Abstract
Contrary to the increasing amount of knowledge regarding the functional roles of glycine-rich RNA-binding proteins (GRPs) in Arabidopsis thaliana in stress responses, the physiological functions of GRPs in rice (Oryza sativa) currently remain largely unknown. In this study, the functional roles of six OsGRPs from rice on the growth of E. coli and plants under cold or freezing stress conditions have been evaluated. Among the six OsGRPs investigated, OsGRP1, OsGRP4, and OsGRP6 were shown to have the ability to complement cold-sensitive BX04 E. coli mutant cells under low temperature conditions, and this complementation ability was correlated closely with their DNA- and RNA-melting abilities. Moreover, OsGRP1 and OsGRP4 rescued the growth-defect of a cold-sensitive Arabidopsis grp7 mutant plant under cold and freezing stress, and OsGRP6 conferred freezing tolerance in the grp7 mutant plant, in which the expression of AtGRP7 was suppressed and is sensitive to cold and freezing stresses. OsGRP4 and OsGRP6 complemented the defect in mRNA export from the nucleus to the cytoplasm in grp7 mutants during cold stress. Considering that AtGRP7 confers freezing tolerance in plants and harbours RNA chaperone activity during the cold adaptation process, the results of the present study provide evidence that GRPs in rice and Arabidopsis are functionally conserved, and also suggest that GRPs perform a function as RNA chaperones during the cold adaptation process in monocotyledonous plants, as well as in dicotyledonous plants.
Keywords: Arabidopsis thaliana, cold stress, glycine-rich RNA-binding protein, RNA chaperone, rice
Introduction
RNA-binding proteins are ubiquitous cellular proteins that regulate gene expression principally at the post-transcriptional level, which involves pre-mRNA splicing, nucleocytoplasmic mRNA transport, mRNA stability and decay, and translation (Dreyfuss et al., 1993; Simpson and Filipowicz, 1996). RNA-binding proteins are characterized by the presence of several conserved motifs and domains, including the RNA-recognition motif (RRM), glycine-rich domain, K homology domain, RGG-box, and zinc-finger motif (Burd and Dreyfuss, 1994; Lorković and Barta, 2002). The RNA-binding proteins that harbour RRMs at the N-terminus and a glycine-rich region at the C-terminus are referred to as the glycine-rich RNA-binding proteins (GRPs). The presence of GRPs in a variety of plant species (Carpenter et al., 1994; Ferullo et al., 1997; Moriguchi et al., 1997; Horvath and Olson, 1998; Aneeta et al., 2002; Stephen et al., 2003; Nomata et al., 2004; Shinozuka et al., 2006) and the regulation of their expression patterns by a number of external stimuli (Sachetto-Martins et al., 2000) have been amply reported. In particular, an increase in the expression of GRP genes under low temperatures has compelled us to hypothesize that GRPs might be involved in the responses of plants to cold stress conditions.
As a component of a focused effort to understand the biological roles of GRPs in plants under stress conditions, the functional roles of AtGRPs in Arabidopsis plants have been extensively investigated under a variety of stress conditions. It is reported that AtGRP2, AtGRP4, and AtGRP7, the three AtGRPs of eight AtGRP family members in Arabidopsis, have different impacts on seed germination, seedling growth, and stress tolerance of Arabidopsis plants under diverse stress conditions (Kwak et al., 2005; Kim et al., 2007a, 2008). In particular, AtGRP2 and AtGRP7 but not AtGRP4 accelerated seed germination and seedling growth under low temperatures and conferred freezing tolerance to Arabidopsis plants. Moreover, it has been shown that AtGRP7 has an RNA chaperone activity during the cold adaptation process in Escherichia coli (Kim et al., 2007b), and confers freezing tolerance via the regulation of mRNA export in the guard cells (Kim et al., 2008). The results reported in this series of founding studies are clearly reflective of GRP's prominent roles in the responses of plants to diverse environmental factors in dicotyledonous plants such as Arabidopsis.
Despite increasing knowledge regarding the functional roles of GRPs in Arabidopsis, our current understanding of the biological functions of GRPs in monocotyledonous plants, including rice, is severely limited. A recent proteomic analysis of RNA-binding proteins revealed the existence and regulation of RNA-binding proteins in dry seeds of rice (Masaki et al., 2008). The rice genome encodes at least six GRPs, which share a high degree of sequence homology in the ribonucleoprotein1 (RNP1) and RNP2 regions, but vary in the length of the C-terminal glycine-rich domain (Table 1; see Supplementary Fig. S1 at JXB online). The six OsGRPs are designated as OsGRP1 to OsGRP6 (Table 1). The results of sequence analyses showed that OsGRP1 (Os01g68790) and OsGRP6 (Os12g31800) are closely related to AtGRP2, and OsGRP3 (Os03g46770) and OsGRP5 (Os05g13630) are the most homologous to AtGRP7. Although the overall homology is relatively low, OsGRP2 (Os03g56020) and OsGRP4 (Os04g33810) are the most homologous to AtGRP3 and AtGRP2/7, respectively (Table 1). Considering that a particular type of AtGRPs confers cold and freezing tolerance to Arabidopsis plants, it is of interest comparatively to assess the roles of OsGRPs on seed germination and seedling growth in plants under low temperature conditions. This article provides compelling evidence that GRPs in rice and Arabidopsis are functionally conserved, and that they perform a function as RNA chaperones during cold adaptation processes in monocotyledonous plants, as well as in dicotyledonous plants.
Table 1.
