RNF-121 is an E3 ligase RING finger protein that is localized to the ER in Caenorhabditis elegans and functions in the UPR and ERAD pathways. The β subunit of the heterodimeric integrin receptor was identified as a substrate for RNF-121, suggesting a link between ERAD and cell adhesion through the regulation of β-integrin.
Abstract
We report on the characterization of RNF-121, an evolutionarily conserved E3 ligase RING finger protein that is expressed in the endoplasmic reticulum (ER) of various cells and tissues in Caenorhabditis elegans. Inactivation of RNF-121 induced an elevation in BiP expression and increased the sensitivity of worms to ER stress. Genetic analysis placed RNF-121 downstream of the unfolded protein response (UPR) regulator protein kinase-like endoplasmic reticulum kinase (PERK). We identify PAT-3::GFP, the β subunit of the heterodimeric integrin receptors, as an RNF-121 substrate; whereas induction of RNF-121 expression reduced the level of PAT-3::GFP in the gonad distal tip cells, inhibition of RNF-121 led to the accumulation of stably bound PAT-3::GFP inclusions. Correspondingly, overexpression of RNF-121 during early stages of gonad development led to aberrations in germline development and gonad migration that overlap with those observed after PAT-3 inactivation. The formation of these gonad abnormalities required functional ER-associated degradation (ERAD) machinery. Our findings identify RNF-121 as an ER-anchored ubiquitin ligase that plays a specific role in the ERAD pathway by linking it to the regulation of the cell adhesion integrin receptors.
INTRODUCTION
Stress conditions in the endoplasmic reticulum (ER) cause the accumulation of misfolded proteins in the ER lumen or membrane. The unfolded-protein response (UPR) is a collection of signaling pathways that regulate the adaptation of cells to ER stress (Schroder and Kaufman, 2005; Ron and Walter, 2007). The general effects of the UPR are a decrease in protein synthesis and translocation to the ER, an increase in retrotranslocation and degradation of misfolded ER-localized proteins by the ER-associated degradation (ERAD) machinery, and an increase in the volume of the ER and in its protein-folding capacity. Severe ER stress can lead to autophagy and also to cell death (Schroder and Kaufman, 2005; Zhang and Kaufman, 2006; Ron and Walter, 2007). The UPR in mammals includes three stress sensors: inositol requirement 1 (IRE1), protein kinase-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) (Schroder and Kaufman, 2005). The UPR is activated not only by external stress but also during development and cellular differentiation, when changes in the physiological state of the cell cause different levels of protein-folding load upon the ER (Zhang and Kaufman, 2006). The UPR in Caenorhabditis elegans is regulated by orthologues of IRE1, PERK and ATF6 (IRE-1, PEK-1 and ATF-6, respectively) and mediates the expression of multiple genes under both normal and ER-stress conditions (Shen et al., 2001; Shen et al., 2005).
The ERAD pathway complements the activity of the UPR molecular chaperones and modifying enzymes in quality control by removing misfolded secretory and membrane proteins, as well as unassembled subunits of multimeric proteins from the ER. The ERAD process starts with the recognition of a substrate in the ER, retrotranslocation from the ER lumen to the cytoplasm through a channel (Meusser et al., 2005; Romisch, 2005; Vembar and Brodsky, 2008), and ubiquitination through a cascade of enzymatic reactions by using ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (Hershko et al., 2000). The polyubiquitin chain is recognized by the CDC48/p97 ATPase complex (CDC48/p97-Ufd1-Npl4) that transports substrates to the proteasome (Vembar and Brodsky, 2008).
E3 ubiquitin ligases in ERAD function as quality-control ligases, recognizing misfolded proteins and subunits that lack their oligomerization partner (Kostova et al., 2007). In yeast, Doa10p and Hrd1p are membrane-associated RING finger ligases that have been found to degrade all studied yeast ERAD substrates (Vashist and Ng, 2004; Carvalho et al., 2006; Denic et al., 2006). Orthologues of Doa10p and Hrd1p have been identified in higher organisms, as have additional ERAD-related cytoplasmic and ER-membrane-bound E3 ligases (Kostova et al., 2007) whose specific in vivo targets and functions are largely unknown. The main mammalian membrane-bound E3 ligases related to ERAD are Hrd1/synoviolin, gp78, TEB4/MARCHVI, and RNF5 (Fang et al., 2001; Amano et al., 2003; Nadav et al., 2003; Kikkert et al., 2004; Chen et al., 2006; Kreft et al., 2006; Omura et al., 2006; Tsai et al., 2007). The C. elegans genome encodes orthologues of Hrd1, gp78, MARCHVI, and RNF5 (named HRD-1, HRDL-1, MARC-6, and RNF-5, respectively) (Sasagawa et al., 2007). Depletion of HRD-1 or MARC-6 causes increased sensitivity of worms to ER stress (Sasagawa et al., 2007). RNF5/RMA1 is a membrane-anchored RING finger ligase that affects the localization and levels of LIM-domain–containing protein in C. elegans and of paxillin in mammalian cells (Matsuda and Nakano, 1998; Matsuda et al., 2001; Didier et al., 2003; Broday et al., 2004, 2007) and has been implicated in ERAD (Younger et al., 2006; Delaunay et al., 2008; Tcherpakov et al., 2009). In C. elegans, RNF-121 resides on the same operon (CEOP3156) as RNF-5. Here, we demonstrate that RNF-121 is an ER-bound E3 ligase that is required to resist ER stress and that its inactivation induces the UPR. We further identify a substrate for RNF-121, PAT-3/β-integrin, and show that PAT-3/β-integrin::GFP levels and accumulation into stable inclusions are dependent on RNF-121 activity and thus offer a new link between ERAD and the regulation of cell adhesion.
MATERIALS AND METHODS
Strains
C. elegans strains were grown at 20°C according to standard protocols. (Brenner, 1974). The wild-type strain used is Bristol N2. Alleles used in this study are as follows: ire-1(v33)II, rnf-121(ok848)III, pek-1(ok275)X, atf-6(ok551)X (received from the Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, MN [CGC]), and rnf-5(tm794)III (received from Mitani Laboratory, Tokyo Japan). The deletion alleles rnf-121(ok848),pek-1(ok275), atf-6(ok551) were isolated by the C. elegans Gene Knockout Project team at Oklahoma Medical Research Foundation (OMRF, Oklahoma City, OK), and strains were outcrossed three to six times. The following transgenes were used: rhIs2[PAT-3::HA::GFP] (Plenefisch et al., 2000), tvEx55[unc-54p::mRFP::SP12] (Kubota et al., 2006), tvEx58[hsp-16p:: mRFP::MANS;rol-6] (this study), zcIs4[hsp-4p::GFP] (Calfon et al., 2002), tvEx33[RNF-121::GFP;rol-6] (this study), tvIs34[RNF-121::GFP;rol-6] (this study), tvEx35[hsp-16p::HA::RNF-121;myo-2p::GFP] (this study), tvEx36[hsp-16p::HA::RNF-121C222AC225A;myo-2p::GFP] (this study), and tvEx48 [hsp-16p::RNF-5; myo-2p::GFP] (this study).
