of Molecular and Cell Biology, Singapore | Byrappa Venkatesh | http://www.imcb.a-star.edu.sg/php/venkatesh.php |
| Instituto Nacional de Cancer, Genetics Division | Hector N Seuanez | http://www.inca.gov.br/ |
| Kunming Institute of Zoology, Chinese Academy of Sciences | Ya-ping Zhang | http://www.kiz.ac.cn/en/ |
| LIRANS Institute, University of Bedfordshire, UK, | David Michael Rawson | http://www.beds.ac.uk/research/lirans/personnel/rawson_d |
| LSU Museum of Natural Science | Frederick H. Sheldon | http://appl003.lsu.edu/natsci/lmns.nsf/$Content/Sheldon?OpenDocument |
| Marine Mammal Institute, Oregon State University | C. Scott Baker | http://mmi.oregonstate.edu/ |
| Monterey Bay Aquarium Research Institute | Robert C. Vrijenhoek | http://www.mbari.org/molecular |
| Museo Nacional de Ciencias Naturales (MNCN), Madrid | David R. Vieites | http://www.vieiteslab.com |
| Museu de Zoologia da Universidade de São Paulo | Hussam Zaher | http://www.mz.usp.br/ |
| Museum of Comparative Zoology, Harvard | Scott Edwards | http://www.mcz.harvard.edu |
| Museum of Vertebrate Zoology, UC Berkeley | Jimmy A. McGuire | http://mvz.berkeley.edu/ |
| Museum Victoria, Australia | Joanna Sumner | http://museumvictoria.com.au/collections-research/our-research/sciences/staff/joanna-sumner/ |
| National Cancer Institute Lab of Genomic Diversity | Stephen J. O'Brien | http://home.ncifcrf.gov/ccr/lgd/ |
| Natural History Museum of Los Angeles County | To be determined | http://www.nhm.org/research |
| Ocean Park Corporation, Hong Kong | Paolo Martelli | http://www.oceanpark.com.hk/ |
| Pontificia Universidade Catolica do Rio Grande do Sul | Sandro Bonatto | NA |
| Royal Ontario Museum (ROM) | Robert W. Murphy | http://labs.eeb.utoronto.ca/murphy/ |
| San Diego Zoo's Institute for Conservation Research | Oliver A. Ryder | http://www.sandiegozoo.org/conservation/about/administrators/oliver_ryder_ph.d/ |
| Smithsonian Institution, National Museum of Natural History | Roy W. McDiarmid | http://vertebrates.si.edu/herps/ |
| Smithsonian Tropical Research Institute | Eldredge Bermingham | http://www.stri.org/ |
| South Australian Museum | Steve Donnellan | http://www.samuseum.sa.gov.au/page/default.asp?site=1&id=1307 |
| Southwest Fisheries Science Center, NMFS | Gabriela Serra-Valente | http://swfsc.noaa.gov/textblock.aspx?Division%20=%20PRD%20=%20229%20=%2012498 |
| Swedish Museum of Natural History | Per Ericson | http://www.nrm.se/en/menu/researchandcollections/departments/vertebratezoology.74_en.html |
| Texas A&M University | William J. Murphy | http://gene.tamu.edu/faculty_pages/faculty_MurphyW.php |
| The Frozen Ark | Olivier Hanotte | http://www.frozenark.org/ |
| The Global Viral Forecasting Initiative | Matthew LeBreton | www.gvfi.org |
| Universidade Federal do Rio de Janeiro, Genetics Dept. | Miquel Moreira & Cibele Bonvicino | http://www.inca.gov.br/conteudo_view.asp?id=414 |
| University of Auckland, New Zealand, School of Biological Sciences | Rochelle Constantine & C. Scott Baker | http://www.sbs.auckland.ac.nz/ |
| University College-Dublin | To be determined | http://www.ucd.ie/research/people/biologyenvscience/dremmacteeling/ |
| University of California, Riverside | To be determined | http://www.biology.ucr.edu/people/faculty/Springer.html |
| University of California, Santa Cruz, Department of Ecology & Evolutionary Biology | Barry Sinervo | http://bio.research.ucsc.edu/∼barrylab/ |
| University of California, Santa Cruz, Mammal Physiology Program | Terrie M. Williams | http://bio.research.ucsc.edu/people/williams/ |
| University of Kansas, Department of Ecology and Evolutionary Biology | Rafe Brown | http://www.nhm.ku.edu/rbrown/ |
| University of Minnesota, Cell Biology & Development | Tony Gamble | http://www.tc.umn.edu/∼gambl007/ |
| University of Montana | Gordon Luikart | http://dbs.umt.edu/research_labs/allendorflab/ |
| University of Sheffield | Terry Burke | http://www.shef.ac.uk/molecol/terry-burke |
| University of Texas at Arlington | Jonathan A. Campbell | http://biology.uta.edu/herpetology |
| Villanova University | Aaron Bauer | http://www.villanova.edu/artsci/biology/ |
| Zoological Institute, Technical University of Braunschweig | Miguel Vences | http://www.mvences.de/ |
| Zoological Museum of Copenhagen, Denmark | Jon Fjeldsa | http://zoologi.snm.ku.dk/english/ |
Tissues in genetic resource collections are stored by different methods, which yield varying results with regard to DNA quality (Edwards et al. 2005). Tissues that are sampled from the field may be left at ambient temperatures for several hours before they are finally frozen in liquid nitrogen and subsequently stored there at or near −80 °C. Nonetheless, many of these tissues still yield high-quality DNA (Brumfield R, LSU, personal communication). In other cases, noncryogenic field buffers are used, although with varying results. In addition to DNA quality, permit and species validation are also important issues to consider (Supplementary Material, Appendix 1). We will follow 4 general guidelines for G10K sample collection:
We seek 20 μg of genomic DNA or about 1 g of frozen tissue for each target species.
Tissues may be field preserved in liquid nitrogen, ethanol, or dimethyl sulfoxide (DMSO). Initial preservation in liquid nitrogen is strongly recommended for new acquisitions when field conditions permit. Transport from the field in liquid nitrogen or dry ice, or the use of dry shippers at liquid nitrogen temperatures is encouraged. In the laboratory, tissues should be stored in −80 °C freezers or in liquid nitrogen. The “gold standard” that would permit cell lines to be harvested is storage in liquid nitrogen of finely minced tissue fragments equilibrated in tissue-culture medium containing DMSO.