Compilation of OsGRPs investigated in this study
| Gene name | Accession no. | Length (aa) | Localizationa | Homology (%)b |
| OsGRP1 | Os01g68790 | 150 | chloroplast | AtGRP2 (60) |
| OsGRP2 | Os03g56020 | 141 | mitochondria/cyto | AtGRP3 (24) |
| OsGRP3 | Os03g46770 | 162 | nucleus | AtGRP7 (73) |
| OsGRP4 | Os04g33810 | 137 | nucleus/mitochondria | AtGRP2/7(29/27) |
| OsGRP5 | Os05g13620 | 104 | nucleus | AtGRP7 (42) |
| OsGRP6 | Os12g31800 | 258 | nucleus/mitochondria | AtGRP2 (53) |
Cellular localization predicted via PSORT (http://psort.ims.u-tokyo.ac.jp) and TargetP (http://www.cbs.dtu.dk/services/TargetP) programs.
The most homologous Arabidopsis counterparts and their sequence homology (%) predicted via ClustalW program.
Materials and methods
Plant materials, stress treatments, and expression analysis
The rice (Oryza sativa) used in this study was of the Dongjin variety. Three-week-old rice seedlings grown in soil at 27±2 °C under a 16/8 h light/dark photocycle were subjected to cold stress at 4 °C for up to 3 d. Total RNA was extracted from the frozen plant samples using the Plant RNeasy extraction kit (Qiagen), and the transcript levels of each gene were determined via real-time RT-PCR with the gene-specific primers provided in Supplementary Table 1 at JXB online. All experimental conditions for real-time RT-PCR and data analysis were essentially as described by Kim et al. (2005). The transcript levels of each gene were measured in both stressed and unstressed control plants at each time point to determine accurately the expression levels of OsGRPs upon cold stress treatment. The experiment was repeated at least three times, and similar results were obtained.
Vector construction and Arabidopsis transformation
To complement grp7 knockout plants with OsGRPs, the coding region of OsGRP cDNA was cloned into the XbaI/BamHI site of the pCambia3301 vector. The transformation of Arabidopsis was conducted via vacuum infiltration using Agrobacterium tumefaciens GV3101. The seeds were harvested and plated on MS medium containing phosphinothricin (50 μg ml−1) as a selection marker. After the further selection of transgenic lines, T2 or T3 lines were utilized for phenotypic investigation. Expression of OsGRP in grp7 mutant plants was analysed via RT-PCR with the gene-specific primer listed in Supplementary Table S1 at JXB online.
Germination and seedling growth assays under low temperatures
The A. thaliana wild-type, grp7 knockout, and transgenic plants used in this work were of the Nossen-0 ecotype. Plants were grown at 23 °C under long-day conditions (16/8 light/dark photocycle) at ∼100 μE m−2 s−1. Arabidopsis plants were grown in either MS medium (Murashige and Skoog, 1962) or soil at 23 °C. The germination assays were conducted on three replicates of 30–40 seeds each, essentially as described by Kim et al. (2005). The seeds were sown on MS medium supplemented with 1.5% sucrose, and the plates were maintained for 3 d at 4 °C in darkness, then transferred to normal growth conditions. To characterize the effects of cold stress on germination, the MS plates were placed in an incubator maintained at 11 °C under white light. A seed was regarded as germinated when the radicle protruded through the seed coat. To determine the effects of cold stresses on the seedling growth of the plants, the seeds were fully germinated for 3 d at normal growth temperature, and the seedlings were transferred to a growth chamber maintained at 8–11 °C.
Electrolyte leakage tests
The electrolyte leakage test was conducted essentially as previously described (Kim et al., 2007a, 2008). Briefly, the fully developed rosette leaves from 4-week-old Arabidopsis plants were placed in a test tube containing 100 μl distilled water, an ice crystal was added to the tube, and the temperature of the water bath was decreased to –10 °C at a rate of 1 °C per 30 min. The conductivity of the solution was measured with a conductivity meter (Cole-Parmer Instrument Co.). The ratio of electrolyte content prior to and after autoclaving was utilized as an indicator for membrane damage after freezing treatment. The experiment was repeated at least five times.
Cold shock test and transcription anti-termination assay in E. coli
The coding region of OsGRP cDNA was cloned into the NdeI/BamHI site of the pINIII vector. For the cold shock test, the cold-sensitive BX04 cells obtained from Dr M Inouye (Xia et al., 2001) were transformed with each vector and grown in Luria-Bertani (LB) medium containing ampicillin and kanamycin. The overnight cultures were subjected to serial dilution, spotted on LB medium, and incubated under low temperature. For transcription anti-termination assay, the E. coli RL211 cells (Landick et al., 1990) that were transformed with each pINIII construct were grown in liquid medium and spotted on LB-carbenicillin plates with or without chloramphenicol.
Nucleic acid-melting assay
The nucleic acid-melting assay was conducted essentially as previously described by Kim et al. (2007b). The molecular beacon was labelled with a fluorophore (tetramethyl rhodamine) and a quencher (dabcyl) as previously described (Phadtare et al., 2002). The recombinant GST-OsGRP fusion proteins were expressed using pGEX-4T-3 vector (Amersham Pharmacia Biosciences) in BL21 DE3 cells, and were purified with glutathione Sepharose 4B resin. Fluorescence measurements were conducted on a Spectra Max GeminiXS spectrofluorometer (Molecular Devices) with excitation and emission wavelengths of 555 nm and 575 nm, respectively.