Transgene Construction
The RNF-121 translational GFP reporter was generated according to O. Hobert (Columbia University) (Hobert, 2002) by amplification of a 5.7-kb genomic fragment upstream of the rnf-121 gene and the entire genomic sequence of rnf-121 (2.4 kb) (amplified from C. elegans genomic DNA) fused to GFP and the 3′ UTR of unc-54 from the pPD95.75 vector (A. Fire, Stanford University School of Medicine). Three extrachromosomal transgenic lines and one integrated transgene were generated and analyzed. The heat-shock inducible construct was generated by cloning the full-length rnf-121 cDNA with an N-terminal hemagglutinin (HA) tag into the pPD49.78 vector that contains the hsp-16-2 promoter site (A. Fire). The heat shock induced RNF-121 RING mutant (C222AC225A) was generated by site directed mutagenesis. The Golgi marker hsp-16p::mRFP::MANS contains 82 amino acids from AMAN-2 (Rolls et al., 2002) under the regulation of the hsp-16-2 promoter fused to monomeric red fluorescent protein (mRFP) at the N terminus. Three independent lines of each construct were analyzed.
In Vitro Ubiquitination
A fragment including the RING finger domain of RNF-121 and the predicted cytoplasmic region upstream the RING domain (amino acids 180-289) was cloned into pGEX 6P-1. RING finger mutants C222AC225A and V224A were generated by site directed mutagenesis of this plasmid. Ubiquitination reactions were performed in 1× ubiquitination buffer (30 mM Tris, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 2 mM ATP, and 1 mM HA-ubiquitin) including 200 ng of E1 (BIOMOL Research Laboratories, Plymouth Meeting, PA), 200 ng of E2 (bacterially expressed and purified His-UBCH5c; David et al., 2010), and 200 ng of GST-RNF-121(180-289) or RING mutants for 1 h at 37°C. The reaction with GST-RNF-5 was incubated for 10 min at 37°C. Reaction products were resolved on 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred onto nitrocellulose membranes. The membranes were probed with anti-HA antibodies.
For in vitro ubiquitination of PAT-3::HA::GFP, worm pellets were incubated with an equal volume of lysis buffer (100 mM NaCl, 50 mM NaF, 50 mM Tris, pH 7.5, 1 mM EGTA, 5% glycerol, 1% Nonidet P40, 1 mM DTT, 10 mM iodoacetamide [added in the extract shown in Supplemental Figure S5], 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin A) on ice followed by sonication (5 s, ×5 times). Samples were centrifuged (15 min at 14,000 rpm), and supernatants were precleared by incubation with protein G beads (Roche Diagnostics, Indianapolis, IN) (30 min at 4°C), followed by incubation with anti-GFP antibody (Roche Diagnostics) (1 h at 4°C). Immunoprecipitation was performed by incubation (2 h at 4°C) with protein G beads (Roche Diagnostics). Beads were washed with 30 mM Tris, pH 7.6, 50 mM NaCl, 5 mM MgCl2, and 1 mM DTT. In vitro ubiquitination reactions were performed as described above with (or without) E1, E2, ATP, and HA-ubiquitin for 20 min at 37°C. Reactions were terminated with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, pH 6.3, and 0.1% Nonidet P40 and washed four times with the lysis buffer. Proteins were solubilized in 3× Laemmli buffer and separated on SDS-PAGE followed by immunoblot analysis with the indicated antibodies.
RNA Interference (RNAi)
A 323-base pair fragment of rnf-121 cDNA (bases 218–540 of the rnf-121 cDNA) was amplified using the primers 5′-ATAGAATGGGCCAACATG and 5′-AACAAGCCACGTGGCCAGGAA and cloned into the L4440 feeding vector (pPD129.36) (A. Fire). RNAi by feeding was carried out as described previously (Fraser et al., 2000). Induction of the bacteria with 1 mM isopropyl β-d-thiogalactoside (IPTG) (Sigma-Aldrich, St. Louis, MO) was performed for 4 h at 37°C. Induced bacteria expressing rnf-121 double-stranded RNA (dsRNA) was seeded onto NMG-RNAi plates containing 1 mM IPTG and 25 μg/ml carbenicillin. Plates were dried at room temperature overnight. Synchronized larval stage 1 (L1) larvae were grown at 20°C on bacteria harboring the L4440 plasmid expressing dsRNA for rnf-121 or control (empty L4440 vector), and the F1 progeny was scored. The der-1 (F25D7.1) RNAi feeding vector was obtained from the J. Ahringer library (Geneservice, Nottingham, United Kingdom). Synchronized L1 larvae of tvEx35 worms were grown at 15°C on RNAi plates, and induction of RNF-121 and analysis were performed as described below.
Tunicamycin Sensitivity Assay
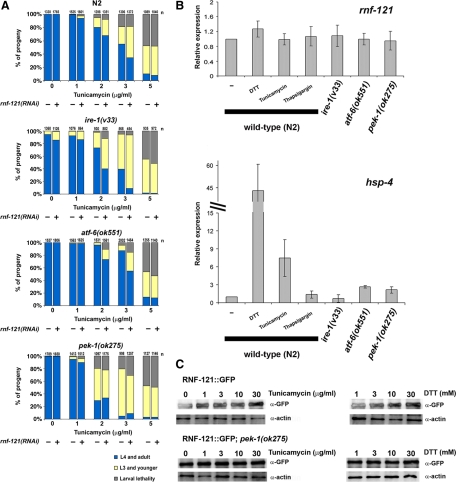
Gravid adults were allowed to lay eggs on plates containing tunicamycin. Analysis of developmental stages was performed 72 h later. At least three independent experiments were performed for each tunicamycin concentration (0–7.5 or 0–5 μg/ml), and three plates for each treatment were analyzed in each experiment; n values above the bars are the total number of embryos. To examine tunicamycin sensitivity of worms treated with RNAi, embryos were transferred to duplicated RNAi feeding plates seeded with rnf-121 dsRNA-producing bacteria or with bacteria containing the empty feeding vector L4440 as a control. Plates were maintained in 20°C, and four P0 larvae were transferred to fresh RNAi plates every day. F1s gravid adults were transferred to RNAi plates containing different amount of tunicamycin. The rnf-121(ok848) worms exhibit higher sensitivity to 2 μg/ml tunicamycin compared with rnf-121(RNAi)-treated worms suggesting incomplete silencing of this gene by RNAi (compare Figure 2A to Figure 3A, top, at 2 μg/ml tunicamycin).
Figure 2.