Tissues should be documented with voucher specimens linked to institutional accession codes when feasible (preserved carcasses are preferred, although photo vouchers are acceptable) and DNA Barcode information will be collected for all specimens (Hebert et al. 2003; Hanner and Gregory 2007; Ratnasingham and Hebert 2007; Field 2008; Field et al. 2008; Borisenko et al. 2009). In the case of rare or endangered species, a tissue sample, locality, identification by a professional zoologist, and DNA Barcode confirmation or listing in the International Species Information System would be acceptable.
All specimens used in the G10K project will be obtained and relocated in accord with national and international statutes regulating the collection, use, and transport of biological specimens.
In addition to samples for DNA extraction, the collection will include 1006 cryopreserved fibroblast cell lines derived from 602 different vertebrate species, primarily mammals, but including representatives of 300 taxa comprising 42 families of nonmammalian amniotes and 1 amphibian species. These resources provide an additional window into the unique cell biology of these species. With the recent development of transformation techniques to create induced pluripotent stem cells from fibroblast lines (Okita et al. 2007; Stadtfeld et al. 2008; Yu 2009; Yusa et al. 2009), the potential of cell-line studies is greatly expanded. Although it is still unclear how well current cell-line generation methods can be extended to all vertebrate clades (Liu et al. 2008; Trounson 2009), we propose to initiate primary fibroblast cell cultures for as many species as possible, with a target of at least 2,000 diverse species, as a corollary outcome of the G10K project. These cell cultures, along with cDNA derived from primary tissues, will provide direct access to gene expression and regulation data in the vertebrate species we catalog and provide a renewable experimental resource to complement the G10K genome sequences. For at least one species of each vertebrate order, we propose to assemble additional genomic resources, including physical maps and a bacterial artificial chromosome (BAC) library, other cell lines, and primary tissues for transcriptome analysis. For these species, we will propose to sequence multiple individuals to assess within-species diversity, including members of both sexes to assess sex-chromosome differences. A resource of this magnitude would help catalyze a much-needed extension of experimental molecular biology beyond the very limited set of model organisms it currently explores.
Integrated analysis and rapid release (genome.gov 2003) of the G10K data represents a substantial informatics challenge, beginning with the construction of a sample tracking database and culminating with the software needed to support a detailed evolutionary analysis of the many terabytes of sequence data (Supplementary Material, Appendices 3 and 4).
The G10K species collection will include tissue/DNA specimens from 5 major organismal groups: mammals, birds, amphibians, nonavian reptiles, and fishes (Table 1, Figure 1). Relevant aspects of each major group compiled by the Taxon committee chairs follow.
Figure 1.
Consensus phylogeny of the major lineages of vertebrates. Topology and divergence dates (Ma) are consensus estimates derived from Hedges and Kumar (2009) and included citations and amended per Benton and Donoghue (2007), Janvier (2006), Maisey (2000), and Sansom et al. (1996). Following the common names of taxon groups in parentheses is the number of living species for that group followed by the number of G10K species with specific biospecimens nominated for G10K whole-genome sequence.
Mammals
Mammals contain a morphologically and behaviorally diverse assemblage of approximately 5400 species from 1200 to 1300 genera distributed in 3 major lineages: monotremes (platypus and echidnas—5 species), marsupials (∼330 species, including the koala, kangaroos, and opossums), and the species-rich eutherian or placental mammals (∼5000 species) (Nowak 1999; Wilson and Reeder 2005), (Table 1, Figure 2).
Figure 2.
Consensus phylogeny of the major lineages of mammals. Topology and dates (Ma) are consensus estimates derived from Hedges and Kumar (2009) and included citations. Following the common names of taxon groups in parentheses is the number of living species for that group, followed by the number of G10K species with specific biospecimens nominated for G10K whole-genome sequence.
The G10K collection contains exemplars of 145 out of the 150 families (Supplementary Material, Appendix 2, mammals). At present, we have access to ∼90% of nonmuroid and nonsciurid rodent genera and nonvespertilionid bat genera. Ultimately, we will target all 1200 to 1300 genera.
Additional sampling will be applied to deeply divergent, and especially endangered, or Evolutionary Distinct and Endangered species (ZSL 2009), currently including all species of Zaglossus (echidna), Cuban and Hispaniolan Solenodon, Malayan Tapir (Tapirus indicus), aardvark (Orycteropus), and others. For fundamental biological investigation, another high priority is to sequence species exhibiting extreme phenotypes, such as deep-sea divers, long-lived species, high-altitude species, and species with distinct sensory modalities, such as echolocation. Our ultimate goal is to include within the collection species spanning the range of brain size, body size, and morphological convergence: aquatic species, gliders, lifespan extremes, nocturnals/diurnals, and social versus solitary species with diverse mating systems and varying levels of paternal care. We will also sample domestic animal species that have undergone recent and rapid evolution and contrast them to their counterpart wild species.
Capturing wide ecological diversity holds great potential for identifying the genomic changes underlying the major mammalian anatomical and behavioral transformations, including the evolution of advanced social and eusocial systems. Determining the genomic infrastructure for extreme physiological responses provides a unique opportunity for understanding the limits of mammalian tissues from resistance to disease to the ability to adapt to environmental disturbance.
Birds
Like eutherian mammals, living birds arose in the mid-Cretaceous (∼100 Ma). Since then, birds have dispersed across the globe and now occupy most of Earth's habitats and ecosystems representing a wide array of lifestyles. At this time, we know very little about the genetic and developmental underpinnings of this biological diversity, as high-quality genome sequences are available for only 2 species, the chicken (Gallus gallus) and zebra finch (Taeniopygia guttata). We expect that many key questions can and will be addressed as additional whole-genome sequences are accumulated and interpreted in the context of an increasingly accurate comparative framework (Hackett et al. 2008).
During recent decades, the avian systematics community has built large collections that house high-quality genetic samples of a substantial portion of avian diversity. These collections provide an essential resource for future genomic analyses of avian structural, functional, and behavioral diversity. With representation from 15 natural history collections distributed globally, the G10K collection includes specimens from 94% of the 34 orders, 91% of the 199 families, 73% of the 2172 genera, and 52% of the 9723 species of birds (Table 1, Figure 3). Every order is represented in multiple biospecimen collections, as are all but 17 families and all but 585 genera, ensuring at least 1 sample of high quality. We plan to sequence both sexes for a number of lineages, including the ratite birds, which like many avian species are externally monomorphic and, additionally, have relatively undifferentiated sex chromosomes.