Poly(A) mRNA in situ localization assay and analysis of cellular localization of OsGRP
Poly(A) mRNA in situ hybridization was conducted essentially as described (Gong et al., 2005; Kim et al., 2008). Briefly, the leaf samples of 2-week-old Arabidopsis plants were fixed in a fixation buffer (120 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 2.7 mM KCl, 0.1% Tween 20, 80 mM EGTA, 5% formaldehyde, 10% DMSO, and 50% heptane). The samples were hybridized in hybridization buffer (Sigma-Aldrich) containing 5 pmol of 45-mer oligo(dT) labelled with fluorescein at the 5′-end at 50 °C in darkness. The samples were immediately observed under an Olympus 1X71 FV500 confocal laser-scanning microscope (Olympus America Inc.) with a 488 nm excitation laser and a 522/DF35 emission filter. Each experiment was repeated at least three times, and similar results were obtained. To determine the cellular localization of OsGRP1 and OsGRP4, the cDNA encoding each protein was fused in-frame with GFP, and the OsGRP1-GFP or OsGRP4-GFP fusion protein was expressed under the control of the CaMV 35S promoter in Arabidopsis. The cellular expression of OsGRP1 and OsGRP4 was investigated via a confocal microscope. Root samples were mounted on microscope slides and observed with a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss, Inc.) equipped with an inverted Zeiss Axiovert 100M microscope. Excitation and emission wavelengths were 488 and 505 to 545 nm, respectively.
Results
Transcript levels and cold stress-responsive expression of OsGRPs in rice
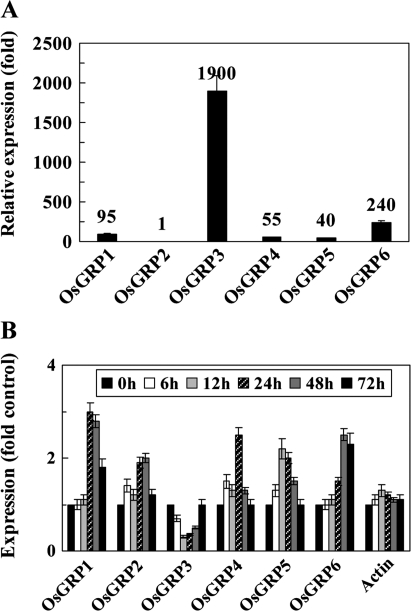
The transcript levels of six OsGRPs in the aerial part of 3-week-old rice seedlings were determined via quantitative real-time RT-PCR analysis with the gene-specific primers listed in Supplementary Table S1 at JXB online. Among the six OsGRPs in rice, OsGRP2 showed the lowest level of expression, and the expression levels of OsGRP1, OsGRP4, OsGRP5, and OsGRP6 were approximately 50–200-fold higher than that of OsGRP2. By comparison, OsGRP3 showed the highest level of expression; its transcript level was approximately 1900-fold higher than that of OsGRP2 (Fig. 1A). The expression patterns of OsGRPs were then assessed in rice under cold stress conditions. An attempt was made first to determine whether cold stress had been adequately applied to the rice by determining the expression of pBC121 (accession no BAA01630) and OsDREB1A (accession no AF300970), both of which are known to be highly induced in rice by chilling treatment (Binh and Oono, 1992; Dubouzet et al., 2003). When the 3-week-old rice seedlings were subjected to cold stress (4 °C), the transcript levels of pBC121 were increased by 2-fold, 4-fold, and 6-fold at days 1, 2, and 3 of cold treatment, respectively, and the transcript level of OsDREB1A was increased by 10-fold at day 1 of the cold treatment (data not shown). While the transcript level of actin was not modulated by cold stress, the transcript levels of each OsGRP in rice were modulated to a different degree by cold stress (Fig. 1B). The levels of OsGRP1, OsGRP2, OsGRP4, OsGRP5, and OsGRP6 expression were increased by more than 2-fold by cold stress, while the transcript level of OsGRP3 was reduced during 48 h of cold treatment, then increased to the pre-stressed levels 3 d after cold treatment. It was noted that the expression of OsGRPs is modulated less significantly by cold stress as compared with that of the Arabidopsis counterpart, in which AtGRPs transcript levels are increased by more than 4-fold by cold stress (Kwak et al., 2005).
Fig. 1.
Expression patterns of OsGRPs. (A) Total RNA was extracted from 4-week-old rice seedlings and the relative expression levels of six OsGRPs were analysed via real-time RT-PCR. OsGRP transcript levels are presented as the relative values to the expression of OsGRP2. (B) Expression patterns of OsGRPs in rice subjected to cold stress (4 °C) for 6, 12, 24, 48, and 72 h were analysed via real-time RT-PCR and presented as the relative expression (fold) of the non-stressed controls. The mean values and standard errors (bar) were obtained from three independent experiments.