Inactivation of rnf-121 increases sensitivity to ER stress and induces the UPR. (A) N2 (wild-type) and rnf-121(ok848) embryos were treated with indicated concentrations of tunicamycin, and the various developmental stages were analyzed after 72-h incubation at 20°C. Animals were categorized into three groups: adults and L4, arrested L3 and younger and dead larvae. Each category is represented by the bar graph as the percentage of the total number of embryos (n = total number of embryos from 9 to 12 plates of 3 independent experiments, indicated above each bar). (B) Elevated basal expression of the hsp-4::gfp transcriptional reporter in rnf-121(RNAi)-treated worms. The hsp-4::gfp transgenic worms were fed with control (vector) or rnf-121(RNAi) bacteria in regular medium at 20°C. Arrow indicates the intestinal anterior cells.
Figure 3.
RNF-121 is regulated by the PEK-1 pathway. (A) N2 (wild-type), ire-1(v33), pek-1(ok275), and atf-6(ok551) mutant worms were treated with control vector (−) or rnf-121(RNAi) (+) and were subjected to the tunicamycin sensitivity assay. The various developmental stages were analyzed after 72-h incubation at 20°C. Animals were categorized into three groups: adults and L4, arrested L3 and younger, and dead larvae. Each category is represented by the bar graph as the percentage of the total number of embryos ([n] indicated above each bar, total number of embryos from 9 to 12 plates of 3–6 independent experiments). (B) Differential expression of rnf-121 and hsp-4 after treatment with DTT, tunicamycin, and thapsigargin and in ire-1, atf-6, and pek-1 mutant worms. Relative quantification of each gene was determined using real-time RT-PCR. Mean values ± SEM from three independent treatments and RNA preparations are shown. Fold regulation was calculated relative to N2 (wild-type) nontreated animals. (C) RNF-121::GFP protein levels in wild-type (top) and pek-1(ok275) mutant worms (bottom) after treatment with tunicamycin and DTT. Total lysates (50 μg) were subjected to immunoblot analysis with anti-GFP and anti-actin antibodies.
For Western analysis, RNF-121::GFP and RNF-121::GFP;pek-1(ok275) worms were treated in M9 buffer supplemented with OP50 bacteria for 4 h at 20°C with the indicated concentrations of tunicamycin or DTT. After treatment, worms were washed three times in M9 buffer, and lysates were prepared using Laemmli buffer. Proteins were separated on SDS-PAGE followed by immunoblot analysis with anti-GFP (Roche Diagnostics) (1:6000) and anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA) (1:2000) antibodies.
Real-Time Polymerase Chain Reaction (PCR) Analysis
Real-time PCR analysis was performed to the mutant strains ire-1(v33), pek-1(ok275), and atf-6(ok551) as well as to wild-type (N2) worms treated with the UPR inducers thapsigargin (5 μM; 4 h), tunicamycin (25 μg/ml; 4 h), and DTT (3 mM; 4 h). At least three biological repeats for each strain and treatment were analyzed. Total RNA was extracted using TRIzol and reverse-transcribed with oligo(dT) and the SuperScript III First-Strand Synthesis kit (Invitrogen, Carlsbad, CA). cDNAs were used as templates for amplification and detection with the SYBR Green dye in an ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA). Relative quantity (RQ) values were calculated using the comparative CT method (ABI Prism SDS software; Applied Biosystems). For endogenous control, we used the small RNA polymerase subunit (F23B2.13). Calibrator samples were prepared from wild-type worms for the UPR mutants and from wild-type worms treated with dimethyl sulfoxide (for thapsigargin and tunicamycin treatments) or water (for DTT treatment).
RNF-121 Induction in Transgenic Worms
Worms were synchronized to L1 and were grown at 15°C. Induction by heat shock was done at 32°C for 30 min at the mid-L2 stage. After heat shock, plates were returned to 15°C. Differential interference contrast (DIC) analysis was performed on adults, and analysis of PAT-3::GFP expression was performed 3 h after heat shock.
Motility Assay
Young adult hermaphrodites were treated with heat shock (32°C; 30 min) and at the indicated time points they were transferred to a drop of M9 medium on an unseeded NGM plate and allowed to acclimate for 30 s. Motility rate was determined by counting the number of body bends in 1-min intervals. A single bend was defined as a sinusoidal movement of the anterior region from left to right and left again. Mean and SE values of body bend per minute were calculated for each group (n = 40).
Microscopy and Quantification
Oocyte maturation and ovulation was observed by time-lapse Nomarski microscopy as described previously (McCarter et al., 1997). Confocal analysis of RNF-121::GFP expression was performed using an LSM 5 EXCITER confocal scanning microscope (Carl Zeiss, Jena, Germany) by using a 63× 1.4 numerical aperture objective lens. Double transgenic worms were recorded using multitracking configuration to prevent signal cross-talk. RNF-121::GFP;mRFP::SP12 (1.1 scan zoom, pinhole 1.4 Airy Unit, 83.2 × 22.3 μm confocal image size) and RNF-121::GFP; mRFP::MANS (2.0 scan zoom, pinhole 1.3 Airy Unit, 47.8 × 14.8 μm confocal image size). Worms were mounted on 4% agarose pads and anesthetized with 0.1% tricaine and 0.01% tetramisole. Mean fluorescence intensities were quantified using LSM 5 EXCITER software. The effect of induced RNF-121 on PAT-3::GFP expression in the body wall muscles was weaker; therefore, quantification was done by normalization of the fluorescence intensity in the distal tip cells (DTC) to the to the fluorescence intensity in the muscle perinuclear region. For fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) analyses, laser power and gain were adjusted so that pixels in the inclusions were under saturation levels. For FRAP experiments, photobleaching was performed by 4× iterative scanning (100% transmission) focused on a region of 1 μm2. Images were acquired (1% transmission) in a single Z-plane by using open pinhole at 10-s intervals. In FLIP experiments, a region outside an inclusion was repeatedly bleached 1× scanning) at 10-s intervals, and images were acquired after each bleach.