Figure 3.
Consensus phylogeny of the major lineages of birds. Topology and dates (Ma) are derived from combined-data tree reported in Hackett et al. (2008), rendered ultrametric by nonparametric rate-smoothing (Sanderson 1997) and scaled to a root age of 119 Ma based on an average of multiple dating studies (van Tuinen et al. 2006). Following the common names of taxon groups in parentheses is the number of living species for that group followed by the number of G10K species with specific biospecimens nominated for G10K whole-genome sequence.
Sampling each genus may result in oversampling of some avian orders and families (such as the extremely diverse passerines and hummingbirds), but we will strive to capture maximal phylogenetic coverage across the avian tree.
Nonavian Reptiles
Nonavian reptile diversity includes snakes, lizards, turtles, crocodilians, and 2 species of tuatara. Because the traditional view of interfamilial relationships (based on morphology) differs appreciably from recent molecular phylogenies and the molecular phylogenies differ from one another, major issues such as the origin of snakes (which are clearly nested within lizards) remain controversial (Fry et al. 2006; Vidal and Hedges 2009). In addition to these uncertainties, the phylogenetic relationships within and among the major groups of reptiles (i.e., families) are often uncertain, for example, among the “colubroid” snakes (Hedges et al. 2009; Zaher et al. 2009) and species-rich assemblages of lizards. Major revisions have occurred within many groups, such as the geckos, where additional families are now recognized (Gamble et al. 2008). Following online databases including the TIGR Reptile Database (Uetz 2009), reptile diversity is distributed among the following groupings: Snakes are divided among 18 families, 484 genera, and 3313 species; lizards comprise 30 families, 499 genera, and 5351 species; and turtle diversity is divided among 13 families, 94 genera, and 313 species (Turtle Taxonomy Working Group 2007). Crocodiles include 23 species divided among 9 genera in 3 taxonomic families. And the 2 species of tuatara are the only extant members of the formerly diverse and widespread Rhyncocephalia. Total reptile diversity therefore includes 65 families, 1087 genera, and 9002 species. The G10K collection has 97%, 69%, and 37% of these, respectively (Table 1, Figure 4). In addition to these DNA and tissue samples, substantial BAC-library resources are available for nonavian reptiles that could facilitate the G10K project (Wang et al. 2006).
Figure 4.
Consensus phylogeny of the major lineages of nonavian reptiles. Topology and dates (Ma) are consensus estimates derived from Hedges and Kumar (2009) and included citations. Following the common names of taxon groups in parentheses is the number of living species for that group followed by the number of G10K species with specific named biospecimens nominated for G10K whole-genome sequence.
Amphibians
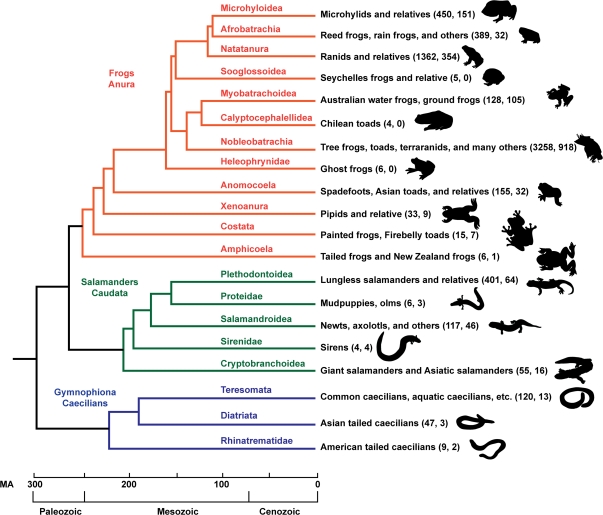
The Class Amphibia is divided into 3 orders: Anura (frogs), Caudata (salamanders), and Gymnophiona (caecilians), derived from a common ancestor 300 Ma and representing the only 3 surviving lineages from a much greater diversity that existed before the Permian extinction 250 Ma (Marjanovic and Laurin 2007). These major clades contain 5811 frog species, 583 salamander species, and 176 caecilian species, respectively (AmphibiaWeb 2009). Amphibian taxonomy is currently in a state of flux, with many new proposed taxonomic changes resulting from molecular phylogenetic analyses. Although controversial, we summarize amphibian diversity and tissue holdings for higher taxonomic groups (Supplemental Material, Appendix 2, amphibians) following the AmphibiaWeb (2009) database. This taxonomy contains 56 families of amphibians shared among the 3 orders, containing a total of 510 genera and 6570 species. The G10K collection contains a total of 1760 species (27%), 301 genera (59%), and 50 families (89%) (Table 1, Figure 5).
Figure 5.
Consensus phylogeny of the major lineages of amphibians. Topology and dates (Ma) are consensus estimates derived from Hedges and Kumar (2009) and included citations. Following the common names of taxon groups in parentheses is the number of living species for that group followed by the list of G10K species with specific named biospecimens nominated for G10K whole-genome sequence.
Amphibians are notorious for their morphological homoplasy due to developmental constraints (Shubin et al. 1995) as well as spectacular adaptive convergences in morphology (Bossuyt and Milinkovitch 2000), behavior, and development, for example, roughly 15 independent evolutionary origins of direct development from an ancestral biphasic life history (Hanken et al. 1997). Perhaps the most striking example is the convergent evolution in toxicity, coloration, and parental care between mantellid frogs of Madagascar and dendrobatid frogs in the Neotropics, as well as repeated parallel evolution of these traits within each of these 2 taxonomic families (Vences et al. 2003; Chiari et al. 2004). Such homoplasies have wreaked havoc on amphibian taxonomy, but offer marvelous opportunities to study the genetic basis of the repeated evolution of complex traits involved in both morphological and behavioral evolution.
