Abstract
Mammalian X chromosome inactivation (XCI) is an essential mechanism to compensate for dosage imbalances between male and female embryos. Although the molecular pathways are not fully understood, heterochromatinization of the Xi requires the coordinate recruitment of multiple epigenetic marks. Using fluorescence in situ hybridization analysis combined with immunocytochemistry, we demonstrate that the chromatin remodeling protein ATRX decorates the chromatids of a single, late replicating X chromosome in female somatic cells and co-localizes with the bona fide marker of the Xi, macroH2A1.2. Chromatin immunoprecipitation using somatic, embryonic stem (ES) cells and trophoblast stem (TS) cells as model for random and imprinted XCI, respectively, revealed that, in somatic and TS cells, ATRX exhibits a specific association with sequences located within the previously described H3K9me2-hotspot, a region 5′ to the X inactive-specific transcript (Xist) locus. While no ATRX-Xi interaction was detectable in undifferentiated ES cells, an enrichment of ATRX was observed after 8 days of differentiation, indicating that ATRX associates with the Xi following the onset of random XCI, consistent with a potential role in maintenance of XCI. These results have important implications regarding a previously described escape from imprinted XCI in ATRX-deficient mice as well as cases of skewed XCI in patients with ATRX syndrome.
Introduction
Large-scale transcriptional silencing of one of the two X chromosomes in female mammals is crucial for dosage compensation between male and female embryos (Huynh and Lee 2005; Lyon 1961). Although a complex phenomenon, recent studies have provided critical insight into the molecular interactions and epigenetic mechanisms leading to the onset of transcriptional repression of an entire X chromosome during early embryonic development and maintenance of the repressed status throughout continuous cell divisions (Brockdorff 2002; Heard and Disteche 2006).
The X-inactive-specific transcript (Xist), a non-coding RNA, plays a pivotal role in mammalian X chromosome inactivation (Penny et al. 1996). During the onset of X inactivation, a massive accumulation of Xist transcripts on the future inactive X chromosome (Xi) occurs in cis, rapidly triggering a series of epigenetic changes that take place on a chromosome-wide basis (Brockdorff 2002; Penny et al. 1996; Wutz and Jaenisch 2000). Notably, a specific region within the Xi located 5′ of the Xist locus (the constitutive H3K9me2-hotspot) has been identified as a potential nucleation center for the coordinate recruitment of repressive histone and chromatin modifications that are essential for the spreading and maintenance of the silenced status on the Xi through the cell cycle (Heard et al. 2001; Rougeulle et al. 2004). As a consequence, facultative heterochromatin on the Xi has been found enriched for several histone modifications such as dimethylation of histone 3 on lysine 9 (H3K9me2), trimethylation of histone 3 on lysine 27 (H3K27me3), ubiquitination of histone 2A (ubH2A), monomethylation of histone 4 on lysine 20, several members of the Polycomb group protein family, and for the unusual core histone macroH2A1.2 (Boggs et al. 2002; Heard et al. 2001; Mermoud et al. 2002; Peters et al. 2002; Plath et al. 2003).
Although a growing list of Xi markers has been identified, our understanding of the epigenetic factors involved in the process of X chromosome inactivation remains incomplete (Brockdorff 2002). For example, additional proteins may prove essential participants within the process, as indicated by the recent identification of PARP protein associations with the inactive X chromosome (Nusinow et al. 2007). Importantly, the factors involved in the spreading of chromosomal silencing and the maintenance or clonal stability of the inactive state in different cell lineages are not clear at present.
The alpha thalassemia/mental retardation X-linked (ATRX) protein is a chromatin remodeling protein belonging to the SWI/SNF2 ATP-dependent helicase family that has been shown to associate with constitutive heterochromatin domains and play a role in the control of DNA methylation at repetitive sequences of the human genome (Gibbons et al. 2000; McDowell et al. 1999). Recent evidence also suggests a role in heterochromatin formation, chromosome alignment at the meiotic spindle, chromosome cohesion in somatic cells, and transcriptional regulation (Baumann et al. 2008; De La Fuente et al. 2004; Gibbons et al. 2000; Kieran et al. 2008; McDowell et al. 1999). Mutations within the coding region of the ATRX gene cause the ATRX-syndrome in human patients, an X-linked genetic disease (Gibbons et al. 1995, 2000), which is characterized by variable combinations of severe mental retardation, characteristic dysmorphic facial features, alpha-thalassemia, seizures, gonadal abnormalities, and sex reversal (Gibbons et al. 1997). Moreover, targeted deletion of ATRX in early preimplantation development results in embryonic lethality due to disorganized extraembryonic tissues and retention of an active paternal X chromosome in extraembryonic tissues in some heterozygous females (Garrick et al. 2006).
Using fluorescence in situ hybridization (FISH) combined with immunochemical detection, we now show that the ATRX protein accumulates at one of the X chromosomes in somatic and trophoblast stem cells. Analysis of replication timing revealed that ATRX associates with the late replicating X chromosome and co-localizes with macroH2A1.2 and ubH2A, two established markers of the inactive X chromosome. Developmental studies using chromatin immunoprecipitation and immuno-FISH in embryonic stem (ES) cells and trophoblast stem (TS) cells demonstrate that the preferential association of ATRX with the Xi is a relatively late event during spontaneous ES cell differentiation. However, ATRX remains stably associated with Xi throughout trophoblast stem cell differentiation. These results identify a novel epigenetic marker of the Xi and place ATRX within the sequence of events, leading toward the heterochromatinization of the inactive X chromosome during mammalian development.
Materials and methods
ES cell culture and differentiation
Female mouse PGK12.1 embryonic stem cells (kindly provided by Dr. Neil Brockdorff) carrying a stable female karyotype with two active X chromosomes (de Napoles et al. 2004; Mermoud et al. 1999, 2002; Silva et al. 2003; Zvetkova et al. 2005) were cultured as described elsewhere (Mermoud et al. 1999, 2002). Briefly, ES cells were plated on 0.1% gelatin-coated culture dishes in ES-Dulbecco's modified essential medium (DMEM) medium (American Type Culture Collection; Manassas, VA, USA) supplemented with 15% fetal bovine serum (FBS, US defined, Hyclone, Logan, UT, USA), 2 mM L-glutamine, 50 U/ml penicillin/streptomycin (Mediatech, Manassas, VA, USA), 100 μM β-mercaptoethanol (Sigma Aldrich, St. Louis, MO, USA), non-essential amino acids (Mediatech), and 10 ng/ml leukemia inhibitory factor (LIF, Stem Cell Technologies, Vancouver, BC, Canada) in the absence of feeder cells. Spontaneous differentiation into ES cell derivatives was induced by the removal of LIF from the culture medium (day 0) and subsequent culture for 5, 8, and 14 days before harvesting for chromosomal analysis or chromatin immunoprecipitation (ChIP).