RESULTS
RNF-121 Is Localized to the ER in C. elegans and Exhibits In Vitro E3 Ubiquitin Ligase Activity
RNF-121 (C16C10.5) is a highly conserved protein with putative transmembrane domains (TMDs) and a RING finger domain. Computer algorithms (PSIPRED, Polyphobius, TMpred, SPLIT, SOSUI, TMHMM2.0, and ConSeq) predict six transmembrane spans and a cytoplasmic loop that includes the RING-H2 domain (C3H2C3 type) (Figure 1A and Supplemental Figure S1A). This structure resembles the predicted structure of human HRD1 (Kikkert et al., 2004). To examine the expression pattern and subcellular localization of RNF-121, we generated a GFP translational fusion including 5.7 kb of the upstream sequence and the full genomic sequence of rnf-121 fused to GFP at its C terminus. The RNF-121::GFP transgene is expressed in intracellular reticular structures as well as in the perinuclear region, suggesting its localization to the ER (Figure 1, B–F). ER membrane localization was confirmed by colocalization with the mRFP::SP12 ER reporter (12-kDa signal peptidase subunit) (Rolls et al., 2002; Kubota et al., 2006; (Figure 1D). SP12 has been shown to be enriched in the rough ER membrane (Vogel et al., 1990); however, in C. elegans neurons it is expressed throughout the ER membrane, both in the cell body and neurites (Rolls et al., 2002). Expression of RNF-121::GFP is also detected in scattered dots that are colocalized with the Golgi reporter α-mannosidase II (Rolls et al., 2002; Figure 1C, arrow; and G). In addition to its localization in the sarcoplasmic reticulum and the ER of the hypodermis (Figure 1, B–D), we suggest that RNF-121 is expressed in the ER membrane of the seam cells (specialized epithelial cells) (Figure 1C), vulva, uterus, spermatheca (Figure 1E), sheath cells (data not shown) and DTCs (Figure 1F), based on the typical pattern of intracellular reticular network in these tissues. This broad expression pattern suggests a physiological role for RNF-121 in ER homeostasis during normal development. To check whether RNF-121 has an intrinsic E3 ligase activity, we tested the activity of the RING-H2 domain in a substrate-independent polyubiquitination assay (Lorick et al., 1999; Yang et al., 2005). Incubation of bacterially expressed GST-RNF-121RING with UBCH5c, in the presence of E1 enzyme, ubiquitin and ATP resulted in the formation of high-molecular-weight ubiquitin conjugates (Figure 1H, top, lane 1). The ligase activity was abolished when Zn2+-liganding residues that are critical for the structure of the RING domain (C222AC225A) were mutated, or when a mutation was introduced in the predicted E2 binding site (V224A) (Brzovic et al., 2003; Katoh et al., 2003; Figure 1H, top, lanes 2 and 3). GST-RNF-5 exhibited high E3 ligase activity under these conditions and was used as a positive control (Figure 1H, top, lane 5). We concluded that RNF-121 is localized to the ER membrane of various tissues and cells in C. elegans and has in vitro ubiquitin ligase activity.
Figure 1.
Topology prediction, expression pattern, and in vitro E3 ligase activity of RNF-121. (A) Schematic representation of the predicted membrane topology of RNF-121. Six TMDs are predicted, the RING finger domain faces the cytosol. The amino acid sequence of the RING finger domain is shown. The cysteine and histidine residues of the RING finger domain are shown in red. The predicted E2 binding site (V224) is labeled in yellow (B and C) RNF-121::GFP is localized to the ER in body wall muscles (mu), hypodermis (hyp), and seam cells (SE). Bar, 10 μm. (D) RNF-121::GFP colocalizes with the mRFP::SP12 reporter. Bar, 5 μm. (E and F) RNF-121::GFP is expressed in the somatic gonad and vulva. Lateral view of midL4 animal. Expression is detected in the vulval cells (vul), spermathecal cells (sp), uterine cells (ut), and the distal tip cell (DTC). Bar, 10 μm. (G) RNF-121::GFP colocalizes with the mRFP::mans reporter. Bar, 5 μm. (H) In vitro self-ubiquitination assays were performed using wild-type GST-RNF-121RING or RING mutant forms of bacterially expressed and purified protein. In lane 4 (−), the reaction includes E1, E2, Ub, and ATP without the E3 ligase. GST-RNF-5 served as a positive control. Arrow indicates the E1 enzyme. Arrowhead indicates the 5% stacking gel-10% running gel boundary. High-molecular-weight smears generated by autoubiquitination of the indicated E3 are labeled with parenthesis. Coomassie-stained gel of the purified glutathione transferase (GST)-tagged expressed proteins is shown (bottom).
Inactivation of RNF-121 Induces ER Stress
rnf-121(ok848) mutant worms (isolated by the C. elegans Gene Knockout Consortium) develop normally and are fertile. The ok848 allele harbors a 1397-bp deletion that spans four of the six predicted TMDs (2–5) and the RING finger domain (Supplemental Figure S1B). To test the possible role of RNF-121 in the cellular response to ER stress, we compared the growth of wild-type (N2) and rnf-121(ok848) mutant worms in a medium containing tunicamycin, an inhibitor of N-glycosylation. At 2 μg/ml tunicamycin 67.6 ± 15.2% of the mutant rnf-121(ok848) worms were arrested at or before the L3 stage, whereas only 22.4 ± 16.5% of N2 (wild-type) animals were arrested (Figure 2A, yellow bar). At 5 μg/ml tunicamycin, we observed major growth arrest and larval lethality in both N2 and rnf-121(ok848) worms. Growth of worms in this concentration of tunicamycin probably leads to accumulation of high levels of unfolded proteins, beyond adaptation capabilities. At the highest tested concentration of tunicamycin (7.5 μg/ml), the distribution was similar and all worms were arrested or died (Figure 2A and Supplemental Table S1). These results suggested that inactivation of RNF-121 induces a low level of ER stress that does not interfere with normal development under optimal growth conditions but has an inhibitory effect on development in response to relatively low levels of exogenous ER stress. Next, we tested whether reduced RNF-121 activity elicits the UPR pathway. The proteins HSP-3 and HSP-4 are the C. elegans orthologues of the mammalian GRP-78/BiP (Shen et al., 2001). We analyzed the expression levels of the hsp-4::gfp transcriptional reporter (Calfon et al., 2002) in rnf-121(RNAi)-treated worms. In control wild-type worms, the hsp-4::gfp reporter displays low fluorescence levels, whereas the expression is strongly induced in the epidermis and intestine upon UPR activation (Calfon et al., 2002). The rnf-121(RNAi)-treated worms exhibit high levels of intestinal GFP expression under normal growth conditions (Figure 2B, bottom, arrow). Higher hsp-4::gfp levels were also observed in the rnf-121(ok848) mutant worms (Supplemental Figure S2). This increase in hsp-4::gfp expression demonstrated that reducing the level of RNF-121 causes UPR activation.