Collectively, amphibians are of global conservation concern, most recently because of a rapid decline in populations and disappearance of species (Mendelson et al. 2006). A chytrid fungus, Batrachochytrium dendrobatidis, has been implicated in these declines (James et al. 2009), but habitat loss, pollutants, pesticides, herbicides, fertilizers, and climatic changes are also factors of concern. In the face of such diversity crises, sequencing many species of amphibians has enormous potential to provide insight into novel antimicrobial compounds, given that many species of frogs harbor a diverse array of such compounds (Zasloff 2002; Vanhoye et al. 2003). The same antimicrobial peptide sequence is rarely recovered from closely related species. Genomic approaches to searching for such antimicrobial diversity using stem cell lines, transcriptomes, and whole-genome sequencing are clearly warranted.
Fishes
Fishes include all nontetrapod vertebrates comprising 1) jawless vertebrates (hagfishes and lampreys, 114 species), 2) chondrichthyans (sharks, rays, and chimaeras, ∼1200 species), 3) actinopterygians (ray-fin fishes, ∼30 000 species), and 4) piscine sarcopterygians (coelacanths, lungfishes, 8 species). Total described diversity comprises approximately 31 500 species (Eschmeyer 1998), but actual diversity is probably greater than 50 000 species. A broad outline of the evolution of these most deeply branching of the vertebrate clades is provided by Stiassny et al. (2004).
Fishes account for nearly 50% of all described living species of vertebrates, exhibiting a vast diversity in their morphology, physiology, behavior, and ecological adaptations and providing an exceptional opportunity to study basic vertebrate biology. Fishes are also important as a food source for human consumption totaling about US $51 billion in trade in 2001 (Tidwell and Allan 2001). In 2006, global capture fisheries were estimated at US $91 billion and global aquaculture (including invertebrates) at US $79 billion (FAO 2008). There is also huge global recreational spending. Fishery activities of all types probably total in excess of US $200 billion per year (FAO 2008). Some 16% of all human protein consumption is fish protein, and about 1 billion people depend on fishes as their major source of protein. Because of the great demand, many groups of fishes are overexploited. Molecular data for commercially important species of fishes, especially those that are currently endangered and those raised by aquaculture, will be valuable in designing strategies for maintaining sustainable stocks and combating disease and other threats.
Fish tissues for the G10K project reside in a number of institutions and are usually curated as parts of formal institutional collections. The total number of species represented by tissue samples is not known precisely, but 6,400 species have been DNA barcoded and collections of new species continue to be added (Wiley E, KU, personal communication). Fresh material from many commonly available species can be obtained easily from fishing boats and the pet-trade industry for both genome and other molecular projects. The G10K project has in hand suitable samples from 62/62 orders (100%), 424/532 families (80%), 1777 of about 4956 genera (36%), and 4246 of about 31 564 named species (13%) (Table 1, Figure 1). We have identified other partner institutions that are anticipated to provide a minimum of 2500 additional species that will be officially incorporated into the project.
The largest known animal genome is that of the marbled lungfish, Protopterus aethiopicus, with a haploid size of 133 pg (about 130 Gbp), followed by the salamanders Necturus lewisi and Necturus punctatus at 120 pg (about 117 Gbp) (Gregory 2009). The genomes are bloated through the activity of transposons that, combined with their enormous size, make genome sequencing and assembly extremely challenging. Although RNA sequencing is one avenue by which we may get direct access to interesting biology in these species, we nevertheless recommend that full-genome sequencing projects be undertaken for large-genome species. There are important questions pertaining to gene regulation, genome structure, and genome evolution that cannot be answered from analysis of transcribed RNA alone.
Discussion
Careful observations of the morphological and functional adaptations in vertebrates have formed the basis of biological studies for a millennium, but it is only recently that we have been able to observe the action of evolution directly at the genetic level. It is not known whether convergent adaptations in independent lineages are often governed by analogous changes in a small number of orthologous genome loci or if macroevolutionary events in separate lineages usually result from entirely idiosyncratic combinations of mutations. The evidence from several recent studies points toward the former hypothesis (Eizirik et al. 2003; Nachman et al. 2003). For example, adaptive hind-limb reduction occurred independently many times in different lineages and even within the same species, just as sticklebacks in different lakes adapted from an oceanic to a freshwater environment (Shapiro et al. 2006). These stickleback adaptations are all traced to independent deletions of the same distal enhancer of the PITX2 development gene, demonstrating remarkable convergent evolution at the genomic level (Kingsley D, HHMI, personal communication). By cataloging the footprints of adaptive evolution in every genomic locus on every vertebrate lineage, the G10K project will provide the power to thoroughly test the “same adaptation, same loci” hypothesis, along with other fundamental questions about molecular adaptive mechanisms.
In the course of this investigation, we will discover the genetic loci governing fundamental vertebrate processes. The study of the evolution of viviparity is an outstanding example. Birds, crocodiles, and turtles all lay eggs, whereas apart from monotremes, mammals are all live bearers. Thus, there was one fundamental transition from oviparity to viviparity in these amniotes, which caused a fundamental reorganization in the developmental program and large-scale change in gene interactions that we are only just beginning to understand. Remarkably, however, nonavian reptiles have over 100 independent evolutionary origins of viviparity (Blackburn 2000). Fish have an equally spectacular variety of such transitions, along with some amphibians, such as the frog genus Gastrotheca, which includes species with placental-like structures (Duellman and Trueb 1986). These many independent instances of the evolution of viviparity afford an extraordinary opportunity to explore the genomics behind this reproductive strategy.
The architecture of sex determination in vertebrates is similarly diverse, with examples of XY, ZW, and temperature-dependent mechanisms. The G10K project thus provides an equally exciting opportunity for dissection of this diversity. In fact, a few vertebrate species have abandoned sex altogether. What happens when an asexual genome descends from an ancestral sexual genome, as has occurred repeatedly in Aspidoscelis lizard lineages? Are the independent parthenogenetic genomes parallel in any way? In one group of lizards, genus Darevskia, the formation of unisexual species is phylogenetically constrained (Murphy et al. 2000), yet in others, for example, Aspidoscelis, it is not. Many species of lizards and snakes are also known to have facultative parthenogenesis: Unmated females produce viable eggs and offspring. Unisexuality also occurs in amphibians and fishes by gynogenesis, hybridogenesis, and in amphibians by kleptogenesis (Bogart et al. 2007). Sequential hermaphrodite fishes can change their sex. Do these parallel convergent changes involve the same genes? The evolution of longevity remains another question of great interest. What mechanisms are responsible for the 2 orders of magnitude differences among vertebrates and what sets the limits for long-lived species found in each of the vertebrate clades? By identifying genomic loci that support different evolutionary innovations such as these, the data from the G10K project will drive fundamental progress in molecular and developmental biology.