TS cell culture and differentiation
Female mouse TS cells with a stable diploid karyotype (a generous gift from Dr. Janet Rossant) were cultured in the presence of fibroblast growth factor (FGF4) and heparin as described previously (Tanaka et al. 1998). Briefly, TS cells were cultured on a feeder layer of mitomycin C-inactivated, male mouse embryonic fibroblasts (MEFs) in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 20% FBS (Hyclone), 1 mM sodium pyruvate, 2 mM L-glutamine, 50 U/ml penicillin/streptomycin (Mediatech), 100 μM β-mercaptoethanol, 25 ng/ml fibroblast growth factor (FGF4), and 1 μg/ml heparin (Sigma). Spontaneous differentiation into TS cell derivatives was induced through removal of FGF4 and heparin from the culture medium (day 0) and subsequent culture for 2, 4, and 6 days prior to chromosome and ChIP analysis.
Derivation and culture of MEFs and primary granulosa cells
Mouse embryonic fibroblasts (MEFs) were obtained from E14.5 embryos of a hybrid C57Bl/6/SJL background according to standard procedures (Abbondanzo et al. 1993) and cultured in DMEM medium (Invitrogen) supplemented with 10% FCS (Hyclone), 1 mM sodium pyruvate, 2 mM L-glutamine, 50 U/ml penicillin/streptomycin (Mediatech), and 100 μM β-mercaptoethanol (Sigma) at 37°C under an atmosphere of 5% O2, 5% CO2, and 90% N2 for two to three passages before harvest and allocation to different experiments. Primary granulosa cells were obtained following mechanical dissection from cumulus–oocyte complexes (COC's) and cultured under similar conditions before harvesting for chromosome analysis.
Immunochemistry and FISH
Metaphase chromosome spreads from undifferentiated ES cells and TS cells and following induction of spontaneous differentiation were prepared after hypotonic treatment in 1% sodium citrate for 20 min following immediate fixation in methanol/acetic acid (3:1). For the analysis of histone modifications, additional chromosome spreads were fixed in 2% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) and 0.15% Triton X-100 (Bio-Rad Laboratories, Hercules, CA, USA). Analysis of chromosomal ATRX localization patterns was conducted using a mouse monoclonal antibody (McDowell et al. 1999) at a (1:5) dilution. The histone variant macroH2A was detected using an anti-rabbit antibody (Costanzi and Pehrson 1998) at a dilution of 1:50 on paraformaldehyde-fixed metaphase spreads, while the subcellular distribution of ubH2A (clone E6C5, Millipore, Billerica, MA, USA; dilution 1:20) was analyzed according to procedures described previously (Fang et al. 2004). Co-localization analysis of ATRX protein with ubH2A was conducted using a rabbit polyclonal ATRX antibody (Santa Cruz Biotech, Santa Cruz, CA, USA) at a dilution of 1:200. Primary antibodies were detected with Alexa Fluor®555 or 488-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Molecular Probes, Eugene, OR, USA) applied for 2 h at room temperature at a dilution of 1:1,000 in a humidified chamber. For the detection of ubH2A, an Alexa Fluor® 488-conjugated anti-mouse IgM secondary antibody was used as described above. Hoechst 33258 DNA stain was used to visualize condensed heterochromatin areas, and cover slips were mounted with Vectashield anti-fading medium (Vector Laboratories, Burlingame, CA, USA). Chromosome analysis was conducted using a Leica DMRE fluorescence microscope, and images were captured using a Leica DFC 350F CCD camera.
Following immunochemistry and image capture of individual metaphase spreads, slides were processed for DNA-FISH analysis using a fluorescein isothiocyanate-conjugated X-chromosome paint (Cambio, Cambridge, UK), according to the manufacturer's specifications and with the following modifications. Briefly, metaphase spreads were denatured in 70% formamide in 2× saline-sodium citrate (SSC) at 80°C for 10 min and subsequently chilled in ice-cold 70% ethanol for 5 min. The X chromosome probe was denatured for 7 min at 75°C and subsequently incubated at 41°C for 1 h. Overnight hybridization was carried out in a humidified chamber at 41°C. Stringency washes were conducted in a solution containing 50% formamide in 2× SSC as previously described (De La Fuente et al. 2004). The position of the X chromosomes was then recorded and correlated to the previously documented chromosomal patterns of ATRX associations.
BrdU incorporation
Mouse embryonic fibroblasts and primary granulosa cells were cultured in the presence of 5-bromo-2′-deoxyuridine (BrdU, Sigma) at a concentration of 50 μg/ml for 4 h. Following removal of BrdU, cells were cultured for an additional 2 h before treatment with 100 ng/ml colchicine (Invitrogen) for 2–3 h. Metaphase chromosomes were fixed in methanol/acetic acid (3:1) and spread as described earlier. Following ATRX immunolocalization and image capture, chromosomal DNA was denatured in 2 N HCl for 20 min and extensively rinsed in PBS. BrdU incorporation was subsequently detected using an anti-BrdU antibody (BrdU, Boehringer Mannheim/Roche, Mannheim, Germany) at a concentration of 2 μg/ml and detected with an Alexa Fluor-488 secondary antibody (Molecular Probes).
ChIP assays
Chromatin immunoprecipitation was performed using the ChIP-IT™ kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, culture dish adherent cells were cross-linked for 10 min with 1% formaldehyde at room temperature and mechanically harvested using a cell scraper. Following nuclear extraction with a dounce homogenizer, chromatin fragments (200–1,000 bp) were prepared by adding ChIP-IT™ enzymatic digestion cocktail for 5 min at 37°C. Chromatin was then pre-cleared using Protein G beads and immunoprecipitated at 4°C overnight. Antibodies against the transcription factor TFIIB (Santa Cruz) and normal pre-immune IgG (Active Motif) were used as positive and negative controls, respectively. Rabbit polyclonal ATRX antibody (Santa Cruz), rabbit polyclonal anti-H3K9me2 (Millipore, Billerica, MA, USA), and mouse monoclonal anti-H3K27me3 (abcam, Cambridge, MA, USA) were used at a concentration of 1.2 μg/μl. Following elution and reversal of crosslinks, one fifth of the precipitated chromatin was subjected to PCR amplification with primers (C21-fwd, 5′-acaggctgtgaaccagagtacc-3′; C21-rev, 5′-acaggaatgtaggattcaccaa-3′) corresponding to a region 58-kb upstream of the Xist transcription start site, a putative nucleation center for the spreading of epigenetic modifications on the future inactive X chromosome (Heard et al. 2001). An aliquot of the enzymatically digested, not immunoprecipitated, chromatin was treated in parallel, diluted 1:10 and used as “input” sample. Conditions for the PCR amplification were as follows: denaturation at 94°C for 3 min, followed by 39 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 1 min, elongation at 72°C for 1 min, and a final elongation step of 72°C for 10 min. Individual band intensity after ChIP was quantified using the Kodak-1D Image Analysis Software (Kodak, Rochester, NY, USA), and data are presented as percent mean net intensity of the input sample from three independent ChIP experiments.