RNF-121 Acts through the PERK/PEK-1 Signaling Pathway
The above-mentioned experiments indicate that depletion of RNF-121 causes ER stress and activates the UPR. To determine whether RNF-121 acts via the UPR pathway, we analyzed sensitivity to ER stress after depletion of RNF-121 in a mutant background of each of the three known ER stress transducers in C. elegans: IRE-1, PEK-1, and ATF-6. The ire-1(v33) and pek-1(ok275) mutant strains are more sensitive to tunicamycin than the wild-type (Figure 3A, − bars; Shen et al., 2001), whereas atf-6(ok551) worms are less sensitive to tunicamycin (Shen et al., 2005), and in our hands are more resistant than the wild type (Figure 3A, − bars). Sensitivity to tunicamycin was increased in ire-1(v33);rnf-121(RNAi), and atf-6(ok551);rnf-121(RNAi) compared with the ire-1(v33) and atf-6(ok551) mutant worms, respectively (Figure 3A, compare − to + bars, and Supplemental Tables S1–S4; p < 0.005). However, rnf-121(RNAi) treatment had no significant effect on tunicamycin sensitivity of pek-1(ok275) mutant worms (p > 0.2) (Figure 3A, bottom, 2–3 μg/ml tunicamycin, and Supplemental Table S5). These results suggest that RNF-121 operates in C. elegans in the same genetic pathway with PEK-1. Because pek-1(ok275) worms are more sensitive to tunicamycin than rnf-121(RNAi)-treated wild-type N2 worms (Figure 3A and Supplemental Tables S1 and S5), this suggests that rnf-121 is located downstream of pek-1 and therefore depletion of rnf-121 did not cause a more severe phenotype in the pek-1(ok275) mutant worms. PERK not only mediates general translational attenuation upon ER stress but also increases the translation of the ATF4 transcriptional activator and thus regulates gene expression during ER stress response (Scheuner et al., 2001; Harding et al., 2003). To determine whether the transcription of rnf-121 is regulated by PEK-1 and the UPR pathway in C. elegans, we performed a real-time PCR analysis of the mutant strains pek-1(ok275), ire-1(v33), and atf-6(ok551), as well as of wild-type worms, treated with the UPR inducers DTT, tunicamycin, and thapsigargin. Although the mRNA levels of hsp-4 were induced upon tunicamycin or DTT treatment and in pek-1(ok275) and atf-6(ok551) mutant backgrounds, and abolished in ire-1(v33) as shown previously (Shen et al., 2001), the levels of rnf-121 mRNA were largely unaffected (Figure 3B). No changes in rnf-121 mRNA levels were detected when any of the UPR mutants were treated with tunicamycin (data not shown). These results indicate that RNF-121 is not regulated on the transcriptional level by PEK-1 and the UPR system. To examine whether RNF-121 protein level is regulated by the PEK-1 pathway, we performed immunoblot analysis of RNF-121::GFP worms treated with tunicamycin and DTT. Although the level of RNF-121 increases after ER stress in wild-type background, it is constantly high in pek-1(ok275) worms (Figure 3C) and does not increase after ER stress. It suggests that PEK-1 pathway activates RNF-121 translation or protein stability under ER stress conditions, whereas inactivation of PEK-1 during development in the pek-1(ok275) mutant may cause ER stress and up-regulation of RNF-121 by an alternative pathway.
Overexpression of RNF-121 Causes Defects in Gonad and Germline Development That Depends on DER-1/Derlin-1
The number of identified ER-membrane located E3-ligases in multicellular organisms is increasing, suggesting specific roles for these E3s as the ability to recognize specific ERAD substrates. We expressed RNF-121 at specific stages of development by using a heat-inducible promoter. Moderate decrease in worm motility was detected following RNF-121 expression in adults (Supplemental Figure S3). We observed defects in normal development only when RNF-121 was induced at a specific developmental stage, the mid-L2 stage, resulting in decreased hermaphrodite fertility. The average number of progeny of hsp-16p::RNF-121 was 55 ± 18 (n = 65) compared with 254 ± 28 (n = 60) in wild-type worms treated with the same heat-shock conditions at the mid-L2 stage (Supplemental Figure S4). To characterize this partial sterility phenotype, we analyzed gonad morphology and oocyte development, maturation, and ovulation in young adult worms. The C. elegans gonad is formed from two symmetrical U-shaped arms, each connected by a spermatheca to a common uterus. At the L2 stage, migration of the hermaphrodite gonads begins, guided by two special somatic cells, the DTCs (Kimble and White, 1981). Germ cells proliferate in the distal region of the gonad and as they move proximally, they enter meiosis and differentiate into sperm or oocyte. Oocyte meiotic maturation occurs at the most proximal gonad, followed by entrance into the spermetheca at ovulation and fertilization (McCarter et al., 1999). After induction of RNF-121 at the mid-L2 stage, the adult gonad arms exhibited defective gametogenesis that resulted in proximal gonads with a small number of oocytes (Figure 4, C–H, 3.6 ± 1.5 oocytes per gonad arm, n = 61) in mid- and late-stage adults compared with 9.6 ± 1.5 oocytes in mid-stage wild-type adults (n = 49) and 9.2 ± 1.3 in mid-stage adults expressing a ligase-inactive RNF-121 mutant (hsp-16::RNF-121C222AC225A; n = 67). DIC and 4,6-diamidino-2-phenylindole (DAPI) analyses revealed delayed development of the oocytes in the proximal gonads (Figure 4, C–E, compared with Figure 4, A and B). In wild-type gonad, progression of nuclei from pachytene into diplotene occurs in the loop region, whereas in the proximal gonad arm oocytes progress to diakinesis (Figure 4B, short arrow). In contrast, pachytene nuclei were found in the hsp-16p::RNF-121 proximal gonads after the loop region, indicating a delay in progression into diplotene (72.6 ± 4.5%; n = 256) (Figure 4E, arrowhead). In addition, the developing oocytes and especially the most proximal oocyte had abnormal elongated shapes (Figure 4, C, D, and F, white outline) and were not rectangular as the wild-type oocytes before maturation (Figure 4A, white outline) (McCarter et al., 1999). Meiotic maturation rates were slower (0.85 ± 0.23 maturations/gonad arm/h; n = 27) relative to the maturation rates in wild-type hermaphrodite that were treated with the same heat-shock conditions (2.65 ± 0.50 maturations/gonad arm/h; n = 16). Abnormal signaling from the sperm to the oocyte or sheath cells that surround the oocyte could contribute to the lower maturation phenotype (McCarter et al., 1999; Miller et al., 2001). Because the sperm in the hsp-16::RNF-121 adults seem abnormal and swollen (Figure 4F, arrow), we examined whether cross to wild-type males will rescue the partial sterility phenotype. Indeed, the wild-type sperm partially rescued the sterility and the number of progeny increased (118 ± 37, n = 31) (Supplemental Figure S4).
Figure 4.
Overexpression of RNF-121 at the L2 stage results in germline and gonad defects. (A and B) DIC and DAPI staining of wild-type adult hermaphrodite gonad arms. Marked are pachytene nuclei (arrowhead), diplotene (long arrow), and oocytes at diakinesis (short arrow). A scheme of the DTC path is shown on the left. (C–E) DIC and DAPI staining of hsp-16p::RNF-121 adult hermaphrodite gonad arms. Marked are pachytene nuclei (arrowhead) and oocytes at diakinesis (short arrow). The first oocyte is outlined. (F) Abnormal sperm in the spermatheca of hsp-16p::RNF-121 adult hermaphrodite (arrow). (G and H) DTCs migration defects in hsp-16p::RNF-121 adults. Bar, 10 μm. (I) Percentage of animals with abnormal gonads. Analysis was performed to the transgenic lines (3 independent lines from each construct) hsp-16p::RNF-121, hsp-16p::RNF-121C222AC225A, and hsp-16p::RNF-121 treated with der-1(RNAi). RNAi of der-1 in wild-type worms (N2 background) was performed as a control. Additional controls are N2 worms and transgenic worms expressing the E3 ligase RNF-5. Worms were treated with heat shock at mid-L2 stage to induce transgene expression (+) or nontreated controls (−). n = total number of worms analyzed in three to five independent experiments (indicated above each bar).