The symphony of vertebrate species that cohabit on our planet attests to underlying life processes with remarkable potential. Genomics reveals a unity behind these life processes that is unrivaled by any other avenue of investigation, exposing the undeniable relatedness and common origin of all species. By revealing genetic vulnerabilities in endangered species and tracking host–pathogen coevolution, genomics also plays an increasing role in sustaining biodiversity and combating emerging infectious diseases. Thus, the information in the genomes of threatened and endangered species revealed by the G10K project will be crucial to conservation efforts (Ryder et al. 2000; O'Brien 2003; Ryder 2005; Kohn et al. 2006; Schwartz et al. 2009). In studying the genomes of recently extinct species as well, molecular aspects of species' vulnerability can be revealed and vital gaps in the vertebrate record restored. In all these ways, the G10K project will engage the public in the quest for the scientific basis of animal diversity and in the application of the knowledge we gain to halt extinctions and improve animal health.
As the printing of the first book by Johannes Gutenberg altered the course of human history, so did the human genome project forever change the course of the life sciences with the publication of the first full vertebrate genome sequence. When Gutenberg's success was followed by the publication of other books, libraries naturally emerged to hold the fruits of this new technology for the benefit of all who sought to imbibe the vast knowledge made available by the new print medium. We must now follow the human genome project with a library of vertebrate genome sequences, a genomic ark for thriving and threatened species alike, and a permanent digital record of countless molecular triumphs and stumbles across some 600 million years of evolutionary episodes that forged the “endless forms most beautiful” that make up our living world.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
American Genetic Association, Gordon and Betty Moore Foundation, NHGRI Intramural Sequencing Center, and UCSC Alumni Association to cost of the Genome 10K workshop; Howard Hughes Medical Institute to D.H.; Gordon and Betty Moore Foundation to S.C.S.; Assembling the Euteleost Tree of Life to E.W.; National Science Foundation (0732819 to E.W., DEB-0640967 and 0543556 to J.A.M., 0817042 to H.B.S., EF0629849 to W.J.M., DEB-0443470 to G.O.); The Global Viral Forecasting Initiative to N.W., B.P., and M.L.; Biomedical Research Council of A*STAR, Singapore to B.V.; Natural Sciences and Engineering Research Council Discovery Grant to R.W.M.; National Basic Research Program of China (973 Program, 2007CB411600), the National Natural Science Foundation of China (30621092), and Bureau of Science and Technology of Yunnan Province to Y.Z.; MCB and SB RAS Programs (A.S.G.); Portuguese-American Foundation for Development, CIBIO, UP, University of Montana [G.L.] and Portuguese Science Foundation [PTDC/CVT/69438/2006; PTDC/BIA-BDE/65625/2006 to G.L].
Supplementary Material
Acknowledgments
We wish to thank R. Fuller and S. Karl for project assistance and our reviewers for helpful comments.
Appendix
Genome 10K Community of Scientists (G10KCOS) Authors
Coordinators and corresponding authors: David Haussler (Howard Hughes Medical Institute, UCSC, CBSE/ITI E2501, University of California, Santa Cruz, Santa Cruz, CA, e-mail: haussler@soe.ucsc.edu); Stephen J. O'Brien (National Cancer Institute, Laboratory of Genomic Diversity, Frederick, MD, e-mail: stephen.obrien@nih.gov); Oliver A. Ryder (San Diego Zoo's Institute for Conservation Research, Escondido, CA, e-mail: oryder@sandiegozoo.org)
Coauthors: committee chairs (alphabetical): F. Keith Barker (University of Minnesota, Department of Ecology, Evolution and Behavior, St Paul, MN); Michele Clamp (The Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, MA); Andrew J. Crawford (Universidad de los Andes, Departamento de Ciencias Biológicas, Carrera 1E No. 18A–10, A.A. 4976, Bogotá, Colombia); Robert Hanner (Biodiversity Institute of Ontario, University of Guelph, Guelph, Ontario, Canada); Olivier Hanotte (The Frozen Ark Project—University of Nottingham, School of Biology, University Park, Nottingham, Nottinghamshire, UK); Warren E. Johnson (National Cancer Institute, Laboratory of Genomic Diversity, Frederick, MD); Jimmy A. McGuire (Museum of Vertebrate Zoology and Department of Integrative Biology, University of California, Berkeley, CA); Webb Miller (The Pennsylvania State University, Biology Department, 208 Mueller Laboratory, University Park, PA); Robert W. Murphy (Royal Ontario Museum, Department of Natural History, 100 Queen's Park, Toronto, ON, Canada); William J. Murphy (Texas A&M University, Department of Veterinary Integrative Biosciences, College Station, TX); Frederick H. Sheldon (Museum of Natural Science and Department of Biological Sciences, Louisiana State University, 119 Foster Hall, Baton Rouge, LA); Barry Sinervo (Department of Ecology and Evolutionary Biology University of California, Santa Cruz, Earth & Marine Sciences A308, Santa Cruz, CA); Byrappa Venkatesh (Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Biopolis, Singapore, Republic of Singapore); Edward O. Wiley (Department of Ecology and Evolutionary Biology, University of Kansas, Natural History Museum and Biodiversity Research Center, Lawrence, KS)