The rice OsGRPs complement the cold sensitivity of E. coli mutant cells
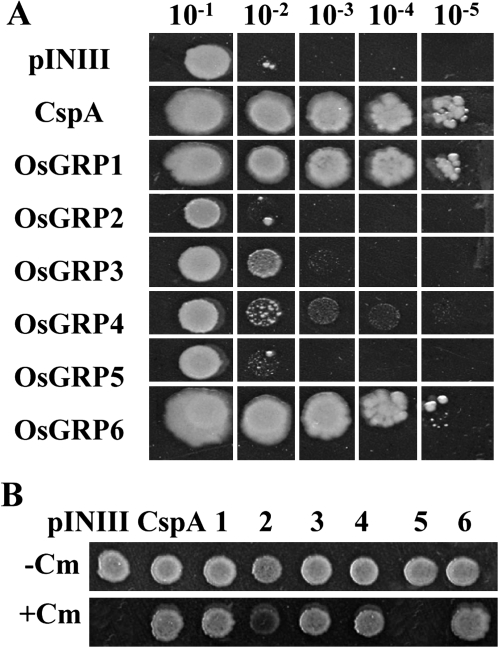
Cold shock proteins (CSPs) in prokaryotes have been determined to function as RNA chaperones during cold stress (Jiang et al., 1997; Phadtare et al., 1999). In an effort to determine whether OsGRPs have RNA chaperone activity during the cold adaptation process, OsGRPs were assessed with regard to their abilities to complement a cold-sensitive BX04 E. coli which lacks four endogenous CSPs and is highly sensitive to cold stress (Xia et al., 2001). When the BX04 cells harbouring each construct were incubated at 37 °C, all cells grew well with no noticeable differences (see Supplementary Fig. S2 at JXB online). By contrast, when the cells were subjected to cold shock at 20 °C, the growths of BX04 cells were significantly different, depending on the expression of each OsGRP (Fig. 2A). The BX04 cells expressing either OsGRP1 or OsGRP6 grew well at low temperatures, and the colony-forming ability of these cells was comparable with that of the positive control cells expressing CspA, a bacterial RNA chaperone. The BX04 cells expressing either OsGRP3 or OsGRP4 grew better than the cells expressing only the pINIII vector, but grew relatively less well than the cells expressing OsGRP1 or OsGRP6. No complementation ability was observed for OsGRP2 and OsGRP5. These results show that specific types of rice OsGRPs are capable of suppressing the cold-sensitivity of E. coli at low temperatures, which indicates that these OsGRPs exhibit an RNA chaperone activity during the cold adaptation process.
Fig. 2.
Complementation ability and transcription anti-termination activity of OsGRPs. (A) The diluted cultures (10−1 to 10−5 dilution) of the BX04 cells harbouring each OsGRP, CspA (positive control), or pINIII vector (negative control) were spotted on LB-agar plates and incubated at 20 °C. The pictures were taken 5 d after incubation. (B) Liquid cultures of RL211 cells harbouring each construct were spotted on LB agar with (+) or without (–) chloramphenicol (Cm), and the cells were grown at 37 °C.
The rice OsGRPs possess DNA- and RNA-melting capabilities
To determine whether the OsGRPs capable of complementing the cold sensitivity of bacterial cells have an RNA chaperone activity, their nucleic acid-melting activities were evaluated in vitro and in vivo. The recombinant GST-OsGRP1, GST-OsGRP4, and GST-OsGRP6 fusion proteins expressed in E. coli were purified (see Supplementary Fig. S3A at JXB online), and their in vitro DNA-melting activities were evaluated by measuring the fluorescence intensity of a molecular beacon. The addition of GST-OsGRP1, GST-OsGRP4, or GST-OsGRP6 proteins resulted in a significant increase in fluorescence, whereas the addition of GST alone resulted in no fluorescence (see Supplementary Fig. S3B at JXB online). The DNA-melting activity of OsGRP1, OsGRP4, and OsGRP6 was comparable to that of CspA. To determine whether OsGRP1, OsGRP4, and OsGRP6 possess RNA-melting activity in vivo, their transcription anti-termination activity was then evaluated in E. coli. An E. coli RL211 strain (Landick et al., 1990) harbours a chloramphenicol resistance gene downstream from a trpL terminator, and serves as an efficient system for the testing of transcription anti-termination activity and RNA-melting activity of putative RNA chaperones (Bae et al., 2000; Phadtare et al., 2002; Nakaminami et al., 2006). As shown in Fig. 2B, the RL211 cells expressing either OsGRP1, OsGRP4, or OsGRP6 grew well on growth media containing chloramphenicol, whereas RL211 cells harbouring OsGRP5 did not grow on chloramphenicol plates. The RL211 cells expressing OsGRP3 which showed partial complementation ability in BX04 cells (Fig. 2A) grew well on growth media containing chloramphenicol. These results show that the OsGRPs capable of complementing the cold sensitivity of bacterial cells harbour nucleic acid-melting activity, which further confirms that these OsGRPs exhibit an RNA chaperone activity during the cold adaptation process.
The rice OsGRPs rescue the growth-defect of cold-sensitive Arabidopsis grp7 mutant plants
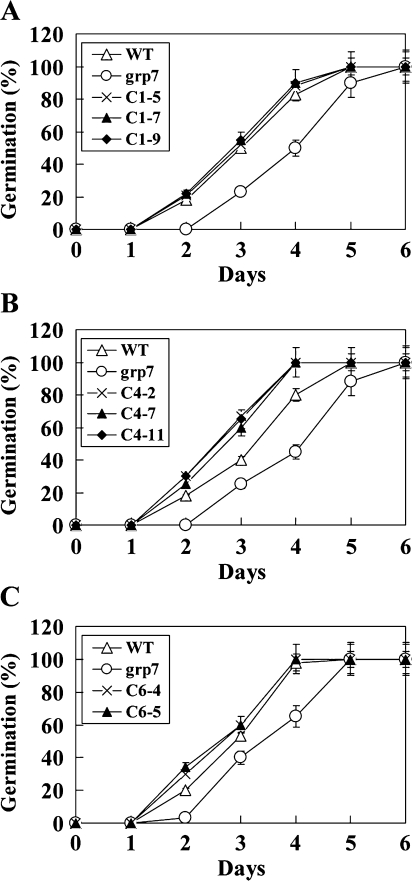
As OsGRPs possess an RNA chaperone activity during the cold adaptation process in E. coli, it is of keen interest to determine whether OsGRPs confer cold or freezing tolerance in plants. The function of OsGRPs was evaluated in plants under cold and freezing stress conditions via a functional complementation of OsGRPs to the cold-sensitive AtGRP7 Arabidopsis mutant plant (grp7). It has been reported that AtGRP7 plays diverse roles in different cellular processes including stress response, circadian regulation, and floral transition (Staiger et al., 2003; Schöning et al., 2007; Kim et al., 2008; Streitner et al., 2008). As AtGRP7 has been shown to exhibit an RNA chaperone function during the cold adaptation process in E. coli (Kim et al., 2007b) and the loss-of-function grp7 mutant was found to be sensitive to cold stress (Kim et al., 2008), the grp7 mutant is a valuable plant system to examine the function and RNA chaperone activity of putative RNA chaperones. The transgenic Arabidopsis plants that ectopically express OsGRP1, OsGRP4, or OsGRP6 were generated, and the expression of OsGRPs in a grp7 background was confirmed via RT-PCR analysis (see Supplementary Fig. S4 at JXB online). Five to six independent transgenic lines were investigated, and similar results were observed. When the plants were grown at normal growth temperature, the germination and seedling growth of grp7 and OsGRP-expressing grp7 plants did not differ noticeably from each other (data not shown). However, when the seeds of the plants were germinated at low temperatures (11 °C), it was apparent that the grp7 mutant expressing either OsGRP1, OsGRP4, or OsGRP6 germinated earlier than the control grp7 did (Fig. 3).