Statistical analysis
Statistical analysis was performed by one-way analysis of variance and comparison of all pairs by Tukey–Kramer Honestly Significant Difference using JMP Start Statistics (SAS Institute, Cary, NC, USA). Variation among replicates is presented as the standard deviation. Differences were considered significant when (P<0.05).
Results
ATRX decorates the late-replicating X chromosome in female somatic cells
ATRX has been previously found associated with constitutive heterochromatin domains in mouse somatic (McDowell et al. 1999) and germ cells (Baumann et al. 2008; De La Fuente et al. 2004) as well as with ribosomal DNA repeats in human metaphase chromosomes (Gibbons et al. 2000). As expected, immunocytological analysis of the chromosomes of female embryonic fibroblasts and primary ovarian granulosa cells revealed a precise co-localization of ATRX with pericentric heterochromatin domains in the majority of chromosomes. However, in these two primary cell cultures, ATRX also revealed a striking association with the chromatids of a single chromosome per metaphase spread (Fig. 1a, bold arrow and inset). Importantly, FISH, using an X chromosome paint probe, revealed that ATRX protein accumulates preferentially at one of the two X chromosomes in 89% (n=68) of the metaphase spreads analyzed. The enrichment of ATRX at one of the X chromosomes is also detectable in interphase nuclei (Fig. 1a, bold arrows, and see Supplemental Figure S2-B, upper panel). However, due to the prominent localization of ATRX to pericentric heterochromatin domains, multiple ATRX signals are detected in addition to that corresponding to the X chromosome.
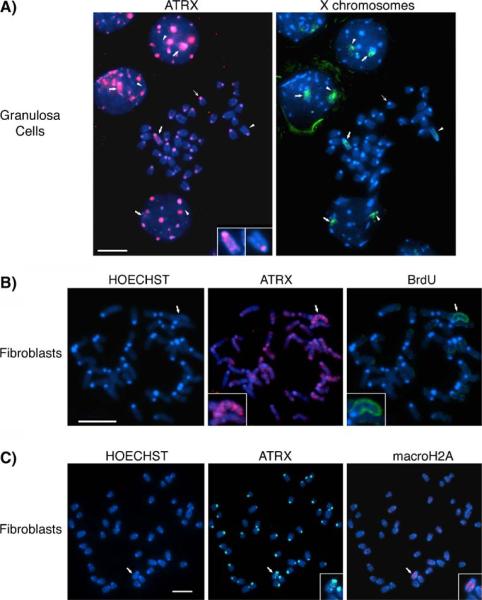
Fig. 1.
Chromosomal localization patterns of ATRX in somatic cells. a Combined fluorescent immunochemistry for ATRX protein (red) and DNA-FISH (green) to determine the position of the X chromosomes on metaphase spreads from a mouse granulosa cell. DNA was visualized by counterstaining with Hoechst 33258. While some autosomes exhibit only a faint banding pattern, a striking accumulation of ATRX on the sister chromatids of one of the two X chromosomes is apparent (bold arrow, inset). The ATRX pattern on the other homolog is indistinguishable from autosomes (arrow head, inset). ATRX protein is found at pericentromeric heterochromatin domains (thin arrow) in autosomes and the sex chromosomes. Notably, ATRX associations are also detectable at the subnuclear domain occupied by one of the two X chromosomes in interphase nuclei (bold arrows, and Supplemental Figure S2-B for nonbiased co-localization analysis). b Analysis of replication timing by BrdU incorporation provides evidence for an association of ATRX protein with the Xi in embryonic fibroblasts. The late replicating inactive X chromosome is clearly labeled with BrdU (green) within the metaphase spread (arrow, inset) and is the only chromosome presenting ATRX protein accumulation (red) along its chromatids (arrow, inset). c Co-immunolocalization of ATRX protein (green) and macroH2A (red), a marker of the inactive X chromosome (Xi), on paraformaldehyde-fixed spreads showing ATRX protein at pericentromeric heterochromatin (inset) and at the chromatids of one chromosome (arrow), which is identified as the Xi by macroH2A co-localization (red, inset). Scale bar=10 μm
In order to determine which of the X chromosomes is marked by ATRX, we set out to distinguish the late replicating (inactive) X chromosome following BrdU incorporation in asynchronously dividing cells (Drouin et al. 1990; Gilbert et al. 1962; Gilbert 2002; Takagi et al. 1982; Zhang et al. 2002). Our analysis revealed that the ATRX protein accumulates on a single, late-replicating X chromosome in 87% (n=94) of female metaphase spreads (Fig. 1b, bold arrow, insets). Moreover, simultaneous analysis of the histone variant macroH2A1.2, a bona fide marker of facultative heterochromatin on the Xi (Costanzi and Pehrson 1998; Costanzi et al. 2000), revealed a precise co-localization of ATRX with macroH2A1.2 throughout the chromatids of the inactive X chromosome (Fig. 1c, bold arrow, insets). Thus, data obtained from the analysis of replication timing and the co-localization with macroH2A1.2 provide the initial evidence for an association of ATRX with the inactive X chromosome in female somatic cells.
ATRX is enriched at X chromosome-specific genomic regions
Previous studies have described the presence of a specific genomic region or hotspot located 5′ to the Xist promoter, which exhibits high levels of lysine methylation on residues 9 (H3K9me2) and 27 (H3K27me3) of histone H3 (Heard et al. 2001; Rougeulle et al. 2004), which might be critical for the recruitment and propagation of a ribonucleoprotein complex containing Xist RNA (Heard et al. 2001; Rougeulle et al. 2004). To determine whether ATRX protein exhibits a direct interaction with X chromosome-specific sequences, we conducted ChIP assays on 14.5 dpc female MEFs analyzing the H3K9me2-hotspot sequences (Fig. 2). Enzymatically digested DNA–protein complexes were immunoprecipitated using antibodies directed against ATRX, H3K9me2, and H3K27me3, while antibodies against the transcription factor (TFIIB) and pre-immune IgG serum served as controls. As expected, precipitation with a pre-immune IgG resulted only in a basal level of amplification. However, both H3K9me2 (62% of the input value) and H3K27me3 (49% of the input value) were found significantly (p<0.05) enriched at X chromosome-specific DNA sequences corresponding to the H3K9me2-hotspot compared against the negative control IgG (Fig. 2). Importantly, immunoprecipitation with an anti-ATRX antibody also revealed a significant enrichment (p<0.05) at these sequences (45% of the input value). In contrast, in embryonic fibroblasts obtained from male embryos carrying only one (active) X chromosome, ATRX protein was not found significantly enriched (16% of input value; Supplemental Figure S1-A). Thus, these results provide novel evidence indicating the presence of a DNA–protein interaction of ATRX with X chromosome-specific sequences in MEFs.