Additional authors (alphabetical) Fred W. Allendorf (Division of Biological Sciences, University of Montana, Missoula, MT); George Amato (Center for Conservative Genetics, American Museum of Natural History, New York, NY); C. Scott Baker (Marine Mammal Institute and Department of Fisheries and Wildlife, Oregon State University, Newport, OR); Aaron Bauer (Department of Biology, Villanova University, Mendel Hall Rm 191, Biology, Villanova, PA); Albano Beja-Pereira (CIBIO, Campus Agrario de Vairao, R., Maonte-Crasto, Vairao, Portugal); Eldredge Bermingham (Smithsonian Tropical Research Institute, PO Box 0843-03092, Balboa, Ancon Panama-Republic Of Panama); Giacomo Bernardi (University of California Santa Cruz, Department of Ecology and Evolutionary Biology, Santa Cruz, CA); Cibele R. Bonvicino (Genetics Division, Instituto Nacional de Câncer, Rua André Cavalcanti, 37, 4o andar, Rio de Janeiro, RJ 20231-050, and Laboratório de Biologia e Parasitologia de Mamíferos Reservatórios Silvestres, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz, Rio de Janeiro, RJ, Brazil); Sydney Brenner (Salk Institute for Biological Studies, PO Box 85800, San Diego, CA, Okinawa Institute of Science and Technology, Okinawa, Japan); Terry Burke (Department of Animal and Plant Sciences, University of Sheffield, Sheffield, UK); Joel Cracraft (American Museum of Natural History, Department of Ornithology, New York, NY); Mark Diekhans (University of California, Santa Cruz, Santa Cruz, CA); Scott Edwards (Harvard University, Department of Organismic and Evolutionary Biology, Cambridge, MA); Per G.P. Ericson (Swedish Museum of Natural History, PO Box 50007, Stockholm, Sweden); James Estes (Department of Ecology and Evolution, Center for Ocean Health, Santa Cruz, CA); Jon Fjelsda (University of Copenhagen, Zoologisk Museum, Universitetsparken 15, Museet—Bygn.11, 2-4-465, Denmark); Nate Flesness (ISIS, International Species Information System, Minneapolis, MN); Tony Gamble (University of Minnesota, Department of Genetics, Cell Biology, Room 6-160 Jackson Hall, Minneapolis, MN); Philippe Gaubert (Muséum National d'Histoire Naturelle, UMR BOREA IRD 207, 43 rue Cuvier—CP 26, Paris, France); Alexander S. Graphodatsky (Institute of Chemical Biology and Fundamental Medicine, Russian Academy of Science, Siberian Branch, Prospect Lavrentieva, 10, Novosibirsk, Novosibirsk Region, Russia); Jennifer A. Marshall Graves (The Australian National University, Research School of Biology, The Australian National University, Canberra, Australia); Eric D. Green (National Human Genome Research Institute, National Institutes of Health, Bldg. 50, Rm. 5222, Bethesda, MD); Richard E. Green (Max-Planck Institute for Evolutionary Anthropology, Leipzig Germany); Shannon Hackett (Field Museum of Natural History, Department of Zoology, Division of Birds, Chicago, IL); Paul Hebert (Biodiversity Institute of Ontario, University of Guelph, Guelph, Ontario, Canada); Kristofer M. Helgen (National Museum of Natural History, MRC 108, Smithsonian Institution, Division of Mammals, PO Box 37012, Washington, DC); Leo Joseph (CSIRO Sustainable Ecosystems—Gungahlin Homestead, Crace ACT 2911, GPO Box 284, Canberra, ACT, Australia); Bailey Kessing (SAIC-Frederick, Inc., National Cancer Institute at Frederick, Laboratory of Genomic Diversity, PO Box B, Frederick, MD); David M. Kingsley (HHMI and Stanford University, Beckman Center B300, Stanford, CA); Harris A. Lewin (Department of Animal Sciences and Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL); Gordon Luikart (Division of Biological Sciences, University of Montana, CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, Universidade do Porto, Portugal, Missoula, MT); Paolo Martelli (Ocean Park, Aberdeen, Hong Kong); Miguel A.M. Moreira (Genetics Division, Instituto Nacional de Cancer, Rua Andre Cavalcante, 37, 4o andar, Rio de Janeiro, RJ, Brazil); Ngan Nguyen (University of California, Santa Cruz, Santa Cruz, CA); Guillermo Ortí (George Washington University, Department of Biological Sciences, 2023 G Street, NW, Washington, D.C. 20052); Brian L. Pike (Global Viral Forecasting Initiative, One Market, Spear Tower, Suite 3574, San Francisco, CA); David Michael Rawson (LIRANS Institute, University of Bedfordshire, 250 Butterfield, Great Marlings, Luton, Bedfordshire, UK); Stephan C. Schuster (Penn State University, 310 Wartik Laboratories, University Park, PA); Héctor N. Seuánez (Genetics Division, Instituto Nacional de Câncer, Universidade Federal do Rio de Janeiro, Rua André Cavalcanti 37, 4o andar, Rio de Janeiro, RJ 20231-050, and Department of Genetics, Universidade Federal do Rio de Janeiro, Cidade Universitária, CCS, Bloco A, Rio de Janeiro, RJ 21949-570, Brazil); H. Bradley Shaffer (Department of Evolution and Ecology and Center for Population Biology, University of California, Davis, CA); Mark S. Springer (Department of Biology, University of California, Riverside, CA); Joshua Michael Stuart (University of California, Santa Cruz, Mail Stop SOE2, Santa Cruz, CA); Joanna Sumner (Museum Victoria, GPO Box 666, Melbourne, Vic., Australia); Emma Teeling (University College Dublin, School of Biology and Environmental Science, Science Centre West, Belfield, Dublin, Ireland); Robert C. Vrijenhoek (Monterey Bay Aquarium Research Institute, Moss Landing, CA); Robert D. Ward (CSIRO Marine and Atmospheric Research, GPO Box 1538, Castray Esplanade, Hobart, Tasmania, Australia); Wesley C. Warren (Genome Sequencing Center, Washington University School of Medicine, St Louis, MO); Robert Wayne (UCLA, Ecology and Evolutionary Biology, Box 951606, 2312 LSB, Los Angeles, CA); Terrie M. Williams (University of California Santa Cruz, Center for Ocean Health—Department of Ecology and Evolutionary Biology, Santa Cruz, CA); Nathan D. Wolfe (Global Viral Forecasting Initiative, One Market, Spear Tower, Suite 3574, San Francisco, CA and Stanford University, Program in Human Biology, Stanford, CA); Ya-Ping Zhang (State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, 32 Jiaochagndong ST, Kunming, Yunnan, China)
Committees:
Mammals Group Members: C. Scott Baker, James Estes, Philippe Gaubert, Jennifer Graves, Alexander Graphodatsky, Kristofer M. Helgen, *Warren E. Johnson, Harris A. Lewin, Gordon Luikart, *William J. Murphy, Stephen J. O'Brien, Oliver A. Ryder, Mark Springer, Emma Teeling, Robert Wayne, Terrie Williams, Nathan Wolfe, Ya-Ping Zhang
Birds Group Members: *F. Keith Barker, Joel Cracraft, Scott V. Edwards, Olivier Hanotte, *Frederick H. Sheldon
Amphibians and Reptiles Group Members: *Andrew J. Crawford, Paolo Martelli, *Jimmy A. McGuire, *Robert W. Murphy, H. Bradley Shaffer, *Barry Sinervo
Fishes Group Members: Fred W. Allendorf, Giacomo Bernardi, Guillermo Orti, David M. Rawson, *Byrappa Venkatesh, Robert C. Vrijenhoek, Robert D. Ward, *Edward O. Wiley