In addition to abnormalities in oocyte development and maturation after induction of RNF-121, we observed gonad migration defects that involved extra turns and abnormal migration paths, as a 45° migration toward the dorsal side followed by a reversed migration to the ventral side and repeated migration toward the dorsal side (Figure 4G) or a reversed migration on the ventral side till the midbody (Figure 4H). These phenotypes depend on the expression of an intact RING finger domain of RNF-121 and could be detected at lower percentage without heat shock treatment, suggesting that low levels of wild-type protein expressed through the basal activity of the heat shock promoter are effective, in contrast to a RING mutant (Figure 4I). To determine the functional relation between the E3 ligase activity of RNF-121 and the ERAD pathway, we induced RNF-121 expression after depletion of the putative retrotranslocation component der-1 (Ye et al., 2004). Depletion of der-1 in wild-type background had no effect on gonad development (Figure 4I). However, analysis of gonads in der-1(RNAi) adult hermaphrodites after induction of RNF-121 suppressed the RNF-121 effect and showed low penetrance of the gonad phenotype (Figure 4I), suggesting that RNF-121 acts with and requires Derlin-1 for its activity in the ERAD pathway. We conclude that ectopic expression of RNF-121 at early stage of gonad development impaired gonad and germline development and that this activity depends on DER-1, probably by contributing to active retrotranslocation of substrates from the ER to the cytosol.
PAT-3/β-Integrin::GFP Is a Substrate for RNF-121
The C. elegans integrin receptors are necessary for normal gonad development and their depletion results in similar phenotypes as described above after overexpression of RNF-121. In C. elegans, there are two α-integrin subunits, INA-1 and PAT-2, and one β-integrin subunit, PAT-3 (Cox et al., 2004). PAT-3 is expressed in the body-wall muscles and the somatic gonad (Hresko et al., 1994; Gettner et al., 1995) and is essential for muscle attachment and organization of the sarcomere during embryogenesis (Williams and Waterston, 1994) and for gonad migration and oocyte development and maturation during postembryonic development (Lee et al., 2001, 2005). In mammalian cells, integrin-β1 is expressed in excess over integrin α-chain, and the excess of noncomplexed β1 is retained in the ER and degraded (Heino et al., 1989; Koivisto et al., 1994). To check whether PAT-3 is an ERAD substrate for RNF-121 E3 ligase activity in vivo, we used the PAT-3/β-integrin::GFP transgenic strain (Plenefisch et al., 2000). The GFP-tagged β subunit expressed through the PAT-3::GFP reporter exhibits a similar pattern as the anti-PAT-3 (MH25) antibody (Francis and Waterston, 1991) with an additional ER membrane localization, probably as a result of high expression levels of the β subunit (Figure 5A). This suggests that the unassembled β subunits are retained in the ER and subjected to degradation through the ERAD pathway. To test the possibility that PAT-3::GFP is degraded by RNF-121, we first measured GFP levels in the PAT-3::GFP;hsp-16p::RNF-121 double transgene after induction of RNF-121 expression during the first phase of gonad migration (mid-L2 stage). PAT-3::GFP levels are high in the ER of the gonad DTCs at this stage of development, whereas fluorescence intensity decreased after induction of RNF-121. The mean ratio of fluorescence intensity DTC/muscle (DTC values were normalized to the fluorescence intensity in the muscle perinuclear region; see Materials and Methods) was lower in the PAT-3::GFP;hsp-16p::RNF-121 worms than in the PAT-3::GFP control strain (0.39 ± 0.07 vs. 0.58 ± 0.11, respectively; n = 50 for each group; p < 0.0001) (Figure 5, A and B). This suggests that RNF-121 is able to ubiquitinate the ER-localized PAT-3::GFP in the DTCs and send it for proteasomal degradation in the cytoplasm. To test whether PAT-3::GFP is ubiquitinated by RNF-121, we performed in vitro ubiquitination assays. To this end, PAT-3::GFP was immunoprecipitated using anti-GFP antibody from rnf-121(ok848); PAT-3::HA::GFP mutant worms that contain a deletion in the rnf-121 coding sequence. The immune-purified PAT-3::GFP was than subjected to in vitro ubiquitination assay using bacterially expressed and purified GST-RNF-121RING in the presence of HA-tagged ubiquitin. This reaction resulted in the formation of high-molecular-weight conjugates that were recognized by antibodies to HA (recognizes both PAT-3 and HA-Ub) as well as antibodies to GFP (Figure 5C). Immune-purified PAT-3::GFP from PAT-3::GFP strain that does not include the rnf-121 deletion also could be ubiquitinated by wild-type RNF-121 but not by a RING finger mutant (Supplemental Figure S5). These data provide direct evidence for the ubiquitination of PAT-3::GFP by RNF-121.
Figure 5.
PAT-3::GFP degradation and accumulation depend on RNF-121 activity. (A) PAT-3::GFP expression in control (top) and in hsp-16p::RNF-121 (bottom). The nuclear envelope (NE) and surrounding reticulum of two body-wall muscle cells and the DTCs are shown in each panel (confocal images). Analysis was performed 3 h after heat shock at the mid-L2 stage. Muscle cell are outlined in red and DTCs in yellow. The NE is labeled with an arrowhead. Bar, 5 μm. (B) Distribution of the ratio of fluorescence intensities (DTC/muscle) in PAT-3::GFP (blue) and PAT-3::GFP; hsp-16p::RNF-121 (black) worms. Intensity values in the DTCs (yellow) are normalized to the intensity values in the nearest muscle nuclei (red) at the same focal plane. Analysis was performed on n = 50 worms from each strain (p < 0.0001) (C) In vitro ubiquitination of PAT-3::GFP immunoprecipitated from rnf-121(ok848); PAT-3::HA::GFP worms. Equal aliquots of immunopurified PAT-3::HA::GFP were used in each reaction in the presence of bacterially expressed and purified GST-RNF-121RING, E2, HA-ubiquitin and ATP, with or without E1 as indicated. Reactions were incubated at 37°C for 20 min and terminated with 8 M urea buffer. Black arrows indicate PAT-3::HA::GFP. Gray arrows indicate a putative spliced-form of PAT-3::HA::GFP. Arrowheads indicate the 5% stacking gel-8% running gel boundary. The ubiquitinated forms of PAT-3::HA::GFP are indicated by parentheses. IP, 25% of the immunoprecipitate used in each ubiquitination reaction. (D) PAT-3::GFP animals treated with control bacteria (empty L4440 vector; top) or rnf-121(RNAi) bacteria (bottom). Worms were grown in normal medium at 20°C. Variable sizes of fluorescent inclusions are shown (arrows) in PAT-3::GFP;rnf-121(RNAi) young adults. (E) Quantitative analysis of inclusion formation in PAT-3::GFP worms. Three independent experiments were performed (n = 150). Mean and SEM values are presented. Control worms were fed with empty L4440 vector. (F) Accumulation of fluorescent inclusions in rnf-121(ok848) worms (top) and immunoblot analysis of GFP and actin levels in total worm lysates (100 μg) from the indicated strains (bottom); rnf-121(ok848) worms were grown in liquid culture at 25°C (G) FRAP analysis of PAT-3::GFP inclusions in muscle cells of rnf-121(RNAi) animals. The boxed area inside the inclusion was photobleached. Example of a representative inclusion is shown (n > 15). (H) FLIP analysis of PAT-3::GFP inclusions in rnf-121(RNAi) animals. The marked region was repeatedly photobleached. Representative analysis is shown (n > 15). (I) PAT-3::GFP animals treated with 2 μg/ml tunicamycin accumulate smaller fluorescent inclusions. Bar, 5 μm.