Fig. 3.
Germination of the wild-type, mutant, and transgenic seeds under cold-stress conditions. Seed germination of the wild-type (WT), grp7 mutants, (A) OsGRP1-expressing lines (C1-5, C1-7, and C1-9), (B) OsGRP4-expressing lines (C4-2, C4-7, and C4-11), and (C) OsGRP6-expressing lines (C6-4 and C6-5) was measured in MS medium at 11 °C, and the germination rate was scored on the indicated days. Mean values and stand errors were obtained from three independent experiments (n=30–40).
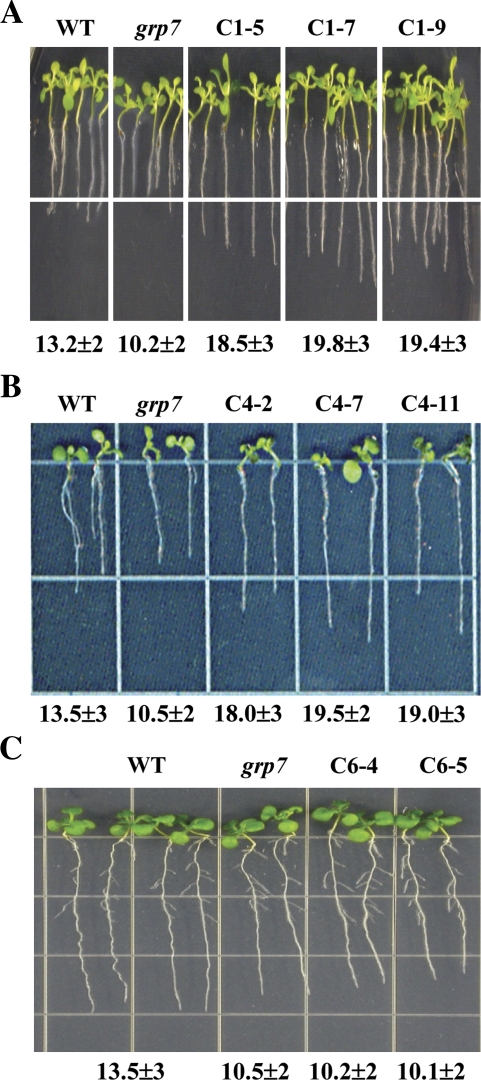
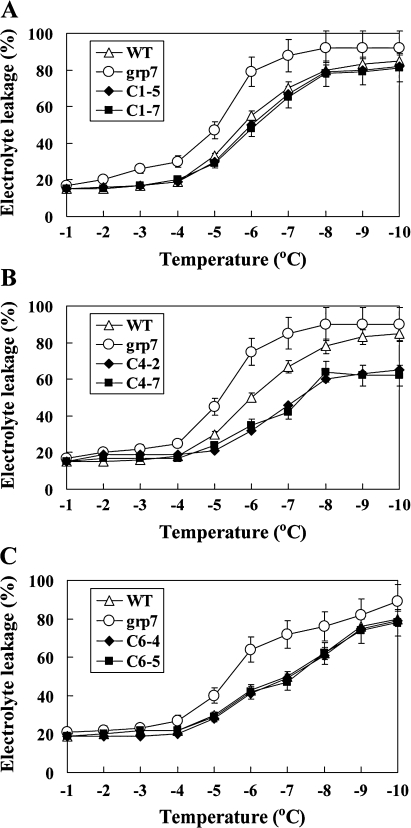
The impact of OsGRP1, OsGRP4, and OsGRP6 on the seedling growth and freezing tolerance of the plants was then examined under cold and freezing stress conditions. When the Arabidopsis plants were grown for 5 weeks at 8 °C, it was observed that the root growth of OsGRP1- or OsGRP4-expressing plants was superior to that of the control plants (Fig. 4). In comparison, no noticeable differences in root growth were observed between the OsGRP6-expressing plants and grp7 plants under low temperature conditions (Fig. 4). The contribution of OsGRP1, OsGRP4, or OsGRP6 to the enhanced freezing tolerance of Arabidopsis plants was further evaluated by measuring electrolyte leakage from the leaves of the plants (Fig. 5). It was shown that the leaves from OsGRP1-, OsGRP4-, or OsGRP6-expressing grp7 plants show much lower rates of electrolyte leakage than was observed in the leaves from grp7 plants. These results demonstrate that OsGRP1-, OsGRP4-, or OsGRP6-expressing grp7 plants are more tolerant to cold and freezing stress, thereby implying that OsGRP1, OsGRP4, and OsGRP6 have the ability to rescue grp7 Arabidopsis plants from cold or freezing damage.