General Policy Group Members: C. Scott Baker, *Adam Felsenfeld, Eric D. Green, *Robert Hanner, *Olivier Hanotte, David Haussler, Paul Hebert, Stephen J. O'Brien, Oliver A. Ryder, Hector N. Seuanez, Ya-Ping Zhang
Analysis Group Members: *Michele Clamp, Mark Diekhans, David Haussler, Bailey Kessing, David M. Kingsley, Harris A. Lewin, *Webb Miller, Ngan Nguyen, Brian L. Pike, Stephan C. Schuster, Joshua M. Stuart, Steve Turner
*Chairs
References
- AmphibiaWeb Information on amphibian biology and conservation. 2009 Available from: http://amphibiaweb.org. [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Baker CV. The evolution and elaboration of vertebrate neural crest cells. Curr Opin Genet Dev. 2008;18:536–543. doi: 10.1016/j.gde.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bell MA, Aguirre WE, Buck NJ. Twelve years of contemporary armor evolution in a threespine stickleback population. Evolution. 2004;58:814–824. doi: 10.1111/j.0014-3820.2004.tb00414.x. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Blackburn DG. Reptilian viviparity: past research, future directions, and appropriate models. Comp Biochem Physiol A Mol Integr Physiol. 2000;127:391–409. doi: 10.1016/s1095-6433(00)00272-5. [DOI] [PubMed] [Google Scholar]
- Bogart JP, Bi K, Fu J, Noble DW, Niedzwiecki J. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome. 2007;50:119–136. doi: 10.1139/g06-152. [DOI] [PubMed] [Google Scholar]
- Borisenko AV, Sones JE, Hebert PDN. The front-end logistics of DNA barcoding: challenges and prospects. Mol Ecol Resour. 2009;9:27–34. doi: 10.1111/j.1755-0998.2009.02629.x. [DOI] [PubMed] [Google Scholar]
- Bossuyt F, Milinkovitch MC. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc Natl Acad Sci U S A. 2000;97:6585–6590. doi: 10.1073/pnas.97.12.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito P, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2009;135:439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- Chiari Y, Vences M, Vieites DR, Rabemananjara F, Bora P, Ramilijaona Ravoahangimalala O, Meyer A. New evidence for parallel evolution of colour patterns in Malagasy poison frogs (Mantella) Mol Ecol. 2004;13:3763–3774. doi: 10.1111/j.1365-294X.2004.02367.x. [DOI] [PubMed] [Google Scholar]
- Collins FS, Morgan M, Patrinos A. The human genome project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Duellman WE, Trueb L. Biology of amphibians. New York: McGraw Hill; 1986. 670 pp. [Google Scholar]
- Edwards S, Birks S, Brumfield R. Future of avian genetic resources collections: archives of evolutionary and environmental history. Auk. 2005;122:979–1284. [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS, O'Brien SJ. Molecular genetics and evolution of melanism in the cat family. Curr Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Erwin TL. How many species are there—revisited. Conserv Biol. 1991;5:330–333. [Google Scholar]
- Eschmeyer WN. Catalog of fishes. San Francisco (CA): California Academy of Sciences; 1998. 3 v. (p. 2905) [Google Scholar]
- FAO . FAO fisheries and aquaculture report. Rome (Italy): Food and Agriculture Organization of the United Nations; 2008. [Google Scholar]
- Field D. Working together to put molecules on the map. Nature. 2008;453:978. doi: 10.1038/453978b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, Vidal N, Norman JA, Vonk FJ, Scheib H, Ramjan SF, Kuruppu S, Fung K, Hedges SB, Richardson MK, et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- Gamble T, Bauer AM, Greenbaum W, Jackman TR. Out of the blue: a novel, trans-Atlantic clade of geckos (Gekkota, Squamata) Zool Scr. 2008;37:355–366. [Google Scholar]
- Gaston KJ. The magnitude of global insect species richness. Conserv Biol. 1991;5:283–296. [Google Scholar]
- genome.gov. Reaffirmation and extension of NHGRI rapid data release policies: large-scale sequencing and other community resource projects [cited 2009 June 28] 2003 Available from: http://www.genome.gov/10506537. [Google Scholar]
- Gregory TR. Animal genome size database. [cited 2009 May 1] 2009 Available from: http://www.genome.com. [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RC, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hanken J, Jennings DH, Olsson L. Mechanistic basis of life-history evolution in anuran amphibians: direct development. Am Zool. 1997;37:160–171. doi: 10.1006/gcen.1998.7111. [DOI] [PubMed] [Google Scholar]
- Hanner RH, Gregory TR. Genomic diversity research and the role of biorepositories. Cell Preserv Technol. 2007;5:93–103. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond Series B-Biological Sciences. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Couloux A, Vidal N. Molecular phylogeny, classification, and biogeography of West Indian racer snakes of the tribe Alsophiini (squamata, dipsadidae, xenodontinae) Zootaxa. 2009;2067:1–28. [Google Scholar]
- Hedges SB, Kumar S. The Timetree of life. New York: Oxford University Press; 2009. 572 pp. [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. The Amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R, Moore A, Dickinson EC. The Howard and Moore complete checklist of the birds of the world. London: Christopher Helm; 2003. p. 1039. [Google Scholar]
- James TY, Litvintseva AP, Vilgalys R, Morgan JA, Taylor JW, Fisher MC, Berger L, Weldon C, du Preez L, Longcore JE. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009;5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier P. Palaeontology: modern look for ancient lamprey. Nature. 2006;443:921–924. doi: 10.1038/443921a. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends Ecol Evol. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Maisey JG. Discovering fossil fishes. Boulder (CA): Westview Press; 2000. 224 pp. [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Marjanovic D, Laurin M. Fossils, molecules, divergence times, and the origin of lissamphibians. Syst Biol. 2007;56:369–388. doi: 10.1080/10635150701397635. [DOI] [PubMed] [Google Scholar]