Fig. 2.
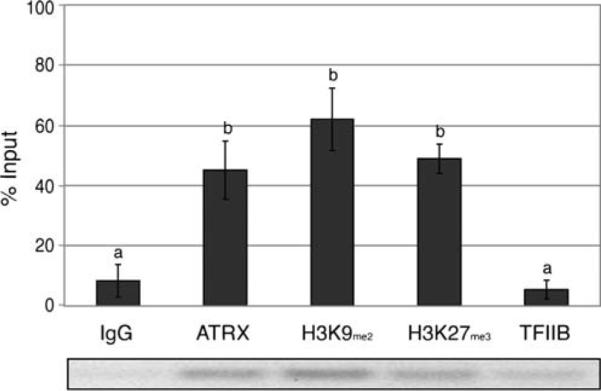
The ATRX protein is preferentially enriched at DNA sequences 5′ of the Xist locus in female embryonic fibroblasts. Chromatin immunoprecipitation (ChIP) assays of X chromosome-specific sequences were performed on primary mouse embryonic fibroblasts using specific antibodies against ATRX, H3K27me3, and H3K9me2 with antibodies against TFIIB and IgG as controls. Amplification of precipitated genomic DNA with an anti-ATRX antibody revealed a significant enrichment of X chromosome-specific DNA fragments corresponding with the constitutive H3K9me2-hotspot located 5′ of the Xist compared to the negative control (IgG), indicating a specific molecular association with the Xi. A representative gel image is shown. Error bars represent the STD of three independent experiments and different superscripts indicate significant differences (p<0.05)
ATRX marks the inactive X chromosome in cells showing imprinted XCI
ATRX protein is highly expressed in trophoblast giant cells of 7.5 dpc embryos (Garrick et al. 2006). Importantly, the extraembryonic lineages of female mammalian embryos, including the trophoblast layer, exhibit a preferential inactivation of the paternally derived X chromosome (Okamoto et al. 2005; Sugawara et al. 1985; Takagi and Sasaki 1975). However, whether ATRX associates with the inactive X chromosome in trophoblast cells is not known. Use of female trophoblast stem cells (Tanaka et al. 1998) thus provides a unique experimental paradigm to determine whether ATRX associates with the paternally derived (inactive) X chromosome during imprinted X-chromosome inactivation.
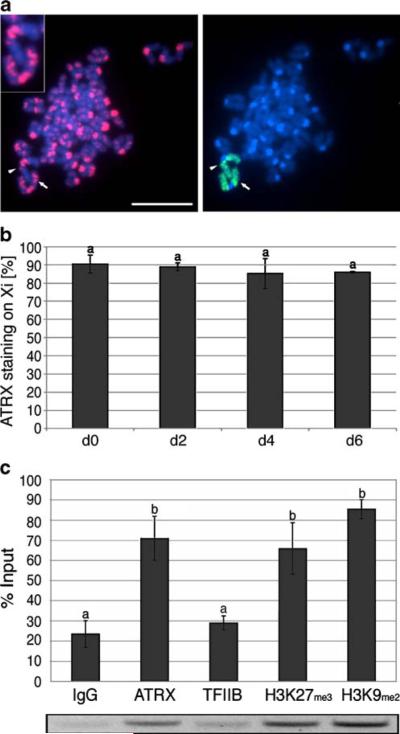
Immuno-FISH analysis revealed that similar to somatic cells, metaphase spreads obtained from undifferentiated trophoblast stem cells exhibit a distinctive localization of ATRX with pericentric heterochromatin in the majority of chromosomes (Fig. 3a, thin arrow). However, a prominent accumulation of ATRX protein on the chromatids of one of the two X chromosomes was also evident in 90% (n=155) of the metaphase spreads analyzed (Fig. 3a, bold arrow and right inset). At this stage, ubiquitination of histone H2A (ubH2A) is transiently associated with the inactive X chromosome in trophoblast stem cells (de Napoles et al. 2004; Fang et al. 2004; Supplemental Figure S2-A). Simultaneous analysis of ATRX (red) and ubH2A (green) revealed the typical association of ATRX with multiple, bright Hoechst-stained heterochromatin foci. In contrast, ubH2A appeared as a single nuclear domain (Supplemental Figure S2-B, lower panel, bold arrow and insets) known to mark the location of the inactive X chromosome (de Napoles et al. 2004; Fang et al. 2004). Notably, co-localization between both ATRX and ubH2A is clearly detectable in some interphase nuclei of undifferentiated trophoblast stem cells (Supplemental Figure S2-B, bold arrow and insets). Moreover, analysis of replication timing to determine the asynchronously replicating (allocyclic) X chromosome in extraembryonic tissues (Sugawara et al. 1985; Takagi and Sasaki 1975) confirmed that, despite a faint banding pattern on the chromatids of autosomes, ATRX (red) is highly enriched throughout the chromatids of a single, allocyclic chromosome per metaphase spread as determined by BrdU incorporation (green; Fig. 3b, bold arrow and insets). These results indicate that during interphase, a small proportion of trophoblast stem cells exhibit a precise co-localization of ATRX with ubH2A. Importantly, during metaphase, ATRX marks an asynchronously replicating (inactive) X chromosome in the majority of cells analyzed, suggesting a stable association of ATRX with the Xi during trophoblast cell mitosis.
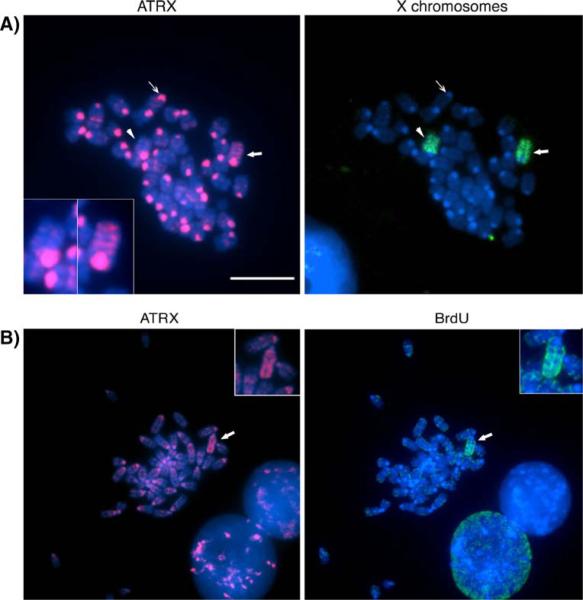
Fig. 3.