To test whether PAT-3 is a main target of RNF-121 E3 ligase activity in the gonad, we analyzed young adults hsp-16::RNF-121;PAT-3::GFP double transgene after RNF-121 induction at the mid-L2 stage (Supplemental Figure S6). The percentage of worms with abnormal gonad was 2.8-fold lower in the double transgene (25.9% in hsp-16::RNF-121;PAT-3::GFP compared with 72.6% in hsp-16::RNF-121; Figure 4I and Supplemental Figure S6), indicating that high levels of PAT-3::GFP partially suppress the gonad defects caused by RNF-121 and suggesting that PAT-3 is a primary substrate of RNF-121 in the gonad.
We next tested PAT-3::GFP expression in rnf-121(RNAi)-treated worms and compared it to wild-type background. Few GFP inclusions (2.7 ± 0.9/worm) could be detected in 31.3 ± 9.0% of the PAT-3::GFP worms (n = 150) (Figure 5, D and E); however, strong increase in fluorescence and accumulation of PAT-3::GFP inclusions (12.9 ± 2.9/worm) was detected in 70.7 ± 18.1% of the rnf-121(RNAi)-treated worms (n = 150) (Figure 5, D and E). In accordance, massive accumulation of inclusions and high GFP levels were detected in PAT-3::GFP;rnf-121(ok848) mutant larvae (Figure 5F). The nature of inclusions and the dynamics of PAT-3::GFP molecules within the inclusions were analyzed by FRAP. Photobleach in a restricted area in the inclusion affected the entire structure implying that the molecules in the inclusion are mobile. However recovery of fluorescence was not detected after 7 min of analysis (as well as 40 min; data not shown) indicating that the misfolded PAT-3::GFP molecules within the inclusion do not exchange with a soluble pool of PAT-3::GFP (Figure 5G). Next, using a complementary FLIP analysis, we could not detect exchange with the cytosolic fraction, implying that the PAT-3::GFP molecules are tightly bound in the inclusions and suggesting that the inclusions are membrane bound (Figure 5H). The inclusion pattern differed from smaller inclusions that appeared after 2 μg/ml tunicamycin treatment, which probably reflected the retention of nonglycosylated PAT-3 precursors in the ER (n = 14) (Figure 5I).
The intense accumulation of PAT-3::GFP inclusions suggests that RNF-121 is essential for the regulated degradation of unassembled PAT-3::GFP subunits in the ER.
DISCUSSION
RNF-121 is a newly identified E3 ligase that is localized to the ER, is essential for larval development during ER stress conditions, and is capable to degrade a type I transmembrane ERAD substrate, PAT-3::GFP. Its predicted structure resembles the predicted structure of Hrd1/Synviolin (Kikkert et al., 2004), harboring six transmembrane domains that may be important for recognition of integral membrane substrates and a RING finger catalytic site exposed to the cytoplasmic ER surface. A difference between the predicted structures of the two proteins is at the C terminus, which contains additional transmembrane domain in RNF-121. The variety of structurally similar ER-related E3 ligases in higher eukaryotes (Kostova et al., 2007) may suggest complementary roles at different stages of the ER stress response. For example, Hrd1 is thought to have a role in the nonadaptive ER stress response that lead cells to apoptosis, because it has been shown that Synovial cells in rheumatoid arthritis overexpress Hrd1/Synviolin and show resistance to ER-stress induced apoptosis (Amano et al., 2003). Alternatively, we demonstrate here that inactivation of RNF-121 is associated with increased ER stress at mild levels of tunicamycin, suggesting a role for RNF-121 in the adaptive response to ER stress. In contrast to Hrd1 and additional ERAD components that are regulated by the UPR on the transcriptional level through the IRE1-XBP1 pathway (Travers et al., 2000; Molinari et al., 2003; Yamamoto et al., 2008), the regulation of RNF-121 is through the PERK/PEK-1 pathway. We detected elevation in RNF-121 protein and not in its mRNA levels upon ER stress. Our data suggest that RNF-121 is regulated by PEK-1 by increased translation or protein stability probably through a tissue-specific downstream target of PEK-1. In the absence of PEK-1, RNF-121 protein levels are constantly high and are not further elevated after ER stress. It suggests that inactivation of PEK-1 causes constant ER stress that results in increased RNF-121 protein levels regulated through an alternative pathway, for example by ATF-6, which was shown to act redundantly to PEK-1 in C. elegans (Shen et al., 2005).
The germline defects that resulted in sterility after induction of RNF-121 at the mid-L2 stage suggest uncontrolled degradation of integrin subunits as well as additional regulators that mature in the ER and function in the developing somatic gonad. Substrates are expected to be expressed in somatic cells because RNF-121 expression is driven by an extrachromosomal array, which is subjected to germline suppression (A. Fire). Therefore, the observed effects are mainly a result of RNF-121 activity in the somatic cells of the gonad or in the body wall muscle cells that assemble the basal laminae on which the distal tip cells migrate. The somatic gonad at the L2 stage includes the two DTCs, the anchor cell, uterine precursor cells, and sheath/spermathecal precursor cells (SS cells) (McCarter et al., 1997). Laser ablation of the SS precursor cells has been shown to result in various germline abnormalities as reduced germline proliferation, defective exit from pachytene stage, feminization of the germline, and endomitotic oocytes (Emo) due to ovulation failure (McCarter et al., 1997). Induction of RNF-121 at this stage caused similar phenotypes as small gonad arms, delay in pachytene exit and abnormal sperm, suggesting damage to the SS precursor cells. Moreover, overexpression of RNF-121 caused severe abnormalities in gonad path that reflect defects in cell migration and therefore in the function of the DTCs. This suggests an association between RNF-121 and regulation of cell–extracellular matrix adhesion, probably by effecting the maturation and exit from the ER of factors required for controlled cell adhesion. Previous support for a possible link between the ERAD process and cell adhesion has been shown for the ubiquitin ligase gp78 that targets the transmembrane metastasis suppressor KAI1 for degradation (Tsai et al., 2007). We focused on PAT-3::GFP as the first identified substrate for RNF-121. Integrin-α and -β subunits are type I transmembrane glycoproteins that must be correctly assembled in the ER, transported to the Golgi, and finally transported to the plasma membrane as the mature assembled heterodimers. In mammalian cells, association of preintegrin-β1-chain with calnexin suggests that it is subjected to the ER protein quality control (Lenter and Vestweber, 1994). The data presented here imply that RNF-121 may trigger ERAD of PAT-3::GFP and therefore participates in the primary steps of substrate recognition as well as its ubiquitination. RNF-121 effect depends on DER-1, providing further support for its function as part of the ERAD machinery. A downstream component in β-integrin regulation is the SCFFbx2 ubiquitin ligase that has been shown previously to bind glycosylated preintegrin-β1 (Yoshida et al., 2002) and may cooperate with RNF-121 in the regulated degradation of nonassembled integrin-β1 after its retrotranslocation from the ER.