Fig. 4.
Cold tolerance of the wild-type, mutant, and transgenic plants. Root length of the wild-type (WT), grp7 mutant, (A) OsGRP1-expressing lines (C1-5, C1-7, and C1-9), (B) OsGRP4-expressing lines (C4-2, C4-7, and C4-11), and (C) OsGRP6-expressing lines (C6-4 and C6-5) was measured in MS medium 5 weeks after incubation at 8 °C.
Fig. 5.
Freezing tolerance of the wild-type, mutant, and transgenic plants. Electrolyte leakage of the leaves taken from the wild-type (WT), grp7 mutant, (A) OsGRP1-expressing lines, (B) OsGRP4-expressing lines, and (C) OsGRP6-expresing lines was measured in a range of temperature from –1 to –10 °C. Mean values and stand errors were obtained from five independent experiments.
OsGRP is involved in mRNA export from the nucleus to the cytoplasm during cold stress
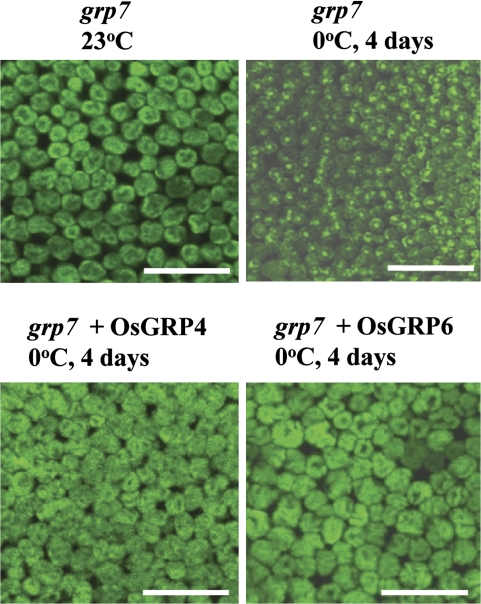
To understand the cellular function of OsGRPs during cold stress, their cellular localization was analysed via confocal microscopy and their potential roles in the export of mRNAs from the nucleus to the cytoplasm were analysed via poly(A) mRNA in situ hybridization assay as previously described (Gong et al. 2005; Kim et al., 2008). The prediction of the cellular localization of OsGRPs using the PSORT and TargetP programs indicates that OsGRP1 and OsGRP4 may be localized to the chloroplasts and the nucleus or mitochondria, respectively (Table 1). Analysis of the cellular localization of OsGRP-GFP fusion proteins showed that OsGRP1 is localized to the chloroplast and OsGRP4 is localized mainly to the nucleus (see Supplementary Fig. S5 at JXB online). Poly(A) mRNA in situ hybridization assay showed that the nuclei of the cells of the grp7 mutant plants grown under normal growth conditions did not show any noticeable fluorescence signals (Fig. 6), which indicates that mRNAs transcribed in the nucleus were efficiently exported to the cytoplasm. When the plants were subjected to 4 d of cold stress at 0 °C, the nuclei of leaf cells in grp7 plants exhibited strong fluorescence signals, indicating that mRNA export was impaired in the mutant cells. Contrary to the strong fluorescent signals in the nuclei of grp7 mutant plant cells under cold stress, no significant fluorescent signal was detected in the nuclei of the cells of OsGRP4- or OsGRP6-expressing grp7 plants under cold stress (Fig. 6). These results show that OsGRP4 and OsGRP6 complement the defect in mRNA export from the nucleus to the cytoplasm in grp7 mutant during cold stress.
Fig. 6.
Poly(A) mRNA in situ localization analysis. Poly(A) mRNA distribution was measured in 2-week-old grp7 mutant, OsGRP4-expressing mutant, and OsGRP6-expressing mutant plants grown at 23 °C and in the plants subjected to cold stress (0 °C) for 4 d. Strong fluorescent signals were detected only in the nuclei of the leaf cells of grp7 plants subjected to cold stress. Bars=100 μm.
Discussion
Despite the increasing amounts of reports evaluating the stress-responsive roles of GRPs in dicotyledonous plants including Arabidopsis, the reports demonstrating the biological functions of GRPs in monocotyledonous plants, including rice, under stress conditions are severely limited. The results of our current study provide clear evidence that, among the six OsGRP family members, OsGRP1, OsGRP4, and OsGRP6 are capable of complementing the growth defect of a cold-sensitive E. coli mutant during cold shock. Interestingly, specific OsGRP family members with E. coli complementation ability during cold stress also confer cold or freezing tolerance in plants. These data are in good agreement with our previous results showing that, in Arabidopsis, AtGRP2 and AtGRP7, which could complement the cold sensitivity of E. coli (Kim et al., 2007b) enhanced the freezing tolerance of Arabidopsis plants (Kim et al., 2007a, 2008), whereas AtGRP4, which had no ability to complement the cold sensitivity of E. coli (Kim et al., 2007b) did not enhance the cold or freezing tolerance of Arabidopsis plants (Kwak et al., 2005). These findings show that the abilities of GRPs to confer cold or freezing tolerance in E. coli and plants are conserved between Arabidopsis and rice.