ATRX marks the inactive X chromosome in trophoblast stem cells. a Indirect immunofluorescence detection of ATRX and DNA-FISH analysis for the X chromosomes on metaphase spreads of trophoblast stem cells. ATRX (red)is present at pericentric heterochromatin domains (thin arrow) in both autosomes and the sex chromosomes (arrowhead and bold arrow). Notably, ATRX exhibits a preferential accumulation at the chromatids of one of the two X chromosomes (see bold arrow and right inset), while the other X chromosome reveals pericentric staining only (arrowhead, left inset). b Asynchronous replication timing, determined by BrdU incorporation (green), identifies the inactive X chromosome (bold arrow and inset) and exhibits a preferential enrichment for ATRX (red)in undifferentiated TS cells (arrow). Scale bars=10 μm
ATRX is a stable marker of the inactive X chromosome throughout trophoblast stem cell differentiation
Several histone modifications, such as ubH2A and H3K27me3, and the polycomb group proteins, EED/Ezh2, exhibit a transient association with the Xi during ES and TS cell differentiation (de Napoles et al. 2004; Fang et al. 2004; Heard et al. 2001; Rougeulle et al. 2004). In order to determine whether the accumulation of ATRX on the Xi is maintained during TS cell differentiation, we determined the patterns of ATRX protein localization in the chromosomes of TS cells on days 2, 4, and 6 of differentiation in vitro (Fig. 4a,b). Similar to undifferentiated TS cells, 89% (n=164) of metaphase spreads on day 2 of spontaneous differentiation displayed ATRX labeling on one of the X chromosomes (bold arrow and insets). Importantly, this immunolocalization pattern persisted in 85% (n=116) and 86% (n=72) of the metaphases analyzed on days 4 and 6 of differentiation, respectively (Fig. 4a,b). Thus, ATRX is a stable mark of the inactive X chromosome throughout early differentiation of TS cells into trophoblast derivatives.
Fig. 4.
ATRX is a stable marker of the inactive X chromosome throughout TS cell differentiation. a Analysis of ATRX immunolocalization patterns (red) and subsequent DNA-FISH analysis (green)of the X chromosomes revealed a stable association of the protein with the Xi (arrow, insets) throughout TS cell differentiation as depicted for day 4 of differentiation. The active X chromosome remains indistinguishable from the autosomes (arrowhead, insets). Scale bars represent 10 μm. b Quantification of ATRX associations with the Xi in TS cell metaphase spreads. Data indicate the mean percentage of diploid metaphase spreads with ATRX association at a single X chromosome at different days (d0–d6) following spontaneous differentiation. Error bars represent the STD of three independent experiments, and different superscripts indicate significant differences (p< 0.05). c ChIP assays were performed on trophoblast stem cells using specific antibodies against ATRX, H3K27me3, and H3K9me2. Precipitated genomic DNA was amplified, and a significant enrichment of DNA fragments encoding the constitutive H3K9me2-hotspot located 5′ of the Xist locus for ATRX and the histone modifications compared to the controls (IgG and TFIIB) was detected. A representative gel image is shown below the graph. Error bars represent the STD of three independent experiments, and different superscripts indicate significant differences (p<0.05)
Next, we determined whether ATRX exhibits a molecular interaction with the H3K9me2-hotspot 5′ of Xist in undifferentiated TS cells. ChIP analysis, demonstrated a significant enrichment (p<0.05) of DNA sequences corresponding to the H3K9me2-hotspot in samples precipitated with the anti-H3K9me2 (85% of the input value) and anti-H3K27me3 (66% of the input value) antibodies when compared to the controls (pre-immune IgG and TFIIB; Fig. 4c). Importantly, immunoprecipitation using anti-ATRX antibodies resulted in a significant enrichment (p<0.05) of DNA sequences corresponding to the H3K9me2-hotspot (71% of the input value). These results are consistent with our cytological findings and confirm the interaction of ATRX protein with DNA sequences specific to Xi in trophoblast stem cells.
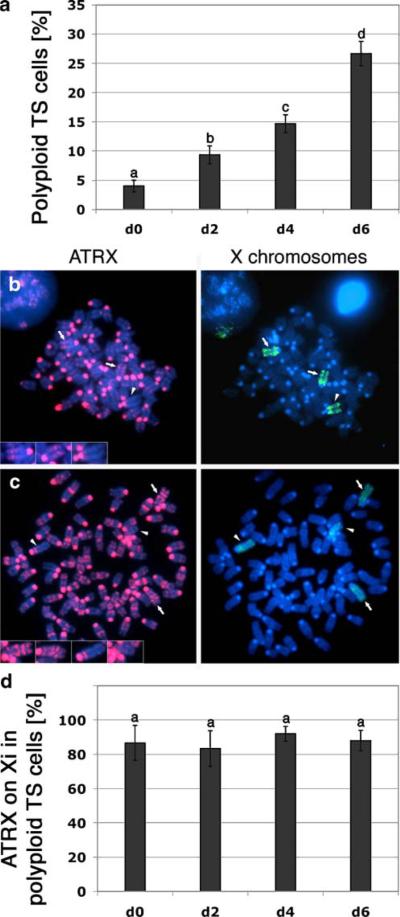
TS cell lines are capable to undergo spontaneous differentiation into trophoblast derivatives in vitro including spongiotrophoblast and polyploid trophoblast giant cells (Tanaka et al. 1998). In our culture system, polyploid metaphase cells increased from 4% (n=325) at day 0 to about 26% (n=269) after 6 days of differentiation (Fig. 5a), indicating that formation of trophoblast giant cells occurs in a fraction of cells within this culture period. Consistent with the inactivation of all X chromosomes in excess of one per diploid genome in somatic cells (Gartler et al. 2006), cells with a hyperdiploid chromosome complement exhibited ATRX association with two (presumably inactive X chromosomes; Fig. 5b; bold arrows and insets), whereas the remaining X chromosome showed ATRX association exclusively at pericentric heterochromatin (arrowhead). Moreover, metaphase spreads obtained from polyploid trophoblast giant cells presented two X chromosomes decorated by ATRX protein along the chromatids (Fig. 5c; bold arrows and insets), while no such accumulation was detectable on the remaining two active X chromosomes (arrowheads and insets). A similar pattern of ATRX accumulation was observed in 85% (n=269) of polyploid giant cell metaphase spreads analyzed (Fig. 5d). These results indicate that the number of X chromosomes which exhibit ATRX labeling correlates with the presence of more than one inactive X chromosome in polyploid trophoblast giant cells and provide further evidence indicating that ATRX is a stable marker of the inactive X chromosome(s) during trophoblast cell differentiation.