- May RM. How many species are there on Earth. Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- Mendelson JR, 3rd, Lips KR, Gagliardo RW, Rabb GB, Collins JP, Diffendorfer JE, Daszak P, Ibanez DR, Zippel KC, Lawson DP, et al. Biodiversity. Confronting amphibian declines and extinctions. Science. 2006;313:48. doi: 10.1126/science.1128396. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- Murphy RW, Fu J, Darevsky IS, Kupriyanova LA, MacCulloch RD. A fine line between sex and unisexuality: the phylogenetic constraints on parthenogenetic lacertid lizards. Zool J Linn Soc. 2000;130:527–549. [Google Scholar]
- Nachman MW, Hoekstra HE, D'Agostino SL. The genetic basis of adaptive melanism in pocket mice. Proc Natl Acad Sci U S A. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak RM. Walker's mammals of the world. 6th ed. Baltimore (MD): Johns Hopkins University Press; 1999. 1936 pp. [Google Scholar]
- O'Brien SJ. Tears of the cheetah: and other tales from the genetic frontier. 1st ed. New York,: Thomas Dunne Books/St. Martin's Press; 2003. xiv, p. 287. [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Osorio J, Retaux S. The lamprey in evolutionary studies. Dev Genes Evol. 2008;218:221–235. doi: 10.1007/s00427-008-0208-1. [DOI] [PubMed] [Google Scholar]
- Parham JW, Bickham JW, Iverson JB, Philippen HD, Rhodin AGJ, Shaffer HB, Spinks PQ, van Dijk PP. 2007. An annotated list of modern turtle terminal taxa, with comments on areas of taxonomic instability and recent change. In: 4 CRMN, (ed) Defining Turtle Diversity: Proceedings of a Workshop on Genetics, Ethics, and Taxonomy of Freshwater Turtles and Tortoises. pp173–1994. [Google Scholar]
- Ratnasingham S, Hebert PD. bold: the barcode of life data system ( http://www.barcodinglife.org) Mol Ecol Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov MN, Tuttle EM, Houck ML, Modi WS, Chemnick LG, Korody ML, Mork EM, Otten CA, Renner T, Jones KC, et al. The value of avian genomics to the conservation of wildlife. BMC Genomics. 2009;10(Suppl. 2):S10. doi: 10.1186/1471-2164-10-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder OA. Conservation genomics: applying whole genome studies to species conservation efforts. Cytogenet Genome Res. 2005;108:6–15. doi: 10.1159/000080796. [DOI] [PubMed] [Google Scholar]
- Ryder OA, McLaren A, Brenner S, Zhang YP, Benirschke K. DNA banks for endangered animal species. Science. 2000;288:275–277. doi: 10.1126/science.288.5464.275. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- Sansom IJ, Smith MM, Smith MP. Scales of thelodont and shark-like fishes from the Ordovician of Colorado. Nature. 1996;379:628–630. [Google Scholar]
- Schwartz M, Luikart G, McKelvey K, Cushman S. Landscape genomics: a brief perspective. In: Huettmann F, Cushman S, editors. Spatial complexity, informatics and wildlife conservation. Tokyo (Japan): Springer; 2009. [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 2006;103:13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, Holland PW. Vertebrate innovations. Proc Natl Acad Sci U S A. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N, Wake DB, Crawford AJ. Morphological variation in the limbs of taricha-granulosa (caudata, salamandridae)—evolutionary and phylogenetic implications. Evolution. 1995;49:874–884. doi: 10.1111/j.1558-5646.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiassny M, Wiley E, Johnson G. Gnathostome fishes. In: Cracraft J, Donoghue M, editors. Assembling the tree of life. New York: Oxford University Press; 2004. pp. 410–429. [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends Ecol Evol. 2003;18:94–101. [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Quoss H, Hartmann J, Eckmann R. Human-induced changes in the reproductive traits of Lake Constance common whitefish (Coregonus lavaretus) J Evol Biol. 2009;22:88–96. doi: 10.1111/j.1420-9101.2008.01622.x. [DOI] [PubMed] [Google Scholar]
- Tidwell JH, Allan GL. Fish as food: aquaculture's contribution. Ecological and economic impacts and contributions of fish farming and capture fisheries. EMBO Rep. 2001;2:958–963. doi: 10.1093/embo-reports/kve236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A. Rats, cats, and elephants, but still no unicorn: induced pluripotent stem cells from new species. Cell Stem Cell. 2009;4:3–4. doi: 10.1016/j.stem.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Uetz P. The TIGR Reptile Database. 2009 Available from: http://www.reptile-database.org/. [Google Scholar]
- Uetz P, Goll J, Hallermann J. Die tigr-reptiliendatenbank. Elaphe. 2007;15:16–19. [Google Scholar]
- van Tuinen M, Stidham M, Hadly TA. Tempo and mode of modern bird evolution observed with large-scale taxonomic sampling. Historical Biol. 2006;18:205–221. [Google Scholar]
- Vanhoye D, Bruston F, Nicolas P, Amiche M. Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur J Biochem. 2003;270:2068–2081. doi: 10.1046/j.1432-1033.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Vences M, Kosuch J, Boistel R, Haddad CFB, La Marca E, Lotters S, Veith M. Convergent evolution of aposematic coloration in Neotropical poison frogs: a molecular phylogenetic perspective. Org Divers Evol. 2003;3:215–226. [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vidal N, Hedges SB. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C R Biol. 2009;332:129–139. doi: 10.1016/j.crvi.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Wang Z, Miyake T, Edwards SV, Amemiya CT. Tuatara (Sphenodon) genomics: bAC library construction, sequence survey, and application to the DMRT gene family. J Hered. 2006;97:541–548. doi: 10.1093/jhered/esl040. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore (MD): Johns Hopkins University Press; 2005. 2 v. (xxxv, p. 2142) [Google Scholar]
- Yu J. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H, Grazziotin FG, Cadle JE, Murphy RW, Moura-Leite JCd, Bonatto SL. Molecular phylogeny of advanced snakes (Serpentes, Caenophidia) with an emphasis on South American xenodontines: a revised classification and descriptions of new taxa. Pap Avuls Zool. 2009;49:115–153. [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- ZSL . EDGE: evolutionary distinct & globally endangered. The Zoological Society of London; 2009. Available from: http://www.edgeofexistence.org/index.php. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.