The molecular mechanism by which OsGRPs confer cold tolerance in plants remains to be clearly elucidated. Because it is believed that GRPs are involved in post-transcriptional gene regulation, including nucleocytoplasmic mRNA transport, mRNA stability, and translation, it appears likely that OsGRPs perform a function in these cellular processes. It has been previously demonstrated that AtGRP2 and AtGRP7, which successfully complemented the growth defect of a cold-sensitive E. coli mutant and enhanced the freezing tolerance of Arabidopsis plants, possess an RNA chaperone activity (Kim et al., 2007a, b). By contrast, AtGRP4, the overexpression of which did not increase cold or freezing resistance in Arabidopsis plants (Kwak et al., 2005), exhibits no RNA chaperone activity (Kim et al., 2007b). Interestingly, OsGRP1, OsGRP4, and OsGRP6, which demonstrated the highest level of complementation ability in cold-sensitive E. coli mutant cells (Fig. 2), share the highest levels of sequence homology with AtGRP2 and AtGRP7 (Table 1). On the basis of these previous observations and current findings, it is proposed that OsGRP1, OsGRP4, and OsGRP6 exhibit RNA chaperone activity during the cold adaptation processes in cells. The complementation ability of OsGRP1, OsGRP4, and OsGRP6 in grp7 mutant plants is indicative of its role as an RNA chaperone in plants.
Considering that the specific types of OsGRPs possess RNA chaperone activity during the cold-adaptation process, the next crucial question is to understand the molecular mechanisms of RNA chaperone action. Several other studies have shown that RNA processing or nucleocytoplasmic transport perform pivotal roles in the responses of plants to cold stress (Lee et al., 2006; Zhu et al., 2007), and a DEAD box RNA helicase, a potential RNA chaperone, is essential for the export of mRNAs from the nucleus to the cytoplasm and cold stress tolerance in Arabidopsis (Gong et al., 2005). It has recently been demonstrated that AtGRP7 confers freezing tolerance in Arabidopsis plants via the regulation of mRNA export from the nucleus to the cytoplasm under cold stress conditions (Kim et al., 2008). The notion that OsGRP4 and OsGRP6, which show the highest level of sequence homology with AtGRP2 and AtGRP7 and that have been determined to be localized to the mitochondria and to both the nucleus and the cytoplasm, respectively (Vermel et al., 2002; Kim et al., 2008), were determined or predicted to be localized to the nucleus or mitochondria (see Supplementary Fig. S5; Table 1) suggest that OsGRP4 and OsGRP6 play a similar function as AtGRP2 or AtGRP7 does. Our current findings demonstrate that OsGRP4 and OsGRP6 are involved in mRNA export from the nucleus to the cytoplasm under cold stress conditions as AtGRP7 does (Fig. 6). It is proposed that OsGRP4 and OsGRP6 confer cold tolerance in rice by functioning as RNA chaperones to regulate mRNA export from the nucleus to the cytoplasm during cold stress. These observations indicate that the molecular mechanism of the action of the GRPs is conserved between Arabidopsis and rice during the cold-adaptation process. The findings that OsGRP1 is localized to the chloroplast and harbours RNA chaperone activity led us to propose that OsGRP1 confers cold tolerance in rice by regulating RNA metabolism in the chloroplast. Considering that numerous nuclear-encoded RBPs take part in chloroplast gene expression by regulating transcription, RNA splicing, RNA editing, RNA degradation and translation (Barkan and Goldschmidt-Clermont, 2000; Schmitz-Linneweber and Small, 2008), it is highly likely that OsGRP1 is one of the nuclear-encoded chloroplast RBPs that play a role during the stress-adaptation process.
The identification of AtGRPs and OsGRPs as RNA chaperones would not only reveal the cellular roles of RNA chaperones during the process of stress adaptation in plants, but might also provide a potential means for the development of stress-tolerant crops. The complementation ability of GRPs in cold-sensitive E. coli and the enhancement of cold and freezing tolerance in GRP-overexpressing Arabidopsis plants suggest that GRPs can be utilized to improve plant performance under stress conditions. It has been demonstrated recently that CspA and CspB, the bacterial RNA chaperones, could be employed to improve the cold or drought stress tolerance of agricultural crops including rice and maize, in which the effective RNA chaperone activity of CSP is critical for providing tolerance to stresses (Castiglioni et al., 2008). The results of this study open new avenues of research involving the use of RNA chaperones as a means to improve plant performance under stress conditions. We are currently attempting to determine whether AtGRPs and OsGRPs harbouring RNA chaperone activity can confer stress tolerance in rice.
In conclusion, the results of the current study provide clear evidence that GRPs in rice and Arabidopsis are functionally conserved, and also suggest that GRPs perform a function as RNA chaperones during the cold-adaptation processes in both monocots and dicots. As our knowledge regarding the cellular functions of GRP family members in the stress responses of monocotyledonous plants remains far from sufficient, our discovery of the conserved functions of specific members of GRPs in Arabidopsis and rice provides new opportunities for the mechanistic examination of their cellular roles during stress adaptation processes. Further studies should be targeted toward a characterization of the GRP-target RNA interactions and the GRP-mediated regulation of RNA metabolism, which is an indispensible step in the formulation of a more comprehensive picture of the cellular functions of GRPs in both monocotyledonous and dicotyledonous plants under stress conditions.
Supplementary data
The following supplementary data are available at JXB online.
Supplementary Table S1. Gene-specific primer pairs used in the RT-PCR experiments
Supplementary Fig. S1. Alignment of amino acid sequences of OsGRPs
Supplementary Fig. S2. Growth of BX04 E. coli mutant cells expressing OsGRPs at 37 °C
Supplementary Fig. S3. Purification of recombinant proteins and nucleic acid-melting activities of OsGRPs
Supplementary Fig. S4. Confirmation of transgenic Arabidopsis plants expressing OsGRP
Supplementary Fig. S5. Cellular localization of OsGRP1 and OsGRP4
Supplementary Material
Acknowledgments
We would like to thank Drs M Inouye and S Phadtare for BX04 mutant cell and pINIII vector and Dr R Landick for E. coli RL211 cell. This work was supported, in part, by a grant from the Korea Science and Engineering Foundation (KOSEF) to the Agricultural Plant Stress Research Center (APSRC, R11-2001-092-04002-0) of Chonnam National University and by a grant (CG2112-1) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the MOST and Rural Development Administration of Republic of Korea.