Fig. 5.
ATRX marks the inactive X chromosomes in polyploid trophoblast giant cells. a The incidence of polyploid metaphases depending on the stage of differentiation was quantified and is displayed as the mean percentage of three independent experiments. Different superscripts indicate significant differences (p<0.05). Error bars represent the STD. b In hyperdiploid metaphases, ATRX marks two of the three X chromosomes (red, bold arrows, insets) in agreement with the rule of one active X chromosome per diploid chromosome set. c Polyploid trophoblast giant cell metaphase presenting ATRX staining on two Xi (red, bold arrows, insets), while there is no such association detectable at the two Xa (arrowheads, insets). d Proportion of trophoblast giant cells showing ATRX protein localization to the inactive X chromosomes in polyploid metaphases in three independent experiments. Different superscripts indicate significant differences (p<0.05). Error bars represent the STD
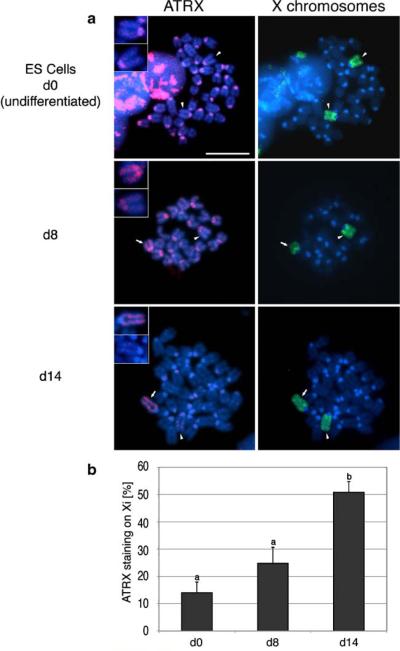
ATRX is a late marker of the Xi in differentiating embryonic stem cells
Consistent with the presence of two active X chromosomes in a previously described female ES cell line (PGK12.1; de Napoles et al. 2004; Mermoud et al. 1999, 2002; Silva et al. 2003; Zvetkova et al. 2005), no significant ATRX labeling was detected in the chromatids of the X chromosomes in 86% (n=211) of the metaphase spreads analyzed from undifferentiated (day 0) ES cells. In fact, ATRX was found localized to pericentromeric heterochromatin in the majority undifferentiated ES cells (Fig. 6 upper panel, arrowheads and insets). A small proportion of ES cells (about 10%; n=164) presented exceptionally low localization of ATRX at pericentric heterochromatin along with a prominent ATRX association at the chromatids of all autosomes (Supplemental Figure S2-C). Interestingly, stable female ES cell lines have recently been demonstrated to show global genomic hypomethylation with a reduction of DNA-methylation levels at pericentric major satellite repeats (Zvetkova et al. 2005). Whether such global epigenetic phenomena relate to the diverging patterns of ATRX immunolocalization patterns observed in a proportion of ES cells in this study remains to be determined.
Fig. 6.
Association of ATRX with the inactive X chromosome is a late event during ES cell differentiation. a In undifferentiated (PGK12.1) ES cell metaphases, ATRX (red) localizes to pericentromeric heterochromatin in all chromosomes. Both X chromosomes (upper panel; arrow and arrowhead) are indistinguishable in regard to ATRX labeling. The association of ATRX with Xi is initially detected following 8 days of spontaneous differentiation (middle panel; arrow and inset) and subsequently increases after 14 days of differentiation (lower panel, arrow, insets). b Quantitative analysis of ATRX associations with the Xi during ES cell differentiation. Data indicate the mean percentage of metaphases with preferential ATRX staining on a single X chromosome in undifferentiated, d8 and d14 differentiated cells. Error bars represent the STD of three independent experiments, and different superscripts indicate significant differences (p<0.05)
However, although onset of XCI could be recognized shortly after LIF removal by the transient subnuclear localization of ubH2A at the Xi at around day 4 (Fang et al. 2004; Supplemental Figure S2-D) an enrichment of ATRX protein at the Xi was detected in about 24% (n=154) and 51% (n=164) of the metaphase spreads only following 8 and 14 days of spontaneous differentiation (Fig. 6a,b). These results indicate that the association of ATRX with Xi is developmentally regulated and that it constitutes a relatively late event during ES cell differentiation.
Next, we determined the ontogeny of any potential associations of ATRX with the H3K9me2-hotspot throughout ES cell differentiation (Fig. 7a). In undifferentiated ES cell cultures (day 0), ATRX levels were not significantly different from those observed using the pre-immune IgG as a negative control (Fig. 7a). However, at this stage and in agreement with previous observations (Fang et al. 2004; Plath et al. 2003; Rougeulle et al. 2004), a significant enrichment (p<0.05) for both H3K9me2 and H3K27me3 was already detectable at the H3K9me2-hotspot 5′ to the Xist locus (Fig. 7a). Although the association of ATRX with this region begins to increase after 5 days of spontaneous differentiation, the values obtained at this stage did not show a significant difference compared with the negative control (IgG). However, consistent with our cytogenetic analysis, ATRX was found significantly enriched at the H3K9me2-hotspot of the inactive X chromosome (49% of the input value; P<0.05) following 8 days of spontaneous differentiation.
Fig. 7.

ATRX is enriched at chromosomal sequences corresponding to the H3K9me2-hotspot on the Xi following spontaneous differentiation of ES cells. Chromatin immunoprecipitation (ChIP) analysis of Xi chromosome-specific sequences on undifferentiated ES cells (d0, white bars) and throughout ES cell differentiation (d5, gray bars) and (d8, black bars) using specific antibodies against ATRX, H3K27me3, and H3K9me2 (antibodies against TFIIB and IgG served as controls). A significant enrichment of DNA fragments encoding sequences within the constitutive H3K9me2-hotspot on the Xi was only observed for samples immunoprecipitated with anti-ATRX antibodies after 8 days of differentiation, while the histone modification H3K9me2 was detectable at all time points investigated. Error bars represent the STD of three independent experiments, and different superscripts indicate significant differences (p<0.05). Representative gel images for each stage of differentiation are displayed below the graph
The levels of H3K9me2 and H3K27me3 remained elevated compared with those observed for the pre-immune IgG group throughout the culture period, suggesting a stable association with Xi. Interestingly, the association of the transcription factor TFIIB with these DNA sequences showed a significant reduction on day 8 of differentiation consistent with the spreading of a transcriptionally repressive chromatin environment (Fig. 7a). Thus, using two independent experimental approaches, we show that ATRX protein associates with the inactive X chromosome relatively late during ES cell differentiation, suggesting that ATRX might not be essential for the onset of X inactivation in ES cells. However, its stable association with Xi during mitosis in differentiating ES cells and trophoblast stem cells indicates a potential involvement in the maintenance of the silenced chromatin status.