In summary, we identified a highly conserved RING finger E3 ligase that is localized to the ER of various tissues in C. elegans and may function as a component of the ERAD machinery in higher organisms. By analysis of its effects on PAT-3::GFP expression, we suggest a link between the ERAD pathway and cell adhesion through the regulation of β-integrin in the ER.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kiyoji Nishiwaki (RIKEN Center for Developmental Biology) for the unc-54p::mRFP::SP12 plasmid, John Plenefisch (University of Toledo) for the rhIs2[PAT-3::HA::GFP] strain, and Rachel Kaminsky and Sharon Sheffy-Levin for technical assistance. We thank the C. elegans Gene Knockout Project team at OMRF and the CGC for providing strains. This study was supported by a Research Career Development Award from the Israel Cancer Research Fund (06-203-RCDA) and by The Israel Science Foundation (grant 980/06) (to L. B.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0774) on March 31, 2010.
REFERENCES
- Amano T., et al. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 2003;17:2436–2449. doi: 10.1101/gad.1096603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L., Hauser C. A., Kolotuev I., Ronai Z. Muscle-epidermis interactions affect exoskeleton patterning in Caenorhabditis elegans. Dev. Dyn. 2007;236:3129–3136. doi: 10.1002/dvdy.21341. [DOI] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Podbilewicz B., Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J. Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P. S., Keeffe J. R., Nishikawa H., Miyamoto K., Fox D., 3rd, Fukuda M., Ohta T., Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. A., Tuskey C., Hardin J. Cell adhesion receptors in C. elegans. J. Cell Sci. 2004;117:1867–1870. doi: 10.1242/jcs.01177. [DOI] [PubMed] [Google Scholar]
- David Y., Ziv T., Admon A., Navon A. The E2 ubiquitin conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A., et al. et al. The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PLoS ONE. 2008;3:e1609. doi: 10.1371/journal.pone.0001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Didier C., et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 2003;23:5331–5345. doi: 10.1128/MCB.23.15.5331-5345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Waterston R. H. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gettner S. N., Kenyon C., Reichardt L. F. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Heino J., Ignotz R. A., Hemler M. E., Crouse C., Massague J. Regulation of cell adhesion receptors by transforming growth factor-beta. Concomitant regulation of integrins that share a common beta 1 subunit. J. Biol. Chem. 1989;264:380–388. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hresko M. C., Williams B. D., Waterston R. H. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh S., Hong C., Tsunoda Y., Murata K., Takai R., Minami E., Yamazaki T., Katoh E. High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides. J. Biol. Chem. 2003;278:15341–15348. doi: 10.1074/jbc.M210531200. [DOI] [PubMed] [Google Scholar]
- Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Koivisto L., Heino J., Hakkinen L., Larjava H. The size of the intracellular beta 1-integrin precursor pool regulates maturation of beta 1-integrin subunit and associated alpha-subunits. Biochem. J. 1994;300:771–779. doi: 10.1042/bj3000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z., Tsai Y. C., Weissman A. M. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft S. G., Wang L., Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J. Biol. Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Sano M., Goda S., Suzuki N., Nishiwaki K. The conserved oligomeric Golgi complex acts in organ morphogenesis via glycosylation of an ADAM protease in C. elegans. Development. 2006;133:263–273. doi: 10.1242/dev.02195. [DOI] [PubMed] [Google Scholar]
- Lee M., Cram E. J., Shen B., Schwarzbauer J. E. Roles for beta pat-3 integrins in development and function of Caenorhabditis elegans muscles and gonads. J. Biol. Chem. 2001;276:36404–36410. doi: 10.1074/jbc.M105795200. [DOI] [PubMed] [Google Scholar]
- Lee M., Shen B., Schwarzbauer J. E., Ahn J., Kwon J. Connections between integrins and Rac GTPase pathways control gonad formation and function in C. elegans. Biochim. Biophys. Acta. 2005;1723:248–255. doi: 10.1016/j.bbagen.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Lenter M., Vestweber D. The integrin chains beta 1 and alpha 6 associate with the chaperone calnexin prior to integrin assembly. J. Biol. Chem. 1994;269:12263–12268. [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Nakano A. RMA1 an Arabidopsis thaliana gene whose cDNA suppresses the yeast secl5 mutation, encodes a novel protein with a RING finger motif and a membrane anchor. Plant Cell Physiol. 1998;39:545–554. doi: 10.1093/oxfordjournals.pcp.a029403. [DOI] [PubMed] [Google Scholar]
- Matsuda N., Suzuki T., Tanaka K., Nakano A. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 2001;114:1949–1957. doi: 10.1242/jcs.114.10.1949. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., Caprioli R. M., Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Nadav E., Shmueli A., Barr H., Gonen H., Ciechanover A., Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- Omura T., et al. A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem. 2006;99:1456–1469. doi: 10.1111/j.1471-4159.2006.04155.x. [DOI] [PubMed] [Google Scholar]
- Plenefisch J. D., Zhu X., Hedgecock E. M. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development. 2000;127:1197–1207. doi: 10.1242/dev.127.6.1197. [DOI] [PubMed] [Google Scholar]
- Rolls M. M., Hall D. H., Victor M., Stelzer E.H.K., Rapoport T. A. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Sasagawa Y., Yamanaka K., Ogura T. ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells. 2007;12:1063–1073. doi: 10.1111/j.1365-2443.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shen X., et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R. E., Sakaki K., Kaufman R. J. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherpakov M., Delaunay A., Toth J., Kadoya T., Petroski M. D., Ronai Z. A. Regulation of ERAD by RNF5-dependent ubiquitination of JAMP. J. Biol. Chem. 2009;284:12099–12109. doi: 10.1074/jbc.M808222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tsai Y. C., et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- Vashist S., Ng D.T.W. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F., Hartmann E., Gorlich D., Rapoport T. A. Segregation of the signal sequence receptor protein in the rough endoplasmic reticulum membrane. Eur. J. Cell Biol. 1990;53:197–202. [PubMed] [Google Scholar]
- Williams B. D., Waterston R. H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Suzuki N., Wada T., Okada T., Yoshida H., Kaufman R. J., Mori K. Human HRD1 promoter carries a functional unfolded protein response element to which XBP1 but not ATF6 directly binds. J. Biochem. 2008;144:477–486. doi: 10.1093/jb/mvn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Lorick K. L., Jensen J. P., Weissman A. M. Expression and evaluation of RING finger proteins. Methods Enzymol. 2005;398:103–112. doi: 10.1016/S0076-6879(05)98010-5. [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H.-Y., Rosser M.F.N., Turnbull E. L., Fan C.-Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang K., Kaufman R. J. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.