References
- Aneeta NS-M, Tuteja N, Sopory SK. Salinity- and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochemical and Biophysical Research Communication. 2002;296:1063–1068. doi: 10.1016/s0006-291x(02)02050-8. [DOI] [PubMed] [Google Scholar]
- Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proceedings of the National Academy of Sciences, USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- Binh LT, Oono K. Molecular cloning and characterization of genes related to chilling tolerance in rice. Plant Physiology. 1992;99:1146–1150. doi: 10.1104/pp.99.3.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiology. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni P, Warner D, Bensen RJ, et al. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiology. 2008;147:446–455. doi: 10.1104/pp.108.118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. HnRNP proteins and the biogenesis of mRNA. Annual Review of Biochemistry. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Ferullo J-M, Vézina LP, Rail J, Laberge S, Nadeau P, Castonguay Y. Differential accumulation of two glycine-rich proteins during cold-acclimation alfalfa. Plant Molecular Biology. 1997;33:625–633. doi: 10.1023/a:1005781301718. [DOI] [PubMed] [Google Scholar]
- Gong Z, Dong C-H, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu J-K. A dead box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. The Plant Cell. 2005;17:256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Olson PA. Cloning and characterization of cold-regulated glycine-rich RNA-binding protein genes from leafy spurge (Euphorbia esula L.) and comparison to heterologous genomic clones. Plant Molecular Biology. 1998;38:531–538. doi: 10.1023/a:1006050208670. [DOI] [PubMed] [Google Scholar]
- Jiang W, Hon Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. Journal of Biological Chemistry. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Kim YO, Kim JS, Kang H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. The Plant Journal. 2005;42:890–900. doi: 10.1111/j.1365-313X.2005.02420.x. [DOI] [PubMed] [Google Scholar]
- Kim JY, Park SJ, Jang B, Jung C-H, Ahn SJ, Goh C-H, Cho K, Han O, Kang H. Functional characterization of a glycine-rich RNA-binding protein2 in Arabidopsis thaliana under abiotic stress conditions. The Plant Journal. 2007 a;50:439–451. doi: 10.1111/j.1365-313X.2007.03057.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung C-H, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in E. coli. Nucleic Acids Research. 2007 b;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. The Plant Journal. 2008;55:455–466. doi: 10.1111/j.1365-313X.2008.03518.x. [DOI] [PubMed] [Google Scholar]
- Kwak KJ, Kim YO, Kang H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. Journal of Experimental Botany. 2005;56:3007–3016. doi: 10.1093/jxb/eri298. [DOI] [PubMed] [Google Scholar]
- Landick R, Stewart J, Lee DN. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes and Development. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu J, Zhu J- K. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. The Plant Cell. 2006;18:1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Barta A. Genomic analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Research. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki S, Yamada T, Hirasawa T, Todaka D, Kanekatsu M. Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnology Letters. 2008;30:955–960. doi: 10.1007/s10529-007-9619-8. [DOI] [PubMed] [Google Scholar]
- Moriguchi K, Sugita M, Sugiura M. Structure and subcellular localization of a small RNA-binding protein from tabacco. The Plant Journal. 1997;6:825–834. doi: 10.1046/j.1365-313x.1997.12010215.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nakaminami K, Karlson D, Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proceedings of the National Academy of Sciences, USA. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomata T, Kabeya Y, Sato N. Cloning and characterization of glycine-rich RNA-binding protein cDNAs in the moss Physcomitrella patens. Plant and Cell Physiology. 2004;45:48–56. doi: 10.1093/pcp/pch005. [DOI] [PubMed] [Google Scholar]
- Phadtare S, Alsina J, Inouye M. Cold-shock response and cold-shock proteins. Current Opinion in Microbiology. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. Journal of Biological Chemistry. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- Sachetto-Martins G, Franco LO, Oliveira DE. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochimica et Biophysica Acta. 2000;1492:1–14. doi: 10.1016/s0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends in Plant Science. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. The Plant Journal. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- Shinozuka H, Hisano H, Yoneyama S, Shimamoto Y, Jones ES, Forster JW, Yamada T, Kanazawa A. Gene expression and genetic mapping analyses of a perennial ryegrass glycine-rich RNA-binding protein gene suggest a role in cold adaptation. Molecular Genetics and Genomics. 2006;275:399–408. doi: 10.1007/s00438-005-0095-3. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Molecular Biology. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek DA, Apel K, Eckstein L. The clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. The Plant Journal. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- Stephen JR, Dent KC, Finch-Savage WE. A cDNA encoding a cold-induced glycine-rich RNA binding protein from Prunus avium expressed in embryonic axes. Gene. 2003;320:177–183. doi: 10.1016/s0378-1119(03)00826-6. [DOI] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. The Plant Journal. 2008;56:239–250. doi: 10.1111/j.1365-313X.2008.03591.x. [DOI] [PubMed] [Google Scholar]
- Vermel M, Guermann B, Delage L, Grienenberger J-M, Maréchal-Drouard L, Gualberto JM. A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proceedings of the National Academy of Sciences, USA. 2002;99:5866–5871. doi: 10.1073/pnas.092019599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Molecular Microbiology. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Dong C-H, Zhu J-K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.