Discussion
Following the accumulation and spreading of Xist RNA on the inactive X chromosome, the establishment and maintenance of a transcriptionally repressive chromatin environment on the Xi requires the coordinate recruitment of multiple chromosome-wide epigenetic modifications (Brockdorff 2002; Masui and Heard 2006; Ng et al. 2007). In the present study, we provide novel evidence indicating that the chromatin remodeling protein ATRX is a mitotically stable epigenetic marker of the Xi in mouse somatic and TS cells. Results from two independent experimental approaches indicate that ATRX becomes preferentially enriched at X chromosome-specific DNA sequences corresponding to the H3K9me2-hotspot 5′ of the Xist locus and throughout the chromatids of the Xi following the onset of random XCI in differentiating ES cells. The association of ATRX with Xi is developmentally regulated and constitutes a relatively late event during ES cell differentiation, suggesting that ATRX might not be essential for the onset of X chromosome inactivation. However, its stable association with Xi during mitosis in differentiating ES cells, TS cells, and somatic cells together with its persistence throughout TS cell differentiation indicate a potential involvement in the maintenance of the silenced chromatin status. We propose that ATRX might be an important component of a large protein complex with a potential involvement in the maintenance of imprinted XCI in extraembryonic tissues as well as the process of heterochromatinization of the Xi during random X chromosome inactivation in the soma. These results contribute to the characterization of the epigenetic landscape of Xi and expand the family of Xi-associated factors by identifying the first chromatin remodeling protein of the SWI/SNF2 family, ATRX, as a component of facultative heterochromatin on the inactive X chromosome.
ATRX associates with the Xi following random XCI in ES cells
During the initial stages of X chromosome inactivation, expression and accumulation of Xist RNA in cis is required for the specific recruitment of repressive chromatin modifications to the Xi (Brown and Willard 1994; Chow and Brown, 2003; Clemson et al., 1996; Csankovszki et al., 1999; Kohlmaier et al., 2004; Penny et al., 1996; Wutz and Jaenisch, 2000). Maintenance of the inactive state however becomes independent of Xist expression during subsequent differentiation stages when epigenetic modifications render the inactive state irreversible and heritable through cell division (Brown and Willard 1994; Csankovszki et al. 1999, 2001; Wutz 2007; Wutz and Jaenisch 2000). The epigenetic mechanisms involved in the maintenance of chromosome-wide silencing during the Xist-independent stage of XCI are not fully understood. However, the initial Xist RNA expression on the Xi has been implicated in the establishment of a `chromosomal memory', the molecular nature of which is still unknown, which, together with heterochromatinization of the entire X chromosome, is required to maintain the inactive state through cell division (Kohlmaier et al. 2004; Wutz 2007).
Analysis of the kinetics of X chromosome inactivation in ES cells indicate that following Xist expression, posttranslational modifications of histones H3/H4 are some of the earliest epigenetic changes on the Xi taking place within 2 days of ES cell differentiation (Brockdorff 2002; Chaumeil et al. 2002). Proteins of the polycomb repressive complex 2 (PRC2), Eed and Ezh2/Enx1, catalyze chromosome-wide trimethylation of H3K27 and are thus found enriched at the Xi within this early developmental window (Czermin et al. 2002; Fischle et al. 2003; Plath et al. 2003; Silva et al. 2003). Importantly, recent studies indicate that the inactive state of Xi is `locked in' as early as 72 h following the induction of ES cell differentiation (Kohlmaier et al. 2004; Wutz and Jaenisch 2000). However, results obtained from immuno-FISH analysis and chromatin immunoprecipitation studies defined the time of ATRX's association with the Xi after 8 days of ES cell differentiation, suggesting that ATRX is an epigenetic mark established following the onset of random XCI. These results place the time of ATRX association with the Xi close to the stage at which the histone variant macroH2A has been observed on the inactive X chromosome (Costanzi and Pehrson 1998; Costanzi et al. 2000; Mermoud et al. 1999) and before the establishment of chromosome-wide DNA methylation changes (Brockdorff 2002; Gilbert and Sharp 1999; Masui and Heard 2006), suggesting that ATRX might be implicated in the maintenance rather than the onset of X chromosome inactivation.
ATRX is a stable marker of Xi in cells showing imprinted X chromosome inactivation
The critical epigenetic factors and or mechanism(s) that are directly responsible for the maintenance of the inactive state through multiple cell divisions during the Xist-independent stage of X chromosome inactivation are not clear at present (Chaumeil et al. 2002; Wutz 2007). Notably, while imprinted X chromosome inactivation initially occurs at the eight-cell stage of preimplantation development (Mak et al. 2002; Okamoto et al. 2004, 2005; Mak et al. 2004), preferential inactivation of the paternal X chromosome persists only in the differentiating extraembryonic lineages of the mammalian embryo (Takagi and Sasaki 1975). Our results indicate that in trophoblast stem cells, ATRX is found co-localized with ubH2A, a transient marker of the Xi (Fang et al. 2004; Mak et al. 2002) during interphase. Importantly, ATRX remains associated with an asynchronously replicating (inactive) X chromosome during trophoblast cell mitosis. Retention of ATRX through mitosis is consistent with a role for ATRX as a bona fide epigenetic mark of the inactive state. Importantly, the number of X chromosomes that exhibit ATRX labeling correlates with the presence of more than one inactive X chromosome in polyploid trophoblast giant cells, providing further evidence indicating that ATRX is a stable marker of the Xi during trophoblast cell differentiation.
Previous studies indicate that most epigenetic modifications associated with the Xi like ubH2A, EED/Ezh2, and H3K27me3 are developmentally regulated and prominent only transiently during the earliest stages of TS cell differentiation (Fang et al. 2004; Plath et al. 2003, 2004). In contrast, ATRX is a stable mark of the Xi during TS cell differentiation in diploid TS cells and during polyploidization, suggesting that ATRX might be involved in the maintenance of imprinted X chromosome inactivation in the trophoblast cell layer, perhaps to facilitate heterochromatinization of the late replicating X chromosome in extraembryonic lineages.
The mechanisms involved in the spreading of ATRX from its initial centromeric localization throughout the entire chromatids of the Xi remain to be determined. However, the ATRX protein has been shown to have a specific interaction with the SET domain (a protein motif common to most histone methyltransferases) of the human EED/Ezh2 complex in yeast two-hybrid screens and in in vitro binding assays (Cardoso et al. 1998), and Ezh2 has previously been shown to interact with ATRX at heterochromatin domains (Cardoso et al. 1998; Kourmouli et al. 2005). In the present study, ATRX was found co-localized with macroH2A1.2 at the Xi in MEFs. In turn, macroH2A1.2 has been found co-localized with well-established early markers of the Xi, like Eed and H3K27me3, in trophoblast stem cells (Kalantry et al. 2006). The polycomb group protein Eed is necessary for the maintenance of XCI in the extraembryonic tissues but not the ICM of the developing conceptus (Kalantry and Magnuson 2006; Kalantry et al. 2006), reflecting major differences in the epigenetic pathways regulating X chromosome inactivation in embryonic and extraembryonic lineages (Kalantry and Magnuson 2006; Kalantry et al. 2006; Mak et al. 2002; Silva et al. 2003; Wang et al. 2001).
Moreover, mice deficient for the EED/Ezh2 complex show defects in the development of secondary trophoblast giant cells similar to ATRX-deficient embryos (Garrick et al. 2006; Wang et al. 2001). In addition, heterochromatin protein 1 (HP1), a well-described molecular partner of ATRX in murine and human cells (Berube et al. 2000; Kourmouli et al. 2005; Le Douarin et al. 1996; McDowell et al. 1999), has been shown to associate with heterochromatin at the Barr body in some human somatic cell types through dimethylation of histone 3 on lysine 9 (Chadwick and Willard 2003). Whether specific interactions between HP1 and ATRX also exist within the context of facultative heterochromatin on the inactive X chromosome, e.g., by providing a specific docking site for ATRX via EED/Ezh2 similar to the HP1/Su(var)3–9 complex (Bannister et al. 2001; Lachner et al. 2001) remains to be determined.
In conclusion, the results presented in this study demonstrate for the first time that ATRX is preferentially enriched at the inactive X chromosome in trophoblast stem cells and female somatic cells. Further studies will be required to determine whether ATRX is strictly necessary for the maintenance of X chromosome inactivation in the extraembryonic tissues of the female embryo or, as most of the epigenetic markers associated with the Xi identified until now, whether ATRX is part of the multiple and at times redundant epigenetic changes that contribute to reinforce the inactive state and heterochromatinization of the Xi through cell division and differentiation.
However, the kinetics of ATRX association with the Xi (this study) and the embryonic phenotype observed in ATRX mutant females (Garrick et al. 2006) suggest a potential role in the maintenance of the inactive state in the extraembryonic lineages. For example, conditional ablation of the ATRX gene disrupts imprinted X chromosome inactivation as indicated by the presence of heterozygous females that retained an active paternal X chromosome in the trophoblast cell layer (Garrick et al. 2006). Importantly, the epigenetic composition of pericentric heterochromatin has a profound effect in the formation of polyploid trophoblast giant cells (Hemberger 2007). Thus, our findings might prove relevant to partially explain the escape from imprinted X inactivation and the severe deficiency of polyploid secondary trophoblast giant cells observed in females carrying an ATRX null allele (Garrick et al. 2006).
Supplementary Material
Acknowledgments
We would like to thank Drs. J. Rossant and N. Brockdorff for kindly providing the TS cell line and the PGK12.1 ES cell line, respectively. We are grateful to Drs. D. Higgs, D. Garrick and J. Pehrson for generous gifts of antibodies and to Drs. M. M. Viveiros and F. Yang for helpful discussions and comments during manuscript preparation. This research was supported by a grant from the National Institute of Child Health and Human Development (NICHD) National Institutes of Health (HD042740) to R. De La Fuente.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00412-008-0189-x) contains supplementary material, which is available to authorized users.
References
- Abbondanzo S, Gadi I, Stewart C. Methods Enzymology. vol. 225. Academic Press; San Diego: 1993. Derivation of embryonic stem cell lines; pp. 803–823. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Baumann C, Schmidtmann A, Muegge K, De La Fuente R. Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia. BMC Mol Biol. 2008;9:29. doi: 10.1186/1471-2199-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brockdorff N. X-chromosome inactivation: closing in on proteins that bind Xist RNA. Trends in Genetics. 2002;18:352–358. doi: 10.1016/s0168-9525(02)02717-8. [DOI] [PubMed] [Google Scholar]
- Brown C, Willard H. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontes M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chow J, Brown C. Forming facultative heterochromatin: silencing of an X chromosome in mammalian females. Cell Mol Life Sci. 2003;60:2586–2603. doi: 10.1007/s00018-003-3121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C, McNeil J, Willard H, Lawrence J. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson J, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros M, Wigglesworth K, Jj E. ATRX, a Member of the SNF2 Family of Helicase /ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Developmental Biology. 2004;272:1–14. doi: 10.1016/j.ydbio.2003.12.012. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endo HM, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Drouin R, Lemieux N, Richer C. Analysis of DNA replication during S-phase by means of dynamic chromosome banding at high resolution. Chromosoma. 1990;99:273–280. doi: 10.1007/BF01731703. [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick BP, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJH, Wood WG, Higgs DR, Gibbons RJ. Loss of atrx affects trophoblast development and the pattern of x-inactivation in extraembryonic tissues. PLoS Genetics. 2006;2:438–450. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM, Varadarajan KR, Luo P, Norwood TH, Canfield TK, Hansen RS. Abnormal X: autosome ratio, but normal X chromosome inactivation in human triploid cultures. BMC Genet. 2006:7. doi: 10.1186/1471-2156-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, et al. Mutations in the transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Gilbert SL, Sharp PA. Promoter-specific hypoacetylation of X-inactivated genes. PNAS. 1999;96:13825–13830. doi: 10.1073/pnas.96.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Muldal S, Lajtha L, Rowley J. Time-sequence of human chromosome duplication. Nature. 1962;195:869–875. doi: 10.1038/195869a0. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of Histone H3 at Lys-9 Is an Early Mark on the X Chromosome during X Inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci. 2007;64:2422–2436. doi: 10.1007/s00018-007-7113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat Rev Genet. 2005;6:410–408. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran R, Seah C, Moulin J, Isaac C, Dick F, Bérubé NG. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180:315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Sun Y-M, van der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem Biophys Res Commun. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen A, Garnier J, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1a and TIF1b in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mak W, Baxter J, Silva J, Newall AE, Otte AP, Brockdorff N. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova T, de Napoles M, Appanah R, Yamanaka S, Otte A, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harbor Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol. 2002;12:247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–39. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Hernández-Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of Histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:313–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Sugawara O, Takagi N, Sasaki M. Correlation between X-chromosome inactivation and cell differentiation in female preimplantation mouse embryos. Cytogenet Cell Genet. 1985;39:210–219. doi: 10.1159/000132137. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis A, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Wutz A. Xist function: bridging chromatin and stem cells. Trends Genet. 2007;23:457–464. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell Biol. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- Zinchuk V, Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr Protoc Cell Biol Chapter. 2008:4. doi: 10.1002/0471143030.cb0419s39. [DOI